Highlights
-
•
Dysregulated and excessive cytokine storm is vital for critical COVID-19 patients.
-
•
Mechanisms of SARS-CoV-2-infected included ACE2 mediated and other pathway response.
-
•
Infected lung epithelial cells and macrophages mainly release cytokines in immunity response.
-
•
Endothelial cells damage and microthrombosis played an important role in COVID-19.
-
•
Targeted cytokine drug therapy might an effective measure to alleviate the inflammation.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Cytokine storm, Pneumonia, Clinical manifestations
Abstract
Coronavirus disease 2019 (COVID-19), caused by the virus designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread widely throughout the world. Despite the strict global outbreak management and quarantine measures that have been implemented, the incidence of COVID-19 continues to rise, resulting in more than 290,000 deaths and representing an extremely serious threat to human life and health. The clinical symptoms of the affected patients are heterogeneous, ranging from mild upper respiratory symptoms to severe pneumonitis and even acute respiratory distress syndrome (ARDS) or death. Systemic immune over activation due to SARS-CoV-2 infection causes the cytokine storm, which is especially noteworthy in severely ill patients with COVID-19. Pieces of evidence from current studies have shown that the cytokine storm may be an important factor in disease progression, even leading to multiple organ failure and death. This review provides an overview of the knowledge on the COVID-19 epidemiological profile, the molecular mechanisms of the SARS-CoV-2-induced cytokine storm and immune responses, the pathophysiological changes that occur during infection, the main antiviral compounds used in treatment strategies and the potential drugs for targeting cytokines, this information is presented to provide valuable guidance for further studies and for a therapeutic reduction of this excessive immune response.
1. Introduction
In December 2019, the emergence of a novel coronavirus-induced pneumonia in Wuhan, China, posed a serious and urgent threat to public health throughout the world [1]. On 11 February 2020, the World Health Organization (WHO) officially renamed 2019‐nCoV as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and designated the disease caused by it as coronavirus disease 2019 (COVID-19). On 30 January 2020, the WHO declared COVID-19 as the sixth public health emergency of international concern, and on March 11, 2020, the WHO classified COVID-19 as a pandemic. Due to the spread of the SARS-CoV-2 virus worldwide, it is currently reported in approximately 200 countries and regions. As of 14 May 2020, the cumulative number of confirmed cases of COVID-19 worldwide has exceeded 4 million, with deaths exceeding 292,046 [2]. The largest number of patients with COVID-19 was observed in the United States, followed by Spain, Russia, the United Kingdom, and Italy. In China, the number of confirmed cases reached 82,933, with fewer new cases being confirmed recently [3]. It is generally believed that the incubation period is two weeks, but there is no unified conclusion. It is the third highly pathogenic coronavirus to rapidly emerge, preceded by severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 [4], which have made public health care institutions around the world face greater challenges.
SARS-CoV-2 has shown a similar pattern of infection and clinical features but an even faster transmission rate [5] when compared with the two previous coronaviruses outbreaks [6]. However, it is of particular attention that acute lung injury (ALI), systemic inflammatory response syndrome (SIRS), and acute respiratory distress syndrome (ARDS) occurred in SARS-CoV- and MERS-CoV-infected patients, as well as in patients with COVID-19 [7]. Cytokines have been found that play a key role in driving the appearance of these clinical features and are also at the core of the development of inflammation [8], [9]. Consistent with the previous findings, patients with severe COVID-19 showed significant increases in cytokines such as IL-2, IL-7, IL-10, GSCF, IP10, MCP-1, MIP1A and TNF-α, with the characteristics of a cytokine storm [10]. When SARS-CoV-2 infects the body, the inflammatory response plays an antiviral role, but a strong cytokine storm due to an unbalanced response can be very damaging to the patients. Therefore, using strategies to effectively suppress cytokine storm is essential for preventing disease deterioration in patients with COVID-19 and for saving patients' lives, which is of great significance for the treatment of critically ill patients and for reducing the mortality rate. In this review, SARS-CoV-2 and the mechanisms by which cytokine storm is induced by the virus will be introduced in detail, including the ways in which cytokines are activated and released, how they cause cell and organ damage, and the therapeutic interventions for preventing and quelling this harmful process.
2. Features of SARS-CoV-2
SARS-CoV-2 is a zoonotic human coronavirus (CoV) closely related to those coronaviruses that cause severe acute respiratory syndrome (SARS-CoV) and Middle East respiratory syndrome (MERS-CoV) [11]. Coronaviruses are a group of positive-sense, single-stranded RNA viruses with the largest genome among known human RNA viruses and a likely ancient origin, named as such because of the envelope spinous processes that resemble a corona [12]. SARS-CoV-2 belongs to the β-coronavirus genus. All coronaviruses have nonsegmented genomes. Two-thirds of the genome consists of two large overlapping open reading frames (orf1a and orf1b), which are translated into 16 nonstructural proteins (nsp1 to nsp16). The remaining genome encodes structural proteins, including nucleoprotein (N), the receptor-binding site spike glycoprotein (S), small envelope glycoprotein (E), and membrane glycoprotein (M) [13]. Coronavirus entry into target cells is mediated by the spike protein. The S protein includes two domains, with the S1 subunit responsible for binding to the receptor and the S2 responsible for fusing the virus and the host cell membrane [14]. Among the coronaviruses that have been identified, the N protein, which is the only protein present in the nucleocapsid and participates in viral replication by binding to RNA, is composed of two separate domains, the N-terminal domain (S1-NTD) and the C-terminal domain (S1-CTD), both of which serve as RNA receptor-binding domains (RBD) [15]. Coronaviruses have been recognized as the causes of mild and severe respiratory tracts diseases in humans and some animals. Compared with the other four low-pathogenicity human viruses (HCoV-NL63, HCoV-229E, HCoV-OC43, and HKU1) that are prone to cause mild cold-like symptoms, the two highly pathogenic coronaviruses, SARS-CoV and MERS-CoV, cause severe respiratory infections in humans that can even progress to fatal multiple organ failure[16]. In light of the published genomic data, the sequence homology between SARS-CoV-2 and SARS-CoV is 79.6% [17], with most of the proteins showing a high homology [18]. In addition, little is known about the origin of SARS-CoV-2, although it was originally thought that the first case were associated with the Huanan Seafood Market in Wuhan [10], and thus these basic but important studies are urgent for rapidly finding the source of SARS-CoV-2 in order to slow the ongoing outbreak.
3. Clinical characteristics of COVID-19 patients
It has been reported that the early clinical manifestations of patients with COVID-19 are mainly fever (98%), cough (82%), shortness of breath and exhaustion, which can rapidly progress to pneumonia [10]. Nausea, vomiting, and diarrhea are uncommon [19]. The respiratory symptoms of COVID-19 are heterogeneous, from mild to severe symptoms with ARDS, accompanied by a generalized weakness and fatigue [20]. Abnormalities are found in computed tomography (CT) images of patients’ chests, primarily ground glass-like opacity areas bilaterally in the lungs of those infected (72%) [20]. Troponin T (TnT) elevation, cardiac dysfunction and arrhythmias are complications in hospitalized COVID-19 patients and are associated with a fatal outcome [21]. Common preexisting diseases in patients may be risk factors for a poor prognosis, including cardiovascular disease (10.5%), hypertension (6%), diabetes (7.3%), chronic respiratory disease (6.3%) and cancers (5.6%) [22], especially in older men. Epidemiological studies have reported that elderly patients are more likely to suffer from critical diseases and that the symptoms in children are often milder. Children under 9 years of age and those aged 10–19 years account for 1% of the total cases [22]. Research from fatal cases in China showed that the majority of the non‐survivors died of multiple organ failure, and most of these cases were in male over 50 years old with noncommunicable chronic diseases [23]. The median times are 5.0 days from the first symptoms to dyspnea and 8.0 days for the development of ARDS [24].
A positive SARS-CoV-2 nucleic acid test can diagnose COVID-19, which can be auxiliary confirmed by CT and specific antibody IgG/IgM tests [25]. The most common abnormalities in the laboratory results include normal or reduced white blood cells, the decreased prevalence of lymphocytes (83.2%) [19], abnormally elevated ALT and AST [26], increased proinflammatory cytokines such as IL-1β, IL-6, and TNF-α, and increases in lactate dehydrogenase (LDH), D-dimer, C-reactive protein (CRP), and procalcitonin (PCT) [10]. In addition, patients' plasma angiotensin II (Ang II) levels are significantly increased, which is related to viral load and lung injury [26]. ICU patients show higher levels of plasma cytokines such as IL-6 [27], D-dimer, fibrinogen, PCT and a prolonged thrombin time [28], suggesting that inflammatory responses play a key role in these injuries and may also be related to the severity of the disease in these patients [29], even being the cause of death. Disseminated intravascular coagulation (DIC) is apparent in most of the patients who have died [30]. Owing to the lack of direct evidence, it is unclear how the process of the inflammatory response involving cytokines fully progresses, but it is certain that the manifested clinical features are directly related to the violent occurrence of inflammation.
4. Emergence and progression of the cytokine storm in COVID-19 patients
The cytokine storm refers to the overproduction of inflammatory cytokines with a wide range of biological activity from a variety of tissues and cells (mainly immune cells), which is due to different infections and a loss of negative feedback on the immune system. In turn, these cytokines drive a positive feedback on other immune cells and continue to recruit them to the sites of inflammation, begetting the exponential growth of inflammation and organ damage. In short, it is the unceasing extreme activation and attack of the autoimmune system. The main cytokines involved are interleukins (IL), interferons (IFN), tumor necrosis factor (TNF), colony stimulating factors (CSF), the chemokine family, growth factors (GF), and others. They are divided into pro-inflammatory factors (such as IL-1β, IL-6, IL-12, TNF, and IFN-γ) and anti-inflammatory factors (such as IL-4, IL-10, IL-13, and TGF-β) based on their functions. The cytokine storm is a crucial cause of ARDS, a systemic inflammatory response, and multiple organ failure [31]. Moreover, the viruses can invade lung epithelial cells and alveolar macrophages to produce viral nucleic acid, which stimulates the infected cells to release cytokines and chemokines, activating macrophages, dendritic cells, and others [15]. Chemokines and cytokines are increasingly released from these cells to attract more inflammatory cells to migrate to the site of inflammation from the blood vessels, thereby cascading the amplification of the inflammatory response.
Acute lung injury (ALI) is a common consequence of cytokine storm in lung tissue and systemic circulation [32]. Recently, the pulmonary pathology of SARS-CoV-2 infection showed that the major changes in the lung tissue is diffuse alveolar damage, alveolar edema and proteinaceous exudates, thickening of alveolar walls, evident desquamation of pneumocytes and hyaline membrane formation, indicative of ARDS [33]. Multinucleated giant cells in the alveolar cavity and inflammatory infiltration of the lymphocytes in the pulmonary mesenchyme have been demonstrated [34]. In addition, the pathological results have confirmed that the number of CD4 + T and CD8 + T cells in the peripheral blood is reduced but that these cells are overactivated. CCR4+/CCR6 + Th17 cells, found to be increased, can have high proinflammatory effects, and CD8 + T cells contain high concentrations of cytotoxic granules, mainly perforin and granulysin, which cause severe immune damage in patients [34]. The damaged alveolar epithelial cells and the extensive phlegm secretion and exudation significantly inhibit the ventilation function of the lungs, leading to hypoxemia, hypotension, and even shock [32]. The existence of disseminated intravascular coagulation is currently reported to be common in deaths from COVID-19[30]. Endothelial cell damage causes coagulation activation and hyperfibrinolysis conditions resulting from the inflammatory progression, which may cause small blood vessel thrombosis, possibly increasing cardiac load and promoting pulmonary embolism [25].
COVID-19 is considered a systemic disease involving a series of other important organs, such as heart, liver and kidney. On the one hand, ACE2, the cellular receptor for the virus, is widely expressed in various organs and tissues, but on the other hand, the damage caused by the viral infection is caused by cytokine storm [24]. The severity and lethality of COVID-19 includes viral damage to the heart muscle and to the blood vessels. Myocardial oxygen demand increases during the state of infection, and the high metabolic rate leads to an increase in myocardial load, which further causes an imbalance between supply and demand [21]. In addition to hypoxia, respiratory distress, metabolic acidosis, fluid or electrolyte disturbances and activated neurohumoral systems after a severe infection may also lead to cardiac arrest and damage and may even induce malignant arrhythmias [25]. Acute kidney injury (AKI) is also an important feature observed in COVID-19 patients. An important autopsy study has reported that patients that died from COVID-19 had significant acute proximal tubule injury, and the presence of clusters of coronavirus particles in podocytes and renal tubular epithelial cells were observed [35]. Activated macrophages and angiotensin II overactivity play a role in AKI and podocyte damage. In addition, hypotension, microvascular damage and contraction, decreased renal perfusion, and hemostatic factors and the related sepsis should be taken into account [36]. Liver damage occurrence may be due to the viral infection or to drug damage during the course of the disease in severe cases, which is yet to be studied [37].
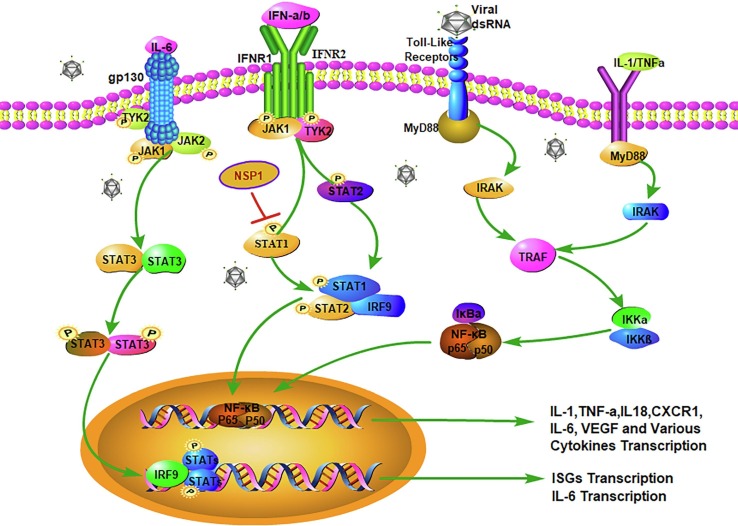
It has long been believed that a well-coordinated and rapid innate immune response is the first line of defense against viral infection. The cytokines synthesized and secreted by immune cells are involved in the induction period and in the effect phases in all inflammatory reactions, but more importantly, activating the initiation of the cytokine transcription mechanism to promote secretion is the key link. When immune cells in the body detect the pathogen-associated molecular pattern (PAMP) from the virus through the pattern recognition receptor (PRR) on the cell membrane, the innate immune response system is immediately activated [38]. Among the PRRs, the most typical is the Toll-like receptor 3(TLR3). TLR3 is a transmembrane receptor, with the extracellular accessory proteins MD-1, MD-2 and RP105 involved in the recognition of the PAMP [39]. Macrophages are the key cells for host defense. TLR3 on macrophages specifically recognizes ds-RNA, the intermediate product of virus replication, followed by the recruitment of signal transfer proteins MyD88, TIRAP, TRAM or TRIF in the cytoplasmic TIR domain. Activation of various kinases (IRAKs, TBK1, and IKKs) and tumor necrosis factor receptor-related factor 6 (TRAF6), according to the different adaptors, eventually lead to activation of the NF-κB, MAPK, or JNK-STAT pathways to promote the transcription of inflammatory cytokines and to produce IFN, IL-1β, IL-6, and others, for coordinating the local or systemic inflammatory responses [40] (Fig. 1 ). IL-1β and IL-6 are the major pro-inflammatory cytokines released during viral infections [41]. IL-1β enhances the inflammatory responses in the bronchi and alveoli in patients with lung injury. At the same time, acute phase proteins from hepatocytes stimulated by IL-1β and IL-6 activate the complement system, and the complement cascade further increases vascular permeability.
Fig. 1.
(a) IL-6 binds to the transmembrane receptor IL-6R to generate the IL-6/IL-6R complex, which induces homodimerization of gp130, leading to activation of the downstream signaling molecules, including TYK2,JAK1,JAK2 and STAT3, promoting IL-6 transcription; (b) the binding of IFN-α/β to the dimer receptor IFNAR activates the JAK-STAT signal transduction pathway, in which STAT1, STAT2 and IRF9 form a complex and then enter the nucleus to initiate the transcription of IFN-stimulated genes (ISG). However, NSP1 can inhibit the IFNs response by blocking STAT1 phosphorylation; (c) TLR3 specifically recognizes the viral ds-RNA followed by the recruitment of signal transfer proteins MyD88, IRAK, IKKε and TRAF6, and eventually the NF-κB complex is activated to promote the transcription of inflammatory cytokines; (d) IL-1 and TNF-α can also activate the NF-κB single pathway to initiate the transcription of inflammatory cytokines. PRR activates the transcription of multiple cytokines and delays IFN-α and -β responses.
IL-6 has always been an important central factor in cytokine storm. In virus-infected lesions, IL-6 can respond to IL-1, TNF-α, or TLR signals, trigger cis-regulatory modules, and activate the NF-κB transcriptional signaling pathway and the binding site of the nuclear factor IL-6 (CAAT/EBPβ) [42]. However, the most important function is that IL-6 binds to the transmembrane receptor IL-6R to generate the IL-6/IL-6R complex. Subsequently, the dimerization of the signal transduction component gp130 is induced by the complex, which activates Janus kinase signal transduction and transcription activators [43] (Fig. 1). High levels of IL-6 can activate the coagulation system and increase vascular permeability, providing conditions for the rapid spread of inflammation [43]. It has been reported that higher levels of IL-6 are present in patients with severe COVID-19, which proves that high levels of IL-6 may cause greater damage to lung tissue [10]. It has been verified in vitro that the SARS-CoV S protein induces upregulation of IL-6 and TNF-α in mouse macrophages through the NF-κB pathway [44]. The cytokine-mediated inflammatory response pathway is a series of intersecting networks, and each one has a degree of redundancy and alternate pathways. The combinations of TLR and ligands initiate a signal cascaded-amplification and lead to the activation of multiple cytokine pathways, which is the main research focus of the current inflammatory responses.
Through the regulation of cytokines and chemokines, conventional lymphocytes (T cells and B cells) differentiate into specific effector cells and localize at the sites of infection. For instance, CD4 + T cells differentiate into Th1 cells and produce IFN-γ to activate macrophages and other types of cells because of the induction of IL-12, thereby triggering the defense against intracellular pathogens. At the same time, IFN-γ can induce the transcription of multiple chemokines. In addition, IL-6 induces the differentiation of CD8 + T cells into cytotoxic T cells, which eliminate viruses by lysing the infected cells. The consumption of cytotoxic T cells may be the cause of the decrease in lymphocytes in most patients with COVID-19 [10]. It has been found that T lymphocytes are a vital source of many chemokines and express multiple molecule receptors [45]. Similarly, neutrophils and macrophages are drawn to regions of injury by IL-8 and MCP-1 by chemotactic influences, respectively, while secreting chemokines to recruit more cells to participate in the battle against pathogens. It is now accepted that each cell can respond to multiple chemokines just by expressing a single type of receptor. It is this complex relationship between chemokines and their receptors that enables chemokines to rapidly replenish in various microenvironments, which thus allows the inflammatory storm to continue to develop [41]. According to a report, in SARS patients, proinflammatory cytokines such as TNF and IL-6 and chemokines IL8, CCL3 (MCP-1), CCL5, CCL2 and CXCL10 were found to be significantly upregulated, while the anti-inflammatory factors such as IL-10 were found to be lacking [31], which showed that the lack of anti-inflammatory factors can cause an imbalance in the inflammatory response and promote cytokine storm.
5. Effects of the cytokine storm resulting from pathogenic SARS-CoV-2 infection
Angiotensin-converting enzyme 2 (ACE2) has been shown to be a cellular receptor for SARS-CoV-2 [46]. Twenty-one mutations have been found in the SARS-CoV-2 spike glycoprotein binding region, suggesting that this coronavirus evolved gradually in adapting to human hosts [47], [48]. Though ACE2 exists in various tissues such as coronary arteries, vascular endothelium, and renal tubular epithelium [49], the macrophages and pulmonary alveolar epithelial cells are the primary targets attacked by the virus [29]. Currently, it is generally believed that all populations are considered susceptible to SARS-CoV-2. In animal models, it has been demonstrated that older rhesus monkeys are more susceptible to SARS-CoV than younger monkeys [50], and the increased expression of ACE2 in the lower respiratory tract induced by smoking may increase sensitivity to SARS-CoV-2. Genetic analysis of ACE2 has shown that expression of a mutant form of ACE2 was higher in East Asian populations, which may indicate that there are differences in SARS-CoV-2 infectivity among different populations [51]. In addition, rapid virus replication causes cell pyroptosis, immune evasion, and cell lysis triggered by anti-Fc antibodies, all of which trigger the mass release of pro-inflammatory cytokines and chemokines. Therefore, exacerbation of COVID-19 patient’s clinical symptoms may be the result of a combination of cytopathic effects caused directly by the virus infection and immunopathology injury caused by a violent cytokine storm.
5.1. ACE2 receptor-mediated inflammatory response
From the molecular modeling structural analysis results of the 2019-nCoV receptor, the receptor-binding domain (RBD) of SARS-CoV-2 shows a more effective interaction with ACE2 compared to that of SARS-CoV [52]. The binding of S protein to ACE2 is the first step for the virus to enter the target cells, which is accomplished by proteolytic cleavage and fusion of the viral and cellular membranes [15]. It is speculated that SARS-CoV-2 might also cause lung tissue injury through the same pathogenic mechanism [53]. On the one hand, when SARS-CoV-2 infects alveolar cells, the S1 and ACE2 transmembrane domains bind to reduce the level of ACE2, resulting in the renin angiotensin system (RAS) tilting towards the ACE-Ang II axis [54]. Meanwhile, the production of Ang II is absolutely or relatively elevated, which causes macrophage infiltration, inducing an increase in cytokines and adhesion molecules, including IL-6, monocyte chemotactic protein 1 (MCP-1), vascular cell adhesion molecule 1 (VCAM-1), selectin E, and others, which cause endothelial dysfunction [55], [56]. In addition, the downregulation of ACE2 reduces the protective effects against acute lung injury [54], leading to increased pulmonary capillary permeability and pulmonary edema, and patients with severe disease may die of respiratory failure. On the other hand, experimental cellular studies in vitro have shown that SARS-CoV S induces shedding of the ACE2 extracellular ectodomain and promotes virus entry into cells through dependence on TNF-α converting enzyme (TACE). However, the function of free sACE2, currently unknown, may also mediate inflammation and tissue injury [57]. In addition, the binding of virus to ACE2 might also be involved in intracellular pathway recognition.
5.2. Cell pyroptosis
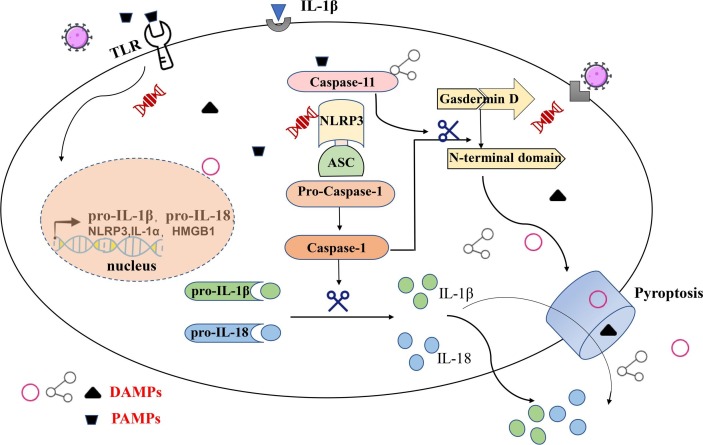
Pyroptosis is a newly identified inflammatory form of programmed cell death, and inflammatory storms caused by SARS-CoV-2 infection may be related to cell pyroptosis. Research evidence from Chen et al. showed that the SARS-CoV viroporin 3a protein activates the NLRP3 (NOD-like receptors protein 3) inflammasome, causing IL-1β production [58]. Reduced cell counts and increased IL-1β in the serum of COVID-19 patients may indicate the activation of cell pyroptosis. When a variety of extracellular PAMPs are recognized by TLRs [59], this triggers the activation of the NF-κB signaling pathway and upregulation of inflammasome-related components, including inactivated NLRP3, proIL-1β, and proIL-18. Subsequently, NLRP3 oligomerizes and it is connected to pro-caspase-1 through the adaptor protein ASC to form a multiprotein complex, thereby activating caspase-1 [60]. Activated caspase-1 recruits and cleaves members of the Gasdermin family such as GSDMD for polymerization in the pathway downstream [61] and simultaneously cleaves the precursors of IL-1β and IL-18 to form active IL-1β and IL-18, which are released into the extracellular environment to recruit more inflammatory cells to aggregate and expand the inflammatory response [62]. In addition, the active cleavage fragment of GSDMD causes extensive cell perforation by inserting into the lipid bilayer, resulting in cell swelling and lysis, which is followed by the release of contents such as the cellular matrix and cytokines [63]. Many endogenous immune molecules are released from the intracellular environment, such as oxidized phospholipids and the cellular matrix, which are known as damage-associated molecular patterns (DAMP). Similar to PAMP, which are known as alarm signals, DAMP can also be recognized by NLRP3, thereby progressively magnifying the inflammatory effects and causing cell pyroptosis [64] (Fig. 2 ). COVID-19 patients often have lymphopenia, but further research is still needed to prove whether it is also related to this mechanism.
Fig. 2.
TLR recognizes extracellular PAMPs and initiates the transcription of NLRP3, proIL-1β, and proIL-18 through the NF-κB signaling pathway, and oligomerized NLRP3, ASC and pro-caspase-1 form a multiprotein complex that activates caspase-1. Activated caspase-1 cleaves the pro-IL-1β and pro-IL-18, forming active IL-1β and IL18, which are released, and simultaneously, the active cleavage fragment of GSDMD causes cell perforation by inserting into the lipid bilayer, resulting in cell swelling and lysis, followed by the release of contents such as the cellular matrix and cytokines. Intracellular signaling mechanism of cell pyroptosis induced by PAMPs.
5.3. Delayed IFN-α and -β response
IFN is a core family in innate antiviral immunity, with type I interferons (IFN-α and -β) being essential, especially for the innate immune response against viruses and other microbial infections. The binding of type I interferon and its dimer receptor (IFNAR) activates the JAK-STAT signal transduction pathway, in which the JAK1 and TYK2 kinases phosphorylate STAT1 and STAT2, which form a complex with IRF9. These complexes enter the nucleus and initiate the transcription of IFN-stimulated genes (ISG) [65]. In vitro studies have found that the rapid replication of SARS-CoV in mice induces a significant but delayed IFN-α/β response, accompanied by a large influx of pathogenic inflammatory mononuclear macrophages (IMM) [66], leading to an increase in lung cytokines and chemokines, vascular leakage and virus-specific T cell apoptosis, further hindering viral clearance. In addition, studies have demonstrated that coronaviruses can rapidly replicate in host cells and encode proteins (NSP1) that antagonize the IFNs response by blocking STAT1 phosphorylation [15], [67]. Meanwhile, structural proteins M and N inhibit the signaling of IFNs by deactivating TRAF3, TBK1/IKKs, and some other mechanisms, respectively [68]. The coronavirus structural and nonstructural proteins cause a delayed response of the IFNs, further amplifying the inflammatory response by promoting viral replication, followed by the increase in the viral PAMPs. In turn, the PAMPs inhibit the delayed IFN signaling and stimulate PRR-induced abnormal inflammatory responses [69]. Above all, it should be clear that these putative antiviral mechanisms have been confirmed in step-by-step studies. Whether they are truly important in infectious viruses and in systemic antagonist pathways needs to be further studied.
5.4. Anti-S IgG-mediated inflammatory response
It is generally considered that antiviral antibodies play a very important role in viral clearance. According to reports, in patients who died from SARS, anti-S neutralizing antibodies (NAb) developed significantly faster (14.7 vs 20 days) and to higher levels than in patients who had recovered [70]. In a SARS-CoV macaque experimental model, after inoculation with an antibody to the S protein, it was found that the anti-S IgG facilitated severe lung injury in the early stages of infection by eliminating the wound-healing macrophage response and TGF-β production, as well as by promoting inflammatory macrophages and the production of factors MCP-1 and IL-8 [71]. This evidence has suggested that anti-S IgG may also play an important role in lung injury caused by acute SARS-CoV-2 infection during the acute infection period [71]. Since the FcR was blockaded, reducing the production of inflammatory cytokines, it is considered that the virus complexed with anti-S IgG may promote cytokine release by binding to Fc receptors on the surface of macrophages or additionally through antibody-dependent cell-mediated cytotoxicity (ADCC) directly lysing the target cells [72]. Whether this complex is associated with antibody-dependent enhancement (ADE) [73] or complement system activation in patients with COVID-19 during viral replication [55] remains to be proven by further studies.
6. Potential antiviral compounds
Corticosteroids are usually used to suppress inflammatory responses, which were the main means of immunomodulatory therapy during the SARS epidemic. However, the early patients were found to have an increased plasma viral load and secondary infections [74]. In some studies, early administration of IFN was beneficial in reducing the viral load and moderately improved clinical performance, and a combination with ribavirin also had a certain therapeutic effect [75]. IFN-λ inhibited the recruitment of inflammatory cells and the production of IL-1β without excessive stimulation of the immune system, which might become a potential therapeutic direction [76].
Remdesivir (RDV) is a nucleotide analog inhibitor of RNA-dependent RNA polymerases with a broad-spectrum antiviral activity, originally designed to target Ebola. RDW was used to treat the first COVID-19 patient in the United States and to reverse lung injury [77]. It has been reported that remdesivir and chloroquine can inhibit SARS-CoV-2 replication in vivo and in vitro [78], and the antiviral protective effect of RDV combined with IFN-β was observed in vivo and in vitro, which is better than that of lopinavir/ritonavir-IFN-β against MERS-CoV [79]. However, its effectiveness and safety still need to be verified in clinical trials. Chloroquine has been shown effective to inhibit SARS-CoV-2 replication in vitro by interfering with ACE2 [78] and can also inhibit the production and release of TNF and IL-6, which suggests that chloroquine can suppress cytokine storm in patients infected with COVID-19 [80]. Hydroxychloroquine shares the same mechanism as chloroquine and was found to be more potent than chloroquine in inhibiting SARS-CoV-2 in vitro [81]. A clinical trial of hydroxychloroquine treatment significantly showed that it was associated with a reduction in viral load in COVID-19 patients and that azithromycin could enhance its effect [82].
The combination of lopinavir and ritonavir increased plasma half-life through the inhibition of cytochrome P450, which has been widely used as an aspartate protease inhibitor to treat HIV infection. Although the effectiveness of these drugs needs to be verified in further trials in COVID-19 patients, the safety of the treatment was confirmed [83]. The results of one study have indicated that arbidol monotherapy may be superior to lopinavir/ritonavir in treating COVID-19 [84]. A new type of RNA-dependent RNA polymerase (RdRp) inhibitor is favipiravir, which was approved for flu treatment. The reason that favipiravir might have a potential antiviral effect on SARS-CoV-2 is that it is converted into an active phosphorylated form (Favipiravir-RTP) in cells and is recognized as a substrate by the viral RNA polymerase, thereby inhibiting RNA polymerase activity [85]. The clinical trial on favipiravir for the treatment of COVID-19 has achieved good results, compared with the lopinavir/ritonavir treatment group, with no significant adverse reactions noted in the favipiravir treatment group [86]. Tocilizumab is an antagonist of IL-6 and has a therapeutic effect on cytokine storm caused by infection. Tocilizumab has been approved for the treatment of severe complications related to SARS-CoV-2 to reduce inflammation in China [87].
In addition, the results from the management of patients with COVID-19 by Xu et al. showed that an artificial liver blood purification system can quickly clear inflammation mediators to suppress cytokine storm, but the repair system may be delayed for excessive clarity [88]. Convalescent plasma transfusion may offer a short-term therapy strategy for susceptible individuals [89]. Although it has shown a good therapeutic effect, its overall safety and the appropriate timing of administration need further study for a rigorous scientific approach in the interest of avoiding harm like ADE [90]. Monoclonal antibody therapy as a potential therapeutic intervention represents the main biological treatment of passive immunotherapy against viral infections. Five critically ill patients with COVID-19 were treated with convalescent plasma, after which their viral loads declined and their clinical conditions improved [91]. The use of these antibodies may be another viable immediate option for emergency prophylaxis and SARS-CoV-2 therapy. Meanwhile, this means that vaccination may be the best choice to control COVID-19, but current vaccine production faces many obstacles. This requires researchers to explore international cooperation. Evidence has revealed that the serine protease TMPRSS2 plays an important role in SARS-CoV-2 entry into cell but that TMPRSS2 inhibitors can prevent entry [46], which may become a treatment option.
Currently, it is unclear which drugs can successfully treat COVID-19. There is not yet complete data from large randomized clinical trials (RCTs) that can provide clinical guidance on the use and dosage of the drugs. Therefore, effective and safe anti-SARS-CoV-2 treatment requires further clinical trials, large-scale randomized controlled studies and comprehensive activities.
7. Conclusion and prospect
In sum, the rapid spread of SARS-CoV-2 has forced all medical institutions worldwide to carry out research, prevention and control. All efforts are being made to slow the spread of COVID-19 to provide better public health recommendations and to develop timely diagnostics, therapeutics and vaccines. The above potential mechanisms are based on two previous outbreaks of coronaviruses and on current partial research [92], with many new mechanisms still unknown. New treatments are being developed based on existing experience in combating viral infections. However, at present, treatment strategies for SARS-CoV-2 infection are only supportive, and more importantly, there are no specific antiviral drugs for COVID-19. Therapeutic interventions targeting these pro-inflammatory cytokines and chemokines may help to alleviate adverse inflammatory responses, such as JAK-STAT pathway inhibition [93].
Studies have found that there may be individual differences in susceptibility to cytokine storm. The innate immune response of healthy people is highly variable, which can be genetically confirmed with the TLR receptor. For example, sepsis patients with single nucleotide polymorphisms (SNPs) of TLR1 hypermorphic variants have been shown to have an association with an increased susceptibility to organ dysfunction, death, and gram-positive bacteremia infection [94]. TLR4 is the major receptor for lipopolysaccharide (LPS); variants of this receptor can make individuals susceptible to sepsis, and genome-wide association studies (GWAS) have linked TLR4 polymorphisms to pathogen susceptibility and disease severity [41]. Future studies will likely reveal potential genetic variations that affect host cytokine storm during infection. In addition, the body’s immune pathological damage is closely related to the viral load, and the degree of the cytokine storm is an essential connection point. If plasma cytokines are monitored dynamically to assess the degree of cytokine storm in a timely and effective way, this may be of great benefit to the care of critically ill patients.
The inflammatory response is an essential part of the immune system for its function; otherwise, pathogens would be difficult to eliminate. SARS-CoV-2 might induce excessive and prolonged cytokine responses, resulting in lung damage and multiple organ failure. To date, most studies have focused on the direct measurement of those cytokines and chemokines in the peripheral blood, but in the context of the rapidly changing cytokine environment after virus infection, we do not have well-rounded understanding of the cause of the vigorous inflammatory response. Although existing studies have shown that during the occurrence of pathogenic hCOV infection, a violent cytokine storm causing immune pathological damage may be a real “killer” in critically ill patients. At the same time, human autopsy and animal models studies have provided some evidence for the pathogenic mechanism of inflammatory cytokines, derived from IMM and neutrophils. However, current studies are limited, and detailed molecular biology principles and broader epidemiology are lacking. Therefore, future studies should not only focus on the identification of specific inflammatory response signaling pathways, in patients and animals infected with hCoV but also include the scientific and effective application for controlling the spread of the virus worldwide.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by Science and Technology Plan Project of Gansu (grant no. 18YF1FA108), Lanzhou University Second Hospital Cuiying Fund Project (grant no. CY2018-MS10), and National Natural Science Foundation of China (grant no. 81560343).
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. New Engl. J. Med. 2020;8:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization, Coronavirus disease (COVID-2019) situation reports, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/, 2020 (accessed 15 May 2020).
- 3.National Health Commission of the People’s Republic of China, Outbreak report, http://www.nhc.gov.cn/xcs/yqtb/202005/d2059dd74f8e4e469c1ad3fb6cf0e3af.shtml/, 2020 (accessed 15 May 2020).
- 4.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;2 doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020 doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Gayle A.A., Wilder-Smith A., Rocklov J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan. China JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., Law K.I., Tang B.S., Hon T.Y., Chan C.S., Chan K.H., Ng J.S., Zheng B.J., Ng W.L., Lai R.W., Guan Y., Yuen K.Y. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;9371:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassar M.S., Bakhrebah M.A., Meo S.A., Alsuabeyl M.S., Zaher W.A. Middle east respiratory syndrome coronavirus (MERS-CoV) infection: epidemiology, pathogenesis and clinical characteristics. Eur. Rev. Med. Pharmacol. Sci. 2018;15:4956–4961. doi: 10.26355/eurrev_201808_15635. [DOI] [PubMed] [Google Scholar]
- 10.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;10223:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan P.K.S., Chan M.C.W. Tracing the SARS-coronavirus. J. Thoracic Dis. 2013:S118–S121. doi: 10.3978/j.issn.2072-1439.2013.06.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Wilde A.H., Snijder E.J., Kikkert M., Van Hemert M.J. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;1–42 doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virology J. 2019;1:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;193–292 doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1–23 doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;3:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;2 doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;4:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Tao, Fan Yongzhen, Chen Ming, Wu Xiaoyan, Zhang Lin, He Tao, Wang Hairong, Wan Jing, Wang Xinghuan, Lu Zhibing. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiology. 2020 doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Zunyou, McGoogan Jennifer M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 23.Du Y., Tu L., Zhu P., Mu M., Wang R., Yang P., Wang X., Hu C., Ping R., Hu P., Li T., Cao F., Chang C., Hu Q., Jin Y., Xu G. Clinical features of 85 fatal cases of COVID-19 from Wuhan: A retrospective observational study. Am. J. Respir. Crit. Care Med. 2020 doi: 10.1164/rccm.202003-0543OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Dawei, Hu Bo, Hu Chang, Zhu Fangfang, Liu Xing, Zhang Jing, Wang Binbin, Xiang Hui, Cheng Zhenshun, Xiong Yong, Zhao Yan, Li Yirong, Wang Xinghuan, Peng Zhiyong. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng Y.-J., Wei Z.-Y., Qian H.-Y., Huang J., Lodato R., Castriotta R.J. Pathophysiological characteristics and therapeutic approaches for pulmonary injury and cardiovascular complications of coronavirus disease 2019, Cardiovascular pathology : the official journal of the Society for. Cardiovasc. Pathol. 2020 doi: 10.1016/j.carpath.2020.107228. 107228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C., Zhang Z., Wang L., Peng L., Chen L., Qin Y., Zhao D., Tan S., Yin L., Xu J., Zhou C., Jiang C., Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury, Science China. Life Sci. 2020 doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y., Tu M., Wang S., Chen S., Zhou W., Chen D., Zhou L., Wang M., Zhao Y., Zeng W., Huang Q., Xu H., Liu Z., Guo L. Clinical characteristics of laboratory confirmed positive cases of SARS-CoV-2 infection in Wuhan, China: A retrospective single center analysis. Travel Med. Infect. Dis. 2020;101606 doi: 10.1016/j.tmaid.2020.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., Akdis C.A., Gao Y.D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020 doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 29.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S.Y. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;3:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thrombosis Haemostasis : JTH. 2020;4:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y., Wang H., Shen H., Li Z., Geng J., Han H., Cai J., Li X., Kang W., Weng D., Lu Y., Wu D., He L., Yao K. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 2003;3:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., Trilling M., Lu M., Dittmer U., Yang D. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020 doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian S., Hu W., Niu L., Liu H., Xu H., Xiao S.Y. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 2020 doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Zhe, Shi Lei, Wang Yijin, Zhang Jiyuan, Huang Lei, Zhang Chao, Liu Shuhong, Zhao Peng, Liu Hongxia, Zhu Li, Tai Yanhong, Bai Changqing, Gao Tingting, Song Jinwen, Xia Peng, Dong Jinghui, Zhao Jingmin, Wang Fu-Sheng. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet Respiratory Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su H., Yang M., Wan C., Yi L.-X., Tang F., Zhu H.-Y., Yi F., Yang H.-C., Fogo A.B., Nie X., Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020 doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batlle D., Soler M.J., Sparks M.A., Hiremath S., South A.M., Welling P.A., Swaminathan S. Acute kidney injury in COVID- 19: emerging evidence of a distinct pathophysiology. J. Am. Soc. Nephrology: JASN. 2020 doi: 10.1681/ASN.2020040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L., Liu J., Lu M., Yang D., Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int.: Official J. Int. Assoc. Study Liver. 2020;5 doi: 10.1111/liv.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;7164:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 39.Lim K.H., Staudt L.M. Toll-like receptor signaling. Cold Spring Harb Perspect Biol. 2013;1 doi: 10.1101/cshperspect.a011247. a011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meylan E., Tschopp J., Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;7098:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 41.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev.: MMBR. 2012;1:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect. Biol. 2014;10 doi: 10.1101/cshperspect.a016295. a016295–a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka T., Narazaki M., Kishimoto T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy. 2016;8:959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 44.Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., Kong L., Fang X., Zheng H., Wu Z., She Y. Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res. 2007;1–2:1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward S.G., Westwick J. Chemokines: understanding their role in T-lymphocyte biology. Biochem. J. 1998;457–70 doi: 10.1042/bj3330457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;1:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen F., Yu H., Guo J., Li Y., Luo K., Huang S. Identification of the hyper-variable genomic hotspot for the novel coronavirus SARS-CoV-2. J. Infect. 2020 doi: 10.1016/j.jinf.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;5:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 50.Smits S.L., De Lang A., Van Den Brand J.M., Leijten L.M., Van I.W.F., Eijkemans M.J., Van Amerongen G., Kuiken T., Andeweg A.C., Osterhaus A.D., Haagmans B.L. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;2 doi: 10.1371/journal.ppat.1000756. e1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discovery. 2020 doi: 10.1038/s41421-020-0147-1. 11–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.C. Y, G. Y, P. Y, Z.Z.J. Biochemicalb.r. Communications, Structure analysis of the receptor binding of 2019-nCoV, 2020. [DOI] [PMC free article] [PubMed]
- 53.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;8:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.-C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;7047:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C., He J., Zhong X., Gan H., Xia Y. CX3CL1/CX3CR1 axis contributes to angiotensin II-induced vascular smooth muscle cell proliferation and inflammatory cytokine production. Inflammation. 2018;3:824–834. doi: 10.1007/s10753-018-0736-4. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X., Yang J., Yu X., Cheng S., Gan H., Xia Y. Angiotensin II-induced early and late inflammatory responses through NOXs and MAPK pathways. Inflammation. 2017;1:154–165. doi: 10.1007/s10753-016-0464-6. [DOI] [PubMed] [Google Scholar]
- 57.Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. U. S. A. 2008;22:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen I.Y., Moriyama M., Chang M.-F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Frontiers Microbiol. 2019:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen I.C., Scull M.A., Moore C.B., Holl E.K., Mcelvania-Tekippe E., Taxman D.J., Guthrie E.H., Pickles R.J., Ting J.P.Y. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;4:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandes-Alnemri T., Wu J., Yu J.W., Datta P., Miller B., Jankowski W., Rosenberg S., Zhang J., Alnemri E.S. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;9:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi J., Gao W., Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 2017;4:245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 62.Man S.M., Karki R., Kanneganti T.-D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017;1:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fink S.L., Cookson B.T. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell. Microbiol. 2006;11:1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- 64.Pandolfi F., Altamura S., Frosali S., Conti P. Key role of DAMP in inflammation, cancer, and tissue repair. Clin. Therapeutics. 2016;5:1017–1028. doi: 10.1016/j.clinthera.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 65.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;8:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;2:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;9:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siu K.-L., Chan C.-P., Kok K.-H., Chiu-Yat Woo P., Jin D.-Y. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 2014;2:141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;5:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L., Zhang F., Yu W., He T., Yu J., Yi C.E., Ba L., Li W., Farzan M., Chen Z., Yuen K.Y., Ho D. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J. Med. Virol. 2006;1:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., Wu T., Cheung K.W., Chan K.H., Alvarez X., Qin C., Lackner A., Perlman S., Yuen K.Y., Chen Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virologica Sinica. 2020 doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tetro J.A. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;S1286–4579(20):30034–30044. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Auyeung T.W., Lee J.S.W., Lai W.K., Choi C.H., Lee H.K., Lee J.S., Li P.C., Lok K.H., Ng Y.Y., Wong W.M., Yeung Y.M. The use of corticosteroid as treatment in SARS was associated with adverse outcomes: a retrospective cohort study. J. Infection. 2005;2:98–102. doi: 10.1016/j.jinf.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discovery. 2016;5:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blazek K., Eames H.L., Weiss M., Byrne A.J., Perocheau D., Pease J.E., Doyle S., Mccann F., Williams R.O., Udalova I.A. IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J. Exp. Med. 2015;6:845–853. doi: 10.1084/jem.20140995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D., Behrens P., Böddinghaus B., Götsch U., Naujoks F., Neumann P., Schork J., Tiarks-Jungk P., Walczok A., Eickmann M., Vehreschild M.J.G.T., Kann G., Wolf T., Gottschalk R., Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. The New Engl. J. Med. 2020;13:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;3:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sheahan T.P., Sims A.C., Leist S.R., Schafer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;1:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;1:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 81.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., TanD W., Liu In. Vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis.: An Official Publ. Infect. Dis. Soc. Am. 2020 doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;105949 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. The New Engl. J. Med. 2020;19:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infection. 2020 doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Y. Furuta, T. Komeno, T. Nakamura, Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase, Proceedings of the Japan Academy. Series B, Physical and Biological Sciences 7 (2017) 449–463. [DOI] [PMC free article] [PubMed]
- 86.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. Experimental treatment with favipiravir for COVID-19: An open-label control study. Engineering (Beijing China) 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Q., Wang Y., Qi C., Shen L., Li J. Clinical trial analysis of 2019-nCoV therapy registered in China. J. Med. Virol. 2020 doi: 10.1002/jmv.25733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu K., Cai H., Shen Y., Ni Q., Chen Y., Hu S., Li J., Wang H., Yu L., Huang H., Qiu Y., Wei G., Fang Q., Zhou J., Sheng J., Liang T., Li L. Management of corona virus disease-19 (COVID- 19): the Zhejiang experience, Zhejiang da xue xue bao. Yi xue ban = J. Zhejiang Univ. Med. Sci. 2020;1 doi: 10.3785/j.issn.1008-9292.2020.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., Van Buskirk C., Grossman B.J., Joyner M., Henderson J.P., Pekosz A., Lau B., Wesolowski A., Katz L., Shan H., Auwaerter P.G., Thomas D., Sullivan D.J., Paneth N., Gehrie E., Spitalnik S., Hod E., Pollack L., Nicholson W.T., Pirofski L.-A., Bailey J.A., Tobian A.A. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Investig. 2020 doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fleming A.B., Raabe V. Current studies of convalescent plasma therapy for COVID-19 may underestimate risk of antibody-dependent enhancement. J. Clin. Virology: The Official Publ. Pan Am. Soc. Clin. Virology. 2020 doi: 10.1016/j.jcv.2020.104388. 104388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. JAMA. 2020 doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ashour H.M., Elkhatib W.F., Rahman M.M., Elshabrawy H.A. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens (Basel, Switzerland) 2020;3:E186. doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seif F., Aazami H., Khoshmirsafa M., Kamali M., Mohsenzadegan M., Pornour M., Mansouri D. JAK inhibition as a new treatment strategy for patients with COVID-19. Int. Arch. Allergy Immunol. 2020;1–9 doi: 10.1159/000508247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wurfel M.M., Gordon A.C., Holden T.D., Radella F., Strout J., Kajikawa O., Ruzinski J.T., Rona G., Black R.A., Stratton S., Jarvik G.P., Hajjar A.M., Nickerson D.A., Rieder M., Sevransky J., Maloney J.P., Moss M., Martin G., Shanholtz C., Garcia J.G., Gao L., Brower R., Barnes K.C., Walley K.R., Russell J.A., Martin T.R. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am. J. Respir. Crit. Care Med. 2008;7:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]