Abstract
ATP13A2 is the autosomal recessive causative gene for juvenile-onset Parkinson’s disease (PARK9, Parkinson’s disease 9), also known as Kufor-Rakeb syndrome. The disease is characterized by levodopa-responsive Parkinsonism, supranuclear gaze palsy, spasticity, and dementia. Previously, we have reported that Atp13a2 deficient medaka fish showed dopaminergic neurodegeneration and lysosomal dysfunction, indicating that lysosome-autophagy impairment might be one of the key pathogeneses of Parkinson’s disease. Here, we established Atp13a2 deficient zebrafish using CRISPR/Cas9 gene editing. We found that the number of TH + neurons in the posterior tuberculum and the locus coeruleus significantly reduced (dopaminergic neurons, 64 % at 4 months and 37 % at 12 months, p < 0.001 and p < 0.05, respectively; norepinephrine neurons, 52 % at 4 months and 40 % at 12 months, p < 0.001 and p < 0.05, respectively) in Atp13a2 deficient zebrafish, proving the degeneration of dopaminergic neurons. In addition, we found the reduction (60 %, p < 0.05) of cathepsin D protein expression in Atp13a2 deficient zebrafish using immunoblot. Transmission electron microscopy analysis using middle diencephalon samples from Atp13a2 deficient zebrafish showed lysosome-like bodies with vesicle accumulation and fingerprint-like structures, suggesting lysosomal dysfunction. Furthermore, a significant reduction (p < 0.001) in protein expression annotated with vesicle fusion with Golgi apparatus in Atp13a2 deficient zebrafish by liquid-chromatography tandem mass spectrometry suggested intracellular trafficking impairment. Therefore, we concluded that Atp13a2 deficient zebrafish exhibited degeneration of dopaminergic neurons, lysosomal dysfunction and the possibility of intracellular trafficking impairment, which would be the key pathogenic mechanism underlying Parkinson’s disease.
Keywords: Parkinson’s disease, ATP13A2, Zebrafish, Trafficking impairment
1. Introduction
Parkinson’s disease (PD) is one of the most common neurodegenerative movement disorders characterized by the loss of dopaminergic neurons and Lewy bodies (Kalia and Lang, 2015). The pathologic mechanisms causing the neurodegeneration in PD remain unclear. Although most cases of PD are sporadic, familial cases were also found in approximately 5–10 % of PD patients (Abeliovich and Gitler, 2016). In recent years, an increasing number of genes has been reported as the key risk factor in PD development (Kalia and Lang, 2015).
ATP13A2 is the recessive causative gene for juvenile-onset PD (PARK9, Parkinson’s disease 9), also known as Kufor-Rakeb syndrome, characterized by levodopa-responsive Parkinsonism, supranuclear gaze palsy, spasticity, and dementia (Najim al-Din et al., 1994; Williams et al., 2005). ATP13A2 is mapped on chromosome 1p36 and contains 29 coding exons encoding a lysosomal type 5 ATPase (Schultheis et al., 2004; Ramirez et al., 2006). ATP13A2 protein localizes in intracellular vesicular compartments including endosomes and lysosomes in neurons (Tan et al., 2011; Podhajska et al., 2012; Matsui et al., 2013a). Although ATP13A2 has been considered a regulator for the lysosome-autophagy pathway (Bento et al., 2016), the molecular function of ATP13A2, and how ATP13A2 contributes to the pathogenesis of PD, remain unclear.
Previously, we have reported that Atp13a2 deficient medaka fish showed dopaminergic neurodegeneration and lysosomal dysfunction specific to cathepsin D (Matsui et al., 2013a). These findings indicated that lysosome-autophagy impairment might lead to dopaminergic neuronal death and might be one of the key pathogeneses of PD. However, the underlying mechanism remains unknown. Here, we established and evaluated Atp13a2 deficient zebrafish, and verified the degeneration of dopaminergic neurons, reduction of cathepsin D protein expression and histological abnormalities of lysosome as previously shown with the medaka fish. Furthermore, we found that the protein expression associated with the vesicle fusion significantly reduced in mutant zebrafish, indicating the possibility that intracellular trafficking impairment might occur in Atp13a2 deficient zebrafish, resulting in neurodegeneration.
2. Materials and methods
2.1. Maintenance of zebrafish
Zebrafish (AB) were raised and maintained under a 14-h light/10-h dark cycle at 28 °C according to standard protocols (M, W., 2000; Kimmel et al., 1995). Starting 5 days post-fertilization, fish were fed brine shrimp at 9:00 a.m. and powdered feed (Kyorin, Himeji, Japan) at 12:00 p.m. (Matsui and Sugie, 2017). Only male fish were used in this study.
2.2. Microinjection and gene editing
Glass capillaries (GD-1; Narishige, Tokyo, Japan) were pulled into microinjection needles by using a vertical needle puller (PC-10; Narishige). These needles were used in an IM-31 microinjector (Narishige) equipped with a YOU-1 micromanipulator (Narishige). To generate Atp13a2 deficient zebrafish, guide RNA (target sequence: GGTCTTGGATCCTTTATGAGGGG, 25 ng/μl) and Cas9 protein (0.6 μg/μl; New England Biolabs, Ipswich, MA) were mixed with phenol red (2%) and co-injected into one-cell stage fish embryos according to previous reports (Hwang et al., 2013; Jinek et al., 2012). The F1 generation and subsequent generations were genotyped using PCR (forward primer: ACCAAACGGGAGTGATGTGT, reverse primer: ACACCCATCTGTACCCCTGA) and direct sequencing (sequencing primer: ACACCCATCTGTACCCCTGA). Heterozygous mutant fish were crossed to obtain homozygous mutant (Atp13a2 deficient) and control fish.
2.3. RT-PCR and real-time PCR of zebrafish atp13a2
atp13a2 mRNA expression levels were evaluated by semi-quantitative RT-PCR and real-time PCR. RNA was extracted from zebrafish brain tissue of mutant and wild type with TRIzol (Life Technologies, Carlsbad, CA). cDNA of each genotype was synthesized from 1 μg template RNA for RT-PCR and 0.5 μg template RNA for real-time PCR using ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs).
RT-PCR was carried out using the following thermocycling program: 95 °C for 120 s; 16, 20, and 24 cycles at 98 °C for 10 s, 52 °C for 30 s, 72 °C for 30 s; and 72 °C for 120 s (GeneAtlas Type G Thermal Cycler; ASTEC, Fukuoka, Japan). Primers used for this reaction were as follows: zebrafish beta-actin-specific primers (forward: CGTGACATCAAGGAGAAGCT, reverse: ATCCACATCTGCTGGAAGGT), zebrafish atp13a2-specific primers (forward: TGGATGCTAAAGGGAACTGTG, reverse: GCAAAGTCCAGACTGCCTTC).
Real-time PCR was carried out using the following thermocycling program: 95 °C for 30 s; 40 cycles at 95 °C for 5 s, 60 °C for 30 s; and 95 °C for 15 s, 60 °C for 30 s, 95 °C for 15 s (Thermal Cycler Dice Real Time System Lite; TaKaRa Bio, Kusatsu, Japan). Primers used for this reaction were as follows: zebrafish beta-actin-specific primers (forward: CGAGCAGGAGATGGGAACC, reverse: CAACGGAAACGCTCATTGC) (McCurley and Callard, 2008), zebrafish atp13a2-specific primers at exon 2 and 3 (forward: GACACTGAACCCCTCATAAAGG, reverse: AGGCCGACAGAAAATACTGC), zebrafish atp13a2-specific primers at exon 17 and 18 (forward: ACAGAGTGCTTGCAGTGATG, reverse: ACACTCATCCTCTGCAGAGAAG).
2.4. Immunostaining of TH + neurons
Immunostaining and counting of the TH + neurons were performed according to previously reported method (Matsui and Sugie, 2017). Adult fish at 4 and 12 months were sacrificed using circulating water of the breeding system containing 0.1 % Tricaine. Brains were removed, explanted, and fixed with 4% paraformaldehyde at 4 °C overnight. Subsequently, specimens were embedded in 2% low-melting agarose and 200 μm axial sections were collected using a PRO7 microslicer (Dosaka EM, Kyoto, Japan). Floating slices were incubated in 10 mM sodium citrate buffer, pH 8.5, at 80 °C for 120 min. After washing with PBS/1% TritonX-100, the sections were blocked with 2% BSA in PBS/1% TritonX-100 buffer for 30 min. These pretreated sections were incubated overnight at 4 °C with a rabbit anti-TH antibody (1:500, AB152; EMD-Millipore, Billerica, MA). After washing with PBS/1% TritonX-100 buffer, the sections were incubated overnight at 4 °C with anti-rabbit IgG conjugated with Alexa Fluor 594 (Life Technologies). The following day, sections were washed in PBS/1% TritonX-100 buffer and analyzed using the A1R + confocal microscope (Nikon, Tokyo, Japan). Five fish at each age were used for counting TH + neurons in the posterior tuberculum and the locus coeruleus.
2.5. Immunoblot analysis
A brain specimen was lysed in RIPA buffer containing 50 mM Tris−HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.1 % SDS, 0.5 % sodium deoxycholate. The samples were separated by SDS-PAGE, and proteins were transferred to a pre-hydrophilized PVDF membrane (EMD Millipore, Billerica, MA, USA). The blotted membrane was immersed in 2% BSA at RT for 30 min. The membrane was then incubated with the appropriate primary antibody (mouse anti-beta-actin antibody, AC-15, Sigma-Aldrich, St. Louis, USA; mouse anti-cathepsin D, BD Biosciences, San Jose, CA), followed by incubation with a horseradish peroxidase (HRP)-conjugated anti-mouse IgG (GE Healthcare, Chicago, IL, USA). Immunoreactive proteins were visualized using Western BLoT Quant HRP Substrate (Takara Bio) and MultiImager II ChemiBox (BioTools, Gunma, Japan). Six fish at 12 months were used for detection of cathepsin D. Densitometry analysis was performed to evaluate the cathepsin D protein expression.
2.6. Transmission electron microscopy
Zebrafish brains at 37 months were fixed with 2 % paraformaldehyde (PFA) and 2 % glutaraldehyde (GA) in 0.1 M cacodylate buffer pH 7.4 at 4 °C overnight. After this fixation the samples were washed 3 times with 0.1 M cacodylate buffer for 30 min each, and were postfixed with 2 % osmium tetroxide (OsO4) in 0.1 M cacodylate buffer at 4 °C for 3 h. The samples were dehydrated in graded ethanol solutions (50 %, 70 %, 90 %, 100 %). The schedule was as follows: 50 % and 70 % for 30 min each at 4 °C, 90 % for 30 min at room temperature, and 3 changes of 100 % for 30 min each at room temperature. After these dehydration processes, the samples were continuously dehydrated in 100 % ethanol at room temperature overnight. The samples were infiltrated with propylene oxide (PO) 2 times for 30 min each and were put into a 70:30 mixture of PO and resin (Quetol-812; Nisshin EM Co., Tokyo, Japan) for 1 h, then they kept the cap of tube open and PO was volatilized overnight. The samples were transferred to a fresh 100 % resin, and were polymerized at 60 °C for 48 h. The polymerized resins were ultra-thin sectioned at 70 nm with a diamond knife using an ultramicrotome (Ultracut UCT; Leica, Vienna, Austria) and the sections were mounted on copper grids. They were stained with 2 % uranyl acetate at room temperature for 15 min, and then were washed with distilled water followed by being secondary-stained with Lead stain solution (Sigma-Aldrich Co., Tokyo, Japan) at room temperature for 3 min. The grids were observed by transmission electron microscope (JEM-1400Plus; JEOL Ltd., Tokyo, Japan) at an acceleration voltage of 100 kV. Digital images were taken with a CCD camera (EM-14830RUBY2; JEOL Ltd., Tokyo, Japan).
2.7. Extraction and digestion of zebrafish brain proteins
Zebrafish brains isolated from two fish of each of wild type and mutant at 28 months were homogenized in 200 μl ice-chilled buffer comprised of T-PER™ Tissue Protein Extraction Reagent (Thermo Fisher Scientific, Massachusetts, USA) and protease inhibitor. Homogenates were centrifuged at 10,000 g for 5 min, and the BCA assay kit (Thermo Fisher Scientific) was used to measure the samples in each supernatant. Extracted proteins were precipitated with cold acetone and resuspended in 8 M Urea/30 mM ammonium bicarbonate. The proteins were then subjected to reductive alkylation with dithiothreitol and iodoacetamide, and trypsinized overnight. The tryptic digestions were purified using C-18 spin column (Thermo Fisher Scientific) and resuspended in 0.1 % formic acid before mass spectrometry analysis.
2.8. Liquid-chromatography tandem mass spectrometry
Protein digests were separated using Nano-LC-Ultra 2D-plus equipped with cHiPLC Nanoflex (Eksigent, Dublin, CA, USA) in trap-and-elute mode, with trap column (200 μm x0.5 mm ChromXP C18-CL 3 μm 120 Å, Eksigent) and analytical column (75 μm x15 cm ChromXP C18-CL 3 μm 120 Å, Eksigent). The separation was carried out with a binary gradient, in which solvent A (0.1 % formic acid/water) and solvent B (0.1 % formic acid/acetonitrile) were used. The gradient program was 2%–33.2% solvent B for 250 min, 33.2%–98% solvent B in 2 min, 98 % solvent B for 5 min, 98 % to 2% solvent B in 0.1 min, and 2% solvent B for 17.9 min, at 300 nL/min. The eluates were infused on-line into the mass spectrometer (TripleTOF 5600+ System with NanoSpray III source and heated interface, SCIEX, Framingham, MA, USA) and ionized in an electrospray ionization-positive mode. Datasets were acquired using an information-dependent acquisition method.
2.9. Protein identification and relative quantification
Acquired datasets were analyzed by ProteinPilot software version 5.0.1 (SCIEX) with the Danio rerio protein database, which was extracted from the NCBI protein library (September 2019) and appended with known common contaminant sequences (SCIEX). Quality of database search was confirmed by the false discovery rate analysis, in which reversed amino acid sequences were used as decoy. Peptide identifications were evaluated by the confidence values that were calculated by ProteinPilot software. Relative abundances of the identified proteins were estimated by Progenesis QI for Proteomics software version 4.1 (Nonlinear Dynamics, Newcastle upon Tyne, UK). All raw data files with wiff format (SCIEX) were imported to generate aggregate and peptide identification results by ProteinPilot, with at least 95 % confidence for assignment. Label-free quantification of proteins were performed by relative quantitation using Hi-N(3) method (Nonlinear Dynamics) (Silva et al., 2006).
2.10. Statistical analysis
Data are expressed as mean ± standard error of the mean (SEM). Densitometry analysis for cathepsin D protein expression was performed using ImageJ 1.43 (NIH, Bethesda, MD) on immunoblot. An analysis of variance (ANOVA) was used to assess the statistical significance of the data. Student's t-test was used to test for statistical significance on the counting of TH + neurons. Value of p < 0.05 was considered statistically significant.
2.11. Gene ontology enrichment analysis
To perform Gene Ontology (GO) enrichment analysis, genes were considered differentially expressed when fold change was higher than 1.5. We conducted GO analysis using “topGO” package (Alexa and Rahnenfuhrer, 2018) with the gene annotation database of D. rerio (v3.8.2) (Carlson, 2019). Fisher’s exact tests were performed to compare the frequency of GO terms in differentially expressed genes with the background frequency.
3. Results
3.1. Generation of an Atp13a2 deficient zebrafish
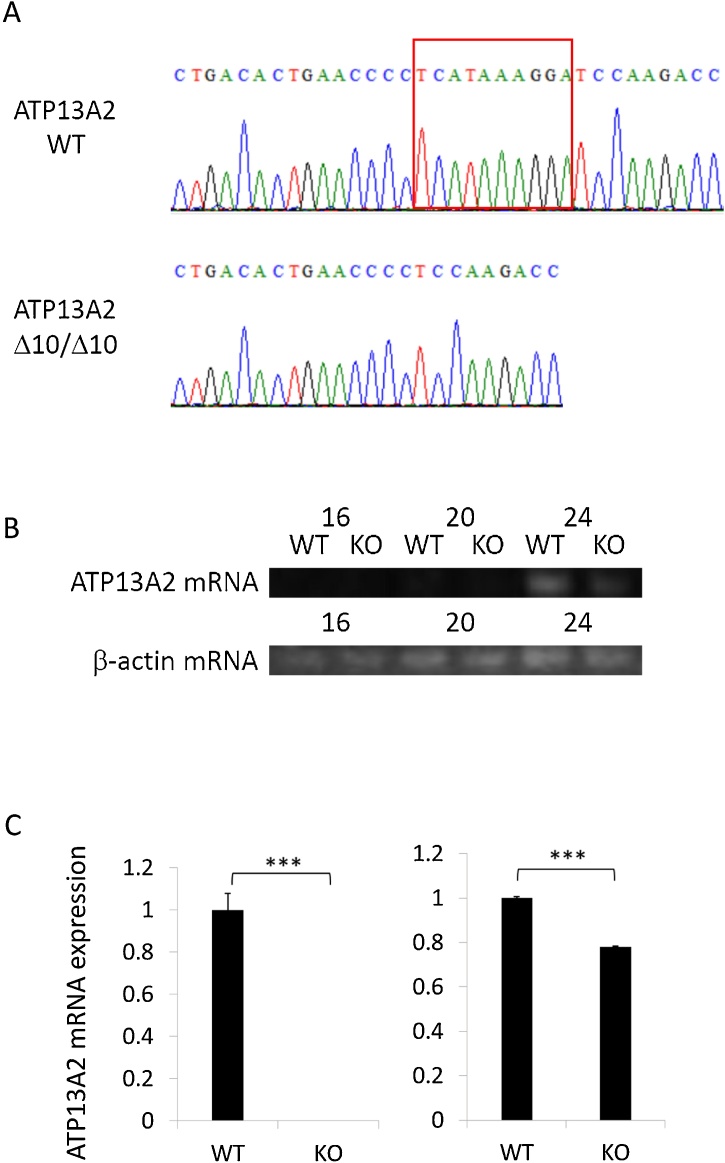
The Atp13a2 deficient zebrafish was generated by introducing a 10 bp deletion in the exon 2 of atp13a2 using the CRISPR/Cas9 gene editing (Fig. 1A). Semi-quantitative RT-PCR at 16, 20, and 24 cycles and real-time PCR showed that mRNA of atp13a2 had decreased in mutant zebrafish (Fig. 1B and 1C). Two types of primers were designed at different positions on the mRNA transcript for real-time PCR. The mRNA containing the deletion site from exon 2 to exon 3 was completely lost (p < 0.001) in mutant zebrafish (Fig. 1C, left), and the mRNA from exon 17 to exon 18 was significantly reduced (22 %, p < 0.001) in mutant zebrafish (Fig. 1C, right). Beta-actin was used as reference to normalize the gene expression. From these results, we confirmed that we have succeeded in generating Atp13a2 deficient zebrafish.
Fig. 1.
Generation of Atp13a2 deficient zebrafish. (A) Sequence data for each genotype. A = green, T = red, G = black, and C = blue. The red square indicates the deletion site. (B) Semi-quantitative RT-PCR at 16, 20, and 24 cycles of atp13a2 mRNA for each genotype. (C) Real-time PCR of atp13a2 mRNA. Two types of primers were designed at different positions on the mRNA transcript. The left is the mRNA containing the deletion sites from exon 2 to exon 3, and the right is the mRNA from exon 17 to exon 18. n = 3 for each genotype. ***p < 0.001. WT indicates wild type and KO indicates Atp13a2 deficient zebrafish (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.2. Atp13a2 deficiency induces degeneration of dopaminergic neurons, cathepsin D deficiency and histological abnormalities of lysosome
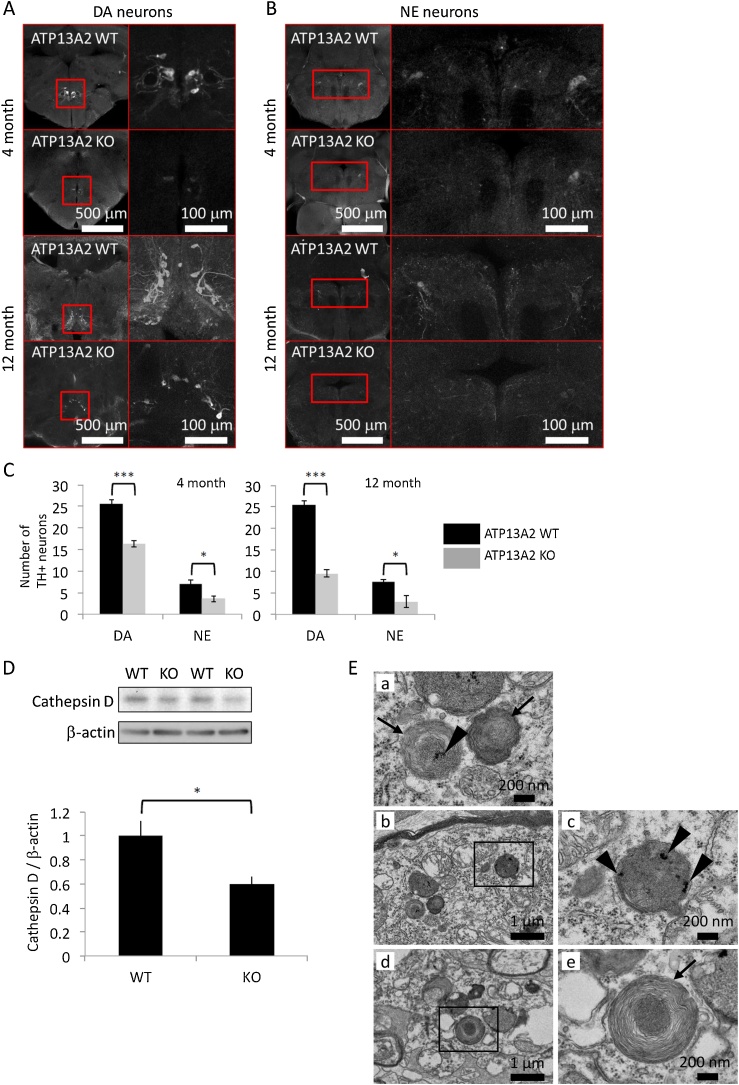
We histologically examined the brain tissue of Atp13a2 deficient zebrafish. We immunostained TH + neurons and counted the number of TH + neurons at 4 and 12 months according to previously reported method (Matsui and Sugie, 2017). The number of TH + neurons in the posterior tuberculum and the locus coeruleus significantly reduced in the mutants at both 4 and 12 months: dopaminergic neurons, 64 % at 4 months and 37 % at 12 months, p < 0.001 and p < 0.05, respectively; norepinephrine neurons, 52 % at 4 months and 40 % at 12 months, p < 0.001 and p < 0.05, respectively (Fig. 2A, 2B and 2C). These findings indicated that Atp13a2 deficient zebrafish showed degeneration of dopaminergic neurons, which was consistent with the pathogenesis of PD.
Fig. 2.
Neuropathology of Atp13a2 deficient zebrafish. (A) Axial sections of the posterior tuberculum containing dopaminergic (DA) neurons of zebrafish brain at 4 and 12 months. Right figures in each section are the enlarged images of the left figures. (B) Axial sections of the locus coeruleus containing norepinephrine (NE) neurons of zebrafish brain at 4 and 12 months. Right figures in each section are enlarged images of the left figures. (C) The number of TH + neurons in the posterior tuberculum and the locus coeruleus. (D) The amount of cathepsin D protein in zebrafish brain. WT indicates wild type and KO indicates Atp13a2 deficient zebrafish. (E) Transmission electron microscopy analysis of the middle diencephalon samples from Atp13a2 deficient zebrafish. Atp13a2 deficiency induces lysosome-like bodies that contain granular deposits (arrowheads) and fingerprint-like structures (arrows). Right figures in each section are the enlarged images of the left figures. Scale bars are indicated in A, B and E. *p < 0.05 and ***p < 0.001.
Next, we measured the amount of cathepsin D protein in zebrafish brain tissue using immunoblot, as we have previously reported that Atp13a2 deficient medaka fish exhibited cathepsin D deficiency. Results confirmed that the amount of cathepsin D protein had reduced (60 %, p < 0.05) in Atp13a2 deficient zebrafish (Fig. 2D).
Next, we performed transmission electron microscopy analysis using middle diencephalon samples from Atp13a2 deficient zebrafish to further investigate the pathological effects of Atp13a2 deficiency. Lysosome-like bodies with vesicle accumulation and fingerprint-like structures were observed in the sections (Fig. 2E), which indicated the lysosomal dysfunction.
3.3. Impaired intracellular trafficking were suggested in Atp13a2 deficient zebrafish
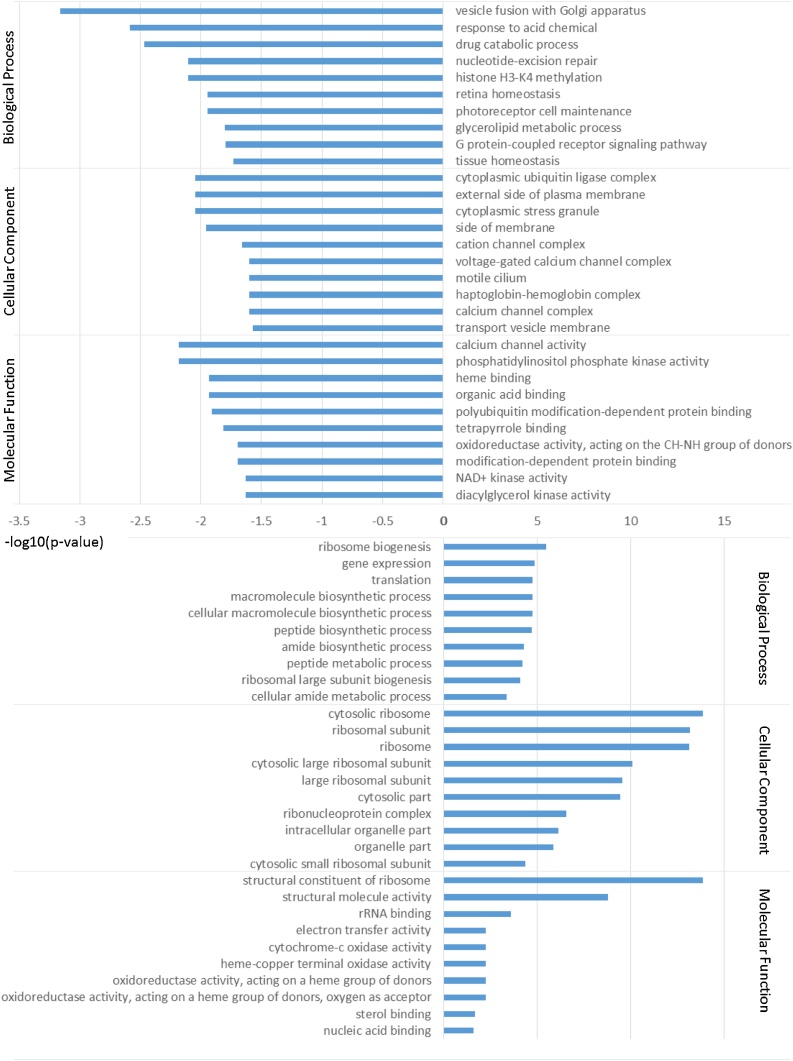
Next, we performed liquid-chromatography tandem mass spectrometry analysis of zebrafish brain tissue samples to elucidate the mechanism of cathepsin D reduction and the degeneration of dopaminergic neurons in Atp13a2 deficient zebrafish. The results of GO analysis were shown in Fig. 3, and the source data of the protein expression changes are presented in Supplementary Table 1. Interestingly, Atp13a2 deficient zebrafish exhibited a significant reduction in protein expression annotated with vesicle fusion of Golgi apparatus (p < 0.001), indicating the possibility of intracellular vesicle trafficking dysfunction. On the other hand, ribosome-associated protein expression increased in Atp13a2 deficient zebrafish, which might suggest the compensation of abnormal protein distribution due to trafficking dysfunction.
Fig. 3.
GO enrichment analysis of zebrafish brain. Protein expression levels in Atp13a2 deficient zebrafish are shown in logarithm compared to wild type. Top 10 GO terms are shown for each category.
From these results, we concluded that Atp13a2 deficient zebrafish exhibited degeneration of dopaminergic neurons and the possibility of intracellular trafficking impairment, which would be the key pathogenic mechanism of PD.
4. Discussion
Previous studies have reported that intracellular trafficking impairment could lead to PD development. Genes such as SNCA, LRRK2, VPS35, DNAJC13, and SYT11, play distinctive roles on various steps of intracellular trafficking such as endocytosis and endosome-lysosome pathways (Abeliovich and Gitler, 2016; Bento et al., 2016; Hunn et al., 2015; Sheehan and Yue, 2019).
We have previously reported that mutations in atp13a2 led to lysosomal dysfunction and abnormal lysosome-related structures in medaka fish. Here, we have shown that not only the reduction of cathepsin D and histological abnormalities of lysosome corroborated previous findings using medaka fish and mice (Matsui et al., 2013a; Kett et al., 2015), but also the possibility of trafficking impairment in Atp13a2 deficient zebrafish by comparing the protein expression with wild type zebrafish.
In this article, we hypothesized that ATP13A2 deficiency might cause trafficking abnormality. Cathepsin D reduction and abnormal lysosomal structures in mutants of both medaka fish (Matsui et al., 2013a) and zebrafish may be due to trafficking abnormalities. ATP13A2 is usually localized in the lysosomal membrane, but a previous study showed that the mutant Atp13a2 localized in the endoplasmic reticulum (Matsui et al., 2013a), which might also be due to trafficking impairment. Bento et al. reported that ATP13A2 interacts with SYT11, which is involved in vesicular fusion, trafficking, and exocytosis (Bento et al., 2016). Although there was no significant change in the protein expression level of SYT11 in our tandem mass spectrometry analysis using zebrafish brain, ATP13A2 dysfunction may have caused the malfunction of SYT11, resulting in trafficking abnormalities. We used cathepsin D as a marker to evaluate lysosome function in this study. Cathepsin D is known to be deeply involved in the neurodegeneration of neuronal ceroid lipofuscinosis, severe neurodegenerative lysosomal storage disease, and a decrease in cathepsin D has been confirmed in the previous report of Atp13a2 deficient medaka fish (Matsui et al., 2013a), so in studies focusing on neurodegeneration, it would be meaningful to assess cathepsin D as a marker of lysosomal dysfunction. However, if abnormality in trafficking is the major pathogenic factor of PARK9, various molecules other than cathepsin D may be affected, contributing to neurodegeneration, as the source data of various protein expression changes in liquid-chromatography tandem mass spectrometry analysis are shown in Supplementary Table 1. Further exploration of affected molecules may contribute to research progress on PD.
In this study, we evaluated the neurodegeneration of Atp13a2 deficient zebrafish by performing the histological analyses of dopaminergic neurons by immunostaining and counting the TH + neurons, and were able to obtain results consistent with the pathology of Parkinson's disease. However, since the human ATP13A2 gene is expressed throughout the brain, the effects of the trafficking abnormalities that may be caused by ATP13A2 deficiency may not be limited to TH + neurons alone. In addition, as liquid-chromatography tandem mass spectrometry analysis was performed using whole brain homogenates from zebrafish, our results may not clearly reflect the degeneration of TH + neurons. Further investigation is necessary to elucidate the detailed mechanism of whether neuronal degeneration due to the potential intracellular trafficking abnormality is specific to TH + neurons in Atp13a2 deficient zebrafish and how it is associated with the pathology of PARK9. In addition, in order to confirm whether the trafficking abnormality actually causes neurodegeneration, additional experiment is required to restore or promote intracellular trafficking with Atp13a2 deficient zebrafish and examine whether neurodegeneration recovers. This is a critical challenge for the future.
Although neurodegeneration such as dopaminergic neuron loss occurred at 4 months, we performed liquid-chromatography tandem mass spectrometry analysis using 28-month-old fish and transmission electron microscopy using 37-month-old fish. This is because Parkinson's disease is a progressive degenerative disease, and it was thought that analysis at an older age would yield results that strongly reflected the pathological condition. As future challenges, it would be worth investigating the changes over time with growth of zebrafish.
Accumulation of inclusion bodies and fingerprint-like structures were observed in Atp13a2 deficient zebrafish in transmission electron microscopy analysis. These abnormal structures have been previously reported in Atp13a2 knockdown cells, Atp13a2 deficient medaka fish, and cathepsin d-deficient mice, and are considered to indicate lysosomal dysfunction (Matsui et al., 2013a; Koike et al., 2000). Fingerprint-like structures are also known to be observed in neuronal ceroid lipofuscinosis (NCL), severe neurodegenerative lysosomal storage diseases (Anderson et al., 2013). Although the detailed pathogenesis of NCL has not yet been elucidated, it may have some overlap with the pathogenesis of PARK9 due to similar histological changes. The detailed mechanism of whether lysosomal dysfunction causes trafficking abnormalities or whether it causes lysosomal dysfunction in Atp13a2 deficient zebrafish remains unclear.
Lewy bodies are aggregates of alpha-synuclein, which is usually degraded by lysosome-autophagy pathway. When lysosomal function is impaired, alpha-synuclein is not degraded, and this could lead to neurodegeneration. Some reports have shown synuclein accumulation in cell models with ATP13A2 deficiency, but there were contradicting reports (Matsui et al., 2013a), in which animal models including medaka fish and mice have shown no synuclein accumulation (Matsui et al., 2013a; Kett et al., 2015). As zebrafish do not have endogenous synuclein, we could not verify synuclein accumulation in the zebrafish model. Further studies in understanding the molecular mechanism related to synuclein accumulation, trafficking abnormalities, and neurodegeneration are necessary.
Recently, zebrafish and medaka fish have been used as animal models for PD. While each fish has its own advantages, there are more researchers who use zebrafish than those who use medaka, and research resources are more abundant in dealing with zebrafish (Matsui et al., 2012). Moreover, we found that medaka fish was less likely to develop the PD phenotype than zebrafish when comparing the previous reports of Pink1 deficient zebrafish and medaka fish (Matsui and Sugie, 2017; Matsui et al., 2013b). Similarly, in Atp13a2 deficient small fish, zebrafish showed more marked degeneration of dopaminergic neurons described in this article than medaka fish (Matsui et al., 2013a).
In summary, we confirmed the degeneration of dopaminergic neurons, the reduction of cathepsin D protein expression and histological abnormalities of lysosome in Atp13a2 deficient zebrafish and described the possibility that these could be caused by trafficking abnormalities. This discovery may lead to the development of new treatments for PD targeting trafficking abnormalities.
Ethics statement
All animal experiments were conducted in compliance with the protocol, which was reviewed and approved by the Institutional Animal Care and Use Committee and by the President of Niigata University (Permit Number: #28 Niigata Univ. Res.367−1 and #28 Niigata Univ. Res.483−1).
CRediT authorship contribution statement
Hiromi Nyuzuki: Conceptualization, Investigation, Writing - original draft. Shinji Ito: Investigation, Writing - review & editing. Keisuke Nagasaki: Supervision. Yohei Nitta: Data curation, Writing - review & editing. Noriko Matsui: Investigation. Akihiko Saitoh: Supervision. Hideaki Matsui: Conceptualization, Methodology, Investigation, Project administration.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
This research was supported by AMED under Grant Number JP19gm6110028, JSPS KAKENHI Grant Numbers JP18955907 and Takeda Science Foundation.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ibror.2020.05.002.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Kalia L.V., Lang A.E. Parkinson’s disease. Lancet. 2015;386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- Abeliovich A., Gitler A.D. Defects in trafficking bridge Parkinson’s disease pathology and genetics. Nature. 2016;539(7628):207–216. doi: 10.1038/nature20414. [DOI] [PubMed] [Google Scholar]
- Najim al-Din A.S., Wriekat A., Mubaidin A., Dasouki M., Hiari M. Pallido-pyramidal degeneration, supranuclear upgaze paresis and dementia: kufor-Rakeb syndrome. Acta. Neurol. Scand. 1994;89(5):347–352. doi: 10.1111/j.1600-0404.1994.tb02645.x. [DOI] [PubMed] [Google Scholar]
- Williams D.R., Hadeed A., al-Din A.S., Wreikat A.L., Lees A.J. Kufor Rakeb disease: autosomal recessive, levodopa-responsive parkinsonism with pyramidal degeneration, supranuclear gaze palsy, and dementia. Mov. Disord. 2005;20(10):1264–1271. doi: 10.1002/mds.20511. [DOI] [PubMed] [Google Scholar]
- Schultheis P.J., Hagen T.T., O’Toole K.K., Tachibana A., Burke C.R., McGill D.L., Okunade G.W., Shull G.E. Characterization of the P5 subfamily of P-type transport ATPases in mice. Biochem. Biophys. Res. Commun. 2004;323(3):731–738. doi: 10.1016/j.bbrc.2004.08.156. [DOI] [PubMed] [Google Scholar]
- Ramirez A., Heimbach A., Grundemann J., Stiller B., Hampshire D., Cid L.P., Goebel I., Mubaidin A.F., Wriekat A.L., Roeper J., Al-Din A., Hillmer A.M., Karsak M., Liss B., Woods C.G., Behrens M.I., Kubisch C. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38(10):1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- Tan J., Zhang T., Jiang L., Chi J., Hu D., Pan Q., Wang D., Zhang Z. Regulation of intracellular manganese homeostasis by Kufor-Rakeb syndrome-associated ATP13A2 protein. J. Biol. Chem. 2011;286(34):29654–29662. doi: 10.1074/jbc.M111.233874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podhajska A., Musso A., Trancikova A., Stafa K., Moser R., Sonnay S., Glauser L., Moore D.J. Common pathogenic effects of missense mutations in the P-type ATPase ATP13A2 (PARK9) associated with early-onset parkinsonism. PLoS One. 2012;7(6):e39942. doi: 10.1371/journal.pone.0039942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H., Sato F., Sato S., Koike M., Taruno Y., Saiki S., Funayama M., Ito H., Taniguchi Y., Uemura N., Toyoda A., Sakaki Y., Takeda S., Uchiyama Y., Hattori N., Takahashi R. ATP13A2 deficiency induces a decrease in cathepsin D activity, fingerprint-like inclusion body formation, and selective degeneration of dopaminergic neurons. FEBS Lett. 2013;587(9):1316–1325. doi: 10.1016/j.febslet.2013.02.046. [DOI] [PubMed] [Google Scholar]
- Bento C.F., Ashkenazi A., Jimenez-Sanchez M., Rubinsztein D.C. The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nat. Commun. 2016;7:11803. doi: 10.1038/ncomms11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M, W . 4th edition. 2000. The Zebrafish Book. [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Matsui H., Sugie A. An optimized method for counting dopaminergic neurons in zebrafish. PLoS One. 2017;12(9) doi: 10.1371/journal.pone.0184363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurley A.T., Callard G.V. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.C., Gorenstein M.V., Li G.Z., Vissers J.P., Geromanos S.J. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell Proteomics. 2006;5(1):144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Hunn B.H., Cragg S.J., Bolam J.P., Spillantini M.G., Wade-Martins R. Impaired intracellular trafficking defines early Parkinson’s disease. Trends Neurosci. 2015;38(3):178–188. doi: 10.1016/j.tins.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan P., Yue Z. Deregulation of autophagy and vesicle trafficking in Parkinson’s disease. Neurosci. Lett. 2019;697:59–65. doi: 10.1016/j.neulet.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kett L.R., Stiller B., Bernath M.M., Tasset I., Blesa J., Jackson-Lewis V., Chan R.B., Zhou B., Di Paolo G., Przedborski S., Cuervo A.M., Dauer W.T. Alpha-Synuclein-independent histopathological and motor deficits in mice lacking the endolysosomal Parkinsonism protein Atp13a2. J. Neurosci. 2015;35(14):5724–5742. doi: 10.1523/JNEUROSCI.0632-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M., Nakanishi H., Saftig P., Ezaki J., Isahara K., Ohsawa Y., Schulz-Schaeffer W., Watanabe T., Waguri S., Kametaka S., Shibata M., Yamamoto K., Kominami E., Peters C., von Figura K., Uchiyama Y. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J. Neurosci. 2000;20(18):6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G.W., Goebel H.H., Simonati A. Human pathology in NCL. Biochim. Biophys. Acta. 2013;1832(11):1807–1826. doi: 10.1016/j.bbadis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Matsui H., Gavinio R., Takahashi R. Medaka fish Parkinson’s disease model. Exp. Neurobiol. 2012;21(3):94–100. doi: 10.5607/en.2012.21.3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H., Gavinio R., Asano T., Uemura N., Ito H., Taniguchi Y., Kobayashi Y., Maki T., Shen J., Takeda S., Uemura K., Yamakado H., Takahashi R. PINK1 and Parkin complementarily protect dopaminergic neurons in vertebrates. Hum. Mol. Genet. 2013;22(12):2423–2434. doi: 10.1093/hmg/ddt095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.