Abstract
Strain Marseille-Q0835T is an aerobic, non-motile and non-spore-forming Gram-positive coccus isolated from the stools of a Burkinabe woman. In this report, we present its phenotypic description including MALDI-TOF mass spectrometry analysis and genome sequencing. Strain Marseille-Q0835T; 2.9768-Mb genome exhibited a 41.9 mol% G+C content and 2699 predicted genes. Considering phenotypic features and comparative genome studies, we propose the strain Marseille-Q0835T as the type strain of Enterococcus burkinafasonensis sp. nov., a new species within the family Enterococcaceae.
Keywords: Culturomics, Enterococcus, Enterococcus burkinafasonensis, gut, new species, taxonogenomics
Introduction
Culturomics strategy is a high-throughput culturing method [1]. This strategy consists of the diversification of culture conditions and uses matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) for identification, to study the human microbiota [[1], [2], [3]]. Culturomics has played a fundamental role in resolving the gaps in 16S rRNA gene-targeted metagenomics [4]. It has been reported that culturomics has contributed up to 66.2% towards updating the repertoire of isolated human bacterial and archaeal species [2]. Taxonogenomics is a concept used for the description of new species that includes phenotypic data, MALDI TOF/MS data and genome sequencing [5,6]. In this study, we report a human gut isolate representative of a novel Enterococcus species purposely named Enterococcus burkinafasonensis.
Isolation and growth conditions
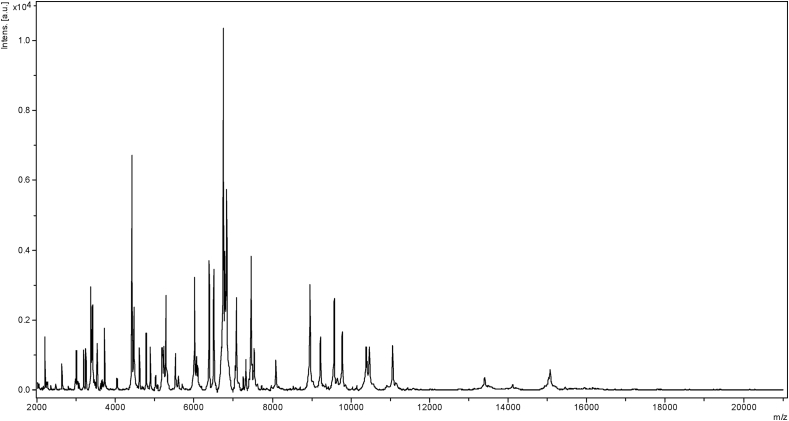
In September 2018, a fresh stool sample was collected from an apparently healthy 28-year-old Burkinabe woman who was admitted for diagnosis check-up in the Regional Tuberculosis Control Centre, Bobo-Dioulasso, Burkina Faso. A stool sample was sent to the collaborative laboratory at IHU in Marseille, France for culturomics analysis, which isolated an unidentified bacterial strain from the stool. The study was validated by the Science and Health Research Ethics Committee of Bobo-Dioulasso, under number (N/Ref.002-2018-CEIRS). The bacterium here referred to as strain Marseille-Q0835 was isolated on Columbia sheep blood agar after a 24-hour incubation under aerobic atmosphere at 37°C and pH 7.5. Purified colonies could not be identified by MALDI-TOF MS. The screening was performed on a Microflex LT spectrometer (Bruker Daltonics, Bremen, Germany), as previously described [7]. The obtained spectra (Fig. 1) were imported into MALDI Biotyper 3.0 software (Bruker Daltonics) and analysed against the main spectra of the bacteria included in the database.
Fig. 1.
MALDI-TOF MS reference mass spectrum. Spectra from 12 individual colonies of Enterococcus burkinafasonensis strain Marseille-Q0835T were compared and a reference spectrum was generated.
16S rRNA gene sequencing
The 16S rRNA gene was sequenced in an attempt to classify this bacterium. Amplification was performed using the primer pair fD1 and rP2 (Eurogentec, Angers, France) and sequencing using the Big Dye® Terminator v1.1 Cycle Sequencing Kit and ABI Prism 3130xl Genetic Analyzer capillary sequencer (Thermofisher, Saint-Aubin, France), as previously described [8]. The 16S rRNA gene nucleotide sequences were assembled and corrected using CodonCode Aligner software (http://www.codoncode.com). BLASTn research was conducted using nucleotide databases for cross-species comparison (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). The search was limited to records that include sequences from type material, and exclude uncultured/environmental sample sequences. The result showed that strain Marseille-Q0835 exhibited a 97.80% sequence identity with Enterococcus gallinarum strain LMG 13129 (GenBank accession number NR_104559.2), the phylogenetically closest species with standing in nomenclature (Fig. 2). We consequently classify strain Marseille-Q0835 as representative of a new species within the genus Enterococcus, family Enterococcaceae, phylum Firmicutes.
Fig. 2.
Phylogenetic tree showing the position of Enterococcus burkinafasonensis strain Marseille-Q0835T relative to other phylogenetically close neighbours, based on the 16S rRNA gene sequences. Pseudomonas aeruginosa ATCC10145 and Escherichia coli strain JCM 1649 AB242910 are used as the outgroup. Sequences were aligned using MUSCLE, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1000 times to generate a majority consensus tree. Only bootstrap values of at least 70 were retained.
Phenotypic characteristics
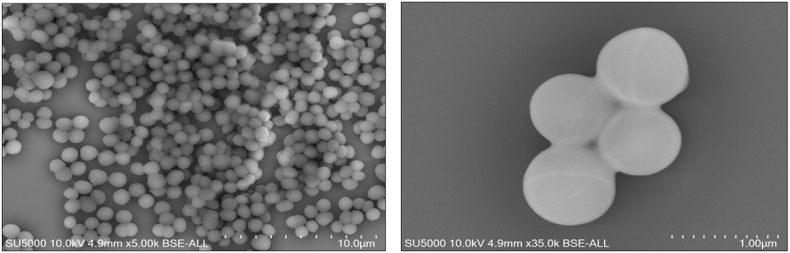
Colonies were smooth, white with entire edges with a mean diameter of 1 mm. The bacterial cells were Gram-positive cocci, non-motile, non-spore forming with a mean diameter of 0.7 μm (Fig. 3). Enterococcus sp. Marseille- Q0835T showed negative catalase and oxidase activities. API 50CH and API ZYM tests (bioMérieux, La Balme les Grottes, France) were performed at 37°C under aerobic conditions and the results are summarized in Table 1. Table 2 compares the characteristics of Enterococcus sp. nov. strain Marseille-Q0835T with other bacterial species (Table 2).
Fig. 3.
Electron micrograph of Enterococcus burkinafasonensis strain Marseille-Q0835T was acquired with a Hitachi SU 5000 Plus tabletop scanning electron microscope.
Table 1.
Phenotypic characterization of Enterococcus burkinafasonensis strain Marseille-Q0835T based on the biochemical tests
| Tests | Results |
|---|---|
| API 50 CH | |
| Control | |
| Glycerol | – |
| Erythol | – |
| d-arabinose | – |
| l-arabinose | + |
| d-ribose | + |
| d-xylose | – |
| l-xylose | – |
| d-adonitol | – |
| Methyl-βd-xylopyranoside | – |
| d-galactose | + |
| d-glucose | + |
| d-fructose | + |
| d-mannose | + |
| l-sorbose | – |
| l-rhammose | + |
| Dulcitom | – |
| Inositol | – |
| d-mannitol | + |
| d-sorbitol | – |
| Methyl-αd-mannopyranoside | – |
| Methyl-αd-glucopyranoside | – |
| N-acetylglucosamine | + |
| Amygdalin | – |
| Arbutin | + |
| Esculin | + |
| Salicin | + |
| d-cellobiose | + |
| d-maltose | + |
| d-lactose | – |
| d-melibiose | – |
| d-saccharose | – |
| d-trehalose | + |
| Inulin | – |
| d-melezitose | – |
| d-raffinose | – |
| Starch | – |
| Glycogen | – |
| Xylitol | – |
| Gentibiose | + |
| d-turanose | – |
| d-lyxose | – |
| d-tagatose | – |
| d-fucose | – |
| l-fucose | – |
| d-arabitol | – |
| l-arabitol | – |
| Potassium gluconate | – |
| Potassium 2-ketogluconate | – |
| Potassium5-Ketogluconate | – |
| API ZYM | |
| Alkaline phosphatase | – |
| Esterase (C4) | – |
| Esterase lipase (C8) | + |
| Lipase (C14) | + |
| Leucine arylamidase | – |
| Valine arylamidase | – |
| Cystine arylamidase | – |
| Trypsin | – |
| α-chymotrypsin | – |
| Acid phosphatase | – |
| Naphthol-AS-BI-phosphohydrolase | + |
| α-galactosidase | + |
| β-galactosidase | + |
| β-glucuronidase | – |
| α-glucosidase | – |
| β-glucosidase | – |
| N-acetyl-β-glucosaminidase | + |
| α-mannosidase | – |
| α-fucosidase | – |
+, positive result; –, negative result.
Table 2.
Differential characteristics of Enterococcus burkinafasonensis strain Marseille-Q0835, Enterococcus timonensis strain Marseille-P2817, Enterococcus hirae strain ATCC 9790, Enterococcus gallinarum strain NBRC 100675, Enterococcus saccharolyticus strain ATCC 43076, Enterococcus casseliflavus strain NBRC 100478 and Enterococcus asini strain ATCC 700915
| Properties |
E. burkinafasonensis |
E. timonensis |
E. hirae |
E. gallinarum |
E. saccharolyticus |
E. casseliflavus |
E. asini |
|---|---|---|---|---|---|---|---|
| Marseille-Q0835 | Marseille-P2817 | ATCC 9790 | NBRC 100675 | ATCC 43076 | NBRC 100478 | ATCC 700915 | |
| Cell diameter (μm) | 0.7 | 0.65–1.1 | na | na | na | na | na |
| Oxygen requirement | Facultative anaerobe | Facultative anaerobe | Facultative anaerobe | Facultative anaerobe | Facultative anaerobe | Facultative anaerobe | Facultative anaerobe |
| Gram stain | Positive | Positive | Positive | Positive | Positive | Positive | Positive |
| Motility | Non-motile | Motile | Non-motile | Non-motile | na | Motile | Non-motile |
| Endospore formation | — | — | na | — | na | na | — |
| Optimum temperature for growth (°C) | |||||||
| Production of: | |||||||
| Alkaline phosphatase | — | + | — | na | + | na | — |
| Catalase | — | — | — | — | — | — | — |
| Oxidase | — | — | na | na | na | na | na |
| α-Glucosidase | — | + | na | na | — | na | na |
| β-Galactosidase | + | + | + | + | — | + | — |
| Acid from: | |||||||
| N-Acetylglucosamine | + | + | + | + | + | + | + |
| l-Arabinose | — | — | — | + | — | + | — |
| d-Ribose | + | — | + | + | — | + | — |
| d-Mannose | + | + | + | + | na | + | — |
| d-Mannitol | + | + | — | + | + | + | — |
| d-Glucose | + | + | + | + | + | + | + |
| d-Fructose | + | + | + | + | + | + | + |
| d-Maltose | + | + | + | + | + | + | + |
| d-Lactose | — | + | + | + | + | + | + |
| G+C content (mol%) | 41.9 | 38.46 | 36.9 | 39.80 | 36.90 | 42.40 | 44.70 |
| Habitat | Human gut | Human lung | Chicken and pig intestines | Intestines of domestic fowl | Fresh broccoli | Plant material | Caecum of donkeys |
Genome sequencing
Genomic DNA was extracted using the EZ1 biorobot (Qiagen, Courtaboeuf, France) with the EZ1 DNA tissue kit and then sequenced on MiSeq technology (Illumina, San Diego, CA, USA) with the Nextera XT Paired end (Illumina), as previously described [9]. The assembly was performed with a pipeline incorporating different softwares (Velvet [10], Spades [11] and Soap Denovo [12]) on trimmed data (Trimmomatic [13]) or raw data. GapCloser was used to reduce assembly gaps. Scaffolds <800 bp and scaffolds with a depth value < 25% of the mean depth were removed [14]. The best assembly was selected by using different criteria (17 scaffolds, 19 contigs). The genome of strain Marseille-Q0835T is 2.9768 Mb long with a 41.9 mol% G+C content and contains 2699 predicted genes.
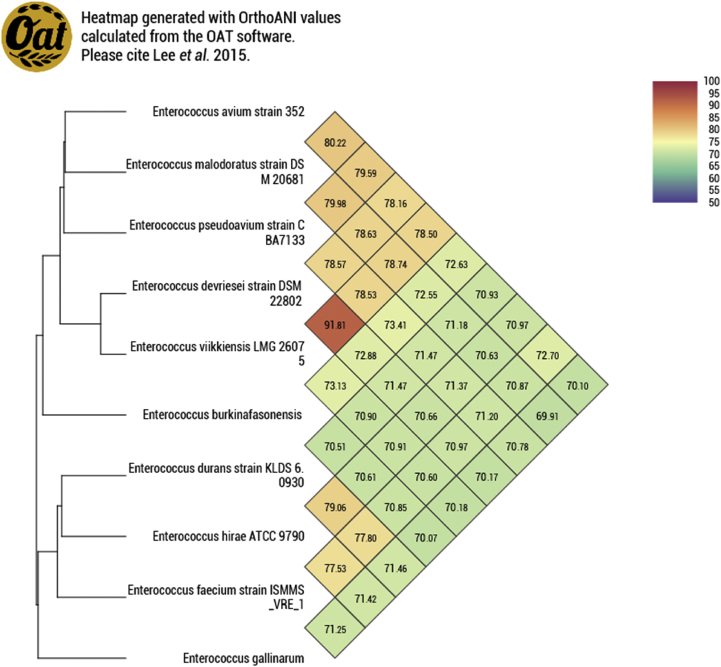
The degree of genomic similarity of Enterococcus sp. Marseille-Q0835T with closely related species was estimated using the OrthoANI software version 0.93.1 (https://www.ezbiocloud.net/tools/orthoani) [15]. Values among closely related species (Fig. 4) ranged from 69.91% for Enterococcus malodoratus strain DSM 20681 and Enterococcus gallinarum strain LMG 13129 to 91.81% for Enterococcus devriesei strain DSM 22802 and Enterococcus viikkiensis strain LMG 26075. When the isolate was compared with these closely related species, values ranged from 70.07% with E. gallinarum strain LMG 13129 to 73.41% with Enterococcus pseudoavium strain CBA7133. These values are lower than the 95% threshold used to discriminate bacterial species [15].
Fig. 4.
Heatmap generated with OrthoANI values calculated using the OAT software between Enterococcus burkinafasonensis strain Marseille-Q0835T and other closely related species with standing in nomenclature.
In silico DNA–DNA hybridization values obtained using the GGDC version 2.0 online tool (http://ggdc.dsmz.de/ggdc.php) are reported in Table 3. For strain Marseille-Q0835T, these values ranged from 20% with E. devriesei strain DSM 22802 to 26.8% with Enterococcus faecium strain ISMMS VRE 1. Such values were lower than the 70% threshold recognized as delineating distinct species [16,17].
Table 3.
Digital DNA–DNA hybridization values obtained by sequence comparison of all studied genomes using GGDC, formula 2a
| Digital DNA–DNA hybridization | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| 1 | 100 | |||||||||
| 2 | 24.10% (21.8%-26.5%) | 100 | ||||||||
| 3 | 22.40% (20.2%-24.9%) | 20.00% (17.8%-22.4%) | 100 | |||||||
| 4 | 25.20% (22.8%-27.7%) | 23.80% (21.5%-26.3%) | 26.40% (24%-28.9%) | 100 | ||||||
| 5 | 40.60% (38.1%-43.1%) | 26.80% (24.4%-29.3%) | 30.00% (27.6%-32.5%) | 25.60% (23.3%-28.1%) | 100 | |||||
| 6 | 25.80% (23.4%-28.2%) | 23.60% (21.3%-26%) | 23.00% (20.7%-25.4%) | 24.80% (22.5%-27.3%) | 28.60% (26.2%-31.1%) | 100 | ||||
| 7 | 24.40% (22.1%-26.9%) | 23.00% (20.7%-25.5%) | 21.30% (19%-23.7%) | 23.90% (21.6%-26.4%) | 23.40% (21.1%-25.8%) | 25.30% (22.9%-27.7%) | 100 | |||
| 8 | 24.20% (21.9%-26.7%) | 20.40% (18.2%-22.9%) | 22.80% (20.5%-25.2%) | 25.40% (23%-27.9%) | 28.90% (26.5%-31.4%) | 24.60% (22.3%-27.1%) | 22.80% (20.5%-25.2%) | 100 | ||
| 9 | 23.90% (21.6%-26.4%) | 22.80% (20.5%-25.3%) | 22.50% (20.3%-25%) | 25.10% (22.7%-27.6%) | 27.80% (25.4%-30.3%) | 26.40% (24%-28.9%) | 27.60% (25.2%-30%) | 23.80% (21.5%-26.2%) | 100 | |
| 10 | 22.70% (20.4%-25.2%) | 20.60% (18.4%-23%) | 46.90% (44.3%-49.4%) | 25.10% (22.8%-27.6%) | 26.40% (24%-28.9%) | 25.20% (22.9%-27.7%) | 24.00% (21.7%-26.5%) | 22.50% (20.2%-25%) | 22.60% (20.3%-25.1%) | 100 |
(1) Enterococcus avium strain 352, (2) Enterococcus burkinafasonensis strain Marseille-Q0835T, (3) Enterococcus devriesei strain DSM 22802, (4) Enterococcus durans strain KLDS 6.0930, (5) Enterococcus faecium strain ISMMS VRE 1, (6) Enterococcus gallinarum strain LMG 13129, (7) Enterococcus hirae strain ATCC 9790, (8) Enterococcus malodoratus strain DSM 20681, (9) Enterococcus pseudoavium strain CBA7133 and (10) Enterococcus viikkiensis strain LMG 26075.
GGDC formula 2: (DNA–DNA hybridization estimates based on identities/high-scoring segment pair length).
Conclusion
Strain Marseille-Q0835T exhibited a 16S rRNA gene sequence divergence <98.65%, DNA–DNA hybridization values < 70% and an OrthoANI value < 95% with its phylogenetically closest species with standing in nomenclature, together with unique phenotypic features. We formally propose strain Marseille-Q0835T as the type strain of the new species named Enterococcus burkinafasonensis.
Description of Enterococcus burkinafasonensis sp. nov.
Enterococcus burkinafasonensis (bur.ki.na.fa.so.nen'sis, L. masc. adj. burkinafasonensis related to Burkina Faso, the name of the country where the sample was collected). The bacterium belongs to the family Enterococcaceae within the phylum Firmicutes. The type strain Marseille-Q0835T (CSUR P0835) was isolated after a 24-hour incubation at 37°C and pH 7.5 in an anaerobic atmosphere of a fresh stool sample collected from a 28-year-old Burkinabe woman. Colonies were smooth, white with entire edges with an average diameter of 1 mm. Bacterial cells were Gram-positive, coccus-shaped, non-motile and non-spore-forming with negative catalase and oxidase activities (see Table 4).
Table 4.
Description of Enterococcus burkinafasonensis sp. nov. strain Marseille-Q0835T
| Type of description | New description |
|---|---|
| Species name | Burkinafasonensis |
| Genus name | Enterococcus |
| Specific epithet | Burkinafasonensis |
| Species status | sp. nov. |
| Species etymology | Enterococcus burkinafasonensis (bur.ki.na.fa.so.nen'sis, L. masc. adj. burkinafasonensis related to Burkina Faso, the name of the country where the sample was collected) |
| Authors | Nina GOUBA, Edmond KUETE YIMAGOU, Yasmine HASSANI, Jamal SAAD, Mustapha FELLAG, Michel DRANCOURT, Maxime Descartes MBOGNING FONKOU |
| Designation of the type strain | Marseille-Q0835 |
| Strain collection number | CSURP0835 |
| 16S rRNA gene accession number | LR746132.1 |
| Genome accession number | CADDWJ010000001.1 |
| Genome status | Whole genome |
| Genome size | 2.9768 Mb |
| GC% | 41.9 |
| Country of origin | Bobo-Dioulasso, Burkina Faso |
| Date of isolation | 04/05/2019 |
| Source of isolation | Human stool sample |
| Growth medium, incubation conditions used for standard cultivation | Growth on Columbia agar supplemented with 5% sheep's blood after 24 hours of incubation under aerobic atmosphere at 37°C and pH 7.5. |
| Gram stain | Positive |
| Cell shape | Coccus |
| Cell size | Mean diameter 0.7 μm |
| Motility | Non-motile |
| Sporulation | Non-sporulating |
| Colony morphology | smooth, white with entire edges with an average diameter of 1 mm. |
| Temperature range | Mesophile |
| Temperature optimum | 37°C |
| Relationship to O2 | Facultative anaerobe |
| O2 for strain testing | Anaerobiosis, microaerophilic, aerobiosis |
| Oxidase | Negative |
| Catalase | Negative |
Using an APIZYM strip, strain Marseille-Q0835T exhibits positive reaction for esterase lipase (C8), lipase (C14), naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase and N-acetyl-β-glucosaminidase, but negative reaction for alkaline phosphatase, esterase (C4), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase and α-fucosidase. Using an API 50CH strip, positive reactions were obtained for d-galactose, d-glucose, d-fructose, l-arabinose, d-ribose, d-mannose, l-rhammose, d-mannitol, N-acetylglucosamine, arbutin, esculin, salicin, d-cellobiose, d-maltose, d-trehalose and gentibiose.
The strain Marseille-Q0835T genome is 2.9768 Mb long, with a G-C content of 41.9%.
Nucleotide sequence accession number
The 16S rRNA gene and genome sequences were deposited in GenBank under accession number LR742708 and NZ_CACSLH000000000, respectively.
Deposit in culture collection
The strain Marseille-Q0835T has been deposited in the French culture collection centre, Collection de Souches de l’Unité des Rickettsies (CSUR), under the number Q0835.
Conflict of interest
None to declare.
Funding sources
This work was funded by the IHU Méditerranée Infection (Marseille, France) and by the French Government under the Investissements d'avenir (Investments for the Future) programme managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03).
Acknowledgements
The authors thank Hitachi Corporation for providing the SU 5000 Plus Tabletop microscope. They also thank Aurelia Caputo from IHU-Méditerranée Infection, Marseille for submitting the genomic sequences to GenBank.
References
- 1.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Bilen M., Dufour J.C., Lagier J.C., Cadoret F., Daoud Z., Dubourg G. The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome. 2018;6:94. doi: 10.1186/s40168-018-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J.-C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Amrane S., Raoult D., Lagier J.-C. Metagenomics, culturomics, and the human gut microbiota. Expert Rev Anti Infect Ther. 2018;16:373–375. doi: 10.1080/14787210.2018.1467268. [DOI] [PubMed] [Google Scholar]
- 5.Fournier P.-E., Lagier J.-C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy D., Mishra A.K., Lagier J.-C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 7.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.-E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 8.Morel A.-S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.-P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 9.Diop A., Khelaifia S., Armstrong N., Labas N., Fournier P.-E., Raoult D. Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb Ecol Health Dis. 2016;27:32049. doi: 10.3402/mehd.v27.32049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol J Comput Mol Cell Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma Oxf Engl. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anani H., Khodor M., Raoult D., Fournier P.-E. Whole-genome sequence of French clinical Olivibacter jilunii strain P8502. Microbiol Resour Announc. 2019;8 doi: 10.1128/MRA.00701-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee I., Ouk Kim Y., Park S.-C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 16.Auch A.F., von Jan M., Klenk H.-P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier-Kolthoff J.P., Auch A.F., Klenk H.-P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]