Abstract
Non-coding RNAs represent a significant proportion of the human genome. After having been considered as ‘junk’ for a long time, non-coding RNAs are now well established as playing important roles in maintaining cellular homeostasis and functions. Some non-coding RNAs show cell- and tissue-specific expression patterns and are specifically deregulated under pathological conditions (e.g. cancer). Therefore, non-coding RNAs have been extensively studied as potential biomarkers in the context of different diseases with a focus on microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) for several years. Since their discovery, miRNAs have attracted more attention than lncRNAs in research studies; however, both families of non-coding RNAs have been established to play an important role in gene expression control, either as transcriptional or post-transcriptional regulators. Both miRNAs and lncRNAs can regulate key genes involved in the development of cancer, thus influencing tumour growth, invasion, and metastasis by increasing the activation of oncogenic pathways and limiting the expression of tumour suppressors. Furthermore, miRNAs and lncRNAs are also emerging as important mediators in drug-sensitivity and drug-resistance mechanisms. In the light of these premises, a number of pre-clinical and early clinical studies are exploring the potential of non-coding RNAs as new therapeutics. The aim of this review is to summarise the latest knowledge of the use of miRNAs and lncRNAs as therapeutic tools for cancer treatment.
Key points
| Non-coding RNAs (especially long non-coding RNAs and microRNAs) have important roles as oncogenic and tumour suppressor molecules. |
| Long non-coding RNAs and microRNAs are attracting increasing interest as therapeutic targets after they have been used widely as biomarkers in the past. |
| One bottle neck is tissue- and cell type-specific delivery and targeting of deregulated non-coding RNAs as well as reducing off-target effects especially innate immune responses. |
Introduction
Non-coding RNAs are a large family of RNAs that are not coding for known proteins. In general, non-coding RNAs can be classified according to their length into small (< 200 nucleotides) and long (> 200 nucleotides) RNAs or according to their function as housekeeping and regulatory RNAs [1]. About 17 categories of non-coding RNA molecules have been identified so far; among them transfer RNAs, ribosomal RNAs, small nucleolar RNAs, endogenous small interfering RNAs, sno-derived RNAs, transcription initiation RNAs, microRNA-offset-RNAs, circular RNAs, vault RNAs, microRNAs (miRNAs), small interfering RNAs (siRNAs), small nuclear RNAs, extracellular RNAs, piwi-interacting RNAs, small Cajal body RNAs, long intergenic non-coding RNAs and long non-coding RNAs (lncRNAs) are known [2–23]. Non-coding RNAs constitute the majority of the human genome and retain fundamental biological properties within cells [24, 25]. Among their functions, they regulate transcription, influence translation of coding genes, are components of the protein synthesis machinery, and regulate each other, e.g. modify ribosomal RNAs, and lncRNAs can counteract miRNAs by sequestering them (miRNA sponges) [17, 26]. Moreover, many physiological processes are regulated by non-coding RNAs, including development, gametogenesis, stress, immune response and tumourigenesis [17, 27]. MicroRNAs may also influence longevity as was shown by analysing the lifespan of the roundworm Caenorhabditis elegans [28]. In this particular case, a loss-of-function mutation in the lin-4 miRNA down-regulated FOXO/DAF-16 and up-regulated lin-14, resulting in a shorter lifespan [27].
Long non-coding RNAs and miRNAs are the most studied classes of non-coding RNAs involved in pathological conditions [13, 29, 30]. This review aims to focus on the different functions of lncRNAs and miRNAs and on their potential utility as therapeutic targets.
MicroRNAs
MicroRNAs are short RNA transcripts of 18–24 nucleotides and they regulate gene expression at the translational level [14]. They are among the most extensively studied and best characterised non-coding RNAs. According to the canonical view, miRNAs are regarded as negative regulators of gene expression that upon binding to the 3ʹ-untranslated region of the target messenger RNA (mRNA) cause a block of translation and/or degradation of the transcript, therefore down-regulating the final protein synthesis rate [31]. However, novel non-conventional mechanisms of action have been suggested for miRNAs that challenge traditional paradigms. For instance, in contrast to the general consensus that miRNA-mRNA interactions occur in the cytoplasm, miR-29b has been demonstrated to localise predominantly in the nucleus [32]. Furthermore, some miRNAs (like miR-10) can exert their effects also by binding to the 5ʹ-untranslated region [33] or like miR-155 to the coding sequence of target mRNAs [34]. Sometimes miRNAs can affect translation by up-regulation mechanisms. It has been shown that miR-369-3p recruits a protein complex to the AU-rich elements in the tumour necrosis factor-alpha mRNA and induces an increase of the tumour necrosis factor-alpha translation rate [35].
A single miRNA can target several mRNAs, thus each miRNA can simultaneously regulate multiple target genes, both within a single pathway or even across different pathways [36–38]. For example, while the miR-17 family targets various components of the transforming growth factor-beta (TGF-β) signalling pathway, including TGF-β receptors and down-stream transducers such as SMADs and cyclin-dependent kinase inhibitor 1A [39], miR-21 suppresses key genes of major tumour suppressive pathways such as p53, TGF-β and mitochondrial apoptosis [40].
MicroRNA expression patterns are tissue specific [41] and often define the physiological nature of the cell [42]. Numerous publications show that altered mRNA expression in the context of several diseases (e.g. cancer, viral diseases, cardiovascular diseases, neurodegenerative and immune-related diseases) is caused by deregulation of miRNA expression [13, 43–48].
MicroRNA dysfunction occurs as a consequence of diverse mechanisms such as promoter methylation, mutations, amplifications or deletions involving miRNA coding regions, transcriptional regulation by proteins (e.g. p53 and c-myc) or lncRNAs, and down-regulation as well as mutations in genes coding for miRNA biogenesis-related enzymes (i.e. Drosha, Exportin 5, Dicer, Argonaute 2) [31, 49–51].
Multiple lines of evidence suggest that tissue- and disease-specific miRNA signatures may be used as biomarkers for diagnostic, prognostic and predictive purposes in the clinic [46, 52–58]. However, the most exciting avenue of current translational research efforts is focusing on their therapeutic potential [59–64]. Indeed, their pivotal role in a huge range of pathological conditions together with the fact that a single miRNA can modulate the expression of several genes across multiple pathways altered in diseases makes them attractive therapeutic targets [65]. Modulation of the expression of miRNAs could be a novel and promising therapeutic modality that can be achieved through administration of either miRNA mimics to restore expression of down-regulated miRNAs or miRNA inhibitors (anti-miRNA) to counteract the up-regulation of pathogenic miRNAs [66]. Research on miRNAs as both targets and therapeutics is proceeding at a rapid pace. In less than 20 years after the discovery of the first miRNA in the nematode C. elegans [67, 68], the first miRNA-based therapeutic entered clinical evaluation in patients with chronic hepatitis C virus (HCV) genotype 1 infection [69]. Owing to a deeper understanding of disease-relevant miRNAs and advances in in-vivo delivery systems, the administration of miRNA-based therapeutics has recently shown to be feasible and safe in humans with encouraging efficacy results in early-phase clinical trials (Table 1) [69–71].
Table 1.
Clinical trials investigating microRNA therapeutics in cancer and other diseases
| Drug name | Type | Disease | Phase | Status | ClinicalTrials.gov identifier |
|---|---|---|---|---|---|
| Miravirsen | Anti-mir122 | HCV | I | Completed | NCT01646489 |
| Miravirsen | Anti-mir122 | HCV | II | Completed | NCT02508090 |
| Miravirsen | Anti-mir122 |
Treatment naïve HCV |
II | Completed | NCT01200420 |
| Miravirsen | Anti-mir122 | HCV | II | Completed | NCT02452814 |
| Miravirsen | Anti-mir122 | HCV | II | Completed | NCT01200420 |
| Miravirsen | Anti-mir122 | HCV | II | Active | NCT01727934 |
| Miravirsen | Anti-mir122 | Refractory HCV | II | Active | NCT01872936 |
| RG-101 | GalNAc-conjugated anti-mir122 | HCV | Ib | Terminated | |
| MesomiR | miR-16 mimic | Malignant pleural mesothelioma, NSCLC | I | Completed | NCT02369198 |
| MRX34 | miR-34 mimic | Primary HCC, melanoma, RCC, SCLC, NSCLC, lymphoma, multiple myeloma | I | Terminated | NCT01829971 |
| MRX34 | miR-34 mimic | Melanoma | I | Terminated | NCT02862145 |
| MRG-106 | Anti-miR155-5p | Cutaneous TCL, mycosis fungoides, CLL, DLBCL, ATLL | I | Recruiting | NCT02580552 |
| MRG-106 | Anti-miR155-5p | Cutaneous TCL, mycosis fungoides | II | Recruiting | NCT03713320 |
| MRG-106 | Anti-miR155-5p | Cutaneous TCL, mycosis fungoides | II | Not yet recruiting | NCT03837457 |
| RG-125 | GalNAc-conjugated anti-miR-103/107 | Type 2 diabetes mellitus and NAFLD | I/IIa | Suspended (sponsor decision) | NCT02826525 |
| MRG-201 | miR-29 mimic | Keloids | I | Completed | NCT02603224 |
| MRG-201 | miR-29 mimic | Keloids | II | Recruiting | NCT03601052 |
| EXTH-61 | Anti-miR-10 | Glioblastoma | I | Recruiting | NCT01849952 |
| MRG-110 | Anti-miR-92 | Small skin wound | I | Completed | NCT03603431 |
| RG-012 | Anti-miR-21 | Alport’s syndrome | I | Active | NCT03373786 |
| RG-012 | Anti-miR-21 | Alport’s syndrome | II | Suspended (sponsor decision) | NCT02855268 |
| pSil-miR200 | miR-200a/c plasmid | Inflammation and osteogenesis after tooth extraction | I | Not yet recruiting | NCT02579187 |
ATLL adult T-cell leukemia/lymphoma, CLL chronic lymphocytic leukemia, DBCLC diffuse large B-cell lymphoma, GalNAc N-acetylgalactosamine, HCC hepatocellular carcinoma, HCV hepatitis C virus, NAFLD non-alcoholic fatty liver disease, NSLC non-small cell lung cancer, RCC renal cell carcinoma, SCLC small-cell lung cancer, TCL T-cell lymphoma
However, although a plethora of miRNA-based compounds have been investigated in preclinical studies, only a minority of these has moved to clinical development. Challenges concerning a proper target selection, stability in body fluids, and specificity of target binding as well as off-target effects remain to be addressed in the future to optimise the in-vivo delivery and efficiency of miRNA-based therapeutics. As miRNAs are implicated in virtually all physiological and pathological processes, a huge therapeutic potential is expected from miRNA-based constructs.
Interestingly, in addition to acting within cells, circulating cell-free miRNAs have been detected in plasma, serum, urine, and many other body fluids and have been shown to act at distant sites within the body [72, 73]. It has been recently shown that miRNAs can be either released by passive leakage from lytic cells or actively secreted via extracellular vesicles (e.g. exosomes) as well as via high-density lipoprotein [74, 75]. Still, another significant proportion of extracellular miRNAs is exported in conjunction with RNA-binding proteins, such as AGO2 and NPM1 [76–78]. Of note, this horizontal transfer of secreted miRNA is emerging as a new form of intercellular communication, by which a donor can influence the gene expression of a recipient cell, both in an autocrine and paracrine manner [79, 80]. More importantly, besides the tremendous contribution to the understanding of mechanisms governing cell-to-cell signalling and miRNA functions, the evidence of a microvesicle-dependent miRNA trafficking system provides intriguing insights for the development of efficient miRNA delivery systems for targeted therapeutics.
Long Non-Coding RNAs
Long non-coding RNAs are constituted by more than 200 nucleotides and have an important role as regulators during development and pathological processes [81–85]. Under physiological conditions, most lncRNAs are transcribed from promoters with low CpG dinucleotide contents and therefore expressed at low levels [86]. An aberrant expression of lncRNAs in several diseases is due to modulation of the chromatin state. Changes in chromatin structure can also be induced by miRNAs; one example is the up-regulated expression of lncRNA MEG3 by miRNA-29 via its inhibition of DNA methyltransferase activity that in turn hampers the methylation of the MEG3 promoter in the context of hepatocellular cancer [87].
Long non-coding RNAs can be localised in the nucleus or cytoplasm [88, 89]. Often lncRNAs have been shown to serve as scaffolds for the assembly of multiple component complexes [90]. Depending on the subcellular localisation of the lncRNAs, they directly regulate gene expression by (a) influencing chromatin modification by binding to chromatin regulatory proteins; (b) regulating splicing and stability of mRNAs; as well as (c) indirectly in transcriptional and post-transcriptional gene expression mechanisms by interacting with other RNAs and proteins [91–93]. For instance, several lncRNAs (e.g. MALAT1, H19, MEG3) are important regulators of the cell cycle via their influence on p21 or p53 [94–96]. On the contrary, some lncRNAs (e.g. H19, PINT, RP11–467J12.4, LincRNA-p21, PANDA) are induced by p53 [97–101].
Several reports have underlined the fundamental interaction between lncRNAs and miRNAs in RNA regulation processes [13, 29, 102, 103]. Long non-coding RNAs can act as miRNA decoys by sequestering miRNAs, thus acting as competing endogenous RNAs and leading to re-expression of miRNA target genes [13, 102, 103]. In addition, lncRNAs can promote gene expression by competing with miRNAs for specific binding sites in the non-coding regions of mRNAs and preventing the transcriptional repression caused by miRNAs [29, 102, 103]. The fact that some lncRNAs can be processed into miRNAs highlights the strong interaction among different classes of non-coding RNAs [13, 29, 102, 103].
Many lncRNAs are expressed in a tissue- and cell type-specific manner; the expression varies between different development stages and they are often deregulated in a disease-specific pattern [104–107] and changes in lncRNA expression has been observed in body fluids such as urine and blood in several pathological conditions [108–110]. Long non-coding RNAs are released either encapsulated in exosomes or inside apoptotic bodies and they are bound to RNA-binding proteins [111, 112]; therefore, they are resistant against RNase degradation and they could be of interest as non-invasive biomarkers [108, 113].
Altered lncRNA expression has been observed in several diseases and especially in cancer [114]. Because of the ability of lncRNAs to interact with DNA, RNA, proteins and their combination, lncRNAs have the potential to contribute to the onset of cancer, modulating virtually all hallmarks of cancer and fostering cancer progression.
Long non-coding RNAs were shown to respond to DNA damage and to be involved in DNA repair; for instance, estrogen-regulated lncRNAs CUPID1 and CUPID2 trigger progression of breast cancer by an inhibition of cell-cycle arrest following DNA damage [115]. In triple-negative breast cancer, lncRNA LINK-A increases the stability of hypoxia-inducible factor 1-alpha [116] and, in oral cancer, lncRNA HIFCAR was identified as a hypoxia-inducible factor 1-alpha co-activator [117]. In both cases, lncRNAs support tumour growth by regulating the Warburg effect. In a hypoxic environment, the expression of some lncRNAs (e.g. lncRNA LET) is also repressed for instance by histone deacetylase 3 activity, resulting in a reduced histone acetylation rate of the lncRNA promoter region [118]. Low lncRNA LET expression is also an important prerequisite for stabilisation of nuclear factor 90 protein that in turn results in increased cancer invasion [118]. In renal tumours, the down-regulation of tumour suppressor lncRNA FILNC1 increases c-Myc expression and in parallel enhances glucose metabolism and lactic acid production [119]. Moreover, epithelial-mesenchymal transformation (EMT) is under the influence of lncRNAs as demonstrated for the up-regulated expression of zinc finger proteins ZEB1 and ZEB2 following loss of lncRNA ATB in hepatocellular carcinoma, which leads to increased distant metastasis [120]. In contrast, hepatitis B virus X protein down-regulates lncRNA hDREH, an inhibitor of EMT in hepatocellular carcinoma [121]. Another lncRNA that promotes EMT and metastasis in various tumours such as liver cancer, pancreatic cancer and breast cancer is HOTAIR [122–124]. Expression of HOTAIR in cancer cells is induced by TGF-β secreted by carcinoma-associated fibroblasts [125]. HOTAIR in turn induces EMT by activating the SMAD cascade signalling in tumour cells [125].
Recently, lncRNAs have been identified as important regulators for the tumour immune micro-environment [126–130], as for instance, the immune escape of liver tumours by lncRNA epidermal growth factor receptor (EGFR)-dependent stimulation of regulatory T-cell differentiation [131]; other reports have evidenced the regulation of macrophage recruiting into bladder cancer by lncRNA LNMAT1 [132] and lncRNA NKILA by enhancing sensitivity of tumour-specific cytotoxic T lymphocytes and type 1 helper T cells to activation-induced cell death and by inhibiting the nuclear factor-κB signalling pathway [133]. All of this activity clearly shows the deep relevance of lncRNAs in the regulation of cancer from initiation to progression but also in the mantainance of the micro-enviroment needed to support cancer invasion and escape to metastatic sites. Long non-coding RNAs are contributing to all levels in cancer development and they could potentially be exploited for treatments.
Use of MicroRNAs and Long Non-Coding RNAs as Clinical Biomarkers
MicroRNA and lncRNA expression patterns are known to vary under pathological conditions. In addition, non-coding RNAs can be detected in tissue samples as well as in all body fluids such as blood (plasma/serum), urine, cerebrospinal fluid or tears [134]. MicroRNAs, especially, are very stable even in RNAse-rich biofluids thus making them potential new biomarkers [73]. Furthermore, miRNAs and lncRNAs are present also in exosomes, which protect them from degradation within body fluids [135]. Therefore, tissues as well as liquid biopsies can be used as starting material for exploring the potential of both non-coding RNA classes as biomarkers and they are already widely used as biomarkers in cardiac and neural diseases as well as cancer [13, 53, 105, 136–138].
An increased level of circulating miR-1 has been shown to be a promising biomarker for acute myocardial infarction [139] and miR-208a/b along with miR-499 have been demonstrated as potential biomarkers of the severity of an infarction and the degree of myocardial damage [140]. A panel of ten miRNAs in plasma has been used to predict the individual risk for future fatal acute myocardial infarction in healthy individuals, resulting in nearly 80% correct risk assessment [141] and furthermore expression levels of lncRNAs (ZFAS1 and CDR1AS) are also able to predict myocardial infarction [142].
In the context of neurological diseases, the power of a diagnostic based on non-coding RNAs present in biological fluid is a great advance given that the disease site is largely inaccessible. Disease-specific neural lncRNAs can be detected in the blood of patients with Alzheimer’s disease [143] and patients with major depressive disorder [144].
MicroRNA-based assays can be used for diagnostic and prognostic purposes in the setting of different cancers; e.g. miRNA expression profiling is clinically used as a tool to distinguish between molecular subtypes of breast cancer [145] and different forms of lung cancer [146]. Another assay composed of 64 miRNAs can detect the primary tissue of origin for 49 tumour types with very high accuracy and is an offered laboratory test [147, 148].
In addition, miRNA- and lncRNA-based assays are often used in combination with existing protein biomarkers to improve the specificity and sensitivity of the diagnosis. One example is given by the lncRNA PCA3-based test from urine samples for the detection of prostate cancer [149], which has US Food and Drug Administration approval. By including lncRNA PCA3 in prostate cancer diagnosis, the sensitivity and accuracy are significantly increased in comparison to using the protein biomarker PSA alone [150, 151]. Furthermore, expression patterns of miRNAs and lncRNAs could be used to monitor therapy success and drug resistance development [13] and therefore they are being tested as biomarkers in several registered clinical studies (ClinicalTrials.gov) to monitor disease progression in patients receiving Food and Drug Administration-approved drugs; e.g. NCT02656589 (prognosis of Herceptin®-treated patients with breast cancer), NCT02471469 (identification of enzalutamide-treated patients with prostate cancer with need of a further surgery) and NCT02243592 (analysing miRNA expression in tissue samples from patients with cancer who are exceptional responders to treatment).
Identification of Deregulated Non-Coding RNAs
The identification of non-coding RNA candidates for designing targeted strategies is the foremost step in developing non-coding RNA-based therapeutics. This requires translational research efforts starting with a patient’s sample analysis to select disease-specific non-coding RNAs more likely to be of biological relevance to a given pathological condition. The advent of high-throughput technologies along with massive parallel sequencing analysis have tremendously facilitated the search for non-coding RNAs, especially for key regulatory miRNAs, which could be targeted. To this end, genome-wide functional screening studies using libraries of hundreds of miRNA mimics or inhibitors are among the most commonly employed approaches [152]. For instance, a recent report described the genome-wide data, including miRNAs, isolated from extracellular vesicles of liver cancer cell lines [153]. On a different approach, several publicly available repositories exist that link genomic, transcriptomic and proteomic data of coding genes and non-coding RNAs targeting these genes, thus assisting researchers in identifying the most biologically relevant lncRNAs and miRNAs in a disease-specific context. In fact, databases such as LncRNADisease [154], lncRNASNP [155], miRTarbase [156] and StarBase [157] contain thousands of experimentally validated and continuously updated lncRNA- or miRNA-target interactions including data generated by cutting-edge techniques such as crosslinking immunoprecipitation [158, 159]. In addition, the matching of these data with those coming from prediction platform such as TargetScan [160] has the potential to further improve the accuracy of the search for relevant non-coding RNAs.
Non-Coding RNA-Based Therapeutics in Cancer: Principles, Promises and Challenges
The ability to target and modulate multiple protein-coding genes at different levels of the same pathways or across different pathways in virtually all pathological processes makes non-coding RNAs, especially miRNAs, therapeutic candidates [64, 104, 105, 161]. This offers several advantages including the simultaneous targeting of various compensatory pathways involved in the development of therapy resistance as well as the chance to hit commonly undruggable proteins via their miRNA regulators.
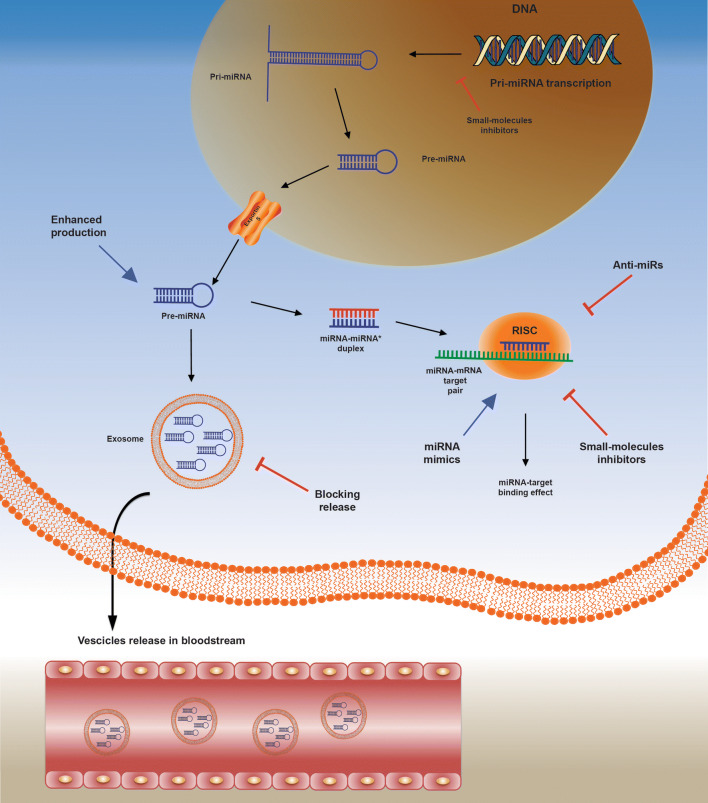
Some promising approaches aimed at manipulating miRNA expression are summarised in Fig. 1. Expression levels of miRNAs can be regulated by anti-miRNAs and miRNA mimics. Anti-miRNAs are synthetic single-stranded RNA molecules that owing to sequence complementary bind and sequester mature miRNAs and by this activity counteract up-regulated miRNAs [61]. In contrast, miRNA mimics are synthetically derived oligonucleotide duplexes, which, upon introduction into the cells, behave similarly to (and therefore mimic) endogenous miRNAs and they are able to restore expression of down-regulated miRNAs [66]. A limitation of this approach could be the ability of cancer cells to overcome miRNA-mediated repression by allowing mutations within miRNA response elements or expressing mRNA isoforms with shorter 3ʹ-untranslated regions lacking miRNA-binding sites by using alternative cleavage and polyadenylation [162–165]. The missing repressive elements in mRNA isoforms will then prevent miRNA mimics binding and result in therapy failure. Another strategy to therapeutically fine tune miRNA levels relies on small-molecule compounds [166, 167] that can perturb several steps of miRNA biogenesis and function mainly through transcriptional regulation [168–170]. For instance, azobenzene was identified as a potent and specific inhibitor of miR-21 expression during a large-scale drug screening on small organic molecules [170]. Azobenzene affects the transcription of the miR-21 gene into pri-miR-21 and is an example for targeting the miRNA synthesis pathway in the nucleus [170].
Fig. 1.
Illustration of the multi-step microRNA (miRNA) biogenesis pathways, miRNA-target messenger RNA (mRNA) interaction and the exosome-mediated miRNA secretion pathway along with relative druggable concentrations. Black arrows refer to enzymatic steps, blue arrows refer to therapeutic approaches enhancing miRNA expression, while red lines refer to therapeutic approaches blocking miRNA functions. For details see the main text
Other small-molecule modifiers act on miRNAs in the cytoplasm as it was shown for the liver-specific miR-122. Some small molecules act either as miR-122 inhibitors (e.g. can be used for blocking HCV replication in liver cells) or as miR-122 activators leading to increased apoptosis in HCC cell lines [171]. Furthermore, DICER and AGO2, two main components interacting with the RISC complex and responsible for the formation of mature miRNAs in the cytoplasm, may be potential targets for interfering with miRNA biosynthesis [172]. In addition, it is possible to block the release of exosomes and other microvesicles [173, 174] that are known carriers of proteins and genetic material including miRNAs between cells [175–177]. Furthermore, the use of vector-based strategies has been successfully adopted both in in-vitro and in-vivo models for targeting miRNAs [178]. This approach is based on the expression of specific lncRNAs acting like sponges or decoys capable of selectively sequestering miRNAs by means of various miRNA-binding sites. As an ultimate result, miRNA down-stream target genes are de-repressed and actively transcribed.
Therapeutic targeting of lncRNA expression can be reached by different approaches (Fig. 2) that have to be adapted to the different sub-cellular localisation of lncRNAs. Nuclear-located lncRNAs can be targeted by chemically modified antisense oligonucleotides (ASOs) resulting in RNAseH-dependent degradation [179]. Furthermore, ASOs or small-molecule inhibitors can be used to inhibit lncRNA-RNA-binding protein interactions [180]. However, the limitations of ASO use has also been recently described. Specifically, it was demonstrated that ASO-mediated cleavage of nascent RNAs can induce premature transcription termination [181]. The introduction of ASOs results in a reduction in polymerase II occupancy down-stream of the transcript cleavage site in an exonuclease XRN2-dependent manner and by this activity influence transcription in general [181]. Nevertheless, it seems possible to target the 3′-end of transcripts with ASOs to reach RNA knock-down without disturbing polymerase II association with the distal gene body [181] and this must be taken into account for therapeutic use of ASOs. Long non-coding RNA expression has been also modulated via steric blocking of the promoter or by using genome-editing techniques such as CRISPR/Cas9 [182–184]. Up-regulation of lncRNA expression can be mediated by knock-down of the relevant corresponding natural antisense transcripts [185]. In contrast, lncRNAs present in the cytoplasm can be degraded with siRNA-based strategies involving the multiprotein complex RISC, RNAse dicer and endonuclease Argonaut2-dependent degradation pathway [122, 186, 187].
Fig. 2.
Illustration of the possibilities to target and alter the expression level of long non-coding RNAs (lncRNAs). a Nuclear localised lncRNAs can be targeted by chemically modified antisense oligonucleotides (ASOs) resulting in RNAseH-dependent degradation. b Antisense oligonucleotides or small-molecule inhibitors can be used to inhibit the lncRNA-RNA-binding protein interactions. c Long non-coding RNA expression can be modulated via steric blocking of the promoter or by using genome-editing techniques such as CRISPR/Cas9 [182–184]. d Up-regulation of lncRNA expression can be mediated by knock-down of the relevant corresponding natural antisense transcripts (NATs). e Long non-coding RNAs present in the cytoplasm can be degraded with small interfering RNA (siRNA)-based strategies involving the multiprotein complex RISC, RNAse dicer and endonuclease Argonaut2-dependent degradation pathway
However, naked oligonucleotides per se, regardless if single or double stranded, display suboptimal pharmacokinetic and pharmacodynamic properties owing to their molecular size (> 6000 Da) and the negatively charged backbone [188]. Indeed, in-vivo stability, cell permeability, tissue-specific targeting and potential off-target effects represent hurdles to be overcome to successfully translate RNA-based compounds from the bench to the bedside.
Chemical Modification and Encapsulation into Delivery Vehicles
One of the greatest challenges related to in-vivo RNA therapeutic delivery to the target tissue is represented by their stability in body fluids. As already mentioned, naked oligonucleotides are subject to degradation by nucleases, especially 2′-hydroxyl-dependent RNAses, both in serum and in the endosomal compartment [189]. To overcome this issue and provide higher stability to RNA therapeutics, two complementary and non-mutually exclusive approaches have been adopted. The first one is based on chemical modifications mostly affecting the 2ʹ-position of the sugar ring of the RNA backbone that include the incorporation of O-methyl and O-methyoxyethyl groups or phosphorothioate-like groups and locked nucleic acid (LNA) modifications (2′,4′-methylene linkage of the ribose) that also improve the binding capacity for complementary sequences [190, 191]. The other strategy involves the adoption of delivery technologies that in addition to protecting RNA from nucleases can also increase the delivery rate to disease-related tissues [192]. Administered antisense oligonucleotides have been found to accumulate and control gene expression in the liver and kidneys of patients but targeting of other tissues was less effective [193, 194].
Often RNA therapeutics are encapsulated into nanoparticle vehicles to overcome these limitations [195]. Among them, lipid-based systems such as neutral lipid emulsions and polymers such as polyethylene glycol and polyethylenimine are the most commonly tested delivery vehicles in both pre-clinical and clinical studies [196, 197]. However, potential immunostimulatory effects and a suboptimal specificity to a targeted disease site need to be overcome to successfully exploit miRNA therapeutics into the clinic.
A strategy to prevent harmful side effects caused by the delivery vehicle is to link RNA therapeutics to a ligand whose receptor is over-expressed on cancerous cells and deliver it directly into tumour cells [198]. Such a targeted delivery was successfully used in in-vitro and in-vivo studies with lung and breast tumour models by fusing miRNAs to folate where FolamiRs were taken up by tumour cells over-expressing the folate receptor in comparison to normal tissue [198–202]. One limitation of this approach is most probably the endocytic uptake of folate derivate and entrapment of FolamiRs into the endosomal compartment [198].
Interestingly, exosomes that are extracellular cell-derived phospholipid nanovescicles horizontally transmitting bioactive molecules to specific recipient cells are increasingly regarded as natural nanocarriers for miRNA- and lncRNA-based therapeutic delivery [203]. Indeed, exosomes are endowed with target-homing specificity, stability and capability of surmounting in-vivo barriers that can lead to specific and efficient delivery of molecules including nucleic acids [204]. In an elegant in-vivo study, human embryonic kidney cells were transfected with the exogenous tumour suppressor let-7a, and after purification from the culture supernatant, let-7a-containing exosomes were systemically administered to mice bearing a EGFR-expressing breast cancer xenograft [205]. The authors were able to show that exosomes loaded with let-7a successfully achieved targeted delivery and tumour regression, providing a proof of principle on the feasibility and the efficacy of these bioengineered exosomes [205]. Despite these encouraging results, before moving to the clinic further research is needed, especially to reduce the immunogenicity of allogeneic exosomes that are enriched in major histocompatibility complex proteins with the risk of immune reactions.
Clinical Development of MicroRNA-Based Therapeutics
The development of miRNA-targeted therapeutics has advanced considerably in recent years, with numerous phase I and II trials currently ongoing across a broad range of pathologic conditions (Table 1).
Locked Nucleic Acid-Antisense Oligonucleotide
As mentioned above, LNA-antisense oligonucleotides are used to repress the expression level of endogenous miRNAs in the cells. The LNA modification increases the RNAse stability of the antisense oligonucleotide and improves the binding capacity for complementary sequences. One of the earliest and at the same time successful example of the translational research efforts behind miRNAs comes from the treatment of HCV infection. In a phase IIa trial, a 15-nucleotide LNA-antisense oligonucleotide complementary to and with high affinity and specificity for the 5ʹ-region of mature miR-122 (Miravirsen®) was administered to 36 patients with chronic HCV genotype 1 infection [69]. The liver-specific miR-122 is (ab-)used by the virus to bind to the 5ʹ-untranslated region of the viral genome and thereby enhance virus replication [69]. In this study, prolonged and dose-dependent reductions in HCV RNA levels were observed, including undetectable viral load in five patients, with no dose-limiting toxicities [69]. This successful therapeutic outcome is based on the fact that no escape mutations in the miR-122-binding sites of the HCV genome were found [69]. Miravirsen had a good safety profile with most common adverse events such as headache and fatigue that were mild in grade and only observed in a few patients [69]. Based on these positive results, other studies testing Miravirsen either alone or combined with other antiviral agents are underway (summarised in Table 1 and [206]). Indeed, longer follow-up and larger patient population are needed to ensure a proper assessment of both efficacy and safety.
In another first-in-human study based on LNA-modified oligonucleotide inhibitor (MRG-106), the onco-miR-155-5p was targeted in patients with mycosis fungoides who are in clinical stage I–III with plaques or tumours on a stable treatment regimen or without any concomitant therapy [207]. During this phase I study, both intratumoral and subcutaneous administration of MRG-106 was evaluated and inhibition of miR-155-5p was well tolerated [207]. In all patients, MRG-106-treated lesions had a Composite Assessment of Index Lesion Severity score reduction of ≥ 50%, which was maintained to the end of the study [207]. One big advantage is the significant pharmacodynamic activity of MRG-106 without requiring additional formulation [207] and by this the risk of unwanted side effects is reduced. Furthermore, the accessibility of cutaneous lesions provides the opportunity to apply MRG-106 directly into the tumour area and this most probably also reduces off-target effects. Biomarker analysis of pre- and post-treatment biopsies showed reductions in the MAPK/ERK, JAK/STAT and PI3K/AKT survival pathways in agreement with the expected MRG-106 mode of action [208]. The simultaneous decreased expression of several signalling pathways associated with the pathogenesis of mycosis fungoides and tumour cell survival is of pivotal importance to minimise tumour escape mechanisms. The possibility to target several survival pathways in parallel underlines the therapeutic power of LNA-based miRNA inhibition. Based on the positive outcome of this clinical trial, a phase II study with MRG-106 has recently started to recruit patients with cutaneous T-cell lymphoma who have confirmed disease progression following treatment with vorinostat.
In the setting of a phase I study, the LNA inhibitor MRG-110 is used to reduce the expression level of the potent anti-angiogenic miR-92 [209]. MRG-110 is expected to accelerate wound healing by improving angiogenesis into the wound area when injected into the skin at the site of the skin wound. Furthermore, MRG-110 has the advantage of reducing off-target effects by applying the inhibitor directly into the area of interest. Inhibition of miR-92 increased angiogenesis and MRG-110 was safe and generally well tolerated when given as 3-weekly intradermal doses. Based on these phase I safety, tolerability, pharmacokinetic, and biomarker data, further clinical studies with MRG-110 are in preparation to evaluate the ability of this miR-92 inhibitor to enhance vascularisation and function in the setting of heart failure.
MicroRNA Mimic
As mentioned above, miRNA mimics are synthetic oligonucleotide duplexes, which, upon introduction into the cells, behave similarly to endogenous miRNAs and they are able to restore expression of down-regulated miRNAs. MRX34, a liposome-formulated miRNA-34 mimic, has been the first miRNA mimic therapeutic to enter a clinical trial in patients with solid tumours including hepatocellular carcinoma, renal cell carcinoma and melanoma [71]. In a phase I, first-in-human, dose-escalation study, MRX34 was administered intravenously twice weekly for 3 weeks within a 28-day cycle (including six total doses of MRX34) in 47 patients with heavily pre-treated advanced solid tumours [71]. The liposomal miR-34 mimic yielded one prolonged therapeutic effect and a partial response was confirmed by means of RECIST criteria in a patient with HCC. Moreover, six patients with stable disease were reported, four of whom lasted more than four cycles [71]. Despite these encouraging preliminary efficacy and safety data, the trial was put on hold by the developing biopharmaceutical company itself because of the occurrence of five immune-related severe adverse events on a longer study follow-up [210]. It is currently unclear whether such adverse events should be attributed to the liposome carrier, the double-stranded oligonucleotides, or both, as they are both known to potentially trigger pro-inflammatory and immune-mediated side effects [210]. Another possible limitation of this approach could be the intravenous administration of the miRNA mimic without any targeting molecule to increase the specific delivery into the tumour.
Following the same approach aimed at restoring the expression of down-regulated miRNAs, a miR-16 mimic recently entered clinical evaluation in patients with pre-treated malignant pleural mesothelioma [70]. MesomiR is the first compound in a clinical trial to use the ‘TargomIR-technology’ [161]. TargomiR delivery vesicles are composed of a miRNA mimic and a targeting molecule (e.g. a specific antibody that recognises a protein on the target cells, in the case of mesomiR, an EGFR antibody is used) surrounded by bacterial-derived minicells [211]. In a first-in-human phase I study, a miR-16 mimic was delivered in TargomiR vesicles to 26 patients with immunohistochemically proven, EGFR-expressing malignant pleural mesothelioma, progressing after standard chemotherapy [70]. MesomiRs were given over a 20-min intravenous infusion either once or twice weekly in a traditional 3 + 3 dose-escalation design in five dose cohorts. Among 22 assessable patients, one (5%) had a partial response and 15 (68%) had stable disease. The administration of MesomiRs was well tolerated and associated with early anti-tumour activity. However, toxicity concerns emerged as cardiac events were reported in five patients [70]. The exact mechanism of the cardiac toxicity remains to be understood and, although a carrier-elicited inflammatory reaction seems the most plausible explanation, a miRNA mimic-related myocardial injury could not be excluded. Pharmacological studies and correlative biomarker analyses embedded in future larger clinical trials are warranted to better understand the pharmacokinetic and pharmacodynamic properties of this new class of therapeutics, thus optimising the delivery of next-generation compounds.
In addition, a phase I study based on a miRNA-29b mimic (MRG-201) was started recently. By using a miRNA-29b mimic, expression of collagen and other proteins involved in fibrous scar formation should be reduced. The therapeutic substance is injected intradermally [210]. The possibility to apply MRG-201 directly into the area of interest will most probably reduce off-target effects. According to first results, collagen expression and the development of fibroplasia in incisional skin wounds have been significantly repressed by the application of MRG-201 and a phase II study to treat patients with a predisposition for keloid formation with MRG-201 is in preparation [212].
Anti-MicroRNA
As mentioned before, anti-miRNA are synthetic single-stranded RNA molecules that, owing to sequence complementary bind and sequester endogenous miRNAs and by this counteract up-regulated miRNAs. MiR-10b is higher expressed in glioblastoma compared with normal brain tissue [213] and therefore the critical function of anti-mir-10b (EXTH-61) in blocking established glioblastoma growth is under investigation in a phase I study [214]. Owing to the fact that miRNAs are able to cross the blood–brain barrier, EXTH-61 could be a further option to overcome the difficulties in treating glioblastoma. However, there is the question on how specific EXTH-61 is taken up by glioblastoma and it will be of interest to see if the anti-miR-10b construct causes off-target effects.
In another phase I/IIa clinical study, 46 patients with type 2 diabetes mellitus and non-alcoholic fatty liver disease receiving metformin monotherapy will be included. A role of miR-103/107 in insulin sensitivity and resistance has been established [215] and, in this study, inhibition of this miRNA via N-acetylgalactosamine conjugated anti-miR-103/107 (RG-125) will be explored. This study was stopped by the company before results were made available.
Following the same approach, anti-miR-122 was used as a conjugate with N-acetylgalactosamine (RG-101) in a multicentre phase Ib study to treat HCV-infected patients who were either treatment naïve or relapsed after interferon-α-based therapy [216, 217]. Twenty-eight patients received one subcutaneous injection of RG-101 and after 4 weeks the viral load was significantly reduced in all patients and the substance was well tolerated. Three patients had undetectable HCV RNA levels even 76 weeks after a single dose of RG-101. In some cases, the observed viral rebound after 12 weeks was associated with the appearance of resistance mediating mutations in miR-122-binding regions in the 5′-untranslated region of the HCV genome [217]. Nevertheless, because of high levels of bilirubin in the blood of the patients, the company decided to terminate the trials based on RG-101. Off-target effects of RG-101 are the inhibition of conjugated bilirubin transport, impaired baseline bilirubin transport and the preferential uptake of RG-101 by hepatocytes resulting in hyperbilirubinemia. This study targets miR-122 in HCV-infected patients like for Miravirsen (described in Sect. 8.1). Locked nucleic acid-antisense-based therapy has an advantage and results in fewer side effects compared with the anti-miRNA strategy. Furthermore, no escape mutations have been observed in patients treated with Miravirsen, but surprisingly, such escape mutations occur during treatment with anti-miR-122 even if both strategies target endogenous miR-122. Considering that both RG-125 as well as RG-101 studies are based on a N-acetylgalactosamine conjugated anti-miRNA, and both have been suspended, it is tempting to speculate that N-acetylgalactosamine conjugation is at least in some part involved in unwanted side effects.
Finally, anti-miR-21 (RG-012) was tested in a phase I study for the efficiency to treat fibrogenetic diseases especially Alport nephropathy [218]. Alport syndrome is a life-threating hereditary kidney disease that currently has no approved therapy [219]. An anti-miRNA-based strategy could be a therapeutic option for rare and orphan diseases such as Alport syndrome for which no drugs for treatment have been developed owing to the small number of patients. One issue of this study is the very limited information on exactly how Alport syndrome progresses, although miR-21 is believed to play a central role in the disease progression. Furthermore, RG-012 is missing a targeting molecule to increase the specific delivery into the kidney and therefore off-target effects are very likely.
Clinical Development of Long Non-Coding RNA-Based Therapeutics
No clinical trials targeting lncRNAs have been started. Nevertheless, the first pre-clinical studies are underway and the first patent for using lncRNA as a therapeutic target has been issued (patent CA 2921457). In several pre-clinical studies, the potential benefit of lncRNA-based therapies for cancer have been demonstrated. Tumour growth and reduction in metastatic sites have been strongly affected by knocking-down lncRNA MALAT1 by a specific ASO in models for breast and lung cancer [179, 220]. Antisense oligonucleotide-mediated down-regulation of lncRN ASChLAP1 has been found to limit tumour formation and metastasis of prostate cancer [221]. Another study using ASO directed against lncRNA USMycN significantly reduced tumour formation in mice with neuroblastoma [222]. The lncRNA-GMAN is highly expressed in gastric cancer cells and is associated with metastatic processes and poor prognosis. By using a CRISPR/Cas9 system targeting lncRNA-GMAN, metastasis of gastric cancer cells was significant reduced and overall survival was improved in a mice model [223].
Furthermore, lncRNAs can also influence the sensitivity of cancer cells towards different types of drugs. Silencing the expression of lncRNA-NEAT1 increased the sensitivity of pre-cancerous cells towards DNA damage-induced cell death and increased lethality of chemotherapy drugs on cancer cells [224]. All these examples underline the significant role of dysregulated lncRNAs in the context of cancer and demonstrate the potential therapeutic power that could result from modifying lncRNA expression.
Of special interest are some clinical phase I/II trials (Table 2) in which the anti-tumour efficiency of a plasmid coding for the A subunit of diphtheria toxin under the control of the lncRNA H19 gene promoter (BC-819) is evaluated in different solid tumours [225–231]. Based on the highly selective tissue expression of lncRNAs such as H19, 22% of the patients with cancer have a complete response and 44% have a partial response [229]. Furthermore, observed side effects are minimal and BC-819 can be considered as safe and well tolerated, thus clinical phase III trials are in preparation [228].
Table 2.
Clinical trials using long non-coding RNA promoter for specific expression of therapeutics in tumour cells
| Drug name | Disease | Phase | Status | ClinicalTrials.gov identifier |
|---|---|---|---|---|
| BC-819 | Unresponsive NMIBC | II | Recruting | NCT03719300 |
| BC-819 | Locally advanced pancreatic adenocarcinoma | IIb | Terminated | NCT01413087 |
| BC-819 | Superficial transitional cell bladder carcinoma | I | Completed | NCT01878188 |
| BC-819 | Advanced ovarian cancer | I/IIa | Completed | NCT00826150 |
| BC-819 | Unresectable pancreatic cancer | II | Completed | NCT00711997 |
| BC-819 | Intermediate-risk superficial bladder cancer | II | Completed | NCT00595088 |
NMIBC non-muscle invasive bladder cancer
Conclusions
The interest in non-coding RNA-based therapeutics was boosted by the first Food and Drug Administration approval of a small-interfering RNA drug in 2018. This small-interfering RNA drug called patisiran targets and degrades the mRNA coding for transthyretin and is used to treat polyneuropathy caused by hereditary transthyretin-mediated amyloidosis [232, 233].
Nevertheless, increasing knowledge about non-coding RNAs especially about lncRNAs is a prerequisite for using the non-coding RNAs as therapeutic targets. Notably, intra- and inter-tumoral heterogeneity are pitfalls in the study of non-coding RNA targets given the tissue-specific interaction. Long non-coding RNAs as well as miRNA expression indeed can vary from one area to another in the same tissue as well as over time; moreover, numerous concomitant conditions can alter the expression levels of non-coding RNAs [234, 235]. Clearly, non-coding RNA-based therapy is still in its infancy. It is very attractive to use these new targets as a further therapeutic option in the context of different diseases because non-coding RNAs are often expressed in a tissue- and cell type-specific manner and their deregulation is disease specific. However, it is also necessary to improve the methods for tissue- and cell type-specific delivery and the targeting of deregulated non-coding RNAs as well as reducing off-target effects especially innate immune responses.
The therapeutic manipulation of miRNAs has gained momentum based on their causative involvement in a broad range of human diseases, ranging from infections to cancer. MicroRNA-targeted therapeutics have recently entered clinical evaluations showing encouraging safety and efficacy in early-phase trials, particularly in patients with HCV infection and malignant pleural mesothelioma [161, 236]. Furthermore, non-coding RNA-based therapies represent an option for life-threatening disease that currently has no approved therapy [219]. A suboptimal in-vivo delivery and emerging side effects could represent drawbacks in successfully translating these compounds to the clinic. A better understanding of their pharmacological properties alongside safer and more efficient delivery technologies will hopefully help realise the tremendous potential of non-coding RNA-based therapeutics.
Authors’ Contributions
MR, NV and JCH conceived and designed the study. MR, AL, MBM and JCH wrote the paper; AL designed the figures; MG and MS summarised the data, analysed the data and created the tables; NV and JCH reviewed and edited the manuscript. All authors read, critically reviewed and approved the final manuscript.
Compliance with Ethical Standards
Funding
No external funding was used in the preparation of this article.
Conflict of Interest
Margherita Ratti, Andrea Lampis, Michele Ghidini, Massimiliano Salati, Milko B. Mirchev and Jens C. Hahne have no conflicts of interest that are directly relevant to the content of this article. Nicola Valeri received speaker honorarium from Bayer, Eli-Lilly, Pfizer and Merck.
References
- 1.Dozmorov MG, Giles CB, Koelsch KA, Wren JD. Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinform. 2013;14(Suppl. 14):S2. doi: 10.1186/1471-2105-14-S14-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med. 2017;21(1):185–192. doi: 10.1111/jcmm.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azlan A, Dzaki N, Azzam G. Argonaute: the executor of small RNA function. J Genet Genom. 2016;43(8):481–494. doi: 10.1016/j.jgg.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Choudhuri S. Small noncoding RNAs: biogenesis, function, and emerging significance in toxicology. J Biochem Mol Toxicol. 2010;24(3):195–216. doi: 10.1002/jbt.20325. [DOI] [PubMed] [Google Scholar]
- 5.Claycomb JM. Ancient endo-siRNA pathways reveal new tricks. Curr Biol. 2014;24(15):R703–R715. doi: 10.1016/j.cub.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 6.de Almeida RA, Fraczek MG, Parker S, Delneri D, O'Keefe RT. Non-coding RNAs and disease: the classical ncRNAs make a comeback. Biochem Soc Trans. 2016;44(4):1073–1078. doi: 10.1042/BST20160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2(12):919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 8.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiger J, Dalgaard LT. Interplay of mitochondrial metabolism and microRNAs. Cell Mol Life Sci. 2017;74(4):631–646. doi: 10.1007/s00018-016-2342-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859(10):1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahne JC, Valeri N. Non-coding RNAs and resistance to anticancer drugs in gastrointestinal tumors. Front Oncol. 2018;8:226. doi: 10.3389/fonc.2018.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 15.Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17(2):93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 16.Langenberger D, Bermudez-Santana C, Hertel J, Hoffmann S, Khaitovich P, Stadler PF. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics. 2009;25(18):2298–2301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- 17.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15(1):39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 20.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15(7):1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 23.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 24.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(1):17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 26.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SS, Lee SV. Non-coding RNAs in Caenorhabditis elegans aging. Mol Cells. 2019;42(5):379–385. doi: 10.14348/molcells.2019.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310(5756):1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 29.Fernandes JCR, Acuna SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Noncoding RNA. 2019;5(1):17. doi: 10.3390/ncrna5010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 33.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA. 2010;107(15):6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 36.Saito T, Saetrom P. MicroRNAs: targeting and target prediction. N Biotechnol. 2010;27(3):243–249. doi: 10.1016/j.nbt.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genom. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13(4):271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30(5):823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68(19):8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 41.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 42.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 43.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer: a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 44.Balatti V, Pekarky Y, Croce CM. Role of microRNA in chronic lymphocytic leukemia onset and progression. J Hematol Oncol. 2015;8:12. doi: 10.1186/s13045-015-0112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 46.Ghidini M, Hahne JC, Frizziero M, Tomasello G, Trevisani F, Lampis A, et al. MicroRNAs as mediators of resistance mechanisms to small-molecule tyrosine kinase inhibitors in solid tumours. Target Oncol. 2018;13(4):423–436. doi: 10.1007/s11523-018-0580-3. [DOI] [PubMed] [Google Scholar]
- 47.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 48.Zhou SS, Jin JP, Wang JQ, Zhang ZG, Freedman JH, Zheng Y, et al. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin. 2018;39(7):1073–1084. doi: 10.1038/aps.2018.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garibaldi F, Falcone E, Trisciuoglio D, Colombo T, Lisek K, Walerych D, et al. Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex. Oncogene. 2016;35(29):3760–3770. doi: 10.1038/onc.2016.51. [DOI] [PubMed] [Google Scholar]
- 50.Gurtner A, Falcone E, Garibaldi F, Piaggio G. Dysregulation of microRNA biogenesis in cancer: the impact of mutant p53 on Drosha complex activity. J Exp Clin Cancer Res. 2016;35:45. doi: 10.1186/s13046-016-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hahne JC, Mirchev M, Kotzev I, Lampis A, Valeri N. Biomarkers for monitoring response to therapies and detection of acquired resistance in advanced gastrointestinal cancers. Front Clin Drug Res. 2017;4:1–73. [Google Scholar]
- 54.Anandappa G, Lampis A, Cunningham D, Khan KH, Kouvelakis K, Vlachogiannis G, et al. miR-31-3p expression and benefit from anti-EGFR inhibitors in metastatic colorectal cancer patients enrolled in the prospective phase II PROSPECT-C trial. Clin Cancer Res. 2019;25(13):3830–3838. doi: 10.1158/1078-0432.CCR-18-3769. [DOI] [PubMed] [Google Scholar]
- 55.Lampis A, Carotenuto P, Vlachogiannis G, Cascione L, Hedayat S, Burke R, et al. MIR21 drives resistance to heat shock protein 90 inhibition in cholangiocarcinoma. Gastroenterology. 2018;154(4):1066–79.e5. doi: 10.1053/j.gastro.2017.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sclafani F, Chau I, Cunningham D, Lampis A, Hahne JC, Ghidini M, et al. Sequence variation in mature microRNA-608 and benefit from neo-adjuvant treatment in locally advanced rectal cancer patients. Carcinogenesis. 2016;37(9):852–857. doi: 10.1093/carcin/bgw073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth EC, Nyamundanda G, Cunningham D, Fontana E, Ragulan C, Tan IB, et al. A seven-gene signature assay improves prognostic risk stratification of perioperative chemotherapy treated gastroesophageal cancer patients from the MAGIC trial. Ann Oncol. 2018;29(12):2356–2362. doi: 10.1093/annonc/mdy407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trevisani F, Ghidini M, Larcher A, Lampis A, Lote H, Manunta P, et al. MicroRNA 193b–3p as a predictive biomarker of chronic kidney disease in patients undergoing radical nephrectomy for renal cell carcinoma. Br J Cancer. 2016;115(11):1343–1350. doi: 10.1038/bjc.2016.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abba ML, Patil N, Leupold JH, Moniuszko M, Utikal J, Niklinski J, et al. MicroRNAs as novel targets and tools in cancer therapy. Cancer Lett. 2017;387:84–94. doi: 10.1016/j.canlet.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 61.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 62.Aslam MI, Patel M, Singh B, Jameson JS, Pringle JH. MicroRNA manipulation in colorectal cancer cells: from laboratory to clinical application. J Transl Med. 2012;10:128. doi: 10.1186/1479-5876-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103(9):919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christopher AF, Kaur RP, Kaur G, Kaur A, Gupta V, Bansal P. MicroRNA therapeutics: discovering novel targets and developing specific therapy. Perspect Clin Res. 2016;7(2):68–74. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 66.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70(18):7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 68.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 69.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 70.van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18(10):1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 71.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35(2):180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 76.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38(20):7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 80.Haimovich G, Ecker CM, Dunagin MC, Eggan E, Raj A, Gerst JE, et al. Intercellular mRNA trafficking via membrane nanotube-like extensions in mammalian cells. Proc Natl Acad Sci USA. 2017;114(46):E9873–E9882. doi: 10.1073/pnas.1706365114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, et al. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28(12):1363–1379. doi: 10.1101/gad.238782.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ounzain S, Pezzuto I, Micheletti R, Burdet F, Sheta R, Nemir M, et al. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J Mol Cell Cardiol. 2014;76:55–70. doi: 10.1016/j.yjmcc.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu Z, Liu X, Liu L, Deng H, Zhang J, Xu Q, et al. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19(4):561–575. doi: 10.2478/s11658-014-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscp Rev RNA. 2018;9(3):e1471. doi: 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun QY, Hao QY, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34(2):142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ribeiro DM, Zanzoni A, Cipriano A, Delli Ponti R, Spinelli L, Ballarino M, et al. Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. 2018;46(2):917–928. doi: 10.1093/nar/gkx1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015;12(10):1094–1098. doi: 10.1080/15476286.2015.1063770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282(34):24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 96.Matouk I, Raveh E, Ohana P, Abu Lail R, Gershtain E, Gilon M, et al. The increasing complexity of the oncofetal H19 gene locus: functional dissection and therapeutic intervention. Int J Mol Sci. 2013;14(2):4298–4316. doi: 10.3390/ijms14024298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci U S A. 2014;111(52):18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14(9):R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-lail R, Fellig Y, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803(4):443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 102.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lopez-Urrutia E, Bustamante Montes LP, Ladron de Guevara Cervantes D, Perez-Plasencia C, Campos-Parra AD. Crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: deciphering molecular mechanisms of master regulators in cancer. Front Oncol. 2019;9:669. [DOI] [PMC free article] [PubMed]
- 104.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hobuss L, Bar C, Thum T. Long non-coding RNAs: at the heart of cardiac dysfunction? Front Physiol. 2019;10:30. doi: 10.3389/fphys.2019.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 108.Martignano F, Rossi L, Maugeri A, Galla V, Conteduca V, De Giorgi U, et al. Urinary RNA-based biomarkers for prostate cancer detection. Clin Chim Acta. 2017;473:96–105. doi: 10.1016/j.cca.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 109.Terracciano D, Ferro M, Terreri S, Lucarelli G, D'Elia C, Musi G, et al. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: new architects in cancer prognostic biomarkers. Transl Res. 2017;184:108–117. doi: 10.1016/j.trsl.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 110.Zhou XY, Yin CQ, Dang YN, Ye F, Zhang GX. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5. [DOI] [PMC free article] [PubMed]
- 111.Li Q, Shao YF, Zhang XJ, Zheng T, Miao M, Qin LJ, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol. 2015;36(3):2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 112.Viereck J, Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res. 2017;120(2):381–399. doi: 10.1161/CIRCRESAHA.116.308434. [DOI] [PubMed] [Google Scholar]
- 113.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114(10):1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 114.Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9(7):1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 115.Betts JA, Moradi Marjaneh M, Al-Ejeh F, Lim YC, Shi W, Sivakumaran H, et al. Long noncoding RNAs CUPID1 and CUPID2 mediate breast cancer risk at 11q13 by modulating the response to DNA damage. Am J Hum Genet. 2017;101(2):255–266. doi: 10.1016/j.ajhg.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shih JW, Chiang WF, Wu ATH, Wu MH, Wang LY, Yu YL, et al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1alpha co-activator driving oral cancer progression. Nat Commun. 2017;8:15874. doi: 10.1038/ncomms15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49(6):1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 119.Xiao ZD, Han L, Lee H, Zhuang L, Zhang Y, Baddour J, et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun. 2017;8(1):783. doi: 10.1038/s41467-017-00902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 121.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57(5):1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 122.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 125.Ren Y, Jia HH, Xu YQ, Zhou X, Zhao XH, Wang YF, et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ss1 secretion. Mol Cancer. 2018;17(1):5. doi: 10.1186/s12943-018-0758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165(7):1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 129.Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015;16(3):318–325. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14(11):1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. doi: 10.1038/ncomms15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen C, He W, Huang J, Wang B, Li H, Cai Q, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun. 2018;9(1):3826. doi: 10.1038/s41467-018-06152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19(10):1112–1125. doi: 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 134.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. The fusion of two worlds: non-coding RNAs and extracellular vesicles: diagnostic and therapeutic implications (review) Int J Oncol. 2015;46(1):17–27. doi: 10.3892/ijo.2014.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nana-Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15(9):445. doi: 10.1186/s13059-014-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sanchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic Acid Ther. 2013;23(1):15–20. doi: 10.1089/nat.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Costa MC, Kurc S, Drozdz A, Cortez-Dias N, Enguita FJ. The circulating non-coding RNA landscape for biomarker research: lessons and prospects from cardiovascular diseases. Acta Pharmacol Sin. 2018;39(7):1085–1099. doi: 10.1038/aps.2018.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391(1):73–77. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 140.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31(6):659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 141.Bye A, Rosjo H, Nauman J, Silva GJ, Follestad T, Omland T, et al. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals: the HUNT study. J Mol Cell Cardiol. 2016;97:162–168. doi: 10.1016/j.yjmcc.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016;6:22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci USA. 2014;111(20):7361–7366. doi: 10.1073/pnas.1405528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu Z, Li X, Sun N, Xu Y, Meng Y, Yang C, et al. Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS ONE. 2014;9(3):e93388. doi: 10.1371/journal.pone.0093388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gilad S, Lithwick-Yanai G, Barshack I, Benjamin S, Krivitsky I, Edmonston TB, et al. Classification of the four main types of lung cancer using a microRNA-based diagnostic assay. J Mol Diagn. 2012;14(5):510–517. doi: 10.1016/j.jmoldx.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 147.Pentheroudakis G, Pavlidis N, Fountzilas G, Krikelis D, Goussia A, Stoyianni A, et al. Novel microRNA-based assay demonstrates 92% agreement with diagnosis based on clinicopathologic and management data in a cohort of patients with carcinoma of unknown primary. Mol Cancer. 2013;12:57. doi: 10.1186/1476-4598-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ferracin M, Pedriali M, Veronese A, Zagatti B, Gafa R, Magri E, et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225(1):43–53. doi: 10.1002/path.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52(6):1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 150.Pal RP, Maitra NU, Mellon JK, Khan MA. Defining prostate cancer risk before prostate biopsy. Urol Oncol. 2013;31(8):1408–1418. doi: 10.1016/j.urolonc.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 151.Wang YH, Ji J, Wang BC, Chen H, Yang ZH, Wang K, et al. Tumor-derived exosomal long noncoding RNAs as promising diagnostic biomarkers for prostate cancer. Cell Physiol Biochem. 2018;46(2):532–545. doi: 10.1159/000488620. [DOI] [PubMed] [Google Scholar]
- 152.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39(16):6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Berardocco M, Radeghieri A, Busatto S, Gallorini M, Raggi C, Gissi C, et al. RNA-seq reveals distinctive RNA profiles of small extracellular vesicles from different human liver cancer cell lines. Oncotarget. 2017;8(47):82920–82939. doi: 10.18632/oncotarget.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gong J, Liu W, Zhang J, Miao X, Guo AY. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015;43:D181–D186. doi: 10.1093/nar/gku1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wessels HH, Lebedeva S, Hirsekorn A, Wurmus R, Akalin A, Mukherjee N, et al. Global identification of functional microRNA-mRNA interactions in Drosophila. Nat Commun. 2019;10(1):1626. doi: 10.1038/s41467-019-09586-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Paraskevopoulou MD, Karagkouni D, Vlachos IS, Tastsoglou S, Hatzigeorgiou AG. microCLIP super learning framework uncovers functional transcriptome-wide miRNA interactions. Nat Commun. 2018;9(1):3601. doi: 10.1038/s41467-018-06046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sclafani F, Chau I, Cunningham D, Peckitt C, Lampis A, Hahne JC, et al. Prognostic role of the LCS6 KRAS variant in locally advanced rectal cancer: results of the EXPERT-C trial. Ann Oncol. 2015;26(9):1936–1941. doi: 10.1093/annonc/mdv285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tan SB, Li J, Chen X, Zhang W, Zhang D, Zhang C, et al. Small molecule inhibitor of myogenic microRNAs leads to a discovery of miR-221/222-myoD-myomiRs regulatory pathway. Chem Biol. 2014;21(10):1265–1270. doi: 10.1016/j.chembiol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 167.Monroig Pdel C, Chen L, Zhang S, Calin GA. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev. 2015;81:104–116. doi: 10.1016/j.addr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang S, Chen L, Jung EJ, Calin GA. Targeting microRNAs with small molecules: from dream to reality. Clin Pharmacol Ther. 2010;87(6):754–758. doi: 10.1038/clpt.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wen D, Danquah M, Chaudhary AK, Mahato RI. Small molecules targeting microRNA for cancer therapy: promises and obstacles. J Control Release. 2015;219:237–247. doi: 10.1016/j.jconrel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47(39):7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132(23):7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 172.Piroozian F, Bagheri Varkiyani H, Koolivand M, Ansari M, Afsa M, AtashAbParvar A, et al. The impact of variations in transcription of DICER and AGO2 on exacerbation of childhood B-cell lineage acute lymphoblastic leukaemia. Int J Exp Pathol. 2019;100(3):184–191. doi: 10.1111/iep.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Essandoh K, Yang L, Wang X, Huang W, Qin D, Hao J, et al. Blockade of exosome generation with GW4869 dampens the sepsis-induced inflammation and cardiac dysfunction. Biochim Biophys Acta. 2015;1852(11):2362–2371. doi: 10.1016/j.bbadis.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells—a new regulatory pathway. Eur J Cancer. 2014;50(5):1025–1034. doi: 10.1016/j.ejca.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 175.Bhome R, Del Vecchio F, Lee GH, Bullock MD, Primrose JN, Sayan AE, et al. Exosomal microRNAs (exomiRs): small molecules with a big role in cancer. Cancer Lett. 2018;420:228–235. doi: 10.1016/j.canlet.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Guay C, Kruit JK, Rome S, Menoud V, Mulder NL, Jurdzinski A, et al. Lymphocyte-derived exosomal microRNAs promote pancreatic beta cell death and may contribute to type 1 diabetes development. Cell Metab. 2019;29(2):348–61.e6. doi: 10.1016/j.cmet.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 177.Kosgodage US, Mould R, Henley AB, Nunn AV, Guy GW, Thomas EL, et al. Cannabidiol (CBD) is a novel inhibitor for exosome and microvesicle (EMV) release in cancer. Front Pharmacol. 2018;9:889. doi: 10.3389/fphar.2018.00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16(11):2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li Z, Li X, Chen X, Li S, Ho IHT, Liu X, et al. Emerging roles of long non-coding RNAs in neuropathic pain. Cell Prolif. 2019;52(1):e12528. doi: 10.1111/cpr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lee JS, Mendell JT. Antisense-mediated transcript knockdown triggers premature transcription termination. Mol Cell. 2020;77(5):1044–54.e3. doi: 10.1016/j.molcel.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Koch L. Functional genomics: screening for lncRNA function. Nat Rev Genet. 2017;18(2):70. doi: 10.1038/nrg.2016.168. [DOI] [PubMed] [Google Scholar]
- 183.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355(6320):7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 186.Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15(8):1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS ONE. 2013;8(10):e77070. doi: 10.1371/journal.pone.0077070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11(2):125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136(4):763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 190.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 191.Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol. 2012;19(8):937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 192.Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44(14):6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sehgal A, Vaishnaw A, Fitzgerald K. Liver as a target for oligonucleotide therapeutics. J Hepatol. 2013;59(6):1354–1359. doi: 10.1016/j.jhep.2013.05.045. [DOI] [PubMed] [Google Scholar]
- 194.Srinivasan SK, Tewary HK, Iversen PL. Characterization of binding sites, extent of binding, and drug interactions of oligonucleotides with albumin. Antisense Res Dev. 1995;5(2):131–139. doi: 10.1089/ard.1995.5.131. [DOI] [PubMed] [Google Scholar]
- 195.Sharma VK, Watts JK. Oligonucleotide therapeutics: chemistry, delivery and clinical progress. Future Med Chem. 2015;7(16):2221–2242. doi: 10.4155/fmc.15.144. [DOI] [PubMed] [Google Scholar]
- 196.Bai ZM, Wei J, Yu CM, Han XS, Qin XF, Zhang CW, et al. Non-viral nanocarriers for intracellular delivery of microRNA therapeutics. J Mater Chem B. 2019;7(8):1209–1225. doi: 10.1039/c8tb02946f. [DOI] [PubMed] [Google Scholar]
- 197.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 198.Orellana EA, Tenneti S, Rangasamy L, Lyle LT, Low PS, Kasinski AL. FolamiRs: ligand-targeted, vehicle-free delivery of microRNAs for the treatment of cancer. Sci Transl Med. 2017;9(401):9327. doi: 10.1126/scitranslmed.aam9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhang Z, Wang J, Tacha DE, Li P, Bremer RE, Chen H, et al. Folate receptor alpha associated with triple-negative breast cancer and poor prognosis. Arch Pathol Lab Med. 2014;138(7):890–895. doi: 10.5858/arpa.2013-0309-OA. [DOI] [PubMed] [Google Scholar]
- 200.Cagle PT, Zhai QJ, Murphy L, Low PS. Folate receptor in adenocarcinoma and squamous cell carcinoma of the lung: potential target for folate-linked therapeutic agents. Arch Pathol Lab Med. 2013;137(2):241–244. doi: 10.5858/arpa.2012-0176-OA. [DOI] [PubMed] [Google Scholar]
- 201.Parker N, Turk MJ, Westrick E, Lewis JD, Low PS, Leamon CP. Folate receptor expression in carcinomas and normal tissues determined by a quantitative radioligand binding assay. Anal Biochem. 2005;338(2):284–293. doi: 10.1016/j.ab.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 202.Yang J, Vlashi E, Low P. Folate-linked drugs for the treatment of cancer and inflammatory diseases. Subcell Biochem. 2012;56:163–179. doi: 10.1007/978-94-007-2199-9_9. [DOI] [PubMed] [Google Scholar]
- 203.Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Syn NL, Wang L, Chow EK, Lim CT, Goh BC. Exosomes in cancer nanomedicine and immunotherapy: prospects and challenges. Trends Biotechnol. 2017;35(7):665–676. doi: 10.1016/j.tibtech.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 205.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ottosen S, Parsley TB, Yang L, Zeh K, van Doorn LJ, van der Veer E, et al. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob Agents Chemother. 2015;59(1):599–608. doi: 10.1128/AAC.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Querfeld C, Pacheco T, Foss FM, Halwani AS, Porcu P, Seto AG, et al. Preliminary results of a phase 1 trial evaluating MRG-106, a synthetic microRNA antagonist (LNA antimiR) of microRNA-155, in patients with CTCL. Blood. 2016;128(22):1829. [Google Scholar]
- 208.Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. 2018;183(3):428–444. doi: 10.1111/bjh.15547. [DOI] [PubMed] [Google Scholar]
- 209.Gallant-Behm CL, Piper J, Dickinson BA, Dalby CM, Pestano LA, Jackson AL. A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound Repair Regen. 2018;26(4):311–323. doi: 10.1111/wrr.12660. [DOI] [PubMed] [Google Scholar]
- 210.Kreth S, Hubner M, Hinske LC. MicroRNAs as clinical biomarkers and therapeutic tools in perioperative medicine. Anesth Analg. 2018;126(2):670–681. doi: 10.1213/ANE.0000000000002444. [DOI] [PubMed] [Google Scholar]
- 211.MacDiarmid JA, Mugridge NB, Weiss JC, Phillips L, Burn AL, Paulin RP, et al. Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer Cell. 2007;11(5):431–445. doi: 10.1016/j.ccr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 212.Gallant-Behm CL, Piper J, Lynch JM, Seto AG, Hong SJ, Mustoe TA, et al. A MicroRNA-29 mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Invest Dermatol. 2019;139(5):1073–1081. doi: 10.1016/j.jid.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 213.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71(10):3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Monroig-Bosque PD, Rivera CA, Calin GA. MicroRNAs in cancer therapeutics: "from the bench to the bedside". Expert Opin Biol Th. 2015;15(10):1381–1385. doi: 10.1517/14712598.2015.1074999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 216.Motavaf M, Safari S, Alavian SM. Targeting microRNA-122: walking on cutting edge of hepatitis C virus infection therapy. Acta Virol. 2014;58(4):301–308. doi: 10.4149/av_2014_04_301. [DOI] [PubMed] [Google Scholar]
- 217.van der Ree MH, de Vree JM, Stelma F, Willemse S, van der Valk M, Rietdijk S, et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: a phase 1B, double-blind, randomised controlled trial. Lancet. 2017;389(10070):709–717. doi: 10.1016/S0140-6736(16)31715-9. [DOI] [PubMed] [Google Scholar]
- 218.Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest. 2015;125(1):141–156. doi: 10.1172/JCI75852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4(121):121. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 222.Liu PY, Erriquez D, Marshall GM, Tee AE, Polly P, Wong M, et al. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J Natl Cancer Inst. 2014;106(7):113. doi: 10.1093/jnci/dju113. [DOI] [PubMed] [Google Scholar]
- 223.Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, et al. Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156(3):676–91.e11. doi: 10.1053/j.gastro.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 224.Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 225.Gofrit ON, Benjamin S, Halachmi S, Leibovitch I, Dotan Z, Lamm DL, et al. DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J Urol. 2014;191(6):1697–1702. doi: 10.1016/j.juro.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 226.Hanna N, Ohana P, Konikoff FM, Leichtmann G, Hubert A, Appelbaum L, et al. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther. 2012;19(6):374–381. doi: 10.1038/cgt.2012.10. [DOI] [PubMed] [Google Scholar]
- 227.Hasenpusch G, Pfeifer C, Aneja MK, Wagner K, Reinhardt D, Gilon M, et al. Aerosolized BC-819 inhibits primary but not secondary lung cancer growth. PLoS ONE. 2011;6(6):e20760. doi: 10.1371/journal.pone.0020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lavie O, Edelman D, Levy T, Fishman A, Hubert A, Segev Y, et al. A phase 1/2a, dose-escalation, safety, pharmacokinetic, and preliminary efficacy study of intraperitoneal administration of BC-819 (H19-DTA) in subjects with recurrent ovarian/peritoneal cancer. Arch Gynecol Obstet. 2017;295(3):751–761. doi: 10.1007/s00404-017-4293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sidi AA, Ohana P, Benjamin S, Shalev M, Ransom JH, Lamm D, et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. J Urol. 2008;180(6):2379–2383. doi: 10.1016/j.juro.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 230.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12(5):607–616. [PubMed] [Google Scholar]
- 231.Sorin V, Ohana P, Gallula J, Birman T, Matouk I, Hubert A, et al. H19-promoter-targeted therapy combined with gemcitabine in the treatment of pancreatic cancer. ISRN Oncol. 2012;2012:351750. doi: 10.5402/2012/351750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Adams D, Gonzalez-Duarte A, O'Riordan WD, Yang CC, Ueda M, Kristen AV, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379(1):11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 233.Yang J. Patisiran for the treatment of hereditary transthyretin-mediated amyloidosis. Expert Rev Clin Pharmacol. 2019;12(2):95–99. doi: 10.1080/17512433.2019.1567326. [DOI] [PubMed] [Google Scholar]
- 234.Petrovic N, Ergun S, Isenovic ER. Levels of microRNA heterogeneity in cancer biology. Mol Diagn Ther. 2017;21(5):511–523. doi: 10.1007/s40291-017-0285-9. [DOI] [PubMed] [Google Scholar]
- 235.Lv D, Wang X, Dong J, Zhuang Y, Huang S, Ma B, et al. Systematic characterization of lncRNAs' cell-to-cell expression heterogeneity in glioblastoma cells. Oncotarget. 2016;7(14):18403–18414. doi: 10.18632/oncotarget.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014;111:53–59. doi: 10.1016/j.antiviral.2014.08.015. [DOI] [PubMed] [Google Scholar]