Abstract
The Northern Wheatear (Oenanthe oenanthe, including the nominate and the two subspecies O. o. leucorhoa and O. o. libanotica) and the Seebohm’s Wheatear (Oenanthe seebohmi) are today regarded as two distinct species. Before, all four taxa were regarded as four subspecies of the Northern Wheatear. Their classification has exclusively been based on ecological and morphological traits, while their molecular characterization is still missing. With this study, we used next-generation sequencing to assemble 117 complete mitochondrial genomes covering O. o. oenanthe, O. o. leucorhoa and O. seebohmi. We compared the resolution power of each individual mitochondrial marker and concatenated marker sets to reconstruct the phylogeny and estimate speciation times of three taxa. Moreover, we tried to identify the origin of migratory wheatears caught on Helgoland (Germany) and on Crete (Greece). Mitogenome analysis revealed two different ancient lineages that separated around 400,000 years ago. Both lineages consisted of a mix of subspecies and species. The phylogenetic trees, as well as haplotype networks are incongruent with the present morphology-based classification. Mitogenome could not distinguish these presumed species. The genetic panmixia among present populations and taxa might be the consequence of mitochondrial introgression between ancient wheatear populations.
Subject terms: Ecology, Evolution, Genetics, Molecular biology, Ecology
Introduction
Maternal inheritance in most taxa (such as birds), rare recombination, and appropriate gene content make mitochondrial DNA (mtDNA) the cornerstone of phylogeographic and taxonomic studies in birds and other organisms1–3. In addition, mtDNA is conservative in size and organization, but nevertheless exhibits comparatively high substitution rates, which makes it a good marker for evolutionary events during the last 20 million years4. Moreover, methods of mtDNA analysis are well established, and do not require high-performance computing power.
Avian mtDNA is a closed circular molecule containing 13 protein coding genes (PCGs), including ATPase subunit 6 (ATP6) and subunit 8 (ATP8), cytochrome c oxidase subunit 1–3 (COX1, COX2 and COX3), cytochrome B (CytB), NADH dehydrogenase subunits 1–6 and 4 l (ND1–6 and ND4l); 2 ribosomal RNAs (rRNAs); 22 transfer RNAs (tRNAs) and a control region (CR). For DNA barcoding, even short stretches of DNA (e.g. <650 base pairs (bp) of cytochrome c oxidase subunits 1 (COX1)) can sometimes effectively delimit species. Accordingly, COX1 has been used as a marker gene and confirmed the classification of >94% of bird species which had previously been classified based on morphology5–10. Nevertheless, single mitochondrial markers are often inappropriate to reliably reconstruct the phylogeography of populations below the species level. Concatenating several mitochondrial gene sequences obtained by traditional Sanger sequencing may improve the situation, but still does not always yield satisfactory results11–13. With the advance of fast moving high-throughput sequencing technologies, the field of phylogeography and population genetics might break the shackles of previous sequencing technologies as they are no longer restricted to a handful of molecular markers. It is now feasible to obtain sequences of complete mitogenomes of large sample sizes to infer the phylogeographic histories of avian species in a short time.
Wheatears (genus Oenanthe) are small passerines in the family Muscicapidae (Old World flycatchers and chats). They are specialized to open habitats and many of them inhabit savannah and desert ecosystems14. To date, the genus Oenanthe sensu stricto comprises 29 species14. The Northern Wheatear (Oenanthe oenanthe) breeds almost across the whole Holarctic. It consists of 3 subspecies, O. o. oenanthe (nominate), O. o. leucorhoa (the “Greenland Wheatear” hereafter), and O. o. libanotica (southern Europe and Asia). In this study, we focused on 3 taxa, O. o. oenanthe, O. o. leucorhoa and O. seebohmi. Breeding sites of O. o. oenanthe range from continental Europe and Great Britain to Siberia and Alaska, while O. o. leucorhoa breeds in Greenland, Iceland and the East of Canada14. During autumn and spring migration, these two subspecies gather at stopover sites in Northern and Western Europe15–18. The sedentary Seebohm’s Wheatear, however, is confined to the Atlas Mountains (Morocco) and had traditionally been included in the Northern Wheatear (O. oenanthe) as a distinctive subspecies (O. o. seebohmi). However, it has recently been separated and considered as a conspecific taxon, O. seebohmi14.
Wheatears exhibit a remarkable complexity and variety of color patterns, while they are rather congeneric with respect to display behavior, foraging, and morphological traits19. Small, but distinctive differences exist between O. o. leucorhoa and O. o. oenanthe. In the field, the 2 subspecies to a certain extent, can be distinguished by wing length and plumage coloration15,20. In line with the current classification as a separate species, O. seebohmi, based on its plumage coloration, can be readily told apart from O. oenanthe. Oenanthe seebohmi can be discriminated by its black throat, its black underwing-coverts and its larger proportion of white on the forehead when compared to other wheatear species. However, species status and particularly the delineation of subspecies within the wheatear genus, have been challenged for decades based on ecological, geographical, morphological and genetic characters19,21–28. In spite of the numerous investigations of the morphology and migration ecology of wheatears18,19,22,29–31, this controversial debate is still ongoing and can only be resolved by molecular data.
In this study, we reconstructed the complete reference mitogenome of the Northern Wheatear and Seebohm’s Wheatear. In order to obtain a comprehensive phylogenetic framework and robust population genetics, we investigated the mitogenome of 117 individuals from 7 breeding populations: Sweden (Ammarnäs, Uppsala, O. o. oenanthe), Norway (O. o. oenanthe), Germany (Norderney, O. o. oenanthe), Alaska (O. o. oenanthe), Iceland (O. o. leucorhoa), and Morocco (O. seebohmi). We compared the resolution power of single mtDNA markers, multiple mtDNA markers and complete mitochondrial genomes for inferring phylogenetic relationships. We further used mitogenomes of Northern Wheatears, which were sampled as passage migrants at two stop-over sites, Helgoland and Crete, to test whether we could infer their origins based on complete mitogenome sequences. Finally, we assessed whether the molecular data is in consensus with the recent morphological and ecological classification of the Northern Wheatear and its closely related species Seebohm’s Wheatear.
Results and Discussion
Mitogenome organization
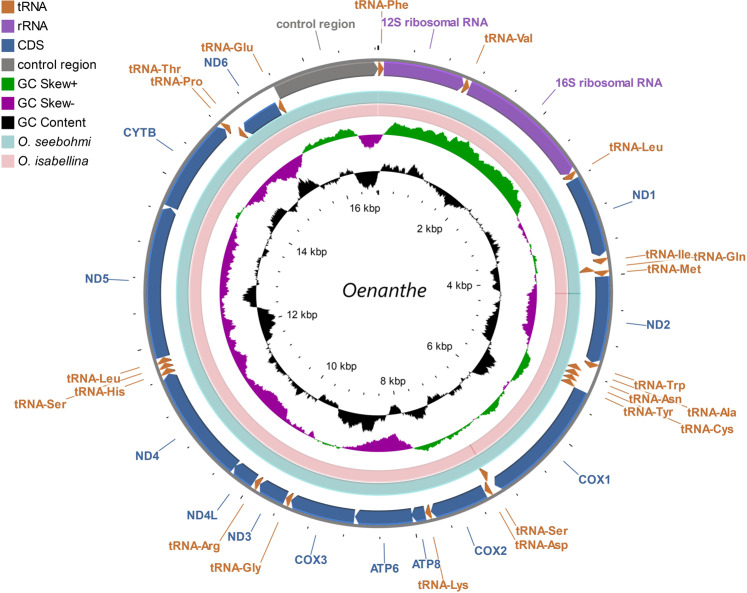
Mitogenomes of 2 subspecies of the Northern Wheatear (O. o. oenanthe and O. o. leucorhoa), 1 congeneric species (O. seebohmi), and the outgroup O. isabellina were assembled de novo and the reads generated by massive parallel sequencing (NGS) of mtDNA were mapped. A mitogenome contraction map (Fig. 1) was built. The total length of the mitochondrial DNA sequence of O. oenanthe ranged from 16,824 to 16,835 bp (Table 1). Except for ND6 and 8 tRNAs, all remaining genes [12 protein-coding genes (PCGs), 2 rRNAs and 14 tRNAs] were located on the heavy DNA strand (H-strand). The arrangement of the whole mitogenome of O. oenanthe, O. seebohmi and O. isabellina were identical to the typical mitogenome of birds, that was initially established in Gallus gallus32. No gene rearrangements and duplications were found in our samples. However, when compared to the O. oenanthe, a few deletions were detected in the mitogenome of O. isabellina inside the non-coding area between ND1 and tRNA-Ile. Another deletion was found in the coding area of tRNA-Trp (Table 1). The overall base compositions of O. oenanthe and O. seebohmi under study were also similar to the related species O. isabellina. The relative abundance of nucleotides was C (32.8%) > A (30.0%) > T (22.9%)> G (14.3%), and the GC skew was negative for the mitogenome.
Figure 1.
Circular map of the mitogenome of Oenanthe oenanthe assembled from NGS data. Features are represented by different color blocks. Arrows indicate the orientation of the gene transcription. The GC content and GC skew were calculated using a sliding window and plotted as the deviation from the average value of the entire sequence. The BLAST comparisons of Oenanthe oenanthe with Oenanthe seebohmi and Oenanthe oenanthe with Oenanthe isabellina are shown in divider rings.
Table 1.
Annotation of assembled mitochondrial genome of Oenanthe oenanthe.
| Product | Start | End | Size | Strand | Name | Anti-code | Start codon | Stop codon |
|---|---|---|---|---|---|---|---|---|
| tRNA-Phe | 1 | 68 | 68 | + | trnF | GAA | ||
| 12S ribosomal RNA | 69 | 1048 | 980 | + | rrnS | − | AAA | TAC |
| tRNA-Val | 1049 | 1118 | 70 | + | trnV | TAC | ||
| 16S ribosomal RNA | 1119 | 2715 | 1597 | + | rrnL | − | TGC | CCC |
| tRNA-Leu | 2716 | 2790 | 75 | + | trnL2 | TAA | ||
| ND1 | 2796 | 3773 | 978 | + | nad1 | − | ATG | AGA |
| tRNA-Ile | 3794 | 3865 | 72 | + | trnI | GAT | ||
| tRNA-Gln | 3871 | 3941 | 71 | − | trnQ | TTG | ||
| tRNA-Met | 3941 | 4009 | 69 | + | trnM | CAT | ||
| ND2 | 4010 | 5049 | 1040 | + | nad2 | − | ATG | TA(A) |
| tRNA-Trp | 5050 | 5120 | 71 | + | trnW | TCA | ||
| tRNA-Ala | 5122 | 5190 | 69 | − | trnA | TGC | ||
| tRNA-Asn | 5195 | 5267 | 73 | − | trnN | GTT | ||
| tRNA-Cys | 5268 | 5334 | 67 | − | trnC | GCA | ||
| tRNA-Tyr | 5334 | 5404 | 71 | − | trnY | GTA | ||
| COX1 | 5406 | 6956 | 1551 | + | cox1 | − | GTG | AGG |
| tRNA-Ser | 6948 | 7022 | 75 | − | trnS2 | TGA | ||
| tRNA-Asp | 7026 | 7094 | 69 | + | trnD | GTC | ||
| COX2 | 7102 | 7785 | 684 | + | cox2 | − | ATG | TAA |
| tRNA-Lys | 7787 | 7854 | 68 | + | trnK | TTT | ||
| ATP8 | 7856 | 8023 | 168 | + | atp8 | − | ATG | TAA |
| ATP6 | 8014 | 8697 | 684 | + | atp6 | − | ATG | TAA |
| COX3 | 8703 | 9486 | 784 | + | cox3 | − | ATG | T |
| tRNA-Gly | 9487 | 9555 | 69 | + | trnG | TCC | ||
| ND3 | 9556 | 9906 | 351 | + | nad3 | − | ATG | TAA |
| tRNA-Arg | 9908 | 9977 | 70 | + | trnR | TCG | ||
| ND4L | 9979 | 10275 | 297 | + | nad4l | − | ATG | TAA |
| ND4 | 10269 | 11646 | 1378 | + | nad4 | − | ATG | T |
| tRNA-His | 11647 | 11717 | 71 | + | trnH | TCG | ||
| tRNA-Ser | 11718 | 11784 | 67 | + | trnS1 | GCT | ||
| tRNA-Leu | 11784 | 11854 | 71 | + | trnL1 | TAG | ||
| ND5 | 11855 | 13672 | 1818 | + | nad5 | − | ATG | AGA |
| CYTB | 13681 | 14823 | 1143 | + | cob | − | ATG | TAA |
| tRNA-Thr | 14827 | 14895 | 69 | + | trnT | TGT | ||
| tRNA-Pro | 14902 | 14971 | 70 | − | trnP | TGG | ||
| ND6 | 14989 | 15507 | 519 | − | nad6 | − | ATG | TAG |
| tRNA-Glu | 15509 | 15580 | 72 | − | trnE | TTC | ||
| control region | 15581 | 16824-16835 | + | CR | − |
The total length of the 13 PCGs was 11,405 bp, representing approximately 67.8% of the entire mitogenome of 3 study taxa. The GC content was 48.1% and 47.1% for the 13 PCG and the complete mitogenome, respectively. The genes coding for individual tRNAs ranged from 67 bp to 75 bp and summed up to 1,547 bp in total length. Since tRNAs play an important role in translating mRNA into protein, they are highly conserved. Accordingly, polymorphisms among birds of the 7 different breeding sites were only detected in 5 out of 22 tRNAs (tRNA-Trp, tRNA-Asn, tRNA-Lys, tRNA-Arg and tRNA-Gly). tRNAs adopted the conventional secondary structure of four-armed cloverleaves and L-shaped tertiary structure33. Owing to the absence of the dihydrouridine arm (D-arm), tRNA-Ser (GCT) did not fold into a cloverleaf conformation. This pattern has been known for other species for decades and is particularly common in animal mitochondrial genomes34.
Phylogeography and population genetics based on the mitogenomic data
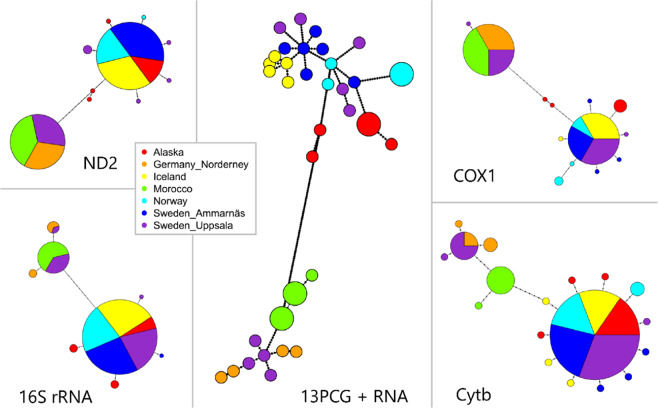
Minimum spanning network analysis of the 13 PCGs, rRNAs and tRNAs from 38 individuals from the 7 breeding populations in Central and Northern Europe (Norway, Germany_Nordeney, Sweden_Ammarnäs and Sweden_Uppsala), Iceland, Alaska and Morocco yielded 34 haplotypes with a maximum of 98 mutation steps (Fig. 2). The highest level of haplotype diversity with maximum mutation steps for single markers was found in ND5 (Fig. S1, Fig. S2). Most of the genetic markers exhibited two haplotype groups (Figs. 2, S2). Exceptions were tRNAs and ATP8 being either highly conservative or extraordinarily short in length (Fig. S2, Table 1). Figure 2 illustrates the minimum spanning networks based on 4 commonly used markers in phylogeography and a concatenated set containing all mitochondrial markers. When looking at the haplotype networks based on the single markers, the Norwegian O. o. oenanthe shared the haplotypes with the other nominate Northern Wheatears from Ammarnäs (Sweden) and Alaska, but also with the “Greenland Wheatears” (O. o. leucorhoa) breeding in Iceland. Hence, this first haplotype group consisted of mixed samples of O. o. oenanthe and O. o. leucorhoa. Similarly, the Moroccan O. seebohmi clustered with the O. o. oenanthe from Uppsala (Sweden) and Norderney (Germany) and constituted the second group of haplotypes. Even when using the full resolution power of all mitochondrial markers, no clear separation among the (sub)species was evident. Accordingly, a limited relationship between the geographic subdivision and haplotype classification was observed. Indeed, these star-like topologies, i.e., a few dominant haplotypes surrounded by less frequent mutations (Fig. 2, S2, Table S1), have often been reported in Eurasian birds11–13.
Figure 2.
Haplotype network for 38 individuals of Oenanthe. Size of the circles represent the frequency of haplotypes. The breeding areas are coded by the colors. Each dot indicates one mutation step. Samples from Morocco are Seebohmi Wheatears (Oenanthe seebohmi); samples from Iceland are Greenland Wheatears (O. o. leucorhoa); samples from Alaska, Germany, Norway and Sweden are nominate wheatears (O. o. oenanthe). The classification of (sub)species are identified by the morphological data.
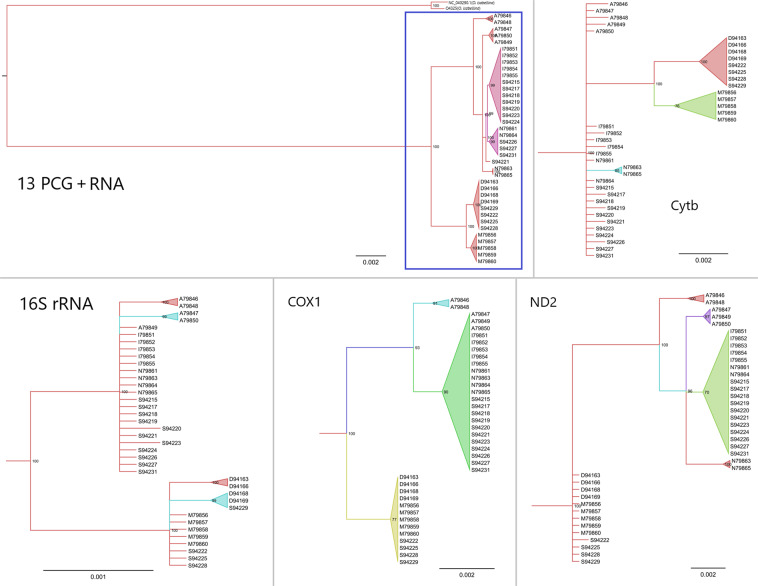
In order to calculate their evolutionary divergence, we firstly estimated the substitutions per site for all the mitochondrial markers of 38 breeding Northern Wheatears. In general, RNA genes were more conservative, resulting in low substitution rates. Among the 13 PCGs, ND2, the most commonly used genetic marker showed the highest substitute rate, followed by ND5, ND6 and Cytb (Fig. S1). On the contrary, other frequently-used barcoding genes (16S rRNA and COX1), exhibited low levels of divergence among the Northern Wheatear populations (Fig. S1). In a next step, we compared the tree topologies of single molecular markers with the phylogenetic tree based on the concatenated set of all mitochondrial markers. We found that all the individual markers could differentiate between O. oenanthe and its sister species O. isabellina, but at the subspecies or population levels, the use of single or a few concatenated markers yielded contradictory subdivisions and shallow trees (Fig. 3, S3). Figure 3 shows inconsistent tree topologies generated from each of four frequently used markers in avian phylogeny studies (16S rRNA, COX1, Cytb and ND2). 16S rRNA and COX1 had reduced substitution rates when compared to ND2 and Cytb, but exhibited high congruence with the concatenated set of markers and separated the two main haplotype clusters.
Figure 3.
MrBayes reconstruction of the Oenanthe oenanthe and it’s outgroup Oenanthe isabellina. Numbers above nodes refer to the support values of Bayesian posterior probability. Sample name indicate the localities by the capital letter, ‘A’ refers to Alaska; ‘D’ refers to Germany; ‘I’ refers to Iceland; ‘M’ refers to Morocco; ‘N’ refers to Norway and ‘S’ refers to Sweden. Samples from Morocco are black Seebohmi Wheatears (Oenanthe seebohmi); samples from Iceland are Greenland Wheatears (O. o. leucorhoa); samples from Alaska, Germany, Norway and Sweden are nominate wheatears (O. o. oenanthe).
For phylogenetic analysis, we followed the most common of the traditional approaches by concatenating two marker genes per analysis, namely ND2 | Cytb and 16S rRNA | COX1. The phylogenetic tree obtained from concatenated ND2 | Cytb was in agreement with the mitogenome results (Fig. S4, Fig. 3, Fig. S3). Furthermore, this combination was much more superior in terms of resolution power, when compared to the tree reconstructions based on the single markers. On the other hand, concatenated 16S rRNA | COX1 was not superior to the individual markers 16S rRNA and COX1, respectively, in terms of resolution (Fig. S4, Fig. 3, Fig. S3). Since our analysis clearly indicated conflicting phylogeographies based on the use of different single or a few concatenated mitochondrial markers (Fig. 3, Fig. S3, Fig. S4), the significance of studies based on traditional approaches should be critically re-examined as they might not mirror actual evolutionary processes. Complete mitogenomes however – as shown in this study – yield potentially enhanced phylogeographic resolution at species and population levels35–37.
In this regard, it is important to mention that high transition-transversion ratios of avian mitochondrial DNA result in a skewed base composition38. For this reason, model selection taking into account codon positions greatly impacts efficiency and accuracy of mtDNA analyses39–41. Accordingly, the comparison of mitochondrial gene trees based on partitioned and non-partitioned sequence data, respectively, (Fig. S4) confirmed that partition strategies can increase accuracy of phylogenetic predictions as well as statistical support42–44.
Can mitogenomes reveal the breeding origin of migrants captured at stop-over sites
Not only for studying phylogenetics, phylogeography and population genetics, the mitochondrial markers have also regarded as logbooks for migration studies. Different mitochondrial lineages can be distinguished by accumulating mutations over time. Based on these mutations, individuals can be assigned to those lineages and their corresponding commons ancestors45. The haplotype of a migrant bird might therefore theoretically predict its geographic origin. In an ideal situation, populations with limited inter-lineage gene flow, contain conservative haplotypes or closely related haplotypic groups corresponding to their geographic localities. The breeding area of migrants could then be recognized by matching to the known breeding population haplotypes45. One of the purposes of this study is to test this scenario. We planned to predict the origins of migrating Northern Wheatears sampled on Helgoland (Germany) and Crete (Greece) through comparing the haplotypes with those of breeding wheatears.
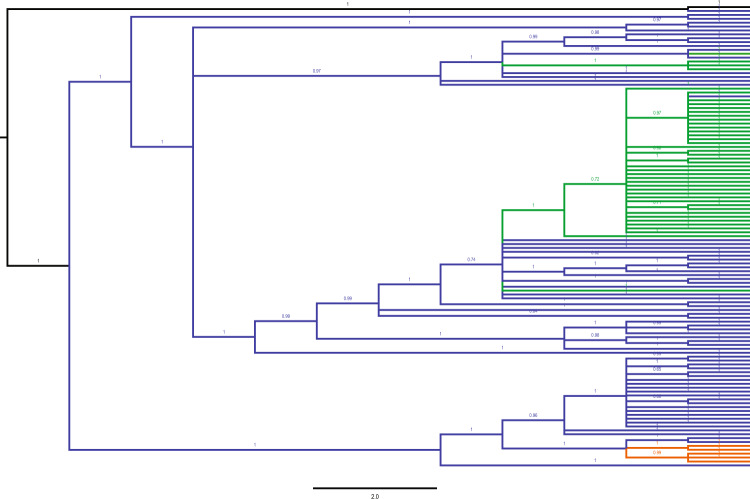
Given the present results, it was very unlikely that the origin of the migrant birds captured on the islands of Helgoland and Crete could be determined, since even birds from different breeding populations could not reliably be separated from each other. However, inadequate sampling46,47 could have contributed to the tree topology of Fig. 3. In order to minimize these effects, we included the Helgoland and Crete birds. These individuals consisted of 2 subspecies O. o. oenanthe and O. o. leucorhoa, that have been identified by morphology and whole genome sequencing (data in preparation). In addition, sequences of the Southern Northern Wheatear (O. o. libanotica)21 obtained from GenBank were included in the analyses. However, the inclusion of additional data did not considerably improve tree topology, and mitogenomic admixture among the subspecies was evident (Figs. 4, S5). This holds true for both molecular classifications based on the single mitochondrial markers and the whole mitogenomes. It was supported by high posterior probabilities implying gene flow among the 3 wheatear taxa (Fig. 3). Moreover, since the breeding birds did not form clear clusters in the haplotype network, the origin of the migrants could not be inferred using haplotype networks either (Fig. S6).
Figure 4.
MrBayes reconstruction based on all the concatenated mitochondrial markers of 3 taxa of Oenanthe and its outgroup. 38 breeding samples and 79 migrant samples (caught in Helgoland and Crete) were included. Numbers above nodes refer to the support values of Bayesian posterior probability. Taxa identifications are color coded based on the field data: outgroup (black), O. o. oenanthe (blue), O. o. leucorhoa (green) and O. seebohmi (orange).
Hypotheses to resolve the conflict between phenotype and haplotype
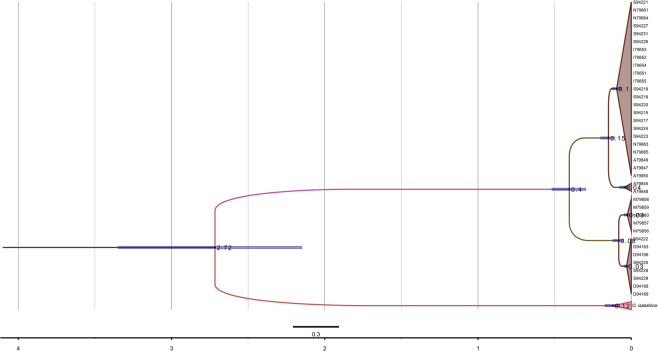
In this study, we successfully detected the footprints of gene flow between ancient Northern Wheatear populations. Mitogenome data identified two clades with about 1% sequence differences, corresponding to an estimate of divergence time of about 400,000 years ago (Figs. 3–5). However, the separation of lineages did not align with the phenotypic differences. Northern Wheatears whose appearance was distinct, were genetically alike, and, vice versa. This finding stands in sharp contrast to recent taxonomy considering the Seebohm’s Wheatear as a separate species.
Figure 5.
Phylogenetic relationships of O. o. oenanthe (blue), O. o. leucorhoa (green) O. seebohmi and O. isabellina based on the mitogenome. The values indicate the split time calculated by BEAST 1.8. The Bayesian posterior probabilities of all the nodes are 1. The blue bars show 95% highest posterior density (HPD) of divergence times.
The explanation might be that Northern Wheatears underwent speciation reversal upon secondary contacts, causing recent introgression of mtDNA across (sub)species boundaries. Recent hybridizations between ancient populations of Morocco and Europe introduced African genetic material into the mitogenome of nominate Northern Wheatears. Due to selective sweep, these new haplotypes have become fixed in a few present populations. This hypothesis is supported by the uniparental inheritance of mtDNA, which results in a lower effective population size and correspondingly in high fixation rates of foreign mitogenomic lineages caused by introgression48. This is in line with multimodal mismatch distributions and non-significant neutrality tests of the 3 taxa (Fig. S7). In addition, the paleoclimatological history corroborates the assumption of speciation reversal. Past climates in the last 2 million years changed in 100,000-year cycles, with alternating warm and cold periods causing the northern hemisphere to repeatedly become inhospitable for most living species49–52. During the glacial periods, large parts of North America, North and West Europe were covered by a thick layer of ice. Only some ice-free Mediterranean areas served as refugia for survivors. For instance, Portugal and Spain, Italy, and the Balkans were three potential refuges during the last glaciation53–55. Consequently, populations of different origins regularly met and exchanged genetic material in the ice-free refugia. They could spread to different locations when ice sheets receded, and suitable new vegetation zones appeared. The hypothesis of cyclical mitochondrial introgression upon secondary contacts could explain that some ancestral mitochondrial polymorphisms are conserved in multiple lineages of recent Northern Wheatear populations and subspecies, although nuclear gene flow is limited (MS in preparation).
Inconsistencies between the mitogenome and morphology-based classifications are not a rare phenomenon. A remarkable example is yellowhammers (Emberiza citrinella) and pine buntings (Emberiza leucocephalos); these two species share similar mitochondrial haplotypes although they are morphologically divergent56. Similar patterns of genetic and phenotypic discordance have also been reported in the other species57–60. In addition to introgression, incomplete lineage sorting (ILS) can lead to non-monophyly and mito-nuclear incongruence, when the species divergence has occurred recently61. Indeed, it is cumbersome to distinguish these two processes because they generate similar genetic signatures62,63. Many recent studies have evaluated the high impact of ILS on the coalescent analysis of reconstructing the speciation process61,64,65. However, we do not consider the ILS as responsible for the unusual mtDNA divergence among these three (sub)species since only part of the nominate populations (O. o. oenanthe) shared haplotypes with the Moroccan populations (O. seebohmi). Therefore, it is more likely that introgressive hybridization (because of cyclic range expansions and reduction during warm and cold periods) determined the haplotype patterns of Northern Wheatears.
Conclusion
Our study is the first one to describe the complete mitogenome of the Northern Wheatear and to reconstruct phylogeographic relationships based on mitochondrial genomes using a large sample size. Our results based on high-quality genetic data differ from the recent subspecies classification derived from morphological traits. The subspecies O. o. oenanthe and O. o. leucorhoa and even O. seebohmi cannot be distinguished by mtDNA. This might be the consequence of recent mitochondrial introgressive hybridizations between the taxa. High resolution markers, such as genome-wide SNPs, should be used, as they could separate these 3 (sub)species (MS in preparation). Although the mitogenome is not congruent with the morphological, ecological and nuclear classification of Northern Wheatears, it can still contribute to deciphering evolutionary processes. A big advantage of mtDNA in phylogeography is its major contribution to detecting recent historical events, which might otherwise have been overlooked if analyses are solely based on nuclear markers.
Materials and methods
Sample collection, DNA extraction and sequencing
In this study, we sampled 117 Northern Wheatears (O. oenanthe) and Seebohm’s Wheatear (O. seebohmi), including samples from birds breeding in Alaska (5), Iceland (5), Norway (4), Germany (4), Sweden (6 in Ammarnäs; 9 in Uppsala), Morocco (5) and samples of actively migrating birds from Greece (6 on Crete) and Germany (73 on Helgoland) (Table S2). As outgroup we used O. isabellina66. All methods carried out in accordance with the relevant guidelines and regulations. In Sweden, birds were captured and marked with permission from the Swedish Bird Ringing Centre. The sampling for Uppsala was approved by the Uppsala animal ethics committee (permit no. C117/8), and for Ammarnäs by the Umeå animal ethics committee M64–05 and M160–11). In Alaska, the sampling was under a license of the U.S. Fish and Wildlife Service (Federal Fish and Wildlife Permits: MB207892-0, MB97904A-0) and the State of Alaska Department of Fish and Game (Permits: 13–103, 14–009). In Germany, the sampling was under license of the German Federal State of Lower Saxony (33.19-42502-04-16/2349) and Schleswig-Holstein (V 244-4829/2017 (33-3/17)). The samples from Greece were provided from IPMB (Institute of Pharmacy and Molecular Biotechnology, Heidelberg University, Germany). Total genomic DNA was extracted from the blood or tissue samples according to the standard phenol-chloroform protocol67. DNA libraries were constructed according to the manufacturer’s instructions (Illumina) with 350 bp insertions. Whole genome sequencing was carried out on the Illumina Novaseq. 6000 platform with a paired-end read length of 150 bp at Berry Genomics company (Beijing, China). The average sequencing depth per individual was 15 folds. Adapter sequencing, low-quality reads were filtered to obtain clean data. We further added sequences obtained from GenBank including more sampling localities (the Netherland, Canada, Iran, Kazakhstan, Mongolia) for investigation (Table S3)21,68,69 6,7,9.
Mitochondrial sequence assembly and gene annotation
Complete mitogenomes were assembled using the MITOBim 1.8 pipeline70, which relies on the baiting and iterative mapping strategy implemented in MIRA471. BLASTn was used to determine the accuracy of the assembled mitogenomes by comparison with the complete mitogenomes of the Collared Flycatcher (Ficedula albicollis) (NC_021621.1) and Isabelline Wheatear (O. isabellina) (NC_040290.1). Protein coding genes, tRNAs and rRNAs were annotated by the MITOS WebServer (http://mitos2.bioinf.uni-leipzig.de/index.py) and blasted against the NCBI database of mitochondrial sequences. The PCG sequences were extracted and concatenated by custom python scripts. tRNA prediction was further performed by tRNAscan-SE v1.3.172. Secondary structures of 16S and 12S RNA were derived using the RNAfold WebServer (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) with minimum free energy (MFE) and partition function. Mitochondrial maps were generated by CGView73. As seen in the chicken Gallus gallus32 and the outgroup O. isabellina66, the highly variable control regions (CR) of all investigated Oenanthe spp. were located between the tRNA-Phe and tRNA-Glu. Since our mitogenome alignments were derived from next-generation sequencing data, coverage became rapidly reduced in the CR. We therefore excluded the CR from downstream phylogeographical analyses to avoid incomplete assemblies or ambiguous alignments.
Phylogenetic analysis
PCG sequences were aligned by MAFFT v7.03774. Neither indels nor interal stop codons were detected in the alignments. SequenceMatrix v.1.7.875 was applied to concatenate the alignments for different gene sets. PartitionFinder v.2.1.176 was applied to determine the best partition arrangement and optimal substitution models of sequence evolution for downstream phylogenetic analyses. Four datasets were pre-defined in the input configuration files (1) 13 PCGs, two rRNAs and combined 22 tRNAs with 15529 residues, (2) 13 PCGs with gene partition, (3) 13 PCGs with codon partition and (4) each single gene with or without partition. The greedy algorithm with unlinked branch lengths estimation and Akaike information criterion with correction (AICc) were used to search for appropriate partitions schemes77.
Phylogenetic trees were constructed based on the best-fit partition schemes suggested by PartitionFinder. Bayesian inference (BI) analysis was applied by MrBayes version 3.2.278. The datasets were run with 2 simultaneous Markov Chain Monte Carlo (MCMC) runs on one cold and three heated chains to confirm the convergence of posterior probability distributions79. Analyses were set to run for 10 million generations with sampling conducted every 1,000 generations until stationarity was reached, i.e., average standard deviation of split frequencies less than 0.01. The initial 25% of the total trees were discarded as burn-in. The speciation time was estimated with the GTR substitution model implemented in BEAST 1.8.380–82. We tested a Yule speciation process with a strict clock model (2.1% per million years for Cytb) and estimated the mutation rate for the remaining markers83. All the analyses were performed in the CIPRES Science Gateway (http://www.phylo.org) and visualization was achieved by FigTree version 1.4.1 (http://beast.bio.ed.ac.uk/FigTree). Population genetic analyses, neutrality tests and mismatch distributions were calculated in ARLEQUIN version 3.5.1.384 and DnaSP version 5.185.
Supplementary information
Acknowledgements
The authors are indebted to Hedwig Sauer-Gürth and Heidi Staudter for lab work support. The authors received financial support from the Deutsche Forschungsgemeinschaft (SCHM 2647/2-1), the Swedish Research Council VR, the County Administrative Board of Västerbotten (LUVRE project) and Ruprecht-Karls-Universität Heidelberg within the funding program Open Access Publishing.
Author contributions
E.W. and M.W. designed with input from D.Z. and M.S.B. the study; D.Z., M.S.B. and F.L. improved the methods for analyses. F.B., H.S., D.A., T.P. and F.L. collected samples; A.H.W. provided infrastructure for bioinformatics analyses. F.B and H.S offered field data. E.W. conducted analyses and wrote the manuscript. M.S.B. and M.W. improved the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Erjia Wang, Email: wang@uni-heidelberg.de.
Michael Wink, Email: wink@uni-heidelberg.de.
Supplementary information
is available for this paper at 10.1038/s41598-020-66287-0.
References
- 1.Avise JC, et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 1987;18:489–522. [Google Scholar]
- 2.Avise, J. C. Phylogeography: the history and formation of species. (Harvard university press, 2000).
- 3.Zink RM, Pavlova A, Drovetski S, Rohwer S. Mitochondrial phylogeographies of five widespread Eurasian bird species. J. Ornitol. 2008;149:399–413. doi: 10.1007/s10336-008-0276-z. [DOI] [Google Scholar]
- 4.Lavrov DV, Pett W. Animal mitochondrial DNA as we do not know it: mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 2016;8:2896–2913. doi: 10.1093/gbe/evw195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aliabadian M, Kaboli M, Nijman V, Vences M. Molecular identification of birds: performance of distance-based DNA barcoding in three genes to delimit parapatric species. PLoS One. 2009;4:e4119. doi: 10.1371/journal.pone.0004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebert PD, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnsen A, et al. DNA barcoding of Scandinavian birds reveals divergent lineages in trans-Atlantic species. J. Ornitol. 2010;151:565–578. [Google Scholar]
- 8.Kerr KC, et al. Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. notes. 2007;7:535–543. doi: 10.1111/j.1471-8286.2007.01670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerr KC, et al. Filling the gap-COI barcode resolution in eastern Palearctic birds. Front. Zool. 2009;6:29. doi: 10.1186/1742-9994-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoeckle MY, Thaler DS. DNA barcoding works in practice but not in (neutral) theory. PLoS One. 2014;9:e100755. doi: 10.1371/journal.pone.0100755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang E, Van Wijk RE, Braun MS, Wink M. Gene flow and genetic drift contribute to high genetic diversity with low phylogeographical structure in European hoopoes (Upupa epops) Mol. Phylogenet. Evol. 2017;113:113–125. doi: 10.1016/j.ympev.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Carneiro de Melo Moura, C. et al. Pliocene origin,ice ages and postglacial population expansion have influenced a panmictic phylogeography of the European Bee-Eater Merops apiaster. J. Divers. 11, 12 (2019).
- 13.Pârâu, L. G., Frias-Soler, R. C. & Wink, M. High Genetic Diversity among Breeding Red-Backed Shrikes Lanius collurio in the Western Palearctic. Diversity11, 10.3390/d11030031 (2019).
- 14.Rising, J. et al. Handbook of the Birds of the World Alive. (Lynx Edicions Barcelona, 2019).
- 15.Schmaljohann H, et al. Proximate causes of avian protandry differ between subspecies with contrasting migration challenges. Behav. Ecol. 2015;27:321–331. [Google Scholar]
- 16.Bairlein F, Eikenaar C, Schmaljohann H. Routes to genes: unravelling the control of avian migration—an integrated approach using Northern Wheatear Oenanthe oenanthe as model organism. J. Ornitol. 2015;156:3–14. doi: 10.1007/s10336-015-1224-3. [DOI] [Google Scholar]
- 17.Bairlein, F. et al. Cross-hemisphere migration of a 25 g songbird. Biol. Lett., rsbl20111223 (2012). [DOI] [PMC free article] [PubMed]
- 18.Bairlein F. Migratory birds under threat. Science. 2016;354:547–548. doi: 10.1126/science.aah6647. [DOI] [PubMed] [Google Scholar]
- 19.Kaboli M, Aliabadian M, Chamani A, Pasquet E, Prodon R. Morphological relationships of the Wheatears (genus Oenanthe) Russ. J. Ecol. 2013;44:251–259. doi: 10.1134/s1067413613030168. [DOI] [Google Scholar]
- 20.Svensson, L. Identification guide to European passerines. (L. Svensson, 1992).
- 21.Aliabadian M, Kaboli M, Prodon R, Nijman V, Vences M. Phylogeny of Palaearctic wheatears (genus Oenanthe)–congruence between morphometric and molecular data. Mol. Phylogenet. Evol. 2007;42:665–675. doi: 10.1016/j.ympev.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Kaboli M, Aliabadian M, Guillaumet A, Roselaar CS, Prodon R. Ecomorphology of the wheatears (genus Oenanthe) Ibis. 2007;149:792–805. [Google Scholar]
- 23.Förschler MI, Khoury F, Bairlein F, Aliabadian M. Phylogeny of the mourning wheatear Oenanthe lugens complex. Mol. Phylogenet. Evol. 2010;56:758–767. doi: 10.1016/j.ympev.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 24.Outlaw RK, Voelker G, Bowie RC. Shall we chat? Evolutionary relationships in the genus Cercomela (Muscicapidae) and its relation to Oenanthe reveals extensive polyphyly among chats distributed in Africa, India and the Palearctic. Mol. Phylogenet. Evol. 2010;55:284–292. doi: 10.1016/j.ympev.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Alaei Kakhki N, et al. Phylogeography of the Oenanthe hispanica-pleschanka-cypriaca complex (Aves, Muscicapidae: Saxicolinae): Diversification history of open-habitat specialists based on climate niche models, genetic data, and morphometric data. J. Zool. Sys. Evol. Res. 2018;56:408–427. doi: 10.1111/jzs.12206. [DOI] [Google Scholar]
- 26.Schweizer M, Shirihai H. Phylogeny of the Oenanthe lugens complex (Aves, Muscicapidae: Saxicolinae): paraphyly of a morphologically cohesive group within a recent radiation of open-habitat chats. Mol. Phylogenet. Evol. 2013;69:450–461. doi: 10.1016/j.ympev.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Panov EN. Comparative ethology and molecular genetics as tools for phylogenetic reconstructions: The example of the genus. Oenanthe. Biol. Bull. 2011;38:809–820. doi: 10.1134/s106235901108005x. [DOI] [Google Scholar]
- 28.Schweizer M, et al. Parallel plumage colour evolution and introgressive hybridization in wheatears. J. Evol. Biol. 2019;32:100–110. doi: 10.1111/jeb.13401. [DOI] [PubMed] [Google Scholar]
- 29.Arizaga J, Schmaljohann H, Bairlein F. Stopover behaviour and dominance: a case study of the Northern Wheatear Oenanthe oenanthe. Ardea. 2011;99:157–165. [Google Scholar]
- 30.Corman A-M, Bairlein F, Schmaljohann H. The nature of the migration route shapes physiological traits and aerodynamic properties in a migratory songbird. Behav. Ecol. Sociobiol. 2014;68:391–402. [Google Scholar]
- 31.Hobson, K. A. & Wassenaar, L. I. Tracking animal migration with stable isotopes. (Academic Press, 2018).
- 32.Desjardins P, Morais R. Sequence and gene organization of the chicken mitochondrial genome: a novel gene order in higher vertebrates. J. Mol. Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 33.Jühling F, et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2008;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe Y, Suematsu T, Ohtsuki T. Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front. Genet. 2014;5:109. doi: 10.3389/fgene.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol. Biol. Evol. 2007;24:269–280. doi: 10.1093/molbev/msl158. [DOI] [PubMed] [Google Scholar]
- 36.Allcock AL, Cooke IR, Strugnell JM. What can the mitochondrial genome reveal about higher-level phylogeny of the molluscan class Cephalopoda? Zool. J. Linn. Soc. 2011;161:573–586. [Google Scholar]
- 37.Cabrera AA, et al. Fin whale (Balaenoptera physalus) mitogenomics: A cautionary tale of defining sub-species from mitochondrial sequence monophyly. Mol. Phylogenet. Evol. 2019;135:86–97. doi: 10.1016/j.ympev.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S. Patterns of nucleotide substitution in mitochondrial protein coding genes of vertebrates. Genetics. 1996;143:537–548. doi: 10.1093/genetics/143.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun EL, Kimball RT. Examining basal avian divergences with mitochondrial sequences: model complexity, taxon sampling, and sequence length. Syst. Biol. 2002;51:614–625. doi: 10.1080/10635150290102294. [DOI] [PubMed] [Google Scholar]
- 40.Duchêne S, Archer FI, Vilstrup J, Caballero S, Morin PA. Mitogenome phylogenetics: the impact of using single regions and partitioning schemes on topology, substitution rate and divergence time estimation. PloS one. 2011;6:e27138. doi: 10.1371/journal.pone.0027138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leavitt JR, Hiatt KD, Whiting MF, Song H. Searching for the optimal data partitioning strategy in mitochondrial phylogenomics: a phylogeny of Acridoidea (Insecta: Orthoptera: Caelifera) as a case study. Mol. Phylogenet. Evol. 2013;67:494–508. doi: 10.1016/j.ympev.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Wang N, Hosner PA, Liang B, Braun EL, Kimball RT. Historical relationships of three enigmatic phasianid genera (Aves: Galliformes) inferred using phylogenomic and mitogenomic data. Mol. Phylogenet. Evol. 2017;109:217–225. doi: 10.1016/j.ympev.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Richards EJ, Brown JM, Barley AJ, Chong RA, Thomson RC. Variation across mitochondrial gene trees provides evidence for systematic error: How much gene tree variation is biological? Syst. Biol. 2018;67:847–860. doi: 10.1093/sysbio/syy013. [DOI] [PubMed] [Google Scholar]
- 44.Kainer D, Lanfear R. The effects of partitioning on phylogenetic inference. Mol. Biol. Evol. 2015;32:1611–1627. doi: 10.1093/molbev/msv026. [DOI] [PubMed] [Google Scholar]
- 45.Wink M. Use of DNA markers to study bird migration. J. Ornitol. 2006;147:234–244. doi: 10.1007/s10336-006-0065-5. [DOI] [Google Scholar]
- 46.Cummings MP, Meyer A. Magic bullets and golden rules: data sampling in molecular phylogenetics. Zoology. 2005;108:329–336. doi: 10.1016/j.zool.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Hillis DM. Inferring complex phytogenies. Nature. 1996;383:130. doi: 10.1038/383130a0. [DOI] [PubMed] [Google Scholar]
- 48.Sloan DB, Havird JC, Sharbrough J. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol. Ecol. 2017;26:2212–2236. doi: 10.1111/mec.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wink M. Bird migration “Out of Africa”: The evolution of bird migration. Der Falke - Journal für Vogelbeobachter. 2014;60:26–30. [Google Scholar]
- 50.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996;58:247–276. [Google Scholar]
- 51.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 52.Finlayson C, et al. Ecological transitions — But for whom? A perspective from the Pleistocene. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2012;329-330:1–9. doi: 10.1016/j.palaeo.2011.04.002. [DOI] [Google Scholar]
- 53.Frenzel, B. Atlas of paleoclimates and paleoenvironments of the Northern Hemisphere. (Geographical Research Institute, Hungarian Academy of Sciences, Budapest, Gustav Fischer Verlag, Stuttgart Jena New York., 1992).
- 54.Lundqvist J, Saarnisto M. Summary of project IGCP-253. Quat. Int. 1995;28:9–18. [Google Scholar]
- 55.Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- 56.Irwin DE, Rubtsov AS, Panov EN. Mitochondrial introgression and replacement between yellowhammers (Emberiza citrinella) and pine buntings (Emberiza leucocephalos)(Aves: Passeriformes) Biol. J. Linn. Soc. 2009;98:422–438. [Google Scholar]
- 57.Semenov GA, Koblik EA, Red’kin YA, Badyaev AV. Extensive phenotypic diversification coexists with little genetic divergence and a lack of population structure in the White Wagtail subspecies complex (Motacilla alba) J. Evol. Biol. 2018;31:1093–1108. doi: 10.1111/jeb.13305. [DOI] [PubMed] [Google Scholar]
- 58.Johnsen A, Kearns AM, Omland KE, Anmarkrud JA. Sequencing of the complete mitochondrial genome of the common raven Corvus corax (Aves: Corvidae) confirms mitogenome-wide deep lineages and a paraphyletic relationship with the Chihuahuan raven C. cryptoleucus. PLoS One. 2017;12:e0187316. doi: 10.1371/journal.pone.0187316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kearns, A. M. et al. Genomic evidence of speciation reversal in ravens. Nat. Commun. 9, 10.1038/s41467-018-03294-w (2018). [DOI] [PMC free article] [PubMed]
- 60.Palacios C, et al. Shallow genetic divergence and distinct phenotypic differences between two Andean hummingbirds: Speciation with gene flow? The Auk. 2019;136:ukz046. [Google Scholar]
- 61.Wang K, et al. Incomplete lineage sorting rather than hybridization explains the inconsistent phylogeny of the wisent. Commun. Biol. 2018;1:169. doi: 10.1038/s42003-018-0176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, et al. Past hybridization between two East Asian long-tailed tits (Aegithalos bonvaloti and A. fuliginosus) Front. Zool. 2014;11:40. doi: 10.1186/1742-9994-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKay BD, Zink RM. The causes of mitochondrial DNA gene tree paraphyly in birds. Mol. Phylogenet. Evol. 2010;54:647–650. doi: 10.1016/j.ympev.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 64.Choleva L, et al. Distinguishing between incomplete lineage sorting and genomic introgressions: complete fixation of allospecific mitochondrial DNA in a sexually reproducing fish (Cobitis; Teleostei), despite clonal reproduction of hybrids. PLoS One. 2014;9:e80641. doi: 10.1371/journal.pone.0080641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He S, et al. An examination of introgression and incomplete lineage sorting among three closely related species of chocolate-dipped damselfish (genus: Chromis) Ecol. Evol. 2019;9:5468–5478. doi: 10.1002/ece3.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S, Luo A, Li G, Li W. Complete mitochondrial genome of the isabelline wheatear Oenanthe isabellina (Passeriformes, Muscicapidae) Mitochondrial DNA Part B. 2016;1:355–356. doi: 10.1080/23802359.2016.1167641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sambrook, J., Fritsch, E. & Maniatis, T. Molecular cloning: A laboratory manual+ Cold Spring Harbor. (New York: Cold spring harbor laboratory press, 1989).
- 68.van Oosten HH, Mueller JC, Ottenburghs J, Both C, Kempenaers B. Genetic structure among remnant populations of a migratory passerine, the Northern Wheatear Oenanthe oenanthe. Ibis. 2016;158:857–867. [Google Scholar]
- 69.Randler C, et al. Phylogeography, pre-zygotic isolation and taxonomic status in the endemic Cyprus Wheatear Oenanthe cypriaca. J. Ornitol. 2012;153:303–312. [Google Scholar]
- 70.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic. Acids. Res. 2013;41:e129–e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chevreux, B., Wetter, T. & Suhai, S. in German conference on bioinformatics. 45-56 (Citeseer).
- 72.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2004;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 74.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaidya G, Lohman DJ, Meier RJC. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics. 2011;27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 76.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 77.Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 78.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 80.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lanave C, Preparata G, Sacone C, Serio G. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- 83.Weir JT, Schluter D. Calibrating the avian molecular clock. Mol. Ecol. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- 84.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Biol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 85.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.