Abstract
Introduction
Obesity is a multifactorial chronic inflammatory disease. Consumption of high energy density (HED) diets is associated with hyperphagia, increased body weight and body fat accumulation, and obesity. Our lab has previously shown that short-term (4 weeks) consumption of a HED diet triggers gut microbiota dysbiosis, gut inflammation, and reorganization of the gut-brain vagal communication.
Objetives
The aim of this study was to investigate the effect of long-term (6 months) consumption of HED diet on body composition, gut microbiome, hepatocellular lipidosis, microglia activation in the nucleus of the solitary tract, and systemic inflammation.
Methods
Male Sprague–Dawley rats were fed a low energy density (LED) diet for 2 weeks and then switched to a HED diet for 26 weeks. Twenty-four-hour food intake, body weight, and body composition were measured twice a week. Blood serum and fecal samples were collected at baseline, 1, 4, 8, and 26 weeks after introduction of the HED diet. Serum samples were used to measure insulin, leptin, and inflammatory cytokines using Enzyme-linked Immunosorbent Assay. Fecal samples were assessed for 16 S rRNA genome sequencing.
Results
HED diet induced microbiota dysbiosis within a week of introducing the diet. In addition, there was significant microglia activation in the intermediate NTS and marked hepatic lipidosis after 4 weeks of HED diet. We further observed changes in the serum cytokine profile after 26 weeks of HED feeding.
Conclusions
These data suggest that microbiota dysbiosis is the first response of the organism to HED diets, followed by increased liver fat accumulation, microglia activation in the brain, and circulating levels of inflammatory markers. To our knowledge, this is the first study to present longitudinal and cross-sectional results on effect of long-term consumption of HED diets on all these parameters in a single cohort of animals.
Subject terms: Feeding behaviour, Neuroscience, Obesity
Introduction
Obesity is a low-grade, chronic inflammatory disease. However, the relationship between inflammation and obesity, and the factors behind obesity-dependent inflammation are not well understood. Typically, inflammation is a transient physiological response. However, the inflammatory state that accompanies the metabolic syndrome observed in most obese individuals is not transient.
Consumption of high energy density (HED) diets triggers microbiota dysbiosis, increased body fat accumulation, and metabolic syndrome1,2. The vast majority of obesity cases result from an imbalance between energy intake and energy expenditure, with intake surpassing expenditure. Excess energy intake leads to enlargement of adipose tissue depositions. Once thought to be inert, adipose tissue is now recognized as a metabolically active endocrine organ3. This enlargement occurs through an increase in the number of adipocytes (adipogenesis) or an increase in the size of existing adipocytes (hypertrophy)4. In the db/db mouse model, there is a high number of adipogenic/angiogenic cell clusters in the early stages of obesity. The number of these cell clusters declines over time and there is an increase in the number of crown-like structures, which are hallmarks of local infiltration of macrophages into tissue surrounding dead adipocytes5. Using three independent adipocyte-specific anti-inflammatory mouse models, Asterholm et al. showed that an acute inflammatory response in adipose tissue is necessary to stimulate adipogenesis as well as proper remodeling and angiogenesis of the extracellular matrix, to allow for healthy adipose tissue expansion6. It would appear that the tonic activation of the innate immune system induced by excess energy intake gradually disrupts the homeostatic state, triggering chronic inflammation.
In the obese state, the production of proinflammatory adipokines induce resident macrophages to change their phenotype from surveillance “M2” to proinflammatory “M1” as well as trigger recruitment of M1 macrophages7,8. In addition, free fatty acids (FFAs) activate toll-like receptor (TLR) 4 in adipose tissue to generate proinflammatory signals9,10. Moreover, deletion of TLR5 triggered a shift in the species composition of the gut microbiota that is associated with development of metabolic syndrome11. Previous work from our laboratory has shown that 4 weeks of HED diet is sufficient to trigger microglia activation in the nucleus of the solitary tract (NTS)12.
Another obesity comorbidity is non-alcoholic fatty liver disease (NAFLD), the most common chronic liver condition in the Western world13. NAFLD is found among people with diabetes (50%) and obesity (76%), and it is almost universal among diabetic people who are morbidly obese14. It encompasses a wide spectrum of liver damage that occurs in people who drink little to no alcohol. Hepatocytes play a primary role in lipid metabolism. FFAs enter the hepatocyte and most FFAs are esterified to form triglycerides (TGs). TGs form complexes with an apolipoprotein to form lipoproteins and then are exported from the hepatocyte. The apolipoproteins are synthesized by the hepatocyte and this is the rate-limiting step in TG export. When consuming a HED diet, the usual cause of hepatic lipidosis is the increased production of TGs, which outpaces apolipoprotein production15.
The aim of this study was to investigate the systemic responses to long-term consumption of a HED diet. Our main goal was to determine the timeline progression of the systemic changes—increased body fat accumulation, development of microbiota dysbiosis, changes in serum of cytokines, development of NAFLD, and increased microglia activation—induced by HED diet consumption. We tested the hypotheses that HED diet consumption induces progressive microbiota dysbiosis and increases circulating levels of leptin, insulin, and proinflammatory cytokines. We also hypothesized that HED diet consumption induces NAFLD and increases microglia activation in the NTS.
Methods
Animals
Male Sprague–Dawley rats (n = 15; ~300 g; Envigo, Indianapolis, IN) were housed individually in conventional polycarbonate shoe-box cages in a temperature-controlled vivarium with ad libitum access to low energy density (LED; 5% fat, 3.25% sucrose) pellets of rat chow (PicoLab rodent diet 20, product #5053, Fort Worth, TX) and water. Rats were maintained on a 12:12-h light: dark cycle with lights on at 0700-h and allowed to acclimate to laboratory conditions for 1 week prior to starting experiments. All animal procedures were approved by the University of Georgia Institutional Animal Care and Use Committee and conformed to National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Food Intake, body weight, and body composition
Following the acclimation period, rats were maintained on LED for an additional 2 weeks and were then switched to a HED diet (45% fat, 20% sucrose, Research Diet #D12451, New Brunswick, NJ). Food intake was measured twice a week as previously reported16. Body weight and body composition were measured weekly using a minispec LF 110 BCA Analyzer (Bruker Corp., The Woodlands, TX). Six rats, chosen randomly, were sacrificed after being on HED diet for 4 weeks (ST-HED, Supplementary Fig. S1). The remaining nine rats were maintained on HED diet for a total of 26 weeks (LT-HED, Supplementary Fig. S1). An additional aged-matched, LED fed group of rats (n = 9, LED26, Supplementary Fig. S1) served as the endpoint controls for the LT-HED group.
Cytokines, leptin, and insulin levels in serum
Blood samples were collected on the last day of LED and 4, 8, and 26 weeks after introduction of the HED diet. The serum was collected and stored at −21 °C. A cytokine array (Rat Cytokine ELISA Kit, cat #EA-4006, Signosis Inc., Sunnyvale, CA) was used to measure levels of cytokines and chemokines. Insulin levels were determined using the Rat Insulin ELISA kit (cat #80-INSRT-E01; ALPCO Diagnostics, Inc., Salem, NH).
Microbiome analysis
Fecal samples were collected following the same timeline as for blood samples mentioned above. Bacterial DNA was extracted from feces using a commercial kit (Quick-DNA Fecal/Soil Microbe Miniprep Kit, cat #D6010, Zymo research, Irvine, CA). High-throughput sequencing was performed using Illumina MiSeq paired-end runs (GGBC, Athens, GA). Amplification targeted the V3–V4 region of the 16 S ribosomal RNA genes using the following primers: S-D-Bact-041-b-S-17 (5’-CCTACGGGNGGCWGCAG-3’) forward and S-D-Bact-0785-a-A-21 (5’-GACTACHVGGGTATCTAATCC-3’)17. Sequences were subsequently trimmed, joined, and quality filtered. To identify Operational Taxonomic Units (OTUs) and to evaluate beta and alpha diversities, we used the Quantitative Insights Into Microbial Ecology (QIIME) software package18. Linear discriminant analysis to identify taxa with differentiating abundance was conducted using the LDA Effect Size (LEfSe) algorithm19. Bacterial abundance was normalized by log-transformation, and statistical analysis and principal component analysis (clustering) were performed using the METAGENassist platform20.
Euthanasia
Rats were anesthetized with CO2 and transcardially perfused with 0.1 M phosphate-buffered saline (PBS; pH 7.4) followed by 4% paraformaldehyde. Hindbrains and liver were harvested, postfixed in 4% paraformaldehyde for 2-h, and immersed in 30% sucrose, 0.1% NaN3 (Sigma-Aldrich; pH 7.4) in PBS and stored at 4 °C until processing.
Microglia activation
Hindbrain samples were cryosectioned (Leica CM1950, Leica Biosystems, Wetzlar, Germany) at 20μm thickness. Sections were incubated overnight with a primary antibody against ionized calcium binding adaptor molecule 1 (Iba-1, Wako Cat#019-19741, RRDI: AB_839504) followed by Alexa-488 secondary antibody to visualize microglia activation as previously described21. Sections were mounted in ProLong (Molecular Probes, OR) and examined under a Nikon 80-I fluorescent microscope. The area fraction of Iba-1 was analyzed using Nikon Elements AR software as previously described16,22.
Hepatic lipidosis
Liver samples were embedded in paraffin and cryosectioned at 4 µm thickness. Tissue sections were stained with hematoxylin and eosin (H&E). In addition, samples for Oil-Red-O (ORO) staining were embedded in optimum cutting temperature compound (VWR Inc., Atlanta, GA), cryosectioned at 7 µm thickness and stained with ORO (Polysciences Inc., Warrington, PA). H&E and ORO stained liver sections were examined microscopically buy a board-certified veterinary pathologist using an Olympus BX41 upright light microscope. Images were captured using an Olympus DP25 digital camera controlled by Olympus cellSense Standard software at ×200 and ×400 original magnification (Olympus, Shinjuku, Japan). A semi-quantitative grading scale (normal [0], minimal [1], mild [2], moderate [3], and marked [4]) was used to express the extent of hepatic lipidosis23. In addition, ORO staining was quantified by Nikon Elements AR Software as previously described16.
Statistical analysis
GraphPad Prism 7 (GraphPad Software, Inc.) was used for statistical analyses. All samples were processed by an experimenter blind to the experimental condition. Data are expressed as mean ± SD and were analyzed using two-tailed t-test or ANOVA followed by Holm–Sidak multiple comparisons test as appropriate. Sample size was determined based on prior studies from our laboratory. To simplify statistical analysis and presentation of results, data for the ST-HED and LT-HED groups were analyzed independently. Alpha value was set at 0.05.
Results
HED diet significantly increased body weight and body fat mass
Caloric intake, body weight, and body fat mass are shown in Fig. 1. In the ST-HED group, the animals significantly increased their caloric intake during the first week after introduction of the HED diet compared to intake of LED diet (Ps < 0.0001) (Fig. 1a, d). Caloric intake decreased to intakes of the LED by week 2 and remained stable. Body weight was significantly higher after one week of HED compared to baseline (Ps < 0.05) and the animals continued gaining weight (Fig. 1b, e). Body fat percent significantly increased after one week on HED diet compared to baseline (Ps < 0.05) and fat deposits continued to grow throughout the experiment (Fig. 1c, f). Caloric intake of the LT-HED group at the end of the study (HED26) was similar to that of aged-matched, LED diet controls (LED26) (Fig. 1g). However, body weight (P = 0.006) (Fig. 1h) and body fat (P < 0.0001) (Fig. 1i) were significantly higher in HED-fed rats compared to LED diet controls.
Fig. 1. High energy density (HED) diet consumption significantly increased body weight and body fat mass.
Shown are mean ± SD kcal consumed (a, d, g), body weight (b, e, h), and body fat mass (c, f, i) for rats fed HED for four weeks (ST-HED; n = 6, top row), rats fed HED for 26 weeks (LT-HED; n = 9, middle row), and endpoint comparison of rats fed LED vs HED for 26 weeks (n = 9 per group, bottom row). Animals significantly increased their caloric intake upon introduction of the HED diet, but caloric intake declined after 1 week and remained stable for the duration of the study in the ST-HED and LT-HED groups. HED diet consumption significantly increased body weight and fat mass. Asterisk indicates statistical significance from week 0. Hash indicates statistical significance from week 1 since start of HED diet, *P < 0.05, **P < 0.01, ***P < 0.0001.
HED diet triggered progressive dysbiosis of the gut microbiota
In the ST-HED group, the rarefaction curve of this analysis indicated >30,000 sequences and >500 OTUs per sample (Supplementary Fig. S2A). In the LT-HED group, the rarefaction curve of this analysis indicated >30,000 sequences and >1500 OTUs per sample (Supplementary Fig. S2B).
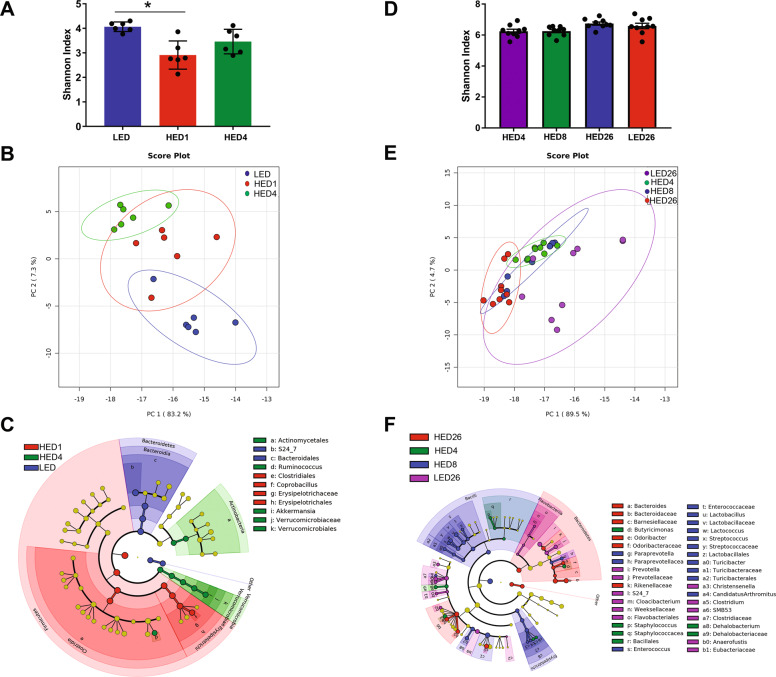
In the ST-HED and LT-HED groups, Firmicutes and Bacteroidetes were the most abundant phyla representing >90% of the bacteria identified. The Shannon index revealed a significant decrease in bacteria diversity after one week of HED diet compared to baseline (P = 0.0146) (Fig. 2a). There were no significant changes observed in bacterial diversity after 4 weeks. Principal Component Analysis (Fig. 2b) showed that at baseline (LED), all animals clustered together. One week after HED diet introduction, the animals clustered away from their baseline (LED) profile. After 4 weeks of HED diet, all animals clustered close to the profile at HED1 and further away from their LED profile.
Fig. 2. Consumption of a high energy density (HED) diet triggered progressive remodeling of the gut microbiota.
The left (a, b, c) and right (d, e, f) columns represent data from the ST-HED (n = 6) and LT-HED (n = 9) groups, respectively. a, d Shannon index shown as mean ± SD for each group and time point. Bacterial diversity was significantly decreased after consuming the HED diet for one week (HED1) compared to baseline (LED). There were no other significant changes observed. b, e Principal Coordinate Analysis showing microbiota from all time points. In the ST-HED group (b), the microbiota of all rats clustered together at baseline (LED). One week after introduction of the HED diet (HED1), the microbiota clustered together and away from their LED profile. At week 4 (HED4), the microbiota clustered together and further away from their LED profile. In the LT-HED group (e), the microbiota of all rats fed the HED clustered together, independent of time point, and away from the microbiota of rats fed LED diet (LED26). c, f Cladogram produced from LDA scores (see Supplementary Fig. S2 for LDA scores). In the ST-HED group (c), at baseline (LED), the microbiota is characterized by abundant members of the Bacteroidetes order Bacteroidales. One week after HED diet introduction (HED1), the microbiota was characterized by abundant members of the Firmicutes order Erysipelotrichales. After 4 weeks of HED diet (HED4), the microbiota was characterized by abundant members of the Actinobacteria order Actinomycetales and Verrucomicrobia order Verrucomicrobiales. In the LT-HED group (f), at HED4 the microbiota was characterized by abundant members of Bacteroidetes order Bacteroidales and Firmicutes orders Bacillales and Clostridiales. Eight weeks after HED diet introduction (HED8), the microbiota was characterized by abundant members of the Bacteroidetes order Bacteroidales and Firmicutes orders Lactobacillales, Turicibacterales, and Clostridiales. After 26 weeks of HED diet (HED26), the microbiota was characterized by abundant members of Bacteroidetes orders Bacteroidales. The microbiota of the LED diet control group (LED26) was characterized by abundant members of Bacteroidetes orders Bacteroidales and Flavobacteriales, and Firmicutes order Clostridiales.
Figure 2c (and Supplementary Fig. S3) represents the microbiota composition of the ST-HED group at each time point. At baseline (LED), the microbiota is characterized by abundant members of the Bacteroidetes order Bacteroidales. One week after HED diet introduction (HED1), the microbiota was characterized by abundant members of the Firmicutes order Erysipelotrichales and after 4 weeks by abundant members of the Actinobacteria order Actinomycetales and Verrucomicrobia order Verrucomicrobiales.
Microbiota composition changed within a week of introduction of HED diet (Fig. 3a). HED diet significantly increased the abundance of Firmicutes (LF 27% vs HED1 86% and HED4 69%, Ps < 0.0001) and decreased the abundance of Bacteroidetes (LED 67% vs HED1 10% and HED4 16%, Ps < 0.0001). There was also a significant increase in abundance of Verrucomicrobia after 4 weeks of HED diet (LED 1% and HED1 0.8% vs HED4 8%, P = 0.04). At the level of family, HED diet significantly increased the abundance of members of Erysipelotrichaceae (LED 3.8% vs HED1 60% and HED4 41%, Ps < 0.0001) of the phylum Firmicutes. Members of the family S24-7 of the phylum Bacteroidetes were significantly depleted by HED diet (LED 69% vs HED1 12% and HED4 17%, Ps < 0.0001). In addition, 1 week of HED diet significantly increased the Firmicutes-to-Bacteroidetes ratio, (LED 0.4 vs HED1 8.7, P = 0.0010). There was no statistically significant difference after 4 weeks on the HED diet; however, the ratio was still higher at HED4 (4.3) than at LED (Fig. 3b).
Fig. 3. Microbial composition of rats fed a high energy density (HED) diet for 4 weeks (ST-HED, n = 6) or 26 weeks (LT-HED, n = 9) and rats fed a low energy density (LED) diet for 26 weeks (LF26, n = 9).
All phylogenetic levels present with abundance >1% are represented. a, c relative abundances of phyla at the family level in the ST-HED group and in the LT-HED. b, d Ratio of Firmucutes to Bacteroidetes in the ST-HED and LT-HED group. In the ST-HED group, HED diet consumption significantly increased the abundance of members of Erysipelotrichaceae (LED 3.8% vs HED1 60% and HED4 41%, Ps < 0.0001) of the phylum Firmicutes. Members of the family S24-7 of the phylum Bacteroidetes were significantly depleted by HED diet consumption (a). In addition, the ratio of Firmicutes to Bacteroidetes was significantly higher at 1 and 4 weeks of HED diet compared to LED (b). In the LT-HED group, compared to LED diet fed rats, HED diet fed rats had significantly higher abundance of members of Bacteroidaceae and Ruminococcaceae, and significantly lower abundance of members of Peptostreptococcaceae and Verrucomicrobiaceae (c). In HED-fed rats, the Firmicutes-to-Bacteroidetes ratio was significantly higher after eight weeks compared to after four and 26 weeks (HED8 11.5 vs HED4 6.4 and HED26 5.7, Ps < 0.01). Compared to LED controls (LED26), HED-fed rats had a significantly lower Firmicutes-to-Bacteroidetes ratio at four and 26 weeks (D). In the legend, following the name of each family, higher taxonomic classifications are indicated by letters in parentheses. Phylum: A, Actinobacteria; B, Bacteroidetes; F, Firmicutes; V, Verrucomicrobia. Class: A, Actinobacteria; B, Bacilli if preceded by F and Bacteroidia if preceded by B; C, Clostridia; E, Erysipelotrichia; F, Flavobacteria; V, Verrucomicrobiae; O, Opitutae. Order: A, Actinomycetales; B, Bacteroidales; C, Clostridiales if preceded by C and Cerasicoccales if preceded by O; E, Erysipelotrichiales; F, Flavobacteriales; L, Lactobacillales; T, Turibacterales; V, Verrucomicrobiales. Asterisk indicates statistical significance *P < 0.05, **P < 0.01, ***P < 0.0001. Data are means ± SD.
In the LT-HED group, the Shannon index showed no significant difference in bacterial diversity (Fig. 2d). Principal Component Analysis showed that all animals fed the HED diet clustered together and away from animals fed LED (LED26) (Fig. 2e).
Figure 2f (and Supplementary Fig. S4) represents the microbiota composition of the LT-HED group at each time point. At HED4, the microbiota was characterized by abundant members of Bacteroidetes order Bacteroidales and Firmicutes orders Bacillales and Clostridiales, at HED8 by abundant members of the Bacteroidetes order Bacteroidales and Firmicutes orders Lactobacillales, Turicibacterales, and Clostridiales, and at HED26 by abundant members of Bacteroidetes orders Bacteroidales. The microbiota of the LED26 group was characterized by abundant members of Bacteroidetes orders Bacteroidales and Flavobacteriales, and Firmicutes order Clostridiales.
At the level of family (Fig. 3c), 4 weeks (HED4) and 8 weeks (HED8) after HED diet introduction there was a significantly higher abundance of Ruminococcaceae compared to after 26 weeks (HED26, P < 0.01) and a significantly lower abundance of Verrucomicrobiaceae compared to after 26 weeks (HED26, P < 0.01). HED diet fed rats had significantly higher abundance of members of Bacteroidaceae (LED26 0.8% vs HED4 14%, HED8 14%, and HED26 16%, Ps < 0.0001) and Ruminococcaceae (LED26 0.8% vs HED4 14% and HED8 14%, Ps < 0.0001) compared to LED fed rats. In addition, HED-diet-fed rats had significantly lower abundance of members of Peptostreptococcaceae (LED26 5% vs HED4 0.6%, HED8 0.9%, and HED26 0.5%, Ps < 0.05) and Verrucomicrobiaceae (LED26 14% vs HED4 1.2%, HED8 3%, and HED26 9%, Ps < 0.01) compared to LED fed rats. In HED-fed rats, the Firmicutes-to-Bacteroidetes ratio was significantly higher at HED8 compared to HED4 and HED26 (HED8 11.5 vs HED4 6.4 and HED26 5.7, Ps < 0.01). Compared to LED26 controls, HED-fed rats had a significantly lower Firmicutes-to-Bacteroidetes ratio at HED4 and HED26 (LED26 13% vs HED4 6% and HED26 6%, Ps < 0.01) (Fig. 3d).
Long-term HED diet significantly changed the cytokine profile in serum
Mean (±SD) serum levels of cytokines (OD) and insulin (ng/ml) are shown in Table 1. In the ST-HED group, the data were compared using paired t-tests. We observed a significant increase in TNFα (P = 0.03) and significant decrease in IL-1α after four weeks on HED diet compared to baseline (P < 0.0001). No other significant changes were observed.
Table 1.
Cytokine/chemokine optical density values ± SD.
| ST-HED | LT-HED | ||||||
|---|---|---|---|---|---|---|---|
| LED | HED4 | LED | HED4 | HED8 | HED26 | LED26 | |
| TNFα, OD | 0.057 ± 0.016 | 0.044 ± 0.011a | 0.049 ± 0.014 | 0.044 ± 0.006b | 0.038 ± 0.003a | 0.057 ± 0.016 | 0.062 ± 0.052 |
| VEGF, OD | 0.029 ± 0.006 | 0.026 ± 0.005 | 0.027 ± 0.005 | 0.025 ± 0.004 | 0.019 ± 0.002 | 0.024 ± 0.005 | 0.022 ± 0.011 |
| FGFβ, OD | 0.016 ± 0.001 | 0.015 ± 0.001 | 0.014 ± 0.003 | 0.028 ± 0.009a c | 0.017 ± 0.007c | 0.037 ± 0.007a | 0.042 ± 0.042 |
| IFNγ, OD | 0.056 ± 0.019 | 0.045 ± 0.009 | 0.055 ± 0.015 | 0.036 ± 0.011 | 0.032 ± 0.005a | 0.031 ± 0.004a | 0.036 ± 0.031 |
| Leptin, OD | 0.049 ± 0.014 | 0.041 ± 0.005 | 0.042 ± 0.002 n = 8 | 0.068 ± 0.022c n = 8 | 0.079 ± 0.071c n = 8 | 0.472 ± 0.092a d n = 8 | 0.089 ± 0.043 n = 8 |
| MCP-1, OD | 0.027 ± 0.007 | 0.022 ± 0.003 | 0.035 ± 0.007 | 0.08 ± 0.040a c | 0.036 ± 0.027c | 0.123 ± 0.039a | 0.103 ± 0.075 |
| SCF, OD | 0.074 ± 0.024 | 0.077 ± 0.038 | 0.064 ± 0.021 | 0.058 ± 0.008c | 0.053 ± 0.010c | 0.124 ± 0.050a d | 0.063 ± 0.032 |
| MIP-1α, OD | 0.019 ± 0.003 | 0.017 ± 0.006 | 0.014 ± 0.002 | 0.074 ± 0.043a | 0.043 ± 0.046c | 0.108 ± 0.019a | 0.089 ± 0.043 |
| IL-1α, OD | 0.059 ± 0.005 | 0.027 ± 0.002a | 0.054 ± 0.022 | 0.058 ± 0.044 | 0.032 ± 0.006c | 0.067 ± 0.026d | 0.024 ± 0.009 |
| IL-1β, OD | 0.069 ± 0.017 | 0.056 ± 0.005 | 0.059 ± 0.021 | 0.050 ± 0.009 | 0.043 ± 0.004 | 0.071 ± 0.028 | 0.054 ± 0.032 |
| IL-5, OD | 0.059 ± 0.018 | 0.047 ± 0.007 | 0.054 ± 0.016 | 0.032 ± 0.009 | 0.048 ± 0.049 | 0.029 ± 0.007a | 0.029 ± 0.019 |
| IL-6, OD | 0.021 ± 0.003 | 0.021 ± 0.007 | 0.018 ± 0.003 | 0.022 ± 0.003 | 0.016 ± 0.003 | 0.031 ± 0.011 | 0.024 ± 0.012 |
| IL-15, OD | 0.051 ± 0.016 | 0.045 ± 0.010 | 0.046 ± 0.011 | 0.035 ± 0.005 | 0.031 ± 0.005a c | 0.046 ± 0.012 | 0.042 ± 0.035 |
| IP-10, OD | 0.029 ± 0.004 | 0.032 ± 0.004 | 0.021 ± 0.007 | 0.021 ± 0.004c | 0.026 ± 0.003 | 0.029 ± 0.004a | 0.027 ± 0.007 |
| Rantes, OD | 1.183 ± 0.179 | 0.898 ± 0.169 | 1.191 ± 0.289 | 0.925 ± 0.317c | 1.042 ± 0.312c | 0.602 ± 0.207a | 0.776 ± 0.252 |
| TGFβ, OD | 0.279 ± 0.065 | 0.187 ± 0.053 | 0.155 ± 0.012 | 0.091 ± 0.034a | 0.198 ± 0.074 | 0.205 ± 0.099d | 0.049 ± 0.009 |
| Insulin, ng/ml | 1.223 ± 0.187 n = 4 | 1.92 ± 0.346 n = 4 | 0.937 ± 0.298 | 1.055 ± 0.185 | 0.888 ± 0.181 | 1.133 ± 0.269 | 1.418 ± 0.486 n = 3 |
For ST-HED, LT-HED, and LED26 n = 6, 9, 9, respectively, unless otherwise stated.
aDifferent from LED.
bDifferent from HED8.
cDifferent from HED26.
dDifferent from LED26.
In the LT-HED group, 4 weeks after introduction of HED diet, we observed a significant increase in FGFβ, MCP-1, MIP-1a, and TGFβ compared to LED (Ps < 0.05). After 8 weeks of HED diet (HED8), IFNγ and IL-15 were significantly decreased compared to LED (Ps < 0.01). TNFα and IL-6 were significantly lower than after four weeks on HED diet (Ps < 0.05). After 26 weeks on HED diet (HED26), FGFβ, Leptin, SCF, and MCP-1 were significantly higher than at LED (Ps < 0.05), HED4 (Ps < 0.01), and HED8 (Ps < 0.05). IP-10 levels were significantly higher than at HED4 (P = 0.0094). MIP-1α, IL-15, and IL-1α were significantly higher than at HED8 (Ps < 0.05). Rantes was significantly lower compared to LED, HED4, and HED8 (Ps < 0.05). IFNγ, IL-5 levels were significantly lower compared to LED (Ps < 0.01). Cross-sectional comparison of the HED26 group to LED26 controls revealed that the HED26 group had significantly higher leptin, SCF, IL-1α, and TGFβ (Ps < 0.01).
HED diet significantly increased microglia activation in the NTS
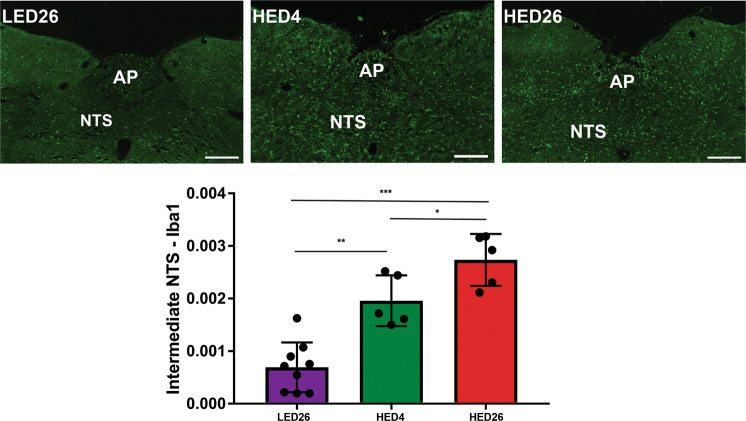
Results of immunostaining against Iba-1 were compared using one-way ANOVA and revealed that compared to LED26 controls, rats fed HED diet for 4 weeks (HED4, P = 0.0005) and 26 weeks (HED26, P < 0.0001) had significantly higher binary area fraction of fluorescent staining against Iba-1 (Fig. 4). In addition, at HED26 binary area fraction was significantly higher than at HED4 (P = 0.0213).
Fig. 4. Consumption of a HED diet significantly increased microglia activation in the intermediate NTS.
Representative sections of intermediate NTS of animals fed a LED diet for 26 weeks (LED26, n = 9), a HED diet for 4 weeks (HED4, n = 5), and a HED diet for 26 weeks (HED26, n = 5) are shown. Binary analysis of the area fraction of Iba-1 immunoreactivity showed that animals fed a HED for four and 26 weeks exhibited significantly more microglia activation than LED fed controls. In addition, microglia activation after 26 weeks of HED diet was significantly higher than after four weeks. Graphs represent mean ± SD Iba-1 intensity. Asterisk indicates statistical *P < 0.05, **P < 0.01, ***P < 0.0001. NTS nucleus tractus solitarius, AP area postrema. Scale bar = 200 μm.
HED diet induced hepatic lipidosis
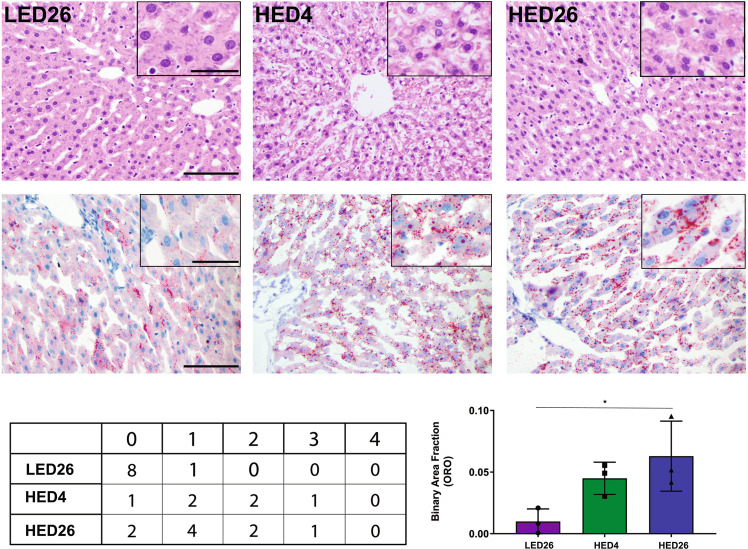
H&E (Fig. 5, top row) and ORO (Fig. 5, middle row) staining showed an increase in rats fed the HED diet (HED4 and HED26) compared to LED26 controls. The quantitative scoring confirmed that while animals fed LED diet (LED26) did not exhibit signs of hepatocellular lipidosis, it was apparent within 4 weeks of introducing the HED diet (HED4). There was no significant difference in hepatocellular lipidosis between the HED4 and HED26 time point. Software analysis revealed a progressive increase in lipid accumulation with LED fed animals showing very little staining (BAF = 0.0099 ± 0.01) compared to animals fed the HED diet for four weeks (BAF = 0.0449 ± 0.01) and 26 weeks (BAF = 0.0629 ± 0.02) (Fig. 5, bottom row table).
Fig. 5. hED diet intake induces hepatic lipidosis.
Representative histopathological images of hematoxylin and eosin stained (top row) and oil-red-o stained (bottom row) hepatic tissue from rats fed a LED diet (left column), rats fed a HED for 4 weeks (middle column), and rats fed a HED for 26 weeks (right column), n = 3 for each group. H&E staining revealed an increase in distinct vacuoles in rats fed the HED diet (HED4 and HED26) compared to LED controls (LED26) (top row). Similarly, ORO staining showed that HED-fed animals exhibited more intensely red granules (HED4 and HED26) than LED controls (LED26) (bottom row). The quantitative scoring (Table) confirmed that hepatocellular lipidosis is apparent after consuming a HED diet for four weeks. We did not observe significant differences in the extent of hepatocellular lipidosis between the ST-HED (HED4) and LT-HED (HED26) groups. LED fed rats did not show signs of hepatocellular lipidosis. Graph represents mean ± SD binary analysis of the area fraction of ORO staining, which further confirms our results. Asterisk indicates statistical significance *P < 0.05.
Discussion
HED diet significantly increased body weight and body fat mass
It is widely recognized that diet is a key factor in the composition, diversity, dynamics, and microbiota-driven host metabolism24. Consistent with prior reports16,25,26, rats fed HED diet did not altered their caloric intake long-term. We observed an initial increase, but the animals adjusted their intake within a few days16,27. In contrast, rats fed HED had significantly higher final body weight and fat mass compared to LED fed animals, despite similar caloric intake. This was previously reported in a study by Lomba et al., which showed that rats fed a high fat diet restricted to the amount of calories consumed by a low fat fed group gained significantly more body weight and white adipose tissue than the low fat fed group28. This phenomenon indicates that HED diets detrimentally affect body weight and fat mass accumulation independent of caloric intake in male Sprague–Dawley rats.
HED diet triggered progressive dysbiosis of the gut microbiota
HED diet consumption induced dynamic fluctuations in the gut microbiota. Consistent with prior reports from our laboratory25, HED introduction triggered a rapid and transient decrease in bacterial diversity. It also induced marked fluctuations in bacterial abundance during the first 4 weeks. Thereafter, we did not observe further fluctuations in bacterial abundance.
We found that HED diet led to a rapid increase in members of the family Erysipelotrichaceae (Firmicutes). These are obligate anaerobes have been associated with consumption of HED, increased adiposity, and inflammation29–31. HED diet also depleted members of the families S24-7 (Bacteroidetes) and Verrucomicrobiaceae (Verrucomicrobia). Members of these families are associated with gut health as they are primarily involved in the fermentation of dietary fibers to produce short-chain fatty acids (SCFAs) and have been shown to be depleted by high fat diets25,32–34. After 4 weeks on the HED diet, we see a blooming of the family Ruminococcaceae (Firmicutes) that persists for the duration of the study. Members of this family are also SCFAs producers and generally associated with gut health33.
Long-term HED diet consumption significantly changed the cytokine profile in serum
In summary, our results revealed that HED induced significant changes in FGFβ, IFNγ, leptin, MCP-1, SCF, MIP-1α, IL-1α, IL-5, IP-10, Rantes, and TGFβ.
Leptin was significantly higher only after 26 weeks on HED diet. Consistent with prior reports35–37, we did not observe changes in insulin. The effect of HED diet on insulin appears to be strain-specific. Studies have reported significant increase in plasma insulin in Long-Evans male rats fed a high fat diet for 70 days38, in WNIN rats after 13 weeks37, and Wistar rats after 18 weeks39.
Our results revealed a significant increase in MCP-1 and MIP-1α after 26 weeks of HED. A similar study by Muralidhar et al. reported no change in MIP-1α and a non-statistically significant increase in MCP-1 after 13 weeks of high fat feeding37. The length of time of HED diet likely underlies the differences observed between the two studies; however, there is an observable trend toward higher levels with HED diet consumption. In addition, we observed a significant decrease in Rantes, consistent with a prior report by Fenton et al. in mice fed a high fat diet for 10 weeks40.
SCF serves as a ligand molecule for the receptor tyrosine kinase c-Kit. Activation of c-Kit is involved in cell migration and survival41. Our results showed a significant increase in SCF after 26 weeks of HED diet consumption.
FGFβ is an endocrine hormone produced by the liver, which is thought to enhance insulin-mediated glucose uptake in the fed state42. IFNγ is an important cytokine for innate and adaptive immune response against viral infections. IP-10 is a chemokine produced in response to IFNγ that acts as a chemoattractant for a host of immune cells43. Our results showed a significant increase in FGFβ and IP-10, and a decrease and IFNγ after 26 weeks of HED diet consumption. However, when compared to aged-matched, LED fed rats, there were no significant differences. In Sprague–Dawley rats, Muralidhar et al. showed that a high fat diet for 13 weeks did not affect plasma levels of IP-1044. It is possible that the changes observed in this study are a result of the aging process and not triggered by the HED diet. In human subjects older than 50 years there is a decrease in IFNγ production from mononuclear cells45.
Interleukin 1α (IL-1α) is an epidermal proinflammatory cytokine46. Interleukin 5 (IL-5) is a key factor in the activation of eosinophils during allergic reactions47. Consistent with prior reports40,48, our data showed that HED diet fed rats had significantly higher levels of IL-1α and a significant decrease in IL-5 after 26 weeks.
Consumption of a HED diet significantly increased microglia activation in the NTS
Reports from our laboratory and others have shown that consumption of a HED diet triggers microglia activation in the nodose ganglia, NTS, and hypothalamus26,49,50. Our data showed that HED diet induced an inflammatory response reflected by microglia activation in the intermediate NTS after 4 weeks. These data further suggest that length of exposure to the HED diet exacerbates this response since microglia activation after 26 weeks of HED diet was significantly higher than after four weeks.
HED diet consumption induced hepatic lipidosis
Our results demonstrated that a HED diet significantly increased intracellular lipid accumulation in the liver. These data are in concert with prior studies in rodents, which report development of hepatic steatosis after 16 weeks of a 40% fat diet51. De Rudder et al., also reported that mice developed hepatic steatosis after only 4 weeks on a 60% fat diet52. Hepatic steatosis has been linked to microbiota dysbiosis53,54. The majority of the nutrient-rich blood supply to the liver comes from the intestines through the portal vein55. Thus, an increase in gut microbes that produce toxic/inflammatory byproducts increases the gut-derived bacterial products entering the liver56. Our data revealed an increase in abundance of members of the family Erysipelotrichaceaea and a study by Spencer et al., showed that levels of these bacteria are directly associated with changes in liver fat in female human subjects57. In addition, we saw an increase in SCFAs producers. The SCFAs acetate, propionate, and butyrate inhibit lipid accumulation in the liver and improve hepatic function in rodents58–60. Given that the abundance of Ruminococcaceae was increased after 26 weeks of HED, it is possible that the presence of these bacteria and their byproducts contributed to prevent the progression of hepatic steatosis, as there was no difference in the degree of steatosis after 4 weeks of HED
In conclusion, we showed that long-term consumption of a HED diet leads to increased adiposity, gut dysbiosis, hepatic steatosis, inflammation in the NTS, and increased systemic levels of inflammatory markers. This study is novel because, to our knowledge, it is the first to present longitudinal and cross-sectional results on the effect of long-term HED diets on all these parameters in a single cohort of animals. Our results suggests that gut dysbiosis starts immediately upon introduction of HED diet. As the liver is overloaded with accumulation of excess fat consumed, hepatic steatosis develops. At the same time, endotoxins produced by the resident gut microbiome damage vagal afferents, which in turn triggers microglia activation in the NTS. Then, cytokines are released from their production site (e.g., adipose tissue) into the systemic circulation. These responses are highly dynamic and play a significant role in the development of obesity.
Supplementary information
Figure S1. Experimental design timeline.
Figure S2. Rarefraction curves by diet group and experimental time point. Data are shown as mean for rats fed a high energy density diet for 4 weeks (A, ST-HED) or 26 weeks (B, LT-HED).
Figure S3. LDA scores used for generation of cladogram (Fig. 3C). Colors designate time point: Blue: LED/baseline, Red: HED1, one week after introduction of HED diet, Green: HED4, four weeks after int
Figure S4. LDA scores used for generation of cladogram (Fig. 3F). Colors designate time point: Purple: LED26, after 26 weeks of LED diet, Green: HED4, four weeks after introduction of HED diet. Blue:
Acknowledgements
This work was supported by The National Institutes of Health grant no. 1R01DC013904.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41387-020-0119-4).
References
- 1.Hildebrandt MA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. e2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de La Serre CB, et al. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299:G440–G448. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stolarczyk E. Adipose tissue inflammation in obesity: a metabolic or immune response? Curr. Opin. Pharmacol. 2017;37:35–40. doi: 10.1016/j.coph.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Jo J, et al. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Computational Biol. 2009;5:e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura S, et al. Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels. Diabetes. 2007;56:1517–1526. doi: 10.2337/db06-1749. [DOI] [PubMed] [Google Scholar]
- 6.Asterholm IW, et al. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab. 2014;20:103–118. doi: 10.1016/j.cmet.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujisaka S, et al. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes. 2009;58:2574–2582. doi: 10.2337/db08-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitseva OI, et al. Inducible Toll‐like receptor and NF‐κB regulatory pathway expression in human adipose tissue. Obesity. 2008;16:932–937. doi: 10.1038/oby.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe Y, Nagai Y, Takatsu K. Activation and regulation of the pattern recognition receptors in obesity-induced adipose tissue inflammation and insulin resistance. Nutrients. 2013;5:3757–3778. doi: 10.3390/nu5093757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berthoud HR, Powley TL. Vagal afferent innervation of the rat fundic stomach: morphological characterization of the gastric tension receptor. J. Comp. Neurol. 1992;319:261–276. doi: 10.1002/cne.903190206. [DOI] [PubMed] [Google Scholar]
- 13.Angulo P. Medical progress-nonalcoholic fatty liver disease. N. Engl. J. Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 14.Adams LA, Angulo P, Lindor KD. Nonalcoholic fatty liver disease. Can. Med. Assoc. J. 2005;172:899–905. doi: 10.1503/cmaj.045232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slauson D. O. & Cooper B. J. Mechanisms of disease: A textbook of comparative general pathology (Mosby Inc., 2002).
- 16.Minaya DM, DiLorenzo PM, Hajnal A, Czaja K. Roux‑en‑Y gastric bypass surgery triggers rapid DNA fragmentation in vagal afferent neurons in rats. Acta Neurobiol. Exp. 2019;79:332–344. [PMC free article] [PubMed] [Google Scholar]
- 17.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1–e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arndt D, et al. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012;40(W1):W88–W95. doi: 10.1093/nar/gks497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallaher ZR, Ryu V, Herzog T, Ritter RC, Czaja K. Changes in microglial activation within the hindbrain, nodose ganglia, and the spinal cord following subdiaphragmatic vagotomy. Neurosci. Lett. 2012;513:31–36. doi: 10.1016/j.neulet.2012.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters JH, Gallaher ZR, Ryu V, Czaja K. Withdrawal and restoration of central vagal afferents within the dorsal vagal complex following subdiaphragmatic vagotomy. J. Comp. Neurol. 2013;521:3584–3599. doi: 10.1002/cne.23374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunt EM, et al. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 2011;53:810–820. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sen T, et al. Diet-driven microbiota dysbiosis is associated with vagal remodeling and obesity. Physiol. Behav. 2017;173:305–317. doi: 10.1016/j.physbeh.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaughn AC, et al. Energy-dense diet triggers changes in gut microbiota, reorganization of gut-brain vagal communication and increases body fat accumulation. Acta Neurobiol. Exp. 2017;77:18–30. doi: 10.21307/ane-2017-033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Lartigue G, de la Serre CB, Espero E, Lee J, Raybould HE. Diet-induced obesity leads to the development of leptin resistance in vagal afferent neurons. Am. J. Physiol. Endocrinol. Metab. 2011;301:E187–E195. doi: 10.1152/ajpendo.00056.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomba A, et al. Weight gain induced by an isocaloric pair-fed high fat diet: a nutriepigenetic study on FASN and NDUFB6 gene promoters. Mol. Genet. Metab. 2010;101:273–278. doi: 10.1016/j.ymgme.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Kaakoush NO. Insights into the role of erysipelotrichaceae in the human host. Front. Cell Infect. Microbiol. 2015;5:1–4. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riva A, et al. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. 2017;19:95–105. doi: 10.1111/1462-2920.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley D, Hughes RJ, Geier MS, Moore RJ. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front. Microbiol. 2016;7:1–13. doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhomah TA, Nishino N, Katsumata E, Wu HM, Tsuruta T. High-fat diet reduces the level of secretory immunoglobulin A coating of commensal gut microbiota. Biosci. Microbiota Food Health. 2019;38:55–64. doi: 10.12938/bmfh.18-027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 34.Evans CC, et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat Diet-induced obesity. PLoS ONE. 2014;9:1–14. doi: 10.1371/journal.pone.0092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borst SE, Conover CF. High-fat diet induces increased tissue expression of TNF-alpha. Life Sci. 2005;77:2156–2165. doi: 10.1016/j.lfs.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Ghibaudi L, Cook J, Farley C, Van Heek M, Hwa JJ. Fat intake affects adiposity, comorbidity factors, and energy metabolism of sprague‐dawley rats. Obes. Res. 2002;10:956–963. doi: 10.1038/oby.2002.130. [DOI] [PubMed] [Google Scholar]
- 37.Mn M, Smvk P, Battula KK, Nv G, Kalashikam RR. Differential response of rat strains to obesogenic diets underlines the importance of genetic makeup of an individual towards obesity. Sci. Rep. 2017;7:9162. doi: 10.1038/s41598-017-09149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woods SC, Seeley RJ, Rushing PA, D’Alessio D, Tso P. A controlled high-fat diet induces an obese syndrome in rats. J. Nutr. 2003;133:1081–1087. doi: 10.1093/jn/133.4.1081. [DOI] [PubMed] [Google Scholar]
- 39.Bravo E, et al. High fat diet-induced non alcoholic fatty liver disease in rats is associated with hyperhomocysteinemia caused by down regulation of the transsulphuration pathway. Lipids Health Dis. 2011;10:1–6. doi: 10.1186/1476-511X-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fenton JI, et al. Diet‐induced adiposity alters the serum profile of inflammation in C57BL/6N mice as measured by antibody array. Diabetes Obes. Metab. 2009;11:343–354. doi: 10.1111/j.1463-1326.2008.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-KIT: from basic science to clinical implications. Physiol. Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 42.BonDurant LD, Potthoff MJ. Fibroblast growth factor 21: a versatile regulator of metabolic homeostasis. Annu Rev. Nutr. 2018;38:173–196. doi: 10.1146/annurev-nutr-071816-064800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metzemaekers M, Vanheule V, Janssens R, Struyf S, Proost P. Overview of the mechanisms that may contribute to the non-redundant activities of interferon-inducible CXC chemokine receptor 3 ligands. Front. Immunol. 2018;8:1–21. doi: 10.3389/fimmu.2017.01970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muralidhar MN, Prasad SMVK, Battula KK, Giridharan NV, Kalashikam RR. Differential response of rat strains to obesogenic diets underlines the importance of genetic makeup of an individual towards obesity. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-09149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abb J, Abb H, Deinhardt F. Age-related decline of human interferon alpha and interferon gamma production. Blut. 1984;48:285–289. doi: 10.1007/BF00320399. [DOI] [PubMed] [Google Scholar]
- 46.Hanel KH, et al. Control of the physical and antimicrobial skin barrier by an IL-31-IL-1 signaling network. J. Immunol. 2016;196:3233–3244. doi: 10.4049/jimmunol.1402943. [DOI] [PubMed] [Google Scholar]
- 47.Sanderson CJ. Interleukin-5, eosinophils, and disease. Blood. 1992;79:3101–3109. [PubMed] [Google Scholar]
- 48.Collins KH, Hart DA, Seerattan RA, Reimer RA, Herzog W. High-fat/high-sucrose diet-induced obesity results in joint-specific development of osteoarthritis-like degeneration in a rat model. Bone Joint Res. 2018;7:274–281. doi: 10.1302/2046-3758.74.BJR-2017-0201.R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waise TMZ, et al. One-day high-fat diet induces inflammation in the nodose ganglion and hypothalamus of mice. Biochem Biophys Res. Commun. 2015;464:1157–1162. doi: 10.1016/j.bbrc.2015.07.097. [DOI] [PubMed] [Google Scholar]
- 50.Yi CX, Tschop MH, Woods SC, Hofmann SM. High-fat-diet exposure induces IgG accumulation in hypothalamic microglia. Dis. Model Mech. 2012;5:686–690. doi: 10.1242/dmm.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen VS, et al. Variation in diagnostic NAFLD/NASH read-outs in paired liver samples from rodent models. J. Pharm. Toxicol. Methods. 2019;101:106651. doi: 10.1016/j.vascn.2019.106651. [DOI] [PubMed] [Google Scholar]
- 52.De Rudder, M. et al. Automated computerized image analysis for the user-independent evaluation of disease severity in preclinical models of NAFLD/NASH. Lab. Invest. 1–14 (2019). [DOI] [PubMed]
- 53.Liu Q, et al. Role and effective therapeutic target of gut microbiota in NAFLD/NASH. Exp. Ther. Med. 2019;18:1935–1944. doi: 10.3892/etm.2019.7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji Y, Yin Y, Li Z, Zhang W. Gut microbiota-derived components and metabolites in the progression of non-alcoholic fatty liver disease (NAFLD) Nutrients. 2019;11:1–16. doi: 10.3390/nu11081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Compare D, et al. Gut-liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 2012;22:471–476. doi: 10.1016/j.numecd.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 56.Baffy G. Potential mechanisms linking gut microbiota and portal hypertension. Liver Int. 2019;39:598–609. doi: 10.1111/liv.13986. [DOI] [PubMed] [Google Scholar]
- 57.Spencer MD, et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishina PM, Freedland RA. Effects of propionate on lipid biosynthesis in isolated rat hepatocytes. J. Nutr. 1990;120:668–673. doi: 10.1093/jn/120.7.668. [DOI] [PubMed] [Google Scholar]
- 59.Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J. Gastroenterol. 2007;13:2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahuri-Arisoylu M, et al. Reprogramming of hepatic fat accumulation and ‘browning’ of adipose tissue by the short-chain fatty acid acetate. Int. J. Obes. 2016;40:955–963. doi: 10.1038/ijo.2016.23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Experimental design timeline.
Figure S2. Rarefraction curves by diet group and experimental time point. Data are shown as mean for rats fed a high energy density diet for 4 weeks (A, ST-HED) or 26 weeks (B, LT-HED).
Figure S3. LDA scores used for generation of cladogram (Fig. 3C). Colors designate time point: Blue: LED/baseline, Red: HED1, one week after introduction of HED diet, Green: HED4, four weeks after int
Figure S4. LDA scores used for generation of cladogram (Fig. 3F). Colors designate time point: Purple: LED26, after 26 weeks of LED diet, Green: HED4, four weeks after introduction of HED diet. Blue: