Abstract
Different dietary nitrogen (N) patterns may have different effects on gut microbiota. To investigate the effects of different crude protein (CP) levels or essential amino acids (EAA) supplementation patterns on the structure and functions of colonic microbiota, 42 barrows (25 ± 0.39 kg) were randomly assigned to 7 dietary treatments including: diet 1, a high CP diet with balanced 10 EAA; diet 2, a medium CP diet with approximately 2% decreased CP level from diet 1 and balanced 10 EAA; diets 3, 4, 5, 6 and 7, low CP diets with 4% decreased CP level from diet 1. Specifically, diet 3 was only balanced for Lys, Met, Thr and Trp; diets 4, 5 and 6 were further supplemented with Ile, Val and Ile + Val on the basis of diet 3, respectively; and diet 7 was balanced for 10 EAA. Results over a 110-d trial showed that reducing the CP level by 2% or 4% dramatically decreased N intake and excretion (P < 0.05) in the presence of balanced 10 EAA, which was not observed when altering the EAA supplementation patterns in low CP diet (−4%). With balanced 10 EAA, 2% reduction in dietary CP significantly reduced Firmicutes-to-Bacteroidetes (F:B) ratio and significantly elevated the abundance of Prevotellaceae NK3B31 (P < 0.05); whereas 4% reduction evidently increased the abundances of Proteobacteria, Succinivibrio and Lachnospiraceae XPB1014 (P < 0.05). Among the 5 low CP diets (−4%), supplementation with Ile, or Val + Ile, or balanced 10 EAA increased F:B ratio and the abundance of Proteobacteria. In addition, the predicted functions revealed that different CP levels and EAA balanced patterns dramatically altered the mRNA expression profiles of N-metabolizing genes, the “N and energy metabolism” pathways or the metabolism of some small substances, such as amino acids (AA) and vitamins. Our findings suggested that reducing the dietary CP levels by 2% to 4% with balancing 10 EAA, or only further supplementation with Ile or Val + Ile to a low protein diet (−4%) reduced the N contents entering the hindgut to various degrees, altered the abundances of N-metabolizing bacteria, and improved the abilities of N utilization.
Keywords: Colonic microbiota, Dietary protein level, Essential amino acid, Pig, 16S high throughput sequencing
1. Introduction
Gut microbiota represents a large and complex microbial community, which is composed of at least 500 to 1,000 species in the mammal (Rajilićstojanović and Vos, 2015). Gut microbiota communities interact with each other and their host, which exerts a vital role in the host physiology and metabolism, including modulation of energy harvest, nutrients metabolism, and immune system development (Willing and Van Kessel, 2010). There is inter-individual variability in the qualitative and quantitative composition of species, taxonomic units, abundance and biodiversity. These heterogeneities are affected by different factors, such as genome, age, sex, geographic location, pathological conditions, pharmacological treatments and daily diet (Walker et al., 2011, Yatsunenko et al., 2012). Among all these factors, changes in the dietary components can quickly alter the composition of microbiota (Walker et al., 2011). Protein is one of the most common and main components in diets, the portion of the dietary nitrogenous compounds escape digestion in the small intestine and enter the large intestine to be further utilized by the hindgut microbiota (mainly at the distal colon) (Gibson et al., 1976).
Gut microbiota can synthesize abundant proteases and peptidases to hydrolyzed proteins and peptides, and they can also catabolize almost all kinds of amino acids (AA) (Gibson et al., 1976, Han et al., 2010). Bacteroides and Propionibacterium have been identified as the predominant proteolytic species in fecal samples, while clostridia, streptococci, Fusobacterium and Lactobacillus are also the frequently seen proteolytic bacteria (Macfarlane et al., 1986, Phillips et al., 1991). Studies revealed that the relative concentrations of different AA available to gut microbiota can greatly affect the overall AA utilization at the community level (Dai et al., 2013).
To ensure the maximum growth performance, nitrogen (N) was usually supplied excessively in the animal diets, which leads to a waste of protein resources and N pollution. Previous studies have shown that the properly reduced dietary protein content and supplementation with crystalline AA reduce N excretion without affecting the growth performance of pigs (Kerr, 2003). Moreover, it is found that the high-protein diet can increase the abundances of Escherichia coli, Clostridium and Clostridium perfringens in the feces of piglets (Opapeju et al., 2009) and rats (Chung et al., 1977). The structural composition of gut microbiota and its metabolites affect the host homeostasis. Therefore, it is necessary to clearly determine the alterations in gut microbiota induced by the low protein diets. At present, several studies have been carried out to examine the effects of low protein diet on gut microbiota, and they mainly focus on different protein contents or protein sources (Torrallardona et al., 2003, Qi et al., 2011, Zhou et al., 2016, Fan et al., 2017). Nonetheless, limited data are available concerning the effects of different EAA supplementation patterns in low protein diet on the gut microbiota of pigs. Therefore, pigs in this study were fed diets containing different dietary protein levels or AA from 25 to 125 kg, and then the composition and functions of colonic microbiota were analyzed through 16S rRNA sequencing and the software package Tax4Fun, respectively.
2. Materials and methods
The experiment was conducted in Metabolism Laboratory of Institute of Animal Nutrition, Sichuan Agricultural University (Ya'an, China). The experimental protocols used in the current study were reviewed and approved by the Animal Care and Use Committee of Sichuan Province (CD-SYXK-2017-015).
2.1. Experimental design and treatments
A total of 42 barrows ([Landrace × Yorkshire] × Duroc; initial body weight = 25 ± 0.39 kg) were randomly assigned to 7 experimental groups based on their body weight for a 110-d trial that was divided into 4 phases (25 to 50 kg, 50 to 75 kg, 75 to 100 kg and 100 to 125 kg). The experimental design was presented in Table 1. Experimental diets were formulated to meet or exceed the nutrient requirements recommended by the NRC (2012), except for the dietary crude protein (CP) contents and AA levels. The 7 diets were as follows. Diet 1 was a high crude protein (HCP) diet with CP levels of 17.5%, 16.0%, 14.5% and 13.0% at the 4 phases, respectively, and balanced for 10 essential amino acids (EAA). Diet 2 was a medium crude protein (MCP) diet, which was based on the NRC (2012) recommendation, with CP levels (approximately 2% lower than those of diet 1) of 15.70%, 13.5%, 12.13% and 10.44% at the 4 phases, respectively, and balanced for 10 EAA. Additionally, the dietary CP levels of diet 3, 4, 5, 6 and 7 (low crude protein [LCP] diets) were 4% lower than those of diet 1. Specifically, diet 3 was only balanced for Lys, Met, Thr and Trp. Diets 4, 5 and 6 were further supplemented with Ile, Val and Ile + Val on the basis of diet 3, respectively. Diet 6 was further supplemented with extra Ile + Val and diet 7 was balanced for 10 EAA. The diets were mainly composed of corn, wheat bran, soybean meal and rapeseed meal (Appendix Tables 1 to 4). The CP and AA contents in the ingredients and experimental diets were analyzed before feeding. All pigs were individually housed into the stainless-steel metabolic crates (1.4 m long × 0.70 m wide × 0.85 m high). Pigs were fed 3 times a day at 08:00, 14:00 and 20:00 and had free access to water throughout the experiment.
Table 1.
Design of experiment.
| Diet | CP level | EAA balanced patterns |
|---|---|---|
| 1 | High CP | 10 EAA1 |
| 2 | Medium CP (−2%) | 10 EAA |
| 3 | Low CP (−4%) | Lys, Thr, Trp and Met |
| 4 | Low CP (−4%) | Lys, Thr, Trp, Met and Ile |
| 5 | Low CP (−4%) | Lys, Thr, Trp, Met and Val |
| 6 | Low CP (−4%) | Lys, Thr, Trp, Met, Val and Ile |
| 7 | Low CP (−4%) | 10 EAA |
CP = crude protein; EAA = essential amino acids.
10 EAA: Lys, Met, Thr, Trp, Val, Ile, Phe, His, Leu and Arg.
2.2. Nitrogen balance study
On the last 4 d of the trial, total feces and urine from all pigs were collected and recorded daily, respectively. Afterwards, the fecal samples from each pig collected on 4 d were mixed, then 500 g of the mixed sample was collected to dry at 65 °C in a forced-draft oven and grounded using a 0.45-mm screen. The final dry samples from all pigs were preserved at −20 °C for further analysis. The urine samples collected on 4 d from each pig were mixed, then 100 mL of the mixed sample was collected and kept at −20 °C. To determine the N balance, the CP contents in diets, as well as fecal and urine samples were analyzed according to methods proposed by the Association of Official Analytical Chemists (2003).
At the end of the trial, all pigs were slaughtered, and approximately 8 g of colonic digesta from each pig was collected in a sterile tube, which was then immediately stored at −80 °C for genomic DNA extraction and metabolite profile detection.
2.3. DNA extraction and sequencing
Approximately 100 mg of colonic contents were weighed, and total genomic DNA was extracted using the stool DNA extraction kit (E.Z.N.A.TM stool DNA isolation kit, D4015-01, Omega Bio-Tek, USA) in accordance with manufacturer instructions. DNA concentration and quality were detected using the Nanodrop Spectrophotometer. Afterwards, the bacterial universal V4 region of the 16S rRNA gene was amplified using the PCR bar-coded primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR reaction system (50 μL) contained 2 × PCR buffer, 1.5 mmol/L MgCl2, 0.4 mmol/L of each dNTP, 1.0 μmol/L of each primer, 0.5 U of KOD-Plus-Neo (TOYOBO) and 10 ng template DNA. The PCR amplification conditions were as follows: initial denaturation at 94 °C for 1 min, followed by 30 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 30 s as well as elongation at 72 °C for 30 s, and then the final extension at 72 °C for 5 min. In addition, the PCR products mixed with 1/6 volume of 6 × loading buffer were loaded onto the 2% agarose gel for detection. Then, samples with a bright main strip of 200 to 400 bp were selected for subsequent experiments. The electrophoresis band was purified using the OMEGA Gel Extraction Kit (Omega Bio-Tek, USA). DNA was quantified using the Qubit 2.0 Fluorometer (Thermo Scientific), and PCR products from different samples were pooled at an equivalent molar amount. The sequencing libraries were generated using the TruSeq DNA PCR-Free Sample Prep Kit in accordance with the manufacturer protocols, and the index codes were added. Moreover, the library quality was assessed by the Qubit 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was subjected to paired-end sequencing (2 × 250 bp) using the Illumina Hiseq apparatus. All reads were uploaded to the National Center for Biotechnology Information (NCBI) and were accessed in the Short Read Archive (SRA) under accession number SRR7585932.
2.4. Detection of metabolite profiles in the colonic contents
Concentrations of main short chain fatty acids (SCFA, including acetate, propionate, butyrate, isobutyrate and branched chain valerate) in the colonic digesta samples were determined by gas chromatography (GC) (GC-14B, Shimadzu, Japan; capillary column: 30 m × 0.32 mm × 0.25 μm film thickness). Briefly, 0.2 g colonic digesta was thawed and suspended into 0.3 mL of methanol (50%). Then, each sample was centrifuged at 10,000 × g and 4 °C for 10 min, and 0.1-mL supernatant was transferred into a 0.5-mL centrifuge tube to mix with 0.02-mL crotonic acid-metaphosphate acid, followed by another 10 min of centrifugation at 15,000 × g and 4 °C. Later, 0.1-mL supernatant was collected and added into 0.7-mL methanol, followed by vortexing and 10 min of centrifugation at 12,000 × g and 4 °C. Afterwards, the supernatant was analyzed by means of GC using a flame ionization detector, with the oven temperature from 100 to 150 °C (N2 was used as the carrier gas at the flow rate of 1.8 mL/min).
The concentration of ammonia nitrogen (NH3–N) in colonic contents was determined through the spectrophotometric approach using the Nessler reagent with yellow color and photometer at the wavelength of 420 nm.
2.5. Analysis of sequencing data
The sequences were analyzed in accordance with the Usearch and QIIME pipeline. The paired-end reads from the original DNA fragments were merged by FLASH4. Then, the sequences were assigned to each sample based on the unique barcode, the low quality reads (with the length of <200 bp and over 2 ambiguous base ‘N’ or the average base quality score of <30) were filtered, and sequences with decayed quality scores (score of <11) were truncated using Trimmomatic36 and Usearch. After identifying the duplicated sequences, all the singletons were discarded, since they might be the unfavorable amplifications and lead to over-estimated diversity. Sequences were then clustered into the operational taxonomic units (OTU) at the identity threshold of 97% using the UPARSE algorithms. Thereafter, the representative sequences were picked and the potential chimeras were removed using the Uchime algorithm. Taxonomy was then assigned using the Silva database and uclust classifier in QIIME. All data were analyzed using R or Python. Subsequently, principal coordinate analysis (PCoA) and LEfSe analysis were carried out using Ape40 package and Python LEfSe package, respectively. The functional profiles of microbial communities were predicted using Tax4Fun.
2.6. Statistical analyses
In this study, each pig was used as an experimental unit. Data normality was analyzed by the Kolmogorov–Smirnov test using SPSS for Windows version 20.0 (SPSS). For normally distributed data that exhibited homogeneity of variance, data on different protein level groups with 10 balanced EAA, and those on different EAA supplementation patterns with the same protein level, were analyzed using the one-way analysis of variance (ANOVA) and Duncan's post hoc test, respectively. The SPSS 20.0 software was adopted for all statistical analyses, and the significance level was set at P < 0.05.
3. Results
3.1. Nitrogen balance of fattening pigs
The N balance data of pigs are shown in Table 2. Balancing 10 EAA and reducing the dietary CP level by 2% or 4% dramatically decreased the N intake and total N excretion (P < 0.05); specifically, the total N excretion decreased by 23.40% and 44.13%, respectively. A 4% reduction in dietary protein levels significantly reduced the N retention but significantly increased the N retention rate (P < 0.05). When the dietary CP level was reduced by 4%, the total N excretion of the group supplemented with Lys, Met, Thr and Trp was higher than that of the further supplemented Ile group, Val group, Val + Ile group and the balanced 10 EAA group by 14.09%, 3.84%, 3.77% and 15.84%, respectively. Similarly, the N retention rate of the group supplemented with Lys, Met, Thr and Trp was lower than that of the other 4 amino acid addition groups by 3.34%, 3.41%, 2.63%, and 7.55%, respectively.
Table 2.
Nitrogen (N) balance of fattening pigs fed low protein diets supplemented with different EAA1.
| Item | HCP |
MCP (−2%) |
LCP (−4%) |
SEM |
P-values2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10 EAA | 10 EAA | Lys, Met, Thr, Trp | Lys, Met, Thr, Trp, Ile | Lys, Met, Thr, Trp, Val | Lys, Met, Thr, Trp, Ile, Val | 10 EAA | P1 | P2 | ||
| N intake, g/d | 59.58x | 49.99y | 42.83 | 40.46 | 44.39 | 43.53 | 44.23z | 1.09 | <0.01 | 0.13 |
| FN, g/d | 6.79x | 6.14xy | 5.79 | 4.97 | 5.63 | 5.43 | 5.21y | 0.15 | 0.04 | 0.26 |
| UN, g/d | 15.35x | 10.82y | 8.54 | 7.59 | 8.17 | 8.38 | 7.15z | 0.57 | <0.01 | 0.82 |
| TN, g/d | 22.14x | 16.96y | 14.33 | 12.56 | 13.80 | 13.81 | 12.37z | 0.66 | <0.01 | 0.54 |
| RN, g/d | 37.44x | 33.03xy | 28.50 | 27.91 | 30.59 | 29.73 | 31.87y | 0.69 | 0.04 | 0.16 |
| N retention rate, % | 62.95y | 65.94y | 66.58 | 68.88 | 68.93 | 68.38 | 72.02x | 0.85 | 0.02 | 0.45 |
| N ABV, % | 70.96y | 75.21y | 77.02 | 78.57 | 78.97 | 78.11 | 81.66x | 0.96 | 0.01 | 0.70 |
EAA = essential amino acids; HCP = high crude protein; MCP = medium crude protein; LCP = low crude protein; FN = fecal nitrogen; UN = urine nitrogen; TN = total nitrogen; RN = retained nitrogen; N ABV = nitrogen apparent biological value.
x, y, z Superscripts in the same row indicate differences among different levels of dietary protein (P < 0.05).
Data are shown as means and standard errors (n = 6, number of replicates).
P1 represents the P-value of ANOVA among the HCP diet, MCP (NRC, 2012) diet and LCP diet under the conditions of 10 EAA balanced; P2 represents the P-value of ANOVA among different amino acid balanced patterns of the LCP diet.
3.2. SCFA and NH3–N concentrations in the colonic contents
As shown in Table 3, in the presence of balanced 10 EAA, reducing the dietary CP level by 2% or 4% apparently decreased the NH3–N concentration in colonic contents (P < 0.05). Nonetheless, there were no significant differences in the concentrations of SCFA, including acetate, propionate, butyrate, isobutyrate and branched chain valerate, among different dietary CP level groups. When the dietary CP level was reduced by 4%, the colonic isobutyrate concentration in pigs fed diet supplemented with Lys, Met, Thr, Trp, Val and Ile was higher than that in pigs fed diets supplemented with Lys, Met, Thr and Trp by 44.44%. No differences in other SCFA and NH3–N concentrations were found between the 5 low protein diet groups.
Table 3.
Colonic microbial metabolism products of fattening pigs fed low protein diets supplemented with different EAA1.
| Item | HCP |
MCP (−2%) |
LCP (−4%) |
SEM |
P-values2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10 EAA | 10 EAA | Lys, Met, Thr, Trp | Lys, Met, Thr, Trp, Ile | Lys, Met, Thr, Trp, Val | Lys, Met, Thr, Trp, Ile, Val | 10 EAA | P1 | P2 | ||
| Acetate, mg/g | 2.03 | 1.98 | 1.84 | 2.25 | 2.33 | 1.98 | 2.39 | 0.07 | 0.21 | 0.20 |
| Propionate, mg/g | 1.18 | 1.0 | 0.92 | 0.95 | 1.08 | 0.81 | 1.14 | 0.05 | 0.61 | 0.39 |
| Butyrate, mg/g | 0.84 | 0.80 | 0.80 | 0.77 | 0.86 | 0.79 | 1.00 | 0.04 | 0.24 | 0.33 |
| Isobutyrate, mg/g | 0.12 | 0.11 | 0.09b | 0.12ab | 0.11ab | 0.13a | 0.12ab | 0.03 | 0.43 | 0.10 |
| BC valerate, mg/g | 0.26 | 0.20 | 0.20 | 0.22 | 0.20 | 0.21 | 0.21 | 0.01 | 0.22 | 0.98 |
| NH3–N, mg/kg | 692.05x | 390.15y | 368.88 | 326.67 | 347.71 | 310.63 | 300.55y | 28.87 | <0.01 | 0.83 |
EAA = essential amino acids; HCP = high crude protein; MCP = medium crude protein; LCP = low crude protein; BC valerate = branched chain valerate; NH3–N = ammonia nitrogen.
x,y,z Superscripts in the same row indicate differences among different levels of dietary protein (P < 0.05).
a,b,c Superscripts in the same row indicated differences among different amino acid balanced patterns of LCP diets groups (P < 0.05).
Data are shown as means and standard errors (n = 6, number of replicates).
P1 represents the P-value of ANOVA among the HCP diet, MCP (NRC, 2012) diet and LCP diet under the conditions of 10 EAA balanced; P2 represents the P-value of ANOVA among different amino acid balanced patterns of the LCP diet.
3.3. Diversity of colonic microbiota
A total of 8,244,072 valid sequences were obtained from all samples after size filtering, quality control and chimera removal, with the mean sequence length of 320.10 ± 13.91 bp/sample. When the overall OTU numbers were classified at a distance level of 0.03 (97% similarity), all the sequences were assigned to 43,374 OTU, which were assigned to 40 bacterial phyla and 676 bacterial genera. As showed in Table 4, no differences were detected in the alpha-diversity indexes, including Shannon, Simpson, ACE and Chao of colonic microbiota, among different dietary protein level or EAA balanced pattern groups.
Table 4.
The alpha-diversity of colonic microbiota of fattening pigs fed low protein diets supplemented with different EAA1.
| Item | HCP |
MCP (−2%) |
LCP (−4%) |
SEM |
P-values2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10 EAA | 10 EAA | Lys, Met, Thr, Trp | Lys, Met, Thr, Trp, Ile | Lys, Met, Thr, Trp, Val | Lys, Met, Thr, Trp, Ile, Val | 10EAA | P1 | P2 | ||
| Chao | 139.86 | 188.86 | 153.57 | 204.69 | 167.55 | 182.22 | 164.81 | 7.98 | 0.32 | 0.47 |
| Ace | 41.66 | 47.80 | 44.62 | 49.33 | 45.35 | 49.13 | 45.62 | 0.87 | 0.23 | 0.31 |
| Shannon | 6.54 | 6.71 | 6.55 | 6.56 | 6.47 | 6.66 | 6.66 | 0.04 | 0.56 | 0.62 |
| Simpson | 0.17 | 0.23 | 0.18 | 0.17 | 0.13 | 0.16 | 0.18 | 0.01 | 0.48 | 0.65 |
EAA = essential amino acids; HCP = high crude protein; MCP = medium crude protein; LCP = low crude protein.
Data are shown as means and standard errors (n = 6, number of replicates).
P1 represents the P-value of ANOVA among the HCP diet, MCP (NRC, 2012) diet and LCP diet under the conditions of 10 EAA balanced; P2 represents the P-value of ANOVA among different amino acid balanced patterns of the LCP diet.
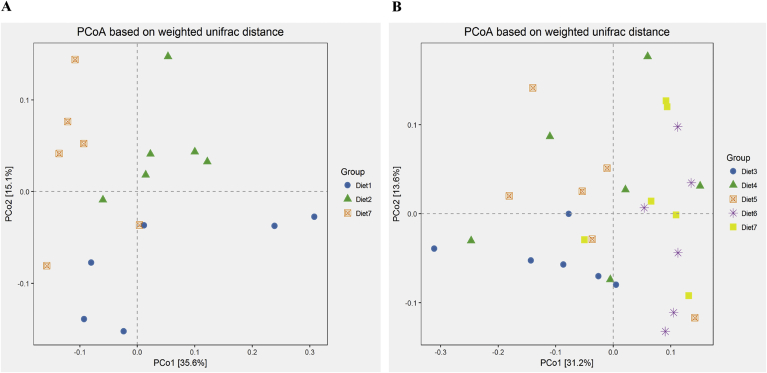
To test the relationships among the colonic bacterial community structures across different dietary protein level diets and EAA balanced pattern diets, PCoA was performed based on the weighted Unifrac distances of the 16S rRNA sequence profiles at OTU level (Fig. 1). The UniFrac distances suggested that both the protein levels and EAA supplementation patterns affected the colonic microbiota. In the presence of balanced 10 EAA, the HCP diet and LCP diet could not be separated in terms of the colonic microbiome, whereas the MCP diet could be separated from the HCP diet and LCP diet. When the dietary CP level was reduced by 4%, the Lys, Met, Thr and Trp supplementation diets were distinctly separated from the further Val + Ile supplementation diet and balanced 10 EAA diet, respectively; meanwhile, the further Val supplementation diet was also separated from the further Val + Ile supplementation diet and the balanced 10 EAA diet.
Fig. 1.
Principal coordinate analysis (PCoA) of bacterial community structures of the colonic microbiota. (A) PCoA of fattening pigs' colonic bacteria among different dietary protein levels groups. Diet 1 was a high protein diet, diet 2 was a medium protein diet (−2%) and diet 7 was a low protein diet (−4%). (B) PCoA of fattening pigs' colonic bacteria among different amino acid supplementation patterns in low protein diet (−4%). Diet 3 was supplemented with Lys, Met, Thr and Trp; diets 4, 5 and 6 were further supplemented with Ile, Val and Ile + Val on the basis of diet 3, respectively; diet 7 was balanced with 10 EAA.
3.4. Community structure of the colonic microbiota
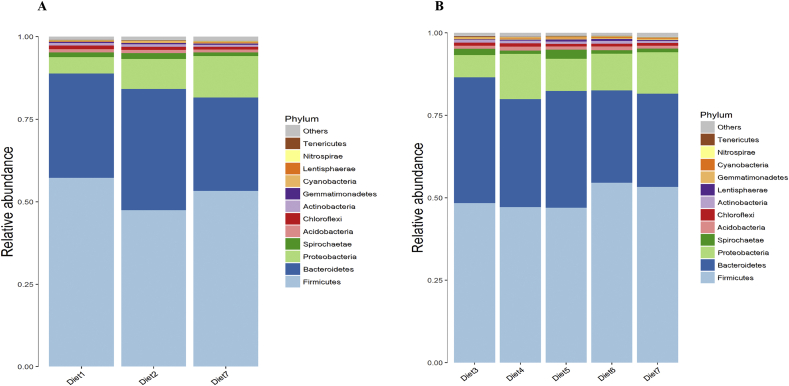
Figure 2 (Appendix Tables 5 and 6) and Fig. 3 (Appendix Tables 7 and 8) showed the overall microbiota compositions in different diets at phylum and genus levels, respectively. As could be observed, Firmicutes, Bacteroidetes, Proteobacteria, Spirochaetae and Acidobacteria were the dominant phyla in all samples, which accounted for over 95% of the total colonic bacterial community. In addition, the dominant genera of all samples were Prevotellaceae NK3B31 group, Anaerovibrio, Prevotella 9, Lachnospiraceae NK4A136 group, Succinivibrio, Lachnospiraceae XPB1014 group, Prevotella 1 and Ruminococcaceae UCG-005. At phylum level, when the dietary CP levels were reduced by 2% and 4% with balanced 10 EAA, the abundances of Firmicutes dramatically decreased from 59.03% to 46.22% (P < 0.05) and 51.78%, respectively, while those of Bacteroidetes markedly altered from 29.46% to 37.88% (P < 0.05) and 28.50%, respectively; furthermore, the decrease in the dietary CP levels significantly increased the abundances of Proteobacteria from 4.70% to 9.26% and 13.75% (P < 0.05), respectively. When the dietary CP levels were reduced by 4%, the abundances of Firmicutes of the Lys, Met, Thr and Trp supplementation group was lower than that of the further Val + Ile supplementation group (P < 0.05) and the balanced 10 EAA group by 14.75% and 10.16%, respectively. Similarly, the abundances of Bacteroidetes in the Lys, Met, Thr and Trp supplementation group were higher than that of the other 2 amino acid addition groups (P < 0.05) by 32.81% and 34.49%, respectively. Meanwhile, the abundance of Actinobacteria in the further Val supplementation group was also reduced.
Fig. 2.
Colonic bacterial community structure on phylum level of fattening pigs. (A) Relative abundance of colonic bacterial of fattening among different dietary protein levels groups. Diet 1 was a high protein diet, diet 2 was a medium protein diet (−2%) and diet 7 was a low protein diet (−4%). (B) Relative abundance of colonic bacterial of fattening among different amino acid supplementation patterns of in low protein diet (−4%). Diet 3 was supplemented with Lys, Met, Thr and Trp; diets 4, 5 and 6 were further supplemented with Ile, Val and Ile + Val on the basis of diet 3, respectively; diet 7 was balanced with 10 EAA.
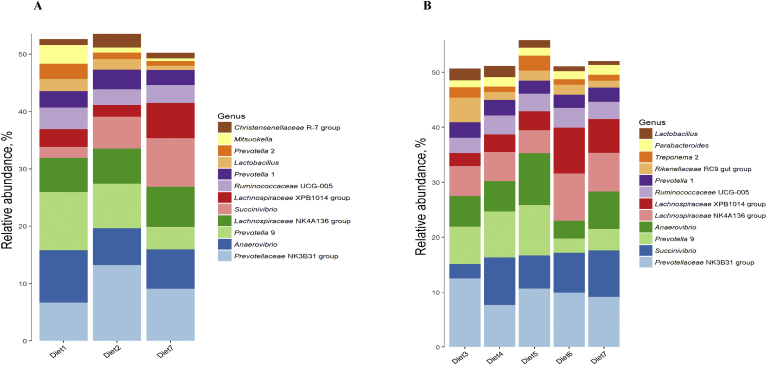
Fig. 3.
Colonic bacterial community structure on genus level of fattening pigs. (A) Relative abundance of colonic bacterial (percentage) of fattening among different dietary protein levels. Diet 1 was a high protein diet, diet 2 was a medium protein diet (−2%) and diet 7 was a low protein diet (−4%). (B) Relative abundance of colonic bacterial (percentage) of fattening among different amino acid supplementation patterns in low protein diet (−4%). Diet 3 supplemented with Lys, Met, Thr and Trp; diets 4, 5 and 6 were further supplemented with Ile, Val and Ile + Val on the basis of diet 3, respectively; diet 7 was balanced with 10 EAA.
At genus level, with balanced 10 EAA, a 2% reduction in the dietary CP level outstandingly enhanced the abundance of Prevotellaceae NK3B31 group (P < 0.05) belonging to Bacteroidetes; whereas a 4% reduction in the dietary CP level markedly elevated the abundances of Succinivibrio and Lachnospiraceae XPB1014 group (P < 0.05), and evidently decreased the abundance of Lactobacillus (P < 0.05). When the dietary CP level was reduced by 4%, compared with the Lys, Met, Thr and Trp supplementation group, the further supplement Ile group and balanced 10EAA group had significantly reduced abundance of Prevotellaceae NK3B31 group (P < 0.05), the group with balanced 10 EAA had notably increased abundance of Succinivibrio (P < 0.05), while other EAA supplementation groups had remarkably reduced abundance of Rikenellaceae RC9 gut group (P < 0.05). The further supplement Val + Ile group had lower abundances of Prevotella 9 and Anaerovibrio than those of the further supplement Val alone group (P < 0.05). At the same time, higher abundances of Lachnospiraceae NK4A136 group and Lachnospiraceae XPB1014 group were detected in the further supplement Val + Ile group (P < 0.05).
3.5. Predicted functions of colonic microbiota
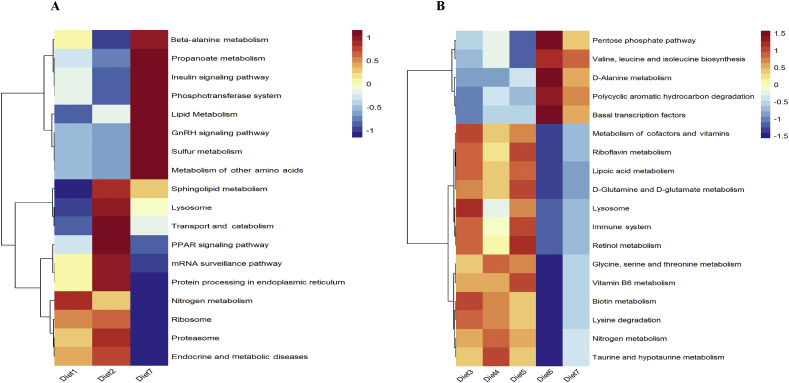
To investigate functional changes of colonic microbial communities, the Tax4FUN software was applied in the 16S rRNA datasets. The results of Tax4FUN were analyzed through the Kruskal–Wallis rank sum test. Among the 47 genes related to N metabolism, 7 differentially expressed genes (DEG) were selected and displayed in the manner of heat map (Fig. 4). As could be observed, a 2% reduction in the dietary CP level markedly down-regulated the mRNA expression of hydroxylamine and glutamate dehydrogenase genes (P < 0.05) with balanced 10 EAA, while a 4% reduction in the dietary CP level remarkably up-regulated that of glutamate synthase genes and nitrogenase gene (P < 0.05). When the dietary CP level was reduced by 4%, the further supplement Val + Ile group and the balanced 10EAA group had down-regulated mRNA expression of denitrification-related genes and carbamate kinase genes whereas up-regulated mRNA expression of glutamate metabolism-related genes (P < 0.05). Tax4Fun analysis also revealed differences in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among various groups. Among the 297 pathways, 18 KEGG Ortholog (KO) profiles with significant differentiation were selected and displayed in the manner of heat map (Fig. 5). With balanced 10 EAA, a 2% or 4% reduction in the dietary CP level mainly dramatically up-regulated these pathways related to N and energy metabolism (P < 0.05) (such as “protein processing in endoplasmic reticulum” and the “insulin signaling pathway”), as well as molecular transport and catabolism (like the “Transport and Catabolism” and “Lysosome”); alternatively, it obviously down-regulated the pathways related to genetic information processing (for instance, the “Ribosome” and “mRNA surveillance pathway”). When the dietary CP level was reduced by 4%, those pathways related to partial AA and glucose metabolism (such as the “Val, Leu and Ile biosynthesis” and the “Pentose phosphate pathway”) were markedly up-regulated in the further Val + Ile supplementation diet group and the balanced 10 EAA diet group; meanwhile, those pathways related to partial AA and vitamin metabolism (like “Glycine, Serine and Thr metabolism” and “vitamin B6 metabolism”) were evidently down-regulated.
Fig. 4.
mRNA expression of genes related to nitrogen metabolism of colonic microbiota. (A) mRNA expression of genes related to nitrogen metabolism of colonic microbiota among different dietary protein levels groups. Diet 1 was a high protein diet, diet 2 was a medium protein diet (−2%) and diet 7 was a low protein diet (−4%). (B) mRNA abundance of genes related to nitrogen metabolism of colonic microbiota among different amino acid supplementation patterns in low protein diet (−4%). Diet 3 supplemented with Lys, Met, Thr and Trp, diets 4, 5 and 6 were further supplemented with Ile, Val and Ile + Val on the basis of diet 3, respectively; diet 7 was balanced with 10 EAA.
Fig. 5.
Predicted function of colonic microbiota. (A) Predicted function of colonic microbiota among different dietary protein levels groups. Diet 1 was a high protein diet, diet 2 was a medium protein diet (−2%) and diet 7 was a low protein diet (−4%). GnRH = gonadotropin-releasing hormone; PPAR = peroxisome proliferators-activated receptors. (B) Predicted function of colonic microbiota among different amino acid supplementation patterns in low protein diet (−4%). Diet 3 supplemented with Lys, Met, Thr and Trp; diets 4, 5 and 6 were further supplemented with Ile, Val and Ile + Val on the basis of diet 3, respectively; diet 7 was balanced with 10 EAA.
4. Discussion
Reducing the dietary CP level is an effective way to decrease N excretion without inhibiting the growth of animals (Kerr, 2003). In this study, with balanced 10 EAA, a 2% or 4% reduction in the dietary CP level markedly reduced N intake and excretion in pigs, which was consistent with results reported from previous studies (Dourmad et al., 1993, Jin et al., 1998). When the dietary CP level was reduced by 4%, the total N excretion in the Lys, Met, Thr, Trp and Ile supplementation group and the 10 EAA balanced group was numerically lower than that in other EAA supplementation groups, which indicated that these 2 diets attained a favorable balanced status of AA, resulting in a high N utilization rate. Results of growth performance (Appendix Table 9) also confirmed this deduction. Our results suggested that both the dietary CP levels and EAA supplementation patterns affected N utilization and excretion, among which, the dietary CP levels had greater influence than the EAA supplementation patterns.
The fermentation of nitrogenous compounds becomes more active from the proximal to distal of large intestine as the pH value increases (Phillips et al., 1991). Similar to carbohydrates, SCFA are also produced during the nitrogenous fermentation process, while the branched chain fatty acids (BCFA), which account for 5% to 10% of total SCFA, are produced exclusively through the microbial metabolism of branched chain AA (Blachier et al., 2007). The amount of BCFA can be used to assess protein fermentation in the large intestine since animals do not contain enzymes that synthesize BCFA. In this study, a 2% or 4% reduction in the protein levels markedly reduced NH3–H concentration in colonic contents, which was consistent with the previous report (Leek et al., 2007). Such results indicated that lowering the dietary CP level reduced the N content entering the hindgut and improved N utilization, and such deduction was confirmed through the N balance results. Besides, no difference was found in SCFA content among different dietary protein levels diets, which was consistent to the previous results (Zhou et al., 2016). However, Fan et al. (2017) reported that the fecal SCFA concentration increased with the dietary protein level increased from 13% to 16%, while the SCFA concentration did not change when the dietary protein level increased from 16% to 21%. Compared with protein fermentation, SCFA was mainly produced by carbohydrate fermentation. Protein and carbohydrate contents in the diets and their utilization efficiencies will affect the amount of nutrients eventually entering the hindgut, which will further affect the production of fermentation products. When the dietary CP level was reduced by 4%, no significant difference in NH3–N content among different EAA supplementation groups was observed. However, the isobutyrate concentration of pigs in further supplement Val + Ile group was markedly higher than that in further supplement Lys, Met, Thr and Trp group. When both Ile and Val are supplemented in diet, these 2 branched chain AA will compete for the same transport system due to their similar branch chain structures (Harper et al., 1970). Our results of growth performance also demonstrated that, Ile was more needed than Val in finishing pigs. Thus, Ile was fully absorbed and utilized, while Val utilization was lower; therefore, more Val entered the hindgut and was utilized by microbes to produce more isobutyrate.
Gut microbiota and their fermented metabolites have potential influence on the host health. In our experiment, with balanced 10 EAA, a 2% or 4% reduction in the dietary CP level had no significant effect on the α diversity index of colonic microbiota in fattening pigs, which was consistent with the finding from Zhou et al. (2016). Nonetheless, different results were obtained from Fan et al. (2017) and Liu et al. (2015). Typically, the inconsistent effects of dietary CP levels on the intestinal microbial diversity might be ascribed to the reduction levels of dietary CP and the duration of experiment.
It was found in our study that, both the dietary CP levels and EAA supplementation patterns altered the structural composition of colonic microbiota, which was also demonstrated by PCoA results. With balanced 10 EAA, a 2% reduction in the dietary CP level markedly reduced Firmicutes to Bacteroidetes (F:B) ratio. On the other hand, previous studies indicate that plant polysaccharides can decrease the F:B ratio (De Filippo et al., 2010). When formulating the low protein diets in this study, the soybean meal was partially replaced by wheat bran and corn. Therefore, there were more fibers and starches in the low protein diets, which could increase the abundance of Bacteroidetes. Further analysis at genus level indicated that, a 2% reduction in the dietary protein remarkably boosted the abundance of Prevotellaceae NK3B31 group belonging to Bacteroidetes. It has been reported that Prevotella spp. can degrade diverse plant polysaccharides, most of which are the fermentable fibers and native cellulose (Chassard et al., 2010). When the dietary protein level was reduced by 4%, the amount of N-containing substances that enter the hindgut sharply decreased; in this way, the N metabolism-associated bacteria will compete for the limited N sources. Proteobacteria and Bacteroidetes are the 2 major N-metabolizing microbial communities (Xie et al., 2013). All ammonia-oxidizing bacteria belong to Proteobacteria (Head et al., 1993, Teske et al., 1994). Therefore, more nitrogenous compounds would be utilized by Proteobacteria in the group with 4% reduction in the dietary protein. Further analysis at genus level also discovered that, a 4% reduction in the dietary protein level evidently enhanced the abundances of Succinivibrio and Lachnospiraceae XPB1014 group. One possible reason is that, the high contents of corn and wheat bran in low protein diet boost the abundances of these 2 bacteria. Succinivibrio is an acetate and succinate producing bacterium which can degrade starch and hemicelluloses (O'Herrin and Kenealy, 1993), while Lachnospiraceae is a kind of soluble polysaccharide-degrading bacterium (Belcheva et al., 2014). Interestingly, when the dietary protein level was reduced by 4%, changes in the abundance of Bacteroidetes were of no statistical significance, which might be related to the fact that the decreases in N metabolism-related bacteria were offset by the increases in plant polysaccharide metabolism-related bacteria.
In this experiment, the colonic microbial diversity was not markedly altered when the dietary CP level was reduced by 4% and different AA were supplemented, however, these treatments changed the abundances of certain bacterial communities. Particularly, in the 3 groups with further Ile and Val + Ile supplementation, as well as balanced 10 EAA, the F:B ratio and the abundance of Proteobacteria increased. Further analysis at genus level also discovered that, the abundance of Anaerovibrio or Lachnospiraceae belonging to Firmicutes was enhanced, while that of Prevotellaceae belonging to Bacteroidetes was reduced in these diets. Prevotella can degrade starches and ferment proteins (Gibson et al., 1989). Results of growth performance and N balance indicated that further Ile and Ile + Val supplementation, as well as balanced 10 EAA, contributed to attaining a balance status of AA, which resulted in a higher N utilization efficiency. A lower fermentation degree of nitrogenous compounds resulted in decreased abundance of Prevotellaceae, whereas a higher fermentation degree of carbohydrates led to increased abundance of Lachnospiraceae. Our experiment also discovered that the lipid metabolism-related bacterium Rikenellaceae RC9 gut group in the Lys, Met, Thr and Trp supplementation group was significantly higher than that in other EAA supplementation groups, which might be related to the fact that the poorly balanced status of AA in these diets not only affected N utilization, but also reduced lipid utilization, resulting in more energy feedstuffs entering the hindgut to be utilized by the Rikenellaceae RC9 gut group (Geurts et al., 2011). Interestingly, we also found that both protein levels and EAA supplementation patterns affected the abundance of Lactobacillus, which suggested that Lactobacillus was sensitive to dietary changes.
Different bacterial community structures are associated with different functional genes and nutrient metabolism networks. Based on the Tax4FUN analysis results, the N metabolism-related genes in gut microbiota were influenced by different dietary CP levels and different EAA supplementation patterns. It was also discovered through comprehensive analysis on the results of N balance and Tax4FUN prediction that, a 2% to 4% reduction in the dietary CP level gradually decreased the N content entering the hindgut and N utilization by microbiota. At the same time, the decomposition of certain AA gradually decreased, and N fixation gradually increased by microbiota. When the dietary CP level was reduced by 4%, those genes related to microbial N metabolism were affected by the different EAA supplementation patterns, especially for the further supplementation of Val + Ile and the balanced 10 EAA. After combining the results of growth performance, N balance and Tax4FUN analysis, it could be concluded that diets with Val + Ile supplementation and balanced 10 EAA had favorable balance state of AA, less N entered the hindgut, and N utilization by microbiota was improved.
In this study, other microbial functions were also detected by Tax4FUN analysis, and the enrichment of “N metabolism” and “energy metabolism” pathways in the low dietary CP diets was notably altered. Decreasing the dietary CP levels not only affected N metabolism, but also impacted the functions of protein-containing transporters and metabolic enzymes. Energy is consumed in N metabolism, as a result, changes in N metabolism also affect energy metabolism, such as lipid metabolism and the insulin signaling pathway. Additionally, it was found in this study that a 4% reduction in the dietary CP level notably down-regulated the pathways related to genetic information processing, which might be associated with polyamines in the intestinal lumen. It has been reported that reducing the amount of N-containing compounds entering the large intestine significantly reduces the concentration of cadaverine and putrescine in the cecal and colon in weaned piglets (Htoo et al., 2007). Polyamines, the positively charged compounds, are essential for the proliferation, differentiation and functions of bacteria and enterocytes (Davila et al., 2013). Overall, a 4% reduction in the dietary CP level had a greater effect on microbial function than that of a 2% reduction. When the dietary CP level was reduced by 4%, different EAA supplementation patterns had no obvious effect on the metabolism of protein and energy, which only mainly affected the metabolism of some small substances, like AA and vitamins.
5. Conclusions
Reducing the dietary CP levels by 2% to 4% with balancing 10 EAA, or only further supplementing Ile, or Ile + Val to a low CP diet (−4%), enhanced the N utilization efficiency in pigs to various degrees, reduced the entering amount of N-containing substances in the hindgut, resulted in the alterations in the abundances of bacteria associated with N metabolism and polysaccharide degradation. Meanwhile, dietary protein levels had greater effect on the microbial N utilization efficiency as well as other metabolic functions in microorganisms than that of AA addition pattern. Furthermore, reduction of the dietary protein level by 4% had greater effect on the alteration in the composition and functions of colonic microbiota than that of 2% reduction in dietary CP level.
Conflict of interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This work was supported by the Science and Technology Support Program of Sichuan Province (2015NZ0042, 2016NZ006) and the National Key R&D Program of China (2018YFD0500605). Thanks for the support of animal experiment base of animal nutrition institute of sichuan agricultural university.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2020.02.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Association of Official Analytical Chemists (AOAC) 17th ed. vol. 17. AOAC; Arlington, VA, USA: 2003. (Official methods of analysis). [Google Scholar]
- Belcheva A., Irrazabal T., J.Robertson S.J., Streutker C., Maughan H., Rubino S J., Moriyama E.H., Copeland J K., Surendra A., Kumar S., Green B., Geddes K., Pezo R C., Navarre W.W., Milosevic M F., Wilson B., Girardin S.E., Wolever T M.S., Edelmann W., Guttman D S., Philpott D.J., Martín A. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- Blachier F., Mariotti F., Huneau J.F., Tomé D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids. 2007;33:547–562. doi: 10.1007/s00726-006-0477-9. [DOI] [PubMed] [Google Scholar]
- Chassard C., Delmas E., Robert Céline, Bernalier-Donadille A. The cellulose-degrading microbial community of the human gut varies according to the presence or absence of methanogens. FEMS Microbiol Ecol. 2010;74:205–213. doi: 10.1111/j.1574-6941.2010.00941.x. [DOI] [PubMed] [Google Scholar]
- Chung K.T., Fulk G.E., Silverman S.J. Dietary effects on the composition of fecal flora of rats. Appl Environ Microbiol. 1977;33:654–659. doi: 10.1128/aem.33.3.654-659.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z.L., Li X.L., Xi P.B., Zhang J., Wu G., Zhu W.Y. L-Glutamine regulates amino acid utilization by intestinal bacteria. Amino Acids. 2013;45:501–512. doi: 10.1007/s00726-012-1264-4. [DOI] [PubMed] [Google Scholar]
- Davila A.M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.H., Sanz Y., Tomé D. Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host. Pharmacol Res. 2013;68:95–107. doi: 10.1016/j.phrs.2012.11.005. [DOI] [PubMed] [Google Scholar]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J.B., Massart S., Collini S., Pieraccini G., Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourmad J.Y., Henry Y., Bourdon D., Quiniou N., Guillou D. Eaap Publication; 1993. Effect of growth potential and dietary protein input on growth performance, carcass characteristics and nitrogen output in growing-finishing pigs. [Google Scholar]
- Fan P., Liu P., Song P., Chen X., Ma X. Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci Rep. 2017;7:43412. doi: 10.1038/srep43412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts L., Lazarevic V., Derrien M., Everard A., Van Roye M., Knauf C., Valet P., Girard M., Muccioli G.G., Francois P., M De Vos W., Schrenzel J., Delzenne N., Cani P.D. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front Microbiol. 2011;2:149. doi: 10.3389/fmicb.2011.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson A.J., Sladen E.G., Dawson, Dawson M.A. Protein absorption and ammonia production: the effects of dietary protein and removal of the colon. Br J Nutr. 1976;35(1):61–65. doi: 10.1079/bjn19760009. [DOI] [PubMed] [Google Scholar]
- Gibson S.A.W., Mcfarlan C., Hay S., Macfarlane G.T. Significance of microflora in proteolysis in the colon. Appl Environ Microbiol. 1989;55:679–683. doi: 10.1128/aem.55.3.679-683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Ma X., Yin J. The effects of lipoic acid on soybean beta-conglycinin-induced anaphylactic reactions in a rat model. Arch Anim Nutr. 2010;64(3):254–264. doi: 10.1080/17450391003625003. [DOI] [PubMed] [Google Scholar]
- Harper A.E., Benevenga N.J., Wohlhueter R.M. Effects of ingestion of disproportionate amounts of amino acids. Physiol Rev. 1970;50:428–558. doi: 10.1152/physrev.1970.50.3.428. [DOI] [PubMed] [Google Scholar]
- Head I.M., Hiorns W.D., Embley T.M., Mccarthy A.J., Saunders J.R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- Htoo J.K., Araiza B.A., Sauer W.C., Rademacher M., Zhang Y., Cervantes M. Effect of dietary protein content on ileal amino acid digestibility, growth performance, and formation of microbial metabolites in ileal and cecal digesta of early-weaned pigs. J Anim Sci. 2007;85(12):3303–3312. doi: 10.2527/jas.2007-0105. [DOI] [PubMed] [Google Scholar]
- Jin C.F., Kim J.H., Han I.K., Bae S.H. Effects of supplemental synthetic amino acids to the low protein diets on the performance of growing pigs. Asian-Australas J Anim Sci. 1998;11:1–7. [Google Scholar]
- Kerr B.J. Proceedings of the 9th international symposium on digestive physiology in pigs, Banff, Alberta, Canadap, 5 May 2003. 2003. Dietary manipulation to reduce environmental impact; pp. 139–158. [Google Scholar]
- Leek A.B.G., Hayes E.T., Curran T.P., Callan J.J., Fearnley S., Dodd V.A., O'Doherty J.V. The influence of manure composition on emissions of odour and ammonia from finishing pigs fed different concentrations of dietary crude protein. Bioresour Technol. 2007;98:3431–3439. doi: 10.1016/j.biortech.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Liu X., Blouin J.M., Santacruz A., Lan A., Andriamihaja M., Wilkanowicz S., Benetti P.H., Tomé D., Sanz Y., Blachier F., Davila A.M. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: the increased luminal bulk connection. Am J Physiol Gastrointest Liver Physiol. 2015;307:G459. doi: 10.1152/ajpgi.00400.2013. [DOI] [PubMed] [Google Scholar]
- Macfarlane G.T., Cummings J.H., Allison C. Protein degradation by human intestinal bacteria. J Gen Microbiol. 1986;132:1647–1656. doi: 10.1099/00221287-132-6-1647. [DOI] [PubMed] [Google Scholar]
- National Research Council (NRC) 11th ed. National Academy Press; Washington, DC, USA: 2012. Nutrient requirements of swine. [Google Scholar]
- O'Herrin S.M., Kenealy W.R. Glucose and carbon dioxide metabolism by Succinivibrio dextrinosolvens. Appl Environ Microbiol. 1993;59:748. doi: 10.1128/aem.59.3.748-755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opapeju F.O., Krause D.O., Payne R.L., Rademacher M., Nyachoti C.M. Effect of dietary protein level on growth performance, indicators of enteric health, and gastrointestinal microbial ecology of weaned pigs induced with postweaning colibacillosis. J Anim Sci. 2009;87:2635–2643. doi: 10.2527/jas.2008-1310. [DOI] [PubMed] [Google Scholar]
- Phillips S.F., Pemberton J.H., Shorter R.G., Talbot I.C. The large intestine: physiology, pathophysiology and disease. Histopathology. 1991;21 [XVIII-XVIII] [Google Scholar]
- Qi H., Xiang Z., Han G., Yu B., Huang Z., Chen D. Effects of different dietary protein sources on cecal microflora in rats. Afr J Biotechnol. 2011;10:3704–3708. [Google Scholar]
- Rajilićstojanović M., De Vos W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2015;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A., Alm E., Regan J.M., Toze S., Rittmann B.E., Stahl D.A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrallardona D., Conde M.R., Badiola I., Polo J., Brufau J. Effect of fishmeal replacement with spray-dried animal plasma and colistin on intestinal structure, intestinal microbiology, and performance of weanling pigs challenged with Escherichia coli K99. J Anim Sci. 2003;81:1220–1226. doi: 10.2527/2003.8151220x. [DOI] [PubMed] [Google Scholar]
- Walker A.W., Ince J., Duncan S.H., Webster L.M., Holtrop G., Ze X., Brown D., Stares M.D., Scott P., Bergerat A., Louis P., McIntosh F., Johnstone A.M., Lobley G.E., Parkhill J., Flint H.J. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing B.P., Van Kessel A.G.V. Host pathways for recognition: establishing gastrointestinal microbiota as relevant in animal health and nutrition. Livest Sci. 2010;133:82–91. [Google Scholar]
- Xie B., Lv Z., Hu C., Yang X., Li X. Nitrogen removal through different pathways in an aged refuse bioreactor treating mature landfill leachate. Appl Microbiol Biotechnol. 2013;97:9225–9234. doi: 10.1007/s00253-012-4623-x. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., DominguezBello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R N., Anokhin A P., Heath A C., Warner B., Reeder J., Kuczynski J., Caporaso G., Lozupone C A., Lauber C., Clemente J.C., Knights D., Knight R., Gordon J I. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Fang L., Sun Y., Su Y., Zhu W. Effects of the dietary protein level on the microbial composition and metabolomic profile in the hindgut of the pig. Anaerobe. 2016;38:61–69. doi: 10.1016/j.anaerobe.2015.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.