Abstract
Disease-specific therapeutic options for critically ill neurological patients are limited. The identification of new preventive, therapeutic, and rehabilitation strategies is of the utmost importance in the field of neurocritical care research. Population genetics offers powerful tools to identify and prioritize biological pathways to be targeted by novel interventions. New treatments with supportive genetic evidence have twice the chances of obtaining final FDA approval compared to those without this support. Large collaborations, public access to data, reproducible science, and innovative analytical methods have exponentially increased the pace of discoveries related to neurocritical care genetics.
Electronic supplementary material
The online version of this article (10.1007/s13311-020-00837-2) contains supplementary material, which is available to authorized users.
Key Words: Neurocritical care, genetics, brain injury, mortality, recovery
Introduction
Patients with neurocritical illness often have severe brain and/or spine injury and an elevated risk of death and severe disability. Therapeutic options are limited for many of the diseases frequently encountered in the Neurosciences Intensive Care Unit (NeuroICU), making the discovery of novel targets for prevention, acute treatment, and rehabilitation a high priority in the field for neurocritical care research. Unfortunately, several treatments aimed to ameliorate acute brain injury in neurocritical care diseases failed to show benefit in clinical trials, despite promising preliminary evidence obtained in observational and animal studies [1–5]. Population genetics offers powerful tools to identify new mechanisms involved in causing the severe neurologic injury that we encounter in the NeuroICU. In this review, we describe the properties that make mutation-disease associations a unique tool for biological discovery, present important concepts needed to appropriately appraise genetic studies, and review concrete examples of the application of population genetics to the field of Neurocritical Care.
Using Genetic Information to Identify New Biological Targets and Pathophysiological Mechanisms
Population genetics offers powerful tools for causal inference. Inherited genetic variation provides numerous “experiments of nature” that connect specific mutations—and underlying genes and cellular pathways—to human disease. Because mutations are randomly distributed during meiosis, mutation-disease associations are immune to confounding by postnatal exposures. In this setting, mutations strongly associated with the diseases encountered in the NeuroICU point to genes, proteins, and cellular pathways involved in these conditions. This notion extends beyond the primary diagnosis that brings the patient to the hospital and applies to mechanisms of secondary brain injury that contribute to the morbidity and mortality regardless of the initial insult. Leveraging genetic analyses to identify new targets and mechanisms constitutes a promising strategy for NeuroICU-related conditions, as common and rare genetic variation explains an important proportion of the variance of several of them (Table 1) [6–10]. For the same reasons, genetic information is likely to become the pillar of precision medicine approaches aimed to identify patients that belong to specific groups of interest, like those that may respond to a specific drug or that are likely to have a poor outcome. Because current genotyping technologies capture information from the entire human genome, it is now possible to agnostically interrogate all possible cellular process and pathways. This is a crucial strength of this approach, as it does not require an a priori hypothesis.
Table 1.
Heritability estimates for diseases commonly managed in the neurocritical care unit
| References | Phenotype | Sample size | Heritability estimate |
|---|---|---|---|
| Bevan et al. [6] | Ischemic stroke |
3752 cases 5972 controls |
38% |
| Devan et al. [7] | Intracerebral hemorrhage |
791 cases 876 controls |
44% |
| Devan et al. [7] | Intracerebral hemorrhage mortality |
791 cases 876 controls |
41% |
| Korja et al. [8] | Subarachnoid hemorrhage |
504 cases 79,644 twin pairs |
41% |
| Speed et al. [9] | Epilepsy |
1258 case 5129 controls |
32% |
| Blum et al. [10] | Guillain-Barré syndrome |
191 cases 639 controls |
15% |
Genetic Analyses Improve the Yield of the Translational Research Cycle
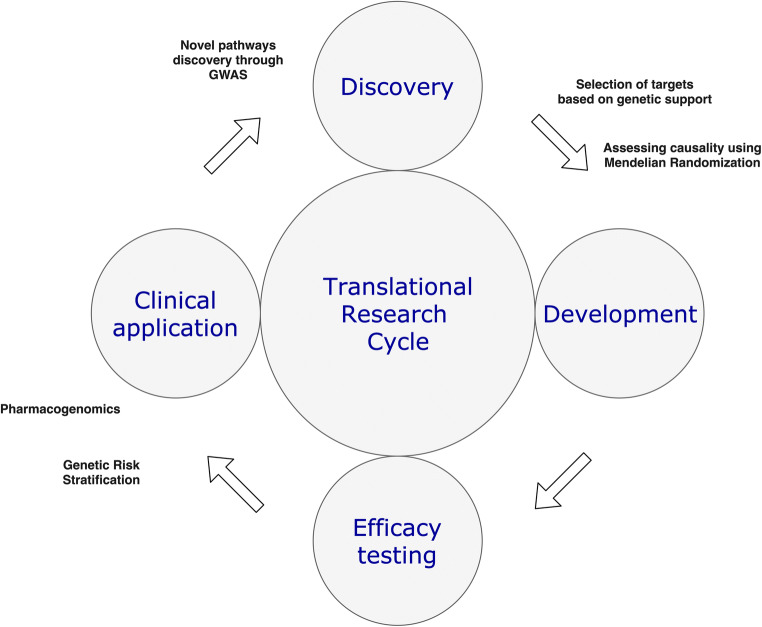
It is challenging to complete the full cycle of clinical research (Fig. 1), from identifying a pathophysiological mechanism to demonstrating the feasibility, safety, and clinical efficacy in humans of using an agent that acts on this pathway. An important proportion of interventions with promising results in preclinical stages fail to show benefit when evaluated in costly and time-consuming clinical studies in humans, a notion called the “Translational Valley of death.” Because of its agnostic approach when identifying novel biological mechanisms in human disease, genetic analyses can significantly reduce the proportion of new treatments that fail in clinical trials. An important study compared clinical interventions with and without genetic support. The investigators utilized the Informa Pharmaprojects database to acquire data on interventions, targets, and indications; and GWASdb and OMIM, two open access resources that collect information on genetic risk factors for complex and Mendelian conditions, respectively, to obtain data on genetic support. After evaluating 19,085 target-indication pairs and 13,855 gene-trait combinations, the authors estimated that selecting genetically supported targets doubles the success rate of the research cycle [11]. Importantly, the continuous increase in number of genetic studies coupled with new methods in statistical genetics, like Mendelian Randomization (see below), will likely increase these estimates.
Fig. 1.
The cycle of translational research. Genetics can contribute to many steps in the cycle. First, genome-wide association studies can identify novel loci and lead to biological pathways implicated in diseases. Secondly, selecting targets based on them having genetic support, and assessing causality through Mendelian randomization can improve the rate of success in the development and efficacy testing of new interventions. Lastly, genetic risk stratification can identify patients that need to be screened or treated, and pharmacogenomics can guide treatment in an individualized manner
Primer on Population Genetics
The field of population genetics studies the distribution of genetic variation across different species, including humans. One important goal is to estimate the proportion of a given trait the can be explained by genetic variation, a concept known as heritability (Table 1). These traits include physiological characteristics, like height and eye color; physiological traits that can become deleterious in the extremes, like blood pressure, blood glucose, and cholesterol levels; and discrete pathological entities. For traits with high genetic contribution, one important additional goal of population genetics is to pinpoint the specific genetic variants that lead to an elevated heritability. The concepts below will facilitate the discussion that follows [12, 13].
Penetrance
Penetrance refers to the proportion of individuals with a specific mutation showing manifestations of the studied condition.
Mendelian Genetic Contribution
A Mendelian pattern of inheritance refers to rare mutations with high penetrance. Diseases with mendelian inheritance are also called monogenic or familial conditions and can be classified between autosomal dominant (one mutated allele is enough to cause the disease) or recessive (both alleles need to have the mutation for the disease to be expressed).
Non-Mendelian Genetic Contribution
A non-Mendelian pattern of inheritance refers to mutations with low penetrance that have a small but statistically significant effect on the risk of expressing disease. Conditions with non-Mendelian patterns of inheritance are usually called complex diseases or traits, meaning that we cannot predict if one individual will have the disease by looking at just one mutation. However, recent advances have made it possible to stratify risk by pooling the contributions of multiple mutations in genetic risk scores.
Heritability
Heritability is a measure of the amount of variation in a trait explained by genetics [14, 15]. It ranges from 0 (genetic variation does not influence the trait) to 1 (genetic variation completely explains the trait). For example, height is a highly heritable trait, with around 80% of the variance explained by genetics. An elevated heritability indicates that the corresponding trait likely has numerous risk factors to be discovered and utilized for treatment development. Heritability can nowadays be estimated using information provided by genome-wide association studies (GWAS). For some traits, the number of identified genetic risk factors does not account for the estimated heritability, leading to “missing” heritability. Although some evidence indicates that this missing heritability is due to shared environment or nonadditive genetic variation, a definitive answer is still lacking.
Minor Allele Frequency
The most commonly studied mutations are single nucleotide polymorphisms (SNPs). SNPs are genetic variants involving only one base pair position of the genome. Most SNPs have only two possible alleles: the major allele and the minor allele, which are carried by the majority and the minority of the population, respectively. This distribution can be expressed by the frequency of the minor allele (MAF), which divides variants into common (MAF > 5%), low-frequency (MAF > 0.5–5%), rare (MAF < 0.5%), and private (minor allele presents only in single families).
Linkage Disequilibrium
Genetic variants usually segregate with other variants located closely in distance. At the population level, this leads to correlation between variants, a phenomenon termed linkage disequilibrium (LD). Conversely, when variants are independent of one another, they are in linkage equilibrium. LD can be measured through a coefficient called r2 which varies from 0 (uncorrelated) to 1 (completely correlated). Researchers use different r2 thresholds to filter out variants, using only uncorrelated SNPs, for example, when developing polygenic risk scores.
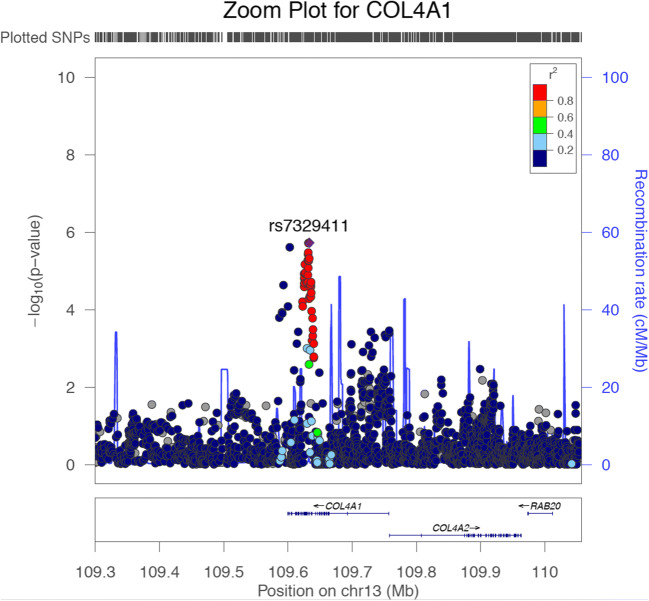
Genomic Locus
A group of highly correlated genetic variants (i.e., in elevated LD) represent an LD block (Fig. 2). In GWAS, these are referred to as susceptibility loci. Association studies can expose these loci, but challenges remain when trying to identify the specific variant causally related to disease.
Fig. 2.
Zoom plot showing a genetic locus
Study Design and Analytical Methods in Population Genetics
Candidate Gene Studies
These studies were the main approach for association testing between genes and diseases before the development of high-throughput genotyping technologies [16–19]. This hypothesis-driven approach builds upon previous data supporting specific biological pathways and allows researchers to evaluate a discrete list of presumed causal SNPs. Nowadays, candidate gene studies are still useful when studying low-frequency mutations or small samples, and they are also a practical option for replication of GWAS results.
Genome-Wide Association Studies
In contrast to candidate gene analyses, genome-wide association studies (GWAS) are hypothesis-free studies that agnostically interrogate millions of SNPs from across the genome for association with a particular disease [20]. Considering the large number of comparisons made, it is standard to use an adjusted significance threshold of p < 5 × 10e−8 to account for the approximately 1 million independent loci found across the human genome. Importantly, the number of genome-wide significant loci identified for many complex traits grows exponentially with sample size.
Sequencing Studies [21]
Next-generation sequencing has revolutionized the amount of data we can produce. With whole-genome sequencing, an important proportion of the 3 billion base pair positions in the human genome can be genotyped and become available for analysis. This technology comes with a new set of statistical and computational challenges, driven by the need to manipulate, curate, and analyze massive amounts of data, and the increasingly small signal-to-noise ratio [22].
Polygenic Risk Scoring Studies [23]
Because the first GWAS was published in 2007 [24], the number of well-described mutation-disease associations has increased dramatically. This growing number of susceptibility loci, including multiple mutations associated with a single trait, allows the creation of scores that summarize the load of mutations related to a specific trait carried by a person. In a simplistic approach, the polygenic risk score can be calculated as the sum of risk alleles for a given person. This score is often a more powerful predictor of disease occurrence than an individual SNP. A more sophisticated approach leverages the effect size of each SNP on the trait of interest as a weight, improving the performance of the scores. Polygenic risk scores can be used for both causal inference and prediction. The latter option is currently being tested in clinical settings [25] and will likely lead to numerous applications in coming years.
Mendelian Randomization Studies [26]
This analytical method constitutes an especial case of instrumental variable analysis, an approach that involves the utilization of a variable that is strongly associated with the exposure and not with the outcome except through the exposure. When the instrument is appropriately chosen and the relevant assumptions are met, this approach yields association results with strong causal implications. In Mendelian Randomization analyses, the instrument is either one mutation or a collection of mutations known to influence the exposure of interest. Because the distribution of alleles occurs randomly before birth, they are immune to confounding by environmental factors encountered throughout life. For example, when investigating causality between high blood pressure and cardiovascular risk, one option would be to randomize patients to blood pressure lowering drugs versus placebo. Another option, leveraging Mendelian Randomization analyses, would be to utilize alleles known to strongly associate with blood pressure reduction. A significant association between these alleles and reduced cardiovascular risk would support a causal link between blood pressure levels and cardiovascular risk.
Accelerated Pace of Discovery in Population Genetics
In the last 10 years, the fields of human and Neurocritical Care genetics research have grown rapidly. This expansion has been the result of a combination of circumstances, including changes in culture, now favoring highly collaborative research environments and public access to data; the creation of large international consortia designed to share ideas and data and pursue harmonization of genetic, clinical, and neuroimaging data with the ultimate goal of achieving massive sample sizes; and the development of novel analytical methods designed to efficiently deal with large-scale data (colloquially referred to as Big Data) [27, 28].
Public access to genetic data is having a particularly profound influence on Neurocritical Care genetics. Genetic studies are complicated, expensive, and, even after completion, require significant amount of resources for quality control, harmonization, and analysis. Public access to genetic data of diseases related to Neurocritical Care is maximizing the return generated by funds destined to this type of research although in parallel increasing the amount of downstream analytical resources utilized to derive new biological insights from a given dataset. Open access resources can be classified into repositories, that aim to store already generated genetic data; large population studies with attached biobanks; disease-specific studies with built-in genomic data intended for public access since conception; and scientific platforms, that combine access to data with a closed computational environment that allow investigators to access and work with data without directly downloading information to their local institutional servers.
Table 2 summarizes open access data resources that are already having a significant impact in the field of Neurocritical Care genetics. The NIH-sponsored Database of Genotypes and Phenotypes (dbGaP) [33] and the Cerebrovascular Disease Knowledge Portal (CDKP) [37], in the USA, and the European Genome-phenome Archive (EGA) [34], in Europe, represent three large and successful examples of open access repositories that seek to make genetic data publicly available. The UK Biobank [29] and the China Kadoorie Biobank [32] constitute very successful examples of large population-based prospective studies providing open access to all collected data, enrolling more than 500,000 individuals each, and gathering a wide range of data types, including genetic, laboratory, electronic health record, and physical measurements. In terms of disease-specific open access studies, the Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH) study [38] has proven an invaluable resource in Neurocritical Care and Stroke research, leading to numerous subanalyses related to ICH outcome and risk [39–44]. The ERICH study has enrolled 6000 extremely well-phenotyped ICH cases and controls, including equal numbers of whites, blacks, and Hispanics. All ICH cases are undergoing whole-genome sequencing. Finally, the American Heart Association Precision Medicine Platform constitutes an example of a scientific platform that offers public access to numerous data sets containing genetic and nongenetic data using a closed computational environment that utilizes Jupyter Notebooks [45] to script, and Amazon Web Services to provide computational support to investigators. Along these lines, the All of Us Research Program [31], scheduled to become operational in the winter of 2020, will provide large amounts of data by enrolling at least 1 million people in the USA from diverse race/ethnicities.
Table 2.
Open access data resources
| Ref. | Name | Website | Resource type | Sample size | Access type |
|---|---|---|---|---|---|
| [29] | UK Biobank | https://www.ukbiobank.ac.uk/ | Observational study | 500,000 | Open |
| [30] | Million Veteran Program | https://www.research.va.gov/mvp/ | Observational study | 1 million | Restricted |
| [31] | All of Us | https://allofus.nih.gov/ | Observational study | 1 million | Open |
| [32] | China Kadoorie Biobank | https://www.ckbiobank.org/ | Observational study | 500,000 | Restricted |
| [33] | dbGAP | https://www.ncbi.nlm.nih.gov/gap/ | Repository | – | Open |
| [34] | EGA | https://ega-archive.org/ | Repository | – | Open |
| [35] | MIMIC-III | https://mimic.physionet.org/ | Database | 61,532 | Open |
| [36] | eICU-CRD | https://eicu-crd.mit.edu/ | Database | 200,000 | Open |
UK = United Kingdom, dbGAP = database of genotypes and phenotypes, EGA = European Genome-phenome Archive, MIMIC-III = Medical Information Mart for Intensive Care-III, eICU-CRD = eICU Collaborative Research Database
In addition to publicly available data, the field of population and medical genetics has significantly benefited from online, open access resources that provide valuable tools for interpretation and analysis. Table 3 summarizes some of the most important of these resources. The UCSC Genome Browser and the Ensembl Genome Browser provide comprehensive genetic information using reference panels. Resources like the Cerebrovascular Disease Knowledge Portal [37], GWAS Catalog [46], and GWAS atlas [47] publicly share summary statistics from published studies. Furthermore, the recently launched Polygenic Score Catalog [48] offers pre-specified and already tested genetic risk scores for several traits, greatly contributing to facilitate and standardize the implementation of PRS analyses. Other useful tools are the Functional Mapping and Annotation of Genome-Wide Association Studies site (FUMA) [49], used to annotate, prioritize, visualize, and interpret GWAS results, LocusZoom [50], an online tool and library to visualize GWAS results at a locus level (Fig. 1).
Table 3.
Online resources
| Ref. | Name | Website | Description |
|---|---|---|---|
| [37] | Cerebrovascular Disease Knowledge Portal | http://www.cerebrovascularportal.org/ | A platform that allows for searching, visualizing and analyzing variations related to cerebrovascular disease. |
| [46] | GWAS Catalog | https://www.ebi.ac.uk/gwas/ | Catalog of GWAS results |
| [47] | GWAS atlas | https://atlas.ctglab.nl/ | Catalog of GWAS results |
| [48] | PGS Catalog | http://www.pgscatalog.org/ | Catalog of Polygenic Risk Scores |
| [49] | FUMA | https://fuma.ctglab.nl/ | Functional Mapping and Annotation of GWAS results |
| [50] | Locus Zoom | http://locuszoom.org/ | Visualization of GWAS results at the locus level |
GWAS = genome-wide association study, PGS = polygenic score, FUMA = functional mapping and annotation
Combining Genetic and Nongenetic Data Sources to Accelerate Discovery
Genetics is one of many contributors to Big Data in clinical research. Other sources include neuroimaging data, electronic health records (natural language data), and biomarkers (e.g., blood pressure monitoring, laboratory data). Analytical methods designed to integrate different types of data constitute a powerful tool in neurocritical care research [51, 52]. Currently, nongenetic data is only utilized to provide the dependent variable for genetic analysis. For example, neuroimaging data on white matter disease is used to conduct GWAS analyses to understand the genetic architecture of cerebral small vessel disease [53]. However, this simplistic utilization of nongenetic data represents only the tip of the iceberg. The integration of different data types via powerful deep learning and artificial intelligence tools has increased the statistical power of genetic analysis, although at the same time broadening the scope of biological questions to be tackled. Along these lines, there have been recent efforts to harmonize and publicly release nongenetic data as well. For example, the Medical Information Mart for Intensive Care-III (MIMIC-III) database is an extensive collection of data of patients admitted to Intensive Care Units at the Beth Israel Deaconess Medical Center [35]. This large dataset has led too several studies using machine learning to develop decision support systems. Inspired by the MIMIC-III project, a joint effort from several centers from across the world led to the eICU Collaborative Research Database [36], another large repository of critical care data available to interested investigators.
Applications to Acute Brain Injury and Recovery in Neurocritical Care
We present a summary of These application to NeuroICU is presented in Table 4
Table 4.
Summary of applications to Neurocritical Care
| Ref. | Year | Disease | Phenotype | Findings |
|---|---|---|---|---|
| [54] | 2018 | TBI | ICP; radiographic edema; functional outcome | rs7105832 and rs2237982 were associated with lower average ICP and decreased radiographic edema. rs2237982 was also associated with favorable 3-month GOS. |
| [55] | 2015 | Ischemic stroke | Infarct size, neurological scores | S1pr2-/- mice had significantly reduced infarct size and improved neurological scores. |
| [56] | 2011 | ICH | Hematoma volume; functional outcome | Each additional ε2 allele associated with 18% increase in the size of hematoma, and 50% increase in the risk of poor outcome. |
| [57] | 2018 | ICH | Hematoma volume; functional outcome | 17p12 is a susceptibility risk locus for hematoma volume, and each additional G allele in rs11655160 is associated with 94% increase in the risk of poor outcome at 3 months. |
| [58] | 2013 | ICH | Hematoma volume; functional outcome | Each additional SD of a polygenic risk score comprising 42 SNPs associated with blood pressure is associated with 29% larger hematoma volume and 71% increased risk of poor functional outcome. |
| [59] | 2019 | Stroke; TBI | Motor and cognitive recovery |
Improvement in motor and cognitive recovery in animal models of stroke and TBI. Patients from an ischemic stroke cohort with naturally occurring loss-of-function mutations in CCR5 had better motor and cognitive recovery months after the event. |
| [60] | 2019 | Ischemic stroke | Functional outcome | Each additional G allele of rs76221407 was associated with a 0.4 increase in mRS. |
| [61] | 2019 | Ischemic stroke | Functional outcome | An intronic variant in the LOC105372028 gene, functionally associated with PPP1R21, a gene implicated in brain plasticity, is associated with functional outcome after ischemic stroke. Several others suggestive associations were also found. |
ICH = intracerebral hemorrhage, TBI = traumatic brain injury, ICP = intracranial pressure, SD = standard deviation, SNPs = single nucleotide polymorphisms, GOS = Glasgow Outcome Scale, mRS = Modified Rankin Scale
Sur1-Trpm4 Influence on Cerebral Edema
The nonselective cation channel formed by the proteins sulfonylurea receptor 1 (SUR1) and transient receptor potential melastatin 4 (TRPM4) constitutes a promising drug target for acute treatment of several conditions encountered in the NeuroICU. The combination of animal, genetic, and clinical trial evidence supporting a role of this pathway in acute brain injury makes SUR1-TRPM4 a prototypic example of new therapy development in our field. Preclinical studies using animal models showed that this channel is not expressed in the normal brain, but it is upregulated in the setting of acute brain injury, when SUR1 associates with TRMP4 causing depolarization and water influx, leading to tissue swelling and cerebral edema [62]. Of note, it is possible to pharmacologically modulate these pathways with sulfonylureas, a commonly used group of anti-diabetic medications.
Importantly for the purposes of this review, genetic studies in humans confirmed a possible role of SUR1-TRPM4 in acute brain injury [54]. A study that evaluated 15 tag SNPs in ABCC8, the gene encoding SUR1, found that 4 of these SNPs were associated with cerebral edema and outcome in patients with severe traumatic brain injury. Specifically, rs7105832 was independently associated with cerebral edema, rs11024286 and rs4148622 with outcome, and rs2237982 with both cerebral edema and outcome. Because these SNPs are intronic, the most likely mechanistic link is via transcriptional function.
The combination of animal and genetic evidence supporting a role of the SUR1-TRPM4 channel in acute brain injury, coupled with the possibility of intervening on this target via existing medications, led to the repurposing of sulfonylureas to treat acute brain injury in several Neurocritical Care diseases. The phase II Glyburide Advantage in Malignant Edema and Stroke (GAMES-RP) randomized clinical trial evaluated the safety and efficacy of intravenous glyburide in a critically ill, acute ischemic stroke population at high risk for brain swelling. The study was stopped early due to funding reasons, but showed that intravenous glyburide was safe in this patient population [63]. Although the trial’s clinical endpoints were negative, subsequent secondary analyses revealed an association between glyburide treatment and reduction in both deaths attributable to malignant edema [64] and water accumulation and mass effect in these patients [65]. Expectations are high for the currently recruiting phase III Cerebral Edema Following Large Hemispheric Infarction (CHARM) study (ClinicalTrials.gov identifier: NCT02864953), with results expected in 2021. This phase 3 randomized clinical trial aims to enroll 680 adult patients with large hemispheric brain infarcts and assign them either to receive intravenous glyburide or placebo, and to evaluate its impact in long-term functional outcome.
Intravenous glyburide is also being evaluated for acute traumatic brain injury. A phase 2 randomized, quadruple-blinded clinical trial is underway (ClinicalTrials.gov NCT03954041), aiming to enroll 160 participants with a traumatic brain contusion. The study’s primary outcome will be the expansion of the qualifying contusion 96 h after starting treatment. Although underpowered to fully evaluate clinical outcome, the trial’s secondary endpoints include 90-day post trauma improvements in Extended Glasgow Outcome Scale and Modified Rankin Scale.
Sphingosine-1-Phosphate Modulates Vascular Integrity
Existing therapies for acute ischemic stroke aim to recanalize obstructed vessels using thrombolytics or mechanical thrombectomy. One limitation of these strategies is the risk of reperfusion injury, mediated by excessive generation of reactive oxygen species that lead to the activation of matrix metalloproteinases (MMP), vasogenic edema and, ultimately, hemorrhagic transformation. Novel promising strategies have been specifically designed to target this pathway. A recent study in mice evaluated the effect knocking out S1PR2 in a standardized animal model of transient cerebral ischemia. S1PR2 encodes sphingosine-1-phospate receptor 2, a member of the G protein-coupled receptors that participates in cell proliferation, survival, and transcriptional activation. The investigators found that animals lacking this gene had reduced infarct size, reduced volume of cerebral edema, and improved neurological scores [55]. The study also showed that the administration of JTE013, a potent and selective S1P antagonist, produced similar results.
The evidence outlined above led to the evaluation of immunomodulatory drugs originally designed for multiple sclerosis in cerebrovascular disease. Fingolimod is an oral sphingosine-1-phosphate–receptor modulator that has shown neuroprotective effects in animal models [66]. This effect could be mediated, at least in part, by reducing brain infiltration of T-lymphocytes [67]. Considering fingolimod’s adverse effects in the cardiovascular system, researchers have also evaluated the more selective S1PR1 modulator RP101075, a drug with fewer off-target effects and a better safety profile. This selective immunomodulatory strategy also displayed protective effects in experimental models of ICH [68]. These results led to a proof-of-concept study in humans that enrolled 23 subjects with ICH and assigned them to standard treatment plus fingolimod administration or standard treatment alone. This small trial showed that the administration of fingolimod was safe, reduced perihematomal edema and improved neurological deficits [69]. This promising evidence ultimately justified a phase-2 randomized clinical trial currently underway (clinicaltrials.gov identifier: NCT03338998). This study randomizes patients to the administration of siponimod or placebo looking for differences in perihematomal edema at day 14 after the event and is expected to be completed by November 2020.
Genetic Determinants of Intracerebral Hemorrhage Volume and Outcome
The volume of the hematoma in the patients with spontaneous, nontraumatic intracerebral hemorrhage (ICH) is the most powerful predictor of outcome in this condition [70]. Importantly for the purposes of this review, a substantial proportion of the variation in this neuroimaging phenotype of radiological severity can be attributed to genetic variation, as expressed by a heritability estimate of ~ 50%. The epsilon 2 variants within APOE, the gene that codes for Apolipoprotein E, constitute one of the genetic risk factors that mediate this elevated heritability. In a study that included ICH patients of European ancestry, each additional e2 allele was associated with a 5.3 mL, or 18%, increase in hematoma volume, with a concomitant 50% increase in risk of poor outcome [56]. Subsequent studies indicated that the effect of APOE epsilon 2 extends beyond admission hematoma volume to also influence the risk of having a spot sign on CT angiogram and of sustaining hematoma expansion [71, 72]. Although the mediating biological mechanism has not been fully elucidated, it is possible that following the rupture of the culprit vessel, the leaking blood injures the already damaged vessels in the vicinity, causing additional bleeding [73]. This paradigm, proposed by C. Miller Fisher and known as the “cascade hypothesis,” serves as a general explanation for the observation that factors that lead to cerebral small vessel disease are associated with both increased risk and larger hematoma size in ICH.
Another genetic determinant of hematoma volume and outcome in intracerebral hemorrhage is the aggregate load of hypertension-related risk alleles. This genetic load can be modeled via polygenic risk scores (see above), a tool that summarizes the genetic contribution of several genetic variants into a single measurement. In a study of 323 ICH patients, a polygenic risk score built with 42 blood pressure-related genetic variants was associated with hematoma volume and poor clinical outcome. In this study, each additional standard deviation of the hypertension-based polygenic risk score was associated with a 29% increase in baseline hematoma volume in patients with deep hemorrhages and 71% increase in the risk of a poor outcome [58]. As for APOE, the exact pathophysiological mechanism mediating this association has not been identified, although the “cascade hypothesis” has also been put forward to explain the observation.
The GWAS framework is now taking its first steps in acute brain injury and clinical severity of ICH. The first genome-wide scan of hematoma volume has been recently completed. The study reported a susceptibility locus located on a narrow genetic region in the short arm of chromosome 17 [57]. The G allele for rs11655160, the top associated variant within this locus, was associated with a 2.6 mL increase in baseline hematoma volume. In line with the known strong correlation between hematoma size and clinical severity and outcome, this same allele was associated with an 83% change in admission Glasgow Coma Scale and an 94% increased risk of poor functional outcome 3 months after the event. The 17p12 locus co-localizes with numerous copy number variants of different size, some compromising half the chromosome, offering a clear next target to pursue to understand the biology underlying this associations.
Recovery After Ischemic Stroke
Significant progress is also happening for genetic studies evaluating recovery after ischemic stroke. A recent study in mice found that knockout animals for CCR5, the gene encoding the C-C chemokine receptor type 5, had improved recovery after stroke and traumatic brain injury. Similarly, a population-based genetic study of stroke demonstrated that patients with naturally occurring loss-of-function mutations in CCR5 had better motor and cognitive recovery months after the event [59]. Of note, CCR5 antagonist drugs are already approved by the FDA [74] for use in the treatment of HIV infection and clinical trials that test this pathway in acute brain injury are expected to be underway soon.
The recent creation of large, international, multidisciplinary research groups focusing on outcome and recovery after stroke now allows a more systematic evaluation of genetic targets. A GWAS meta-analysis undertaken by the Genetics of Ischaemic Stroke Functional Outcome (GISCOME) network found an intronic variant in LOC105372028, a widely expressed gene in the brain that codes for a long noncoding RNA molecule that regulates the expression of KLRAQ1. The latter encodes a regulatory subunit of protein phosphatase 1 (PP1), a protein with putative regulatory roles in processes such as learning, memory and neuroplasticity [61]. Functional experiments to confirm to delineate a possible therapeutic target for recovery are warranted. Another recent meta-analysis of GWAS focusing on stroke recovery identified several low-frequency variants within PATJ, a gene encoding the PALS1-Associated Tight Junction Protein [60]. The large effect size of these variants, in the order of 40% increase in poor functional outcome, makes of this pathway an appealing target for follow-up functional experiments.
Novel Analytical Tools Based on Machine Learning and Artificial Intelligence
Analytical methods based on machine learning, deep learning and artificial intelligence are rapidly changing all aspects of Big Data analysis. One application of these approaches to population genetics involve fine mapping, the process of learning more about a genetic locus identified via genetic studies. Genetic loci identified by GWAS are often located in noncoding regions of the genome, either intronic or intergenic. As discussed above (see primer on population genetics), each newly discovered locus contains several genetic variants in high linkage disequilibrium. The most important next step after finding a novel genome-wide locus is to pinpoint the causal variant that mediates the observed association. Analytical methods based on artificial intelligence and deep learning are increasingly utilized for these purposes (Table 5). Several models based on these tools have been developed and trained to predict the effect of noncoding variants on the expression of other genes and well as protein and pathways function. One example is DeepSea [75], a tool that aims to prioritize regulatory variants using a deep-learning model trained to predict chromatin effects of sequence alterations.
Table 5.
Resources for artificial intelligence and machine learning
| Type | Name | Website | Description |
|---|---|---|---|
| Educational | FastAI | https://www.fast.ai/ | Free courses about machine learning and deep learning |
| Coursera | https://www.coursera.org/ | Free and paid courses and specializations about machine learning, deep learning, genomics, among others. | |
| Udacity | https://www.udacity.com/ | Free and paid courses and nanodegrees about machine learning and deep learning | |
| Frameworks | Tensorflow (Google) | https://www.tensorflow.org/ | Most popular industry framework, recently released v2.0 |
| Keras | https://keras.io/ | High level framework, works on top of tensorflow | |
| Pytorch (Facebook) | https://pytorch.org/ | Most popular research framework | |
| Fastai | https://docs.fast.ai/ | Library simplifying training neural networks using modern best practices. Works on top of pytorch. | |
| Cloud Infrastructure | Google Colab | https://colab.research.google.com/ | Free access to jupyter notebooks with limited-time GPUs |
| Google Cloud | https://cloud.google.com/ | Paid access to Google Cloud infrastructure ($300 credit) | |
| FloydHub | https://www.floydhub.com/ | Paid GPU access | |
| Gradient | https://gradient.paperspace.com/ | Paid GPU access | |
| Onepanel | https://www.onepanel.io/ | Paid GPU access | |
| Azure | https://azure.microsoft.com/ | Paid access to Microsoft Azure Cloud infrastructure | |
| SageMaker | https://aws.amazon.com/sagemaker/ | Cloud platform for machine learning based on AWS | |
| AWS EC2 | https://aws.amazon.com/ec2/ | Paid access to Amazon Web Services Cloud Platform |
AWS EC2 = Amazon Web Services Elastic Compute Cloud
From Genetic Associations to Biological Mechanisms
The genetic studies described above yielded important associations between specific genetic variants and a number of relevant diseases and traits in the field of Neurocritical Care. After identifying the causal variant responsible for each association, it is crucial to precisely delineate the biological pathway that mediates the observed allele–outcome correlations, a process that requires the combination of translational research strategies like gene editing, cellular models and animal models. The successful completion of this process requires the assembly of multidisciplinary teams that include genetic epidemiologists, statistical geneticists, bench researchers, and clinicians. Increasingly, these collaborative networks are created around broad topics with the goal of evaluating multiple pathways and targets, as opposed to ad hoc collaborations intended to pursue one specific questions. As an example, the NIH/NINDS-sponsored Stroke Preclinical Assessment Network (SPAN) will facilitate the evaluation of up to 6 promising neuroprotective drugs or interventions to be given prior to, or at the time of, reperfusion in experimental models of ischemic stroke.
Crucial Next Steps
The intersection of Neurocritical Care and genetics will only grow in coming years. The successful application of genetic advances in Neurocritical Care depend on a number of factors. The creation of new collaborations to bring together clinicians, investigators, and institutions invested in Neurocritical Care research will be crucial. One important goal for these collaborations will be to integrate genetic data with the massive amounts of other data types produced in the Neuroscience Intensive Care Unit, including clinical information, laboratory results, neuroimages, EEG, vital signs, and multimodality monitoring. Public and fast access to these data will constitute an important catalyzer to accelerate the pace of scientific discovery and increase the overall yield of these endeavors. As these impactful changes happen, it will be important to protect our patients’ privacy [76].
Electronic Supplementary Material
(PDF 507 kb)
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Funding
GJF is supported by the National Institute on Aging (K76AG59992), the National Institute of Neurological Disorders and Stroke (R03NS112859), the American Heart Association (18IDDG34280056), a Yale Pepper Scholar Award (P30AG021342), and the Neurocritical Care Society Research Fellowship.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mayer SA, Brun NC, Begtrup K, Broderick J, Davis S, Diringer MN, et al. Efficacy and safety of recombinant activated factor VII for acute intracerebral hemorrhage. N Engl J Med. 2008;358(20):2127–37. doi: 10.1056/NEJMoa042991. [DOI] [PubMed] [Google Scholar]
- 2.Robertson CS, Hannay HJ, Yamal JM, Gopinath S, Goodman JC, Tilley BC, et al. Effect of erythropoietin and transfusion threshold on neurological recovery after traumatic brain injury: A randomized clinical trial. JAMA - J Am Med Assoc. 2014;312(1):36–47. doi: 10.1001/jama.2014.6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baharoglu MI, Cordonnier C, Salman RAS, de Gans K, Koopman MM, Brand A, et al. Platelet transfusion versus standard care after acute stroke due to spontaneous cerebral haemorrhage associated with antiplatelet therapy (PATCH): a randomised, open-label, phase 3 trial. Lancet [Internet]. 2016;387(10038):2605–13. Available from: 10.1016/S0140-6736(16)30392-0 [DOI] [PubMed]
- 4.Sprigg N, Flaherty K, Appleton JP, Salman RAS, Bereczki D, Beridze M, et al. Tranexamic acid for hyperacute primary IntraCerebral Haemorrhage (TICH-2): an international randomised, placebo-controlled, phase 3 superiority trial. Lancet [Internet]. 2018;391(10135):2107–15. Available from: 10.1016/S0140-6736(18)31033-X [DOI] [PMC free article] [PubMed]
- 5.Schmidt-Pogoda A, Bonberg N, Koecke MHM, Strecker J-K, Wellmann J, Bruckmann N-M, et al. Why most acute stroke studies are positive in animals but not in patients. Ann Neurol. 2019;1–12. [DOI] [PubMed]
- 6.Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NLM, Jackson C, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43(12):3161–7. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- 7.Devan WJ, Falcone GJ, Anderson CD, Jagiella JM, Schmidt H, Hansen BM, et al. Heritability estimates identify a substantial genetic contribution to risk and outcome of intracerebral hemorrhage. Stroke. 2013;44(6):1578–83. doi: 10.1161/STROKEAHA.111.000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korja M, Silventoinen K, McCarron P, Zdravkovic S, Skytthe A, Haapanen A, et al. Genetic epidemiology of spontaneous subarachnoid hemorrhage: Nordic twin study. Stroke. 2010;41(11):2458–62. doi: 10.1161/STROKEAHA.110.586420. [DOI] [PubMed] [Google Scholar]
- 9.Speed D, O’Brien TJ, Palotie A, Shkura K, Marson AG, Balding DJ, et al. Describing the genetic architecture of epilepsy through heritability analysis. Brain. 2014;137(10):2680–9. doi: 10.1093/brain/awu206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blum S, Ji Y, Pennisi D, Li Z, Leo P, McCombe P, et al. Genome-wide association study in Guillain-Barré syndrome. J Neuroimmunol [Internet] 2018;323(July):109–14. doi: 10.1016/j.jneuroim.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet [Internet]. 2015;47(8):856–60. Available from: 10.1038/ng.3314 [DOI] [PubMed]
- 12.Burton P. Genetic Epidemiology 1 Key concepts in genetic epidemiology. Lancet -London-. 2005;1(9489):941–51. doi: 10.1016/S0140-6736(05)67322-9. [DOI] [PubMed] [Google Scholar]
- 13.Beyene J, Pare G. Statistical genetics with application to population-based study design: A primer for clinicians. Eur Heart J. 2014;35(8):495–500. doi: 10.1093/eurheartj/eht272. [DOI] [PubMed] [Google Scholar]
- 14.Polderman TJC, Benyamin B, De Leeuw CA, Sullivan PF, Van Bochoven A, Visscher PM, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet [Internet]. 2015;47(7):702–9. Available from: 10.1038/ng.3285 [DOI] [PubMed]
- 15.Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet [Internet]. 2019;51(9):1339–48. Available from: 10.1038/s41588-019-0481-0 [DOI] [PubMed]
- 16.Jorgensen TJ, Ruczinski I, Kessing B, Smith MW, Shugart YY, Alberg AJ. Hypothesis-driven candidate gene association studies: Practical design and analytical considerations. Am J Epidemiol. 2009;170(8):986–93. doi: 10.1093/aje/kwp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat Protoc. 2007;2(10):2492–500. doi: 10.1038/nprot.2007.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: Boosting disease gene discovery. Nat Rev Genet. 2012;13(8):523–36. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- 19.Rebbeck TR, Spitz M, Wu X. Assessing the function of genetic variants in candidate gene association studies. Nat Rev Genet. 2004;5(8):589–97. doi: 10.1038/nrg1403. [DOI] [PubMed] [Google Scholar]
- 20.Bush WS, Moore JH. Chapter 11: Genome-Wide Association Studies. PLoS Comput Biol. 2012;8(12). [DOI] [PMC free article] [PubMed]
- 21.Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, et al. Sequencing studies in human genetics: Design and interpretation. Nat Rev Genet. 2013;14(7):460–70. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khera A V., Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet [Internet]. 2018;50(9):1219–24. Available from: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed]
- 24.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulit SL, Weng LC, McArdle PF, Trinquart L, Choi SH, Mitchell BD, et al. Atrial fibrillation genetic risk differentiates cardioembolic stroke from other stroke subtypes. Neurol Genet. 2018;4(6):1–8. doi: 10.1212/NXG.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grover S, Del Greco FM, Stein CM, Ziegler A. Mendelian randomization. Methods Mol Biol. 2017;1666(19):581–628. doi: 10.1007/978-1-4939-7274-6_29. [DOI] [PubMed] [Google Scholar]
- 27.Eraslan G, Avsec Ž, Gagneur J, Theis FJ. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet [Internet]. 2019;20(7):389–403. Available from: 10.1038/s41576-019-0122-6 [DOI] [PubMed]
- 28.Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. A primer on deep learning in genomics. Nat Genet [Internet]. 2019;51(1):12–8. Available from: 10.1038/s41588-018-0295-5 [DOI] [PMC free article] [PubMed]
- 29.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol [Internet]. 2016;70:214–23. Available from: 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed]
- 31.Sankar PL, Parker LS. The Precision Medicine Initiative’s All of Us Research Program: An agenda for research on its ethical, legal, and social issues. Genet Med. 2017;19(7):743–50. doi: 10.1038/gim.2016.183. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Chen J, Collins R, Guo Y, Peto R, Wu F, et al. China Kadoorie Biobank of 0.5 million people: Survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol. 2011;40(6):1652–66. doi: 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong KM, Langlais K, Tobias GS, Fletcher-Hoppe C, Krasnewich D, Leeds HS, et al. The dbGaP data browser: A new tool for browsing dbGaP controlled-access genomic data. Nucleic Acids Res. 2017;45(D1):D819–26. doi: 10.1093/nar/gkw1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lappalainen I, Almeida-King J, Kumanduri V, Senf A, Spalding JD, Ur-Rehman S, et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat Genet [Internet]. 2015;47(7):692–5. Available from: 10.1038/ng.3312 [DOI] [PMC free article] [PubMed]
- 35.Johnson AEW, Pollard TJ, Shen L, Lehman LWH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:1–9. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O. The eICU collaborative research database, a freely available multi-center database for critical care research. Sci Data. 2018;5:1–13. doi: 10.1038/sdata.2018.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford KM, Gallego-Fabrega C, Kourkoulis C, Miyares L, Marini S, Flannick J, et al. Cerebrovascular disease knowledge portal an open-access data resource to accelerate genomic discoveries in stroke. Stroke. 2018;49(2):470–5. doi: 10.1161/STROKEAHA.117.018922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo D, Rosand J, Kidwell C, Mccauley JL, Osborne J, Brown MW, et al. The ethnic/racial variations of intracerebral hemorrhage (ERICH) study protocol. Stroke. 2013;44(10):120–5. doi: 10.1161/STROKEAHA.113.002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leasure AC, Sheth KN, Comeau M, Aldridge C, Worrall BB, Vashkevich A, et al. Identification and Validation of Hematoma Volume Cutoffs in Spontaneous, Supratentorial Deep Intracerebral Hemorrhage. Stroke. 2019;50(8):2044–9. doi: 10.1161/STROKEAHA.118.023851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh KB, Woo D, Sekar P, Osborne J, Moomaw CJ, Langefeld CD, et al. Untreated Hypertension: A Powerful Risk Factor for Lobar and Nonlobar Intracerebral Hemorrhage in Whites, Blacks, and Hispanics. Circulation. 2016;134(19):1444–52. doi: 10.1161/CIRCULATIONAHA.116.024073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marini S, Lena UK, Crawford KM, Moomaw CJ, Testai FD, Kittner SJ, et al. Comparison of genetic and self-identified ancestry in modeling intracerebral hemorrhage risk. Front Neurol. 2018;9(JUL):1–9. doi: 10.3389/fneur.2018.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Torres A, Murphy M, Kourkoulis C, Schwab K, Ayres AM, Moomaw CJ, et al. Hypertension and intracerebral hemorrhage recurrence among white, black, and hispanic individuals. Neurology. 2018;91(1):E37–44. doi: 10.1212/WNL.0000000000005729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CJ, Brown WM, Moomaw CJ, Langefeld CD, Osborne J, Worrall BB, et al. Alcohol use and risk of intracerebral hemorrhage. Neurology. 2017;88(21):2043–51. doi: 10.1212/WNL.0000000000003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah M, Birnbaum L, Rasmussen J, Sekar P, Moomaw CJ, Osborne J, et al. Effect of Hyperosmolar Therapy on Outcome Following Spontaneous Intracerebral Hemorrhage: Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH) Study. J Stroke Cerebrovasc Dis [Internet]. 2018;27(4):1061–7. Available from: 10.1016/j.jstrokecerebrovasdis.2017.11.013 [DOI] [PMC free article] [PubMed]
- 45.Kluyver T, Ragan-kelley B, Pérez F, Granger B, Bussonnier M, Frederic J, et al. Jupyter Notebooks—a publishing format for reproducible computational workflows. Position Power Acad Publ Play Agents Agendas. 2016;87–90.
- 46.Buniello A, Macarthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–12. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian D, Wang P, Tang B, Teng X, Li C, Liu X, et al. GWAS Atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2019;1–6. [DOI] [PMC free article] [PubMed]
- 48.Lambert SA, Jupp S, Abraham G, Parkinson H, Danesh J, MacArthur JAL, et al. The Polygenic Score (PGS) Catalog: a database of published PGS to enable reproducibility and uniform evaluation. [Internet]. 2019. Available from: www.pgscatalog.org
- 49.Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun [Internet]. 2017;8(1):1–10. Available from: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed]
- 50.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2011;27(13):2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu PY, Cheng CW, Kaddi CD, Venugopalan J, Hoffman R, Wang MD. Omic and Electronic Health Record Big Data Analytics for Precision Medicine. IEEE Trans Biomed Eng. 2017;64(2):263–73. doi: 10.1109/TBME.2016.2573285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hulsen T, Jamuar SS, Moody AR, Karnes JH, Varga O, Hedensted S, et al. From big data to precision medicine. Front Med. 2019;6(MAR):1–14. doi: 10.3389/fmed.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung J, Marini S, Pera J, Norrving B, Jimenez-Conde J, Roquer J, et al. Genome-wide association study of cerebral small vessel disease reveals established and novel loci. Brain. 2019;3176–89. [DOI] [PMC free article] [PubMed]
- 54.Jha RM, Koleck TA, Puccio AM, Okonkwo DO, Park SY, Zusman BE, et al. Regionally clustered ABCC8 polymorphisms in a prospective cohort predict cerebral oedema and outcome in severe traumatic brain injury. J Neurol Neurosurg Psychiatry. 2018;1152–62. [DOI] [PMC free article] [PubMed]
- 55.Kim GS, Yang L, Zhang G, Zhao H, Selim M, McCullough LD, et al. Critical role of sphingosine-1-phosphate receptor-2 in the disruption of cerebrovascular integrity in experimental stroke. Nat Commun. 2015;6. [DOI] [PMC free article] [PubMed]
- 56.Biffi A, Anderson CD, Jagiella JM, Schmidt H, Kissela B, Hansen BM, et al. APOE genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: A genetic association study. Lancet Neurol [Internet]. 2011;10(8):702–9. Available from: 10.1016/S1474-4422(11)70148-X [DOI] [PMC free article] [PubMed]
- 57.Marini S, Devan WJ, Radmanesh F, Miyares L, Poterba T, Hansen BM, et al. 17p12 Influences Hematoma Volume and Outcome in Spontaneous Intracerebral Hemorrhage. Stroke. 2018;49(7):1618–25. doi: 10.1161/STROKEAHA.117.020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falcone GJ, Biffi A, Devan WJ, Brouwers HB, Anderson CD, Valant V, et al. Burden of blood pressure-related alleles is associated with larger hematoma volume and worse outcome in intracerebral hemorrhage. Stroke. 2013;44(2):321–6. doi: 10.1161/STROKEAHA.112.675181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joy MT, Ben Assayag E, Shabashov-Stone D, Liraz-Zaltsman S, Mazzitelli J, Arenas M, et al. CCR5 Is a Therapeutic Target for Recovery after Stroke and Traumatic Brain Injury. Cell [Internet]. 2019;176(5):1143-1157.e13. Available from: 10.1016/j.cell.2019.01.044 [DOI] [PMC free article] [PubMed]
- 60.Mola-Caminal M, Carrera C, Soriano-Tárraga C, Giralt-Steinhauer E, Díaz-Navarro RM, Tur S, et al. PATJ Low Frequency Variants Are Associated with Worse Ischemic Stroke Functional Outcome: A Genome-Wide Meta-Analysis. Circ Res. 2019;124(1):114–20. doi: 10.1161/CIRCRESAHA.118.313533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Söderholm M, Pedersen A, Lorentzen E, Stanne TM, Bevan S, Olsson M, et al. Genome-wide association meta-analysis of functional outcome after ischemic stroke. Neurology. 2019;92(12):e1271–83. doi: 10.1212/WNL.0000000000007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jha RM, Molyneaux BJ, Jackson TC, Wallisch JS, Park SY, Poloyac S, et al. Glibenclamide produces region-dependent effects on cerebral edema in a combined injury model of traumatic brain injury and hemorrhagic shock in mice. J Neurotrauma. 2018;35(17):2125–35. doi: 10.1089/neu.2016.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheth KN, Elm JJ, Molyneaux BJ, Hinson H, Beslow LA, Sze GK, et al. Safety and efficacy of intravenous glyburide on brain swelling after large hemispheric infarction (GAMES-RP): a randomised, double-blind, placebo-controlled phase 2 trial. Lancet Neurol [Internet]. 2016;15(11):1160–9. Available from: 10.1016/S1474-4422(16)30196-X [DOI] [PubMed]
- 64.Kimberly WT, Bevers MB, von Kummer R, Demchuk AM, Romero JM, Elm JJ, et al. Effect of IV glyburide on adjudicated edema endpoints in the GAMES-RP Trial. Neurology. 2018;91(23):e2163–9. doi: 10.1212/WNL.0000000000006618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vorasayan P, Bevers MB, Beslow L, Sze G, Molyneaux BJ, Hinson H, et al. Intravenous Glibenclamide Reduces Water Uptake and Mass Effect in Large Hemispheric Infarction (GAMES-RP Study) Stroke. 2019;50(Suppl_1):1–7. doi: 10.1161/STROKEAHA.119.026036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu L, Barfejani AH, Qin T, Dong Q, Ayata C, Waeber C. Fingolimod exerts neuroprotective effects in a mouse model of intracerebral hemorrhage. Brain Res [Internet]. 2014;1555(2014):89–96. Available from: 10.1016/j.brainres.2014.01.048 [DOI] [PMC free article] [PubMed]
- 67.Rolland WB, Lekic T, Krafft PR, Hasegawa Y, Altay O, Hartman R, et al. Fingolimod reduces cerebral lymphocyte infiltration in experimental models of rodent intracerebral hemorrhage. Exp Neurol [Internet]. 2013;241(1):45–55. Available from: 10.1016/j.expneurol.2012.12.009 [DOI] [PMC free article] [PubMed]
- 68.Sun N, Shen Y, Han W, Shi K, Wood K, Fu Y, et al. Selective sphingosine-1-phosphate receptor 1 modulation attenuates experimental intracerebral hemorrhage. Stroke. 2016;47(7):1899–906. doi: 10.1161/STROKEAHA.115.012236. [DOI] [PubMed] [Google Scholar]
- 69.Fu Y, Hao J, Zhang N, Ren L, Sun N, Li YJ, et al. Fingolimod for the treatment of intracerebral hemorrhage: A 2-arm proof-of-concept study. JAMA Neurol. 2014;71(9):1092–101. doi: 10.1001/jamaneurol.2014.1065. [DOI] [PubMed] [Google Scholar]
- 70.Pinho J, Costa AS, Araújo JM, Amorim JM, Ferreira C. Intracerebral hemorrhage outcome: A comprehensive update. J Neurol Sci. 2019;398(July 2018):54–66. doi: 10.1016/j.jns.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 71.Brouwers HB, Biffi A, Ayres AM, Schwab K, Cortellini L, Romero JM, et al. Apolipoprotein e genotype predicts hematoma expansion in lobar intracerebral hemorrhage. Stroke. 2012;43(6):1490–5. doi: 10.1161/STROKEAHA.111.643262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brouwers HB, Biffi A, McNamara KA, Ayres AM, Valant V, Schwab K, et al. Apolipoprotein e genotype is associated with ct angiography spot sign in lobar intracerebral hemorrhage. Stroke. 2012;43(8):2120–5. doi: 10.1161/STROKEAHA.112.659094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fisher CM. Hypertensive Cerebral Hemorrhage. Demonstration of the Source of Bleeding. J Neuropathol Exp Neurol. 2003;62(1):104–7. doi: 10.1093/jnen/62.1.104. [DOI] [PubMed] [Google Scholar]
- 74.Woollard SM, Kanmogne GD. Maraviroc: A review of its use in hivinfection and beyond. Drug Des Devel Ther. 2015;9:5447–68. doi: 10.2147/DDDT.S90580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12(10):931–4. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Price WN, Cohen IG. Privacy in the age of medical big data. Nat Med [Internet]. 2019;25(1):37–43. Available from: 10.1038/s41591-018-0272-7 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 507 kb)