Abstract
Assembly-line polyketide synthases generate natural products that have led to many live-saving drugs. The use of E. coli as a heterologous host for reconstituting these enormous and complex enzymatic machines has and will continue to be a critical strategy for understanding them. Here, we concisely summarize successful examples in exploiting E. coli for assembly-line polyketide biosynthesis as well as offer examples of new challenges in which this approach is primed to tackle.
Highlights
-
•
Understanding assembly-line PKSs is often challenging in natural hosts.
-
•
E. coli is a robust host for engineered biosynthesis of polyketides.
-
•
E. coli will play a vital role in current challenges like deciphering orphan PKSs.
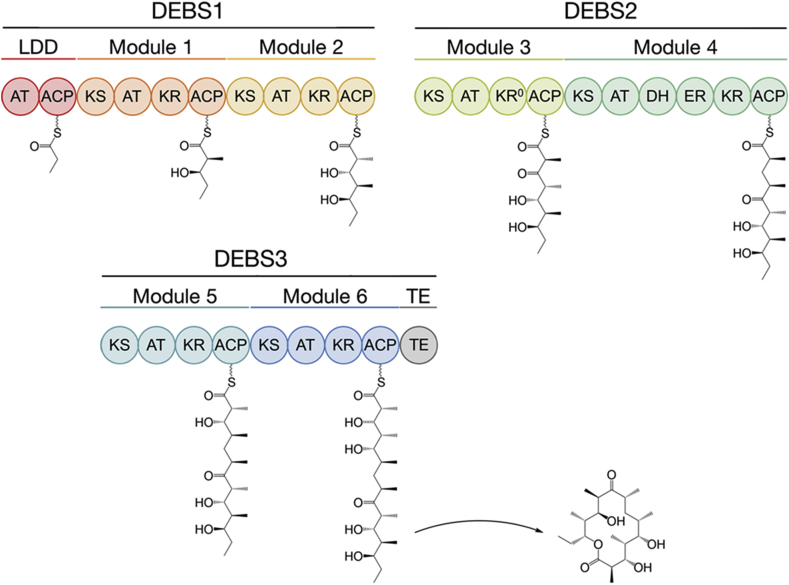
Polyketide synthases (PKSs) manufacture truly remarkable natural products. Polyketides are structurally diverse and possess important medicinal and agricultural capabilities that include but are not limited to: antibacterial, anticancer, antiparasitic, immunosuppressant, herbicidal, insecticidal and fungicidal activities. Assembly-line PKSs are a subset of type I PKSs and are exceptionally complex molecular machines. For example, the extensively studied 6-deoxyerythronolide B synthase (DEBS) is a colossal homodimer (its molecular weight exceeds 2 MDa) that elaborately channels reactive intermediates through 22 distinct enzymatic domains to build 6-deoxyerythronolide B (6-dEB), a precursor of the antibiotic erythromycin A (Walsh, 2004; Khosla et al., 2007; Cortes et al., 1990; Donadio et al., 1991) (Fig. 1).
Fig. 1.
The 6-Deoxyerythronolide B Synthase (DEBS). Enzymatic domains: KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; ER, enoylreductase; ACP, acyl carrier protein; and TE, thioesterase.
Unfortunately, assembly-line PKSs often are housed in poorly characterized and/or difficult to culture microbes. Further, many of the biosynthetic gene clusters (BGCs) involve with these PKSs are not expressed in laboratory conditions (Rutledge and Challis, 2015). Although game-changing tools such as the CRISPR/Cas system are being repurposed for Streptomyces – a treasure trove for assembly-line PKSs – (Cobb et al., 2015), bacteria like Streptomyces have slow growth rates, are labor- and time-intensive to genetically manipulate and have cluttered metabolic backgrounds. On the other hand, model strains of Escherichia coli are obvious “user-friendly” workhorses. They are easy to culture, grow fast and have a wealth of genetic tools. Importantly, their metabolic backgrounds are free from native complex natural product biosynthetic pathways, which lowers the potential for metabolic ‘cross-talk’ with heterologously introduced systems. Despite E. coli’s advantages, only a handful of assembly-line PKSs have been either fully or largely reconstituted in E. coli. Here in this brief perspective, we summarize the most successful case – DEBS – of using E. coli as a heterologous host for assembly-line polyketide biosynthesis. Finally, we propose examples of future challenges and opportunities that researchers exploiting E. coli as a tool may consider as avenues to further our understanding of these breathtaking enzymes.
Following the accomplishments of manipulating DEBS in both its native producer host Saccharopolyspora erythraea (Marsden et al., 1998) as well as Streptomyces hosts (Kao et al., 1994; Xue et al., 1999), the stage was set for its reconstituted biosynthesis in E. coli. Pfeifer et al. expressed the three large DEBS polypeptides (their molecular weights each surpass 300 kDa, and their genes are each longer than 10 kb) in a highly engineered E. coli BL21(DE3) derivative named BAP1 (Pfeifer et al., 2001) and synthesized 6-dEB at titers of 20 mg/L in E. coli. BAP1 encodes Bacillus subtilis Sfp, a phosphopantetheinyl transferase that phosphopantetheinylates the seven acyl carrier protein domains in DEBS. To elevate intracellular concentrations of propionyl-CoA and methylmalonyl-CoA, Pfeifer et al. a) deleted the propionate catabolism genes prpRBCD, b) boosted expression of the propionyl-CoA synthetase PrpE and c) co-expressed Streptomyces coelicolor propionyl-CoA carboxylase, which generates methylmalonyl-CoA from propionyl-CoA. Lau et al. achieved titers that approached Streptomyces hosts (1.1 g/L) by optimizing the medium composition and fermentation conditions (Lau et al., 2004). Installing DEBS in E. coli enabled the engineers to rationally modify specific parts of DEBS to fabricate novel natural polyketides in vivo, a long-standing goal in natural products research. To manufacture a benzoate-containing analog of 6-dEB, Pfeifer et al. replaced the loading module of DEBS with the loading module from the rifamycin polyketide synthase, which accepts a benzoate starter unit. To completely produce erythromycin A in E. coli, Zhang et al. impressively expressed 17 additional genes that are responsible for deoxysugar biosynthesis, macrolide tailoring, and resistance, which led to the biosynthesis of erythromycin A at titers of 0.6 mg/L (Zhang et al., 2010). P8/1-OG (an intermediate of rifamycin biosynthesis) (Watanabe et al., 2003) and epothilone C and D (Mutka et al., 2006) are two other working examples of engineered assembly-line polyketide biosynthesis in E. coli. While these two studies did not reconstitute the entire assembly-line, they are notable for utilizing chaperones and protein dissection to enhance the expression of massive PKS polypeptides: the 530 kDa RifA and 765 kDa EpoD proteins were each split into two polypeptides. Even assembly-line systems that are predominantly composed of non-ribosomal peptide synthetase (NRPS) domains such as the yersiniabactin NRPS/PKS can be successfully expressed in E. coli (Pfeifer et al., 2003).
Engineering erythromycin A biosynthesis in E. coli represents a high-water mark for PKS research. However, one could argue that the small number of pathways targeted for engineered assembly-line polyketide biosynthesis in E. coli suggests that E. coli is a poor “universal host”. Certainly, as seen in the previous examples, E. coli requires nontrivial engineering for the correct post-translational modifications of PKSs, sufficient precursor pools and efficient translation/folding of enormous polypeptides. Notwithstanding these difficulties, E. coli is poised to continue as a top heterologous host for assembly-line PKSs. Here are three – while not exhaustive – examples of challenges and opportunities in which E. coli should remain in the priority list for assembly-line PKS enzymologists.
First, trans-acyltransferase (trans-AT) assembly-line PKSs represent almost 38% of all assembly line PKSs according to a recent survey (O’Brien et al., 2014). Unlike cis-AT PKSs such as DEBS, trans-AT PKSs need AT domains encoded outside of the core polyketide synthase genes that interact noncovalently with the megasynthase. However, the understanding of trans-AT PKSs has significantly lagged behind that of cis-AT PKSs (Helfrich and Piel, 2016). trans-AT PKSs are particularly challenging to study because they frequently originate in poorly understood microbes as well as possess a wide array of unorthodox biosynthetic components. As of this writing, no trans-AT assembly-line PKS has been reconstituted in E. coli; therefore, the lessons learned from the first fruitful attempts will be invaluable to researchers that desire to tap into the advantages that E. coli holds as a heterologous host.
Second, genomic sequencing has revealed large numbers (far surpassing known assembly-line PKS BGCs) of cryptic or “orphan” assembly-line PKS BGCs associated with no known polyketide products (O’Brien et al., 2014). This recent surge provides a vast resource for pharmaceutical and agrochemical natural products research. However, scientists must endeavor to decipher this embarrassment of riches in a timely fashion. Though platforms such as antiSMASH 4.0 (Blin et al., 2017), PRISM 3 (Skinnider et al., 2017) and TransATor (Helfrich et al., 2019) offer useful polyketide structural predictions, there is no substitute for interrogating PKSs and their repertoire of products either in vitro or in vivo. For example, the NOCardiosis-Associated Polyketide (NOCAP) synthase, an orphan assembly-line PKS found in clinical strains of the actinomycete Nocardia, exhibits iterative activity in one of its modules, a behavior which cannot be presently predicted in silico (Kuo et al., 2016). E. coli provides a way to quickly move from assembly-line PKS genes to natural products. E. coli’s aforementioned sizeable genetic toolkit, fast growth rate and uncluttered metabolic background should enable rapid and straightforward cloning of these clusters, polyketide biosynthesis and metabolic profiling. As in the case with trans-AT assembly-line PKSs, no assembly-line PKS identified in silico has been immediately “de-orphanized” by reconstitution in E. coli; this achievement would represent a new method of natural products discovery.
Third, although there have been major advances in the structural analysis of individual PKS domains, multidomain fragments and even intact modules (Robbins et al., 2016; Dutta et al., 2014; Edwards et al., 2014; Li et al., 2018), we know little about how these multienzyme machines behave as a whole in a cell. Transmission electron microscopy of B. subtilis cells revealed that the bacillaene hybrid PKS/NRPS fascinatingly assembles into a single organelle-like membrane-associated complex with approximate mass of 10–100 MDa (Straight et al., 2007). It would be most interesting to establish how many assembly-line PKSs organize into “megacomplexes”. Can microscopists study PKS complex dynamics with live-cell imaging, as oppose to electron microscopy which is limited to fixed cells? Do PKSs retain the ability to assemble as a megacomplex in a heterologous host like E. coli? If so, then pathways previously engineered to operate in E. coli could be easily retrofitted with a variety of fluorescent proteins or tags; therefore, permitting the monitoring of these enzymes with live-cell imaging tools that require fluorophores such as fluorescence or super-resolution microscopy. Once tagged, these proteins can be interrogated “coarsely” in the context of megacomplexes or “finely” in the context of domain-domain interactions with techniques such as FRET (Fluorescence Resonance Energy Transfer).
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgements
This research was supported by the National Institutes of Health (NIH R01 GM087934 and NIH F32 GM123637 to K. P. Y.).
References
- Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J., Kautsar S.A., Suarez Duran H.G., de Los Santos E.L.C., Kim H.U., Nave M., Dickschat J.S., Mitchell D.A., Shelest E., Breitling R., Takano E., Lee S.Y., Weber T., Medema M.H. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb R.E., Wang Y., Zhao H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 2015;4:723. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J., Haydock S.F., Roberts G.A., Bevitt D.J., Leadlay P.F. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348:176. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- Donadio S., Staver M.J., McAlpine J.B., Swanson S.J., Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- Dutta S., Whicher J.R., Hansen D.A., Hale W.A., Chemler J.A., Congdon G.R., Narayan A.R.H., Hakansson K., Sherman D.H., Smith J.L., Skiniotis G. Structure of a modular polyketide synthase. Nature. 2014;510:512. doi: 10.1038/nature13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A.L., Matsui T., Weiss T.M., Khosla C. Architectures of whole-module and bimodular proteins from the 6-deoxyerythronolide B synthase. J. Mol. Biol. 2014;426:2229. doi: 10.1016/j.jmb.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich E.J.N., Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016;33:231. doi: 10.1039/C5NP00125K. [DOI] [PubMed] [Google Scholar]
- Helfrich E.J.N., Ueoka R., Dolev A., Rust M., Meoded R.A., Bhushan A., Califano G., Costa R., Gugger M., Steinbeck C., Moreno P., Piel J. Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat. Chem. Biol. 2019;15:813. doi: 10.1038/s41589-019-0313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.M., Katz L., Khosla C. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science. 1994;265:509. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- Khosla C., Tang Y., Chen A.Y., Schnarr N.A., Cane D.E. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem. 2007;76:195. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- Kuo J., Lynch S.R., Liu C.W., Xiao X., Khosla C. Partial in vitro reconstitution of an orphan polyketide synthase associated with clinical cases of nocardiosis. ACS Chem. Biol. 2016;11:2636. doi: 10.1021/acschembio.6b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J., Tran C., Licari P., Galazzo J. Development of a high cell-density fed-batch bioprocess for the heterologous production of 6-deoxyerythronolide B in Escherichia coli. J. Biotechnol. 2004;110:95. doi: 10.1016/j.jbiotec.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Li X., Sevillano N., La Greca F., Deis L., Liu Y.C., Deller M.C., Mathews I.I., Matsui T., Cane D.E., Craik C.S., Khosla C. Structure-function analysis of the extended conformation of a polyketide synthase module. J. Am. Chem. Soc. 2018;140:6518. doi: 10.1021/jacs.8b02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden A.F., Wilkinson B., Cortés J., Dunster N.J., Staunton J., Leadlay P.F. Engineering broader specificity into an antibiotic-producing polyketide synthase. Science. 1998;279:199. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- Mutka S.C., Carney J.R., Liu Y., Kennedy J. Heterologous production of epothilone C and D in Escherichia coli. Biochemistry. 2006;45:1321. doi: 10.1021/bi052075r. [DOI] [PubMed] [Google Scholar]
- O’Brien R.V., Davis R.W., Khosla C., Hillenmeyer M.E. Computational identification and analysis of orphan assembly-line polyketide synthases. J. Antibiot. 2014;67:89. doi: 10.1038/ja.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B.A., Admiraal S.J., Gramajo H., Cane D.E., Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- Pfeifer B.A., Wang C.C., Walsh C.T., Khosla C. Biosynthesis of Yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl. Environ. Microbiol. 2003;69:6698. doi: 10.1128/aem.69.11.6698-6702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins T., Liu Y.C., Cane D.E., Khosla C. Structure and mechanism of assembly line polyketide synthases. Curr. Opin. Struct. Biol. 2016;41:10. doi: 10.1016/j.sbi.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge P.J., Challis G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015;13:509. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- Skinnider M.A., Merwin N.J., Johnston C.W., Magarvey N.A. Prism 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017;45:W49. doi: 10.1093/nar/gkx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight P.D., Fischbach M.A., Walsh C.T., Rudner D.Z., Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:305. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.T. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303:1805. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Rude M.A., Walsh C.T., Khosla C. Engineered biosynthesis of an ansamycin polyketide precursor in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9774. doi: 10.1073/pnas.1632167100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Q., Ashley G., Hutchinson C.R., Santi D.V. A multiplasmid approach to preparing large libraries of polyketides. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11740. doi: 10.1073/pnas.96.21.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang Y., Wu J., Skalina K., Pfeifer B.A. Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem. Biol. 2010;17:1232. doi: 10.1016/j.chembiol.2010.09.013. [DOI] [PubMed] [Google Scholar]