Abstract
Cadmium (Cd) is a widespread pollutant that is toxic to living organisms. Previous studies have identified certain WRKY transcription factors, which confer Cd tolerance in different plant species. In the present study, we have identified 29 Cd-responsive WRKY genes in Soybean [Glycine max (L.) Merr.], and confirmed that 26 of those GmWRKY genes were up-regulated, while 3 were down-regulated. We have also cloned the novel, positively regulated GmWRKY142 gene from soybean and investigated its regulatory mechanism in Cd tolerance. GmWRKY142 was highly expressed in the root, drastically up-regulated by Cd, localized in the nucleus, and displayed transcriptional activity. The overexpression of GmWRKY142 in Arabidopsis thaliana and soybean hairy roots significantly enhanced Cd tolerance and lead to extensive transcriptional reprogramming of stress-responsive genes. ATCDT1, GmCDT1-1, and GmCDT1-2 encoding cadmium tolerance 1 were induced in overexpression lines. Further analysis showed that GmWRKY142 activated the transcription of ATCDT1, GmCDT1-1, and GmCDT1-2 by directly binding to the W-box element in their promoters. In addition, the functions of GmCDT1-1 and GmCDT1-2, responsible for decreasing Cd uptake, were validated by heterologous expression in A. thaliana. Our combined results have determined GmWRKYs to be newly discovered participants in response to Cd stress, and have confirmed that GmWRKY142 directly targets ATCDT1, GmCDT1-1, and GmCDT1-2 to decrease Cd uptake and positively regulate Cd tolerance. The GmWRKY142-GmCDT1-1/2 cascade module provides a potential strategy to lower Cd accumulation in soybean.
Keywords: transcription factor, WRKY, cadmium stress, CDT1-likes, soybean
Introduction
Cadmium (Cd) is a non-essential metallic trace element that is toxic to both plants and animals. Recently, global climate change and rapid industrialization have contributed to an increase in Cd deposition in soil, leading to a major global threat to crop productivity and human health (Das et al., 1997; Moulis and Thévenod, 2010; Clemens et al., 2013; Okereafor et al., 2020; Zheng et al., 2020). The cultivation of cadmium tolerant crops and reduction of cadmium concentration in the edible parts of plants are solutions that could potentially alleviate the risks to human health (Grant et al., 1998; Gill and Tuteja, 2011; Tang et al., 2017; Zhao and Huang, 2018). To this effect, it is necessary to understand the mechanisms of Cd tolerance in plants and to investigate important genes encoding Cd tolerance.
Plants have evolved a set of versatile adaptive mechanisms to cope with Cd stress. These mainly involve the use of enzymatic and non-enzymatic antioxidants, the extrusion of Cd across the plasma membrane, the restriction of Cd movement to roots by mycorrhizas, and finally, the sequestration of metals in metabolically inactive parts such as root cell walls and leaf vacuoles (Dong et al., 2007; Hasan et al., 2009; Nagajyoti et al., 2010; Gallego et al., 2012; Rizwan et al., 2017; Shanying et al., 2017; Kumar et al., 2019; Sheng et al., 2019; Zhang Z. H. et al., 2019). Considerable progress has recently been made in understanding the molecular mechanisms of Cd accumulation in plants. Several key genes encoding metal transporters, chelator proteins, antioxidant enzymes, defensin genes and transcription factors have been reported to participate in Cd detoxification and tolerance in plants (Zhu et al., 1999a, b; Wu et al., 2012; Sasaki et al., 2014; Chen et al., 2016; Yang et al., 2016; Cai et al., 2017; Luo et al., 2019a, b; Mekawy et al., 2020). Of these functional proteins, Cysteine (Cys) -rich proteins are considered the most important Cd chelator proteins and generally have relatively low molecular weights (4–14 kDa) with a high ratio of cysteine residues (Song et al., 2004; Kuramata et al., 2009). Since the characterization of the first Cd-binding Cys-rich membrane protein from horse kidneys in 1957 (Margoshes and Vallee, 1957), a number of Cd-binding Cys-rich genes have been identified in plants, including AtPcr1 (Song et al., 2004), ThMT3 (Yang et al., 2011), OsDEP1 (Kunihiro et al., 2013), CnMT1 and CnMT2 (Palacios et al., 2014), and OsMT-3a (Mekawy et al., 2020). Particularly, DcCDT1 (Digitaria ciliaris cadmium tolerance 1) and its homologues (AtCDT1 and OsCDT1) are specific to higher plants; they are unique and distinctive in both their peptide lengths (49–60 amino acids) and in their arrangement of Cys residues in the consensus sequence of CL-(Y/F)-A-(C/T)-X5-CC-(F/C)-CCYE-(T/K)-C-(E/K)-C-(CLDCL or delete)-CCCC (Kuramata et al., 2009; Matsuda et al., 2009). Transgenic A. thaliana plants or yeast cells overexpressing DcCDT1, OsCDT1, or AtCDT1 consistently displayed a Cd tolerant phenotype and accumulated lower amounts of Cd. Moreover, 5 DcCDT1 homologs in rice (OsCDT1 – 5) were up-regulated to varying degrees by Cd treatment (Kuramata et al., 2009). However, the mechanism of CDT1 regulation by Cd stress remains to be elucidated.
Transcription factors (TFs) are potentially the core components in the regulatory networks of Cd detoxification and tolerance owing to their functions as key regulators of the Cd stress response via their control on downstream gene expression. Most types of transcription factors regulating Cd detoxification and tolerance have been identified in plants, including metal response element transcription factors (MTF) (Smirnova et al., 2000; Solis et al., 2002; Lin et al., 2017), basic helix-loop-helix (bHLH) transcription factors (Wu et al., 2012; Yao et al., 2018), myeloblastosis (MYB) transcription factors (Hu et al., 2017; Xu et al., 2018; Zhang P. et al., 2019), ethylene responsive factors (ERF) (Yi et al., 2004; Tang et al., 2005; Lin et al., 2017), SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factors (Chen et al., 2018), Zn-regulated (Zip) transporters (Liu et al., 2019; Wu et al., 2019), and WRKY transcription factors (Yang et al., 2016; Hong et al., 2017; Han et al., 2019; Sheng et al., 2019). The WRKY proteins are a superfamily of transcription factors with up to 100, 90, and 170 representatives in Arabidopsis, Rice (Oryza sativa L.) and Soybean (Glycine max. L), respectively (Eulgem et al., 2000; Song et al., 2016; Xu et al., 2016; Yang et al., 2017). The first gene encoding WRKY transcription factor in plants was identified more than 20 years ago (Ishiguro and Nakamura, 1994; Rushton et al., 1995, 1996), and substantial progress in this area of research has since been achieved. Generally, WRKY proteins, composed of a WRKY domain (WRKYGQK) and a novel zinc-finger-like motif (Cx4–5Cx22–23HxH or Cx7Cx23HxC) at the N-terminal, can specifically recognize and bind the cis-acting W-box elements (TTGACC/T) of downstream genes (Eulgem et al., 2000; Eulgem and Somssich, 2007; Rushton et al., 2010). Several studies have found that WRKY TFs participate in the response to Cd stress by regulating the expression of downstream target genes such as AtWRKY12 (Han et al., 2019), AtWRKY13 (Sheng et al., 2019), ThWRKY7 (Yang et al., 2016), and CaWRKY41 (Dang et al., 2019). However, little is known about the role of the soybean WRKY TFs in Cd tolerance.
Soybean is important oil crops and the plant proteins resources widely grown around the world. Heavy metal contamination is an important factor that seriously inhibits soybean growth and threatens the human health. A better understanding of how soybean responds to heavy metal contamination would lay the foundations for developing effective countermeasures. Using the comprehensive mRNA transcriptome of soybean under Cd stress, 29 Cd-responsive WRKY genes were identified and confirmed by qRT-PCR analyses. Of these, GmWRKY142 was selected to verify the function of Cd tolerance, due to its higher expression in root under normal conditions, as well as its strong up-regulation under Cd stress. We demonstrated that GmWRKY142 overexpression in A. thaliana and soybean hair roots resulted in increased Cd tolerance and decreased Cd accumulation. Further analysis indicated that GmWRKY142 directly activated ATCDT1, GmCDT1-1, and GmCDT1-2 expression to enhance Cd tolerance. In summary, the GmWRKY142-GmCDT1-1/2 cascade module is potentially useful for the production of soybean with tolerance to Cd stress along with decreased Cd accumulation in their edible parts.
Materials and Methods
Plant Materials and Growth Conditions
The soybean cultivar “Huaxia 7” was used to perform RNA sequencing (RNA-seq) analyses, mRNA expression and phenotype profiling. Plump seeds of a similar size were surface sterilized in 75% alcohol followed by germination for 3 days in sterile vermiculite. Seedlings of a similar size were transplanted to modified 1/2 Hoagland solution (pH 5.8) containing the following macronutrients KNO3 (2.5 mM), Ca(NO3)2⋅4H2O (2.5 mM), MgSO4⋅7H2O (3.5 mM), and KH2PO4 (0.5 mM), and the micronutrients Fe-EDTA (50 μM), H3BO3 (7.5 μM), (NH4)6Mo7O24 (2.5 μM), MnCl2 (1.25 μM), ZnSO4 (1 μM), and CuSO4 (0.5 μM) (Hoagland and Arnon, 1950; Dang et al., 2019) with or without Cd, in an illuminated growth incubator (Model GXZ-300D; Ningbo, China) under a 16 h light/8 h dark photoperiod, for subsequent RNA-seq, gene cloning, and transcriptional expression experiments. For the Cd dose-response experiment, seedlings were subjected to modified 1/2 Hoagland solution containing 0, 5, 10, 15, 25, or 50 μM of CdCl2 for 2 h. During the time-course experiment, seedlings were subjected to modified 1/2 Hoagland solution with 25 μM CdCl2 for 0, 1, 2, 4, 6, 8, 12, or 24 h. Seedlings cultured in modified 1/2 Hoagland solution with 0 or 25 μM of CdCl2 for 4 h were sampled and subjected to RNA-seq analysis (LC-Bio, Hangzhou, China), according to the vendor’s protocol. All the experiments were performed with three independent biological replicates.
All A. thaliana seeds including the wild-type (WT) ecotype Col-0, and transgenic plants were surface-sterilized and transferred to sterilized matrix soil (Jiffy, Oslo, Norway). Following vernalization in the dark at 4°C for 3 days, the seeds were cultivated in an illuminated growth incubator (Model GXZ-300D, Ningbo, China) under a 16 h light / 8 h dark photoperiod at 24°C.
RNA-Seq and Bioinformatics Analysis
RNA-seq analysis was performed by LC-Bio company (Hangzhou, China). For the transcriptomic analysis of Cd stress in soybean, 3-day old seedlings were cultured in modified 1/2 Hoagland solution with 0 or 25 μM of CdCl2 for 4 h were sampled and subjected to RNA-seq analysis. For the Arabidopsis thaliana, 2-week old seedlings of both the WT (Col-0) and the GmWRKY142-OX line (Line 6) under non-stress conditions were used for transcriptomic analysis. Following the manufacturer’s procedure, total RNA was extracted from the samples using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, United States, STRN10-1KT) and mRNA was enriched and fragmented into shorter fragments by mixed with fragmentation buffer. First-strand cDNA was synthesized from fragmented mRNA using random hexamer primer. End repair of the double-stranded cDNAs was performed using T4 polynucleotide kinase, T4 DNA polymerase and DNA polymerase I Klenow fragment. Then, T4 DNA ligase was used to ligate the fragments to adapters. The available fragments were selected and then enriched by PCR amplification. The constructed libraries were qualified and quantified using an Agilent 2100 Bioanaylzer and the ABI StepOnePlus Real-Time PCR System and then sequenced via Illumina NovaseqTM 6000. The obtained raw reads were then cleaned by removing the low-quality reads and/or adaptor sequences. The clean reads were mapped to reference sequences using SOAPaligner/SOAP2 (Li et al., 2009). The reference genomes and genes set of Soybean and Arabidopsis thaliana were downloaded from the NCBI (National Center for Biotechnology Information) site1, 2. The gene expression levels were calculated using the reads per kilobase per million reads method according to (Mortazavi et al., 2008). Subsequently, based on sequence homology, the differentially expressed genes by gene ontology terms3 were imported into Blast2GO, a software package that retrieves GO terms, allowing gene functions to be determined and compared. To further understand the biological pathways in which the differentially expressed genes are involved, the differentially expressed genes were compared against the KEGG database4. Based on reports of the soybean WRKY gene family from previous studies and differentially expressed genes annotation in this study, probable WRKY transcription factors in soybean were identified and named according to (Yu et al., 2016) (Supplementary Table S3). The sequences for both the soybean and Arabidopsis thaliana used in the present study can be downloaded from the Phytozome database (Version 12)5 according the gene numbers.
RNA Isolation and qRT-PCR Analysis
Total RNA was isolated from samples using the Plant Total RNA Purification Kit (TR02-150, GeneMarkbio, Taiwan, China). The first-strand cDNA was reverse transcribed by PrimeScriptTM RT reagent Kit with a gDNA Eraser kit (RR047, Takara Bio, Shiga, Japan). qRT-PCR analysis was carried out using TB GreenTM Premix Ex TaqTM II (RR820, Takara Bio) and CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). In all experiments, qRT-PCR analyses were performed as triplicates on three different RNA samples isolated independently from each tested condition. Relative gene expression levels were detected using the 2–△ △ Ct algorithm (Livak and Schmittgen, 2001) normalized to the expression of the Actin genes GmACT3 (GenBank: AK285830.1) or AtActin2 (GenBank: AT3g18780). Three independent biological replicates were tested in each experiment for the qRT-PCR analysis. Primers used are listed in Supplementary Table S1.
DNA and cDNA Clones, Vector Construction, and Plant Transformation
The cDNA sequences of GmWRKY142, GmCDT1-1 and GmCDT1-2 genes were isolated using specific primers (Supplementary Table S1) and inserted into the Xba I and Sac I sites of the pTF101.1 vector, using the ClonExpress® II One Step Cloning Kit (C112, Vazyme, Nanjing, China). The resulting plasmid was mobilized into Agrobacterium strains GV3101 and K599 by heat shock, and subsequently used to transform Arabidopsis and soybean hairy roots according to the Agrobacterium-mediated floral dip (Clough and Bent, 1998) and Agrobacterium rhizogenes-based transformation (Matthews and Youssef, 2016) methods, respectively. The complete coding sequence of GmWRKY142 was cloned into the pCambia 1302, pGBKT7, pGADT7, and pGreenII 62-SK binary vectors using the ClonExpress® II One Step Cloning Kit (C112, Vazyme) for the determination of subcellular localization, transcriptional activation activity assay, Yeast one-hybrid assay and Dual LUC assay, respectively. The genomic DNA of Huaxia 7 and WT plants were extracted using the Plant Genomic DNA Kit (DP305, TIANGEN, Beijing, China). Upstream sequences spanning 2,000 bp from the start codons of the ATCDT1, GmCDT1-1, and GmCDT1-2 genes were analyzed to determine the W-box elements. The original ATCDT1, GmCDT1-1, and GmCDT1-2 promoter fragments containing a genuine W-box element were amplified by PCR, while mutated W-box elements were identified by a change in the W-box sequence into “TAAAAT.” Either original or mutated fragments were cloned into the pAbAi vector using the ClonExpress® II One Step Cloning Kit (C112, Vazyme) and used as baits in yeast one-hybrid assays. Details of primers used in vector construction are listed in Supplementary Table S1.
Subcellular Localization
Subcellular localization was investigated by transiently overexpressing 35S:GmWRKY142-GFP in tobacco (Nicotiana tabacum) leaves via Agrobacterium-mediated transformation (Sparkes et al., 2006). Nuclear dye (DAPI) and GFP fluorescence signals were observed using confocal scanning microscopy (Model LSM780, Zeiss, Jena, Germany).
Yeast One-Hybrid Assay
This assay was performed using the MATCHMAKER® Gold Yeast One-Hybrid Library Screening System (Clontech) and the YEASTMAKERTM Yeast Transformation System 2 (Clontech). The full-length coding sequence of GmWRKY142 was cloned from cDNA and inserted into the effector construct pGADT7. A DNA fragments that consisted of two W-box motifs (TATGCTTTA GCTGGAATTGACTTCACCAGGTTTGACCTTACAGGTAGG TAGTTGAGT) or their mutants (ATATGCTTTAGCTGGAA TAAAATTCACCAGGTTAAAACTTACAGGTAGGTAGTTGA GT) were synthesized and introduced into the upstream region of the mini-promoter of AurR (pAbAi), which were termed pAbAi-Wbox and pAbAi-mWbox, respectively. The promoter sequences of GmCDT1-1, GmCDT1-2, and AtCDT1 were amplified and introduced into the upstream region of the mini-promoter of AurR, respectively. Prey and reporter vectors were co-transformed into the yeast strain Y1H Gold. Cells were grown in SD/-Leu liquid media to an OD600 of 0.1 and diluted 10-fold with normal saline. For each dilution, a volume of 7 μL was spotted on SD/-Leu media plates containing either 0 or 150 ng mL–1 AbA in order to test the strength of the interaction. Plates were incubated for 3–4 days at 30°C.
Cd Tolerance Assay
Twenty-five-day-old plants grown in pots (length × width × height = 7.0 × 7.0 × 7.6 cm) were placed in trays and saturated either with 1/10 Hoagland solution (as control) or with 1/10 Hoagland solution containing 500 μM of CdCl2 solution for 10 days. Pots were then transferred to trays containing normal culture medium to promote plant recovery for 12–15 days, after which the plants’ fresh weight and height were measured. Cd content was also measured using an atomic absorption spectrometer (Model AA-6800, Shimadzu Corporation, Japan). The experiment was performed with three independent biological replicates.
Statistical Analysis
All data were analyzed using GraphPad Prism® 5 (Version 5.01, GraphPad Software, Inc., United States) for calculating mean and standard deviation. At least three biological replicates were included in the data, and all data were analyzed using ANOVA or Duncan’s test for the determination of the significant differences with SPSS 21 (IBM Corp, 2012).
Results
The GmWRKY Family of Genes Responds to Cd Stress in Soybean
To identify Cd-responsive WRKY family members in soybean, genome wide transcriptomic analysis was conducted with Huaxia 7 roots exposed to Cd stress. By comparing the transcriptome expression patterns between control and Cd treated groups, a total of 5,285 differentially expressed genes (DEGs) were identified in soybean plants under Cd stress (Supplementary Table S2). Based on reports from previous studies, the soybean genome contains at least 170 WRKY family members (Song et al., 2016; Yang et al., 2017). However, genome wide transcriptomic analysis revealed that only 29 genes were up- or down-regulated more than 1.5-fold in roots in response to Cd stress (Table 1). Of these, 26 GmWRKY genes in the roots were up-regulated, while 3 were down-regulated. Furthermore, qRT-PCR analyses of all 29 Cd-responsive WRKY genes confirmed their differential expression in response to Cd stress (Table 1). It should be noted that GmWRKY142 registered a higher fragments per kilobase of transcript per million mapped reads (FPKM) under normal conditions, as well as being strongly up-regulated by Cd stress, which prompted our interest in further research.
TABLE 1.
GmWRKY family genes respond to Cd stress.
| Gene ID of NCBI | Gene numbers | Gene name |
Fragments Per kilobase per million (FPKM) |
Fold change (FC) | Log2(FC) | qRT-PCR | |||||
| CK_1 | CK_2 | CK_3 | Cd_1 | Cd_2 | Cd_3 | ||||||
| 100781603 | Glyma.15G186300 | GmWRKY146 | 3.89 | 3.23 | 2.47 | 1.13 | 0.68 | 0.69 | 0.26 | –1.94 | -4.33 ± 0.23 |
| 100788213 | Glyma.09G129100 | GmWRKY95 | 1.27 | 1.29 | 0.91 | 0.61 | 0.10 | 0.22 | 0.27 | –1.90 | -4.89 ± 0.52 |
| 100783661 | Glyma.19G221700 | GmWRKY184 | 10.66 | 6.40 | 10.14 | 3.12 | 2.35 | 3.11 | 0.32 | –1.66 | -3.52 ± 0.34 |
| 100127370 | Glyma.01G224800 | GmWRKY7 | 2.42 | 0.97 | 0.90 | 3.14 | 5.27 | 4.05 | 2.90 | 1.54 | 2.35 ± 0.84 |
| 100127423 | Glyma.03G220100 | GmWRKY25 | 2.65 | 1.32 | 3.06 | 8.96 | 7.07 | 4.43 | 2.91 | 1.54 | 2.58 ± 0.09 |
| 100802124 | Glyma.15G110300 | GmWRKY142 | 33.19 | 30.90 | 27.40 | 76.42 | 104.34 | 92.06 | 2.98 | 1.58 | 4.02 ± 0.52 |
| 100127367 | Glyma.01G128100 | GmWRKY4 | 2.18 | 2.36 | 0.82 | 5.55 | 6.03 | 4.63 | 3.02 | 1.60 | 2.36 ± 0.07 |
| 100127421 | Glyma.11G163300 | GmWRKY114 | 11.68 | 10.37 | 13.61 | 31.62 | 46.88 | 34.56 | 3.17 | 1.66 | 3.67 ± 0.10 |
| 100170682 | Glyma.18G213200 | GmWRKY172 | 0.12 | 0.48 | 1.41 | 2.66 | 1.44 | 2.53 | 3.30 | 1.72 | 2.48 ± 0.12 |
| 100797998 | Glyma.12G097100 | GmWRKY115 | 0.93 | 0.67 | 0.12 | 1.93 | 2.32 | 1.74 | 3.48 | 1.80 | 3.47 ± 0.25 |
| 100127387 | Glyma.03G256700 | GmWRKY28 | 1.32 | 0.41 | 0.61 | 2.45 | 3.76 | 2.27 | 3.61 | 1.85 | 3.41 ± 0.33 |
| 100807966 | Glyma.07G023300 | GmWRKY68 | 3.93 | 2.48 | 5.31 | 12.84 | 13.75 | 15.90 | 3.62 | 1.86 | 4.08 ± 0.41 |
| 732588 | Glyma.15G003300 | GmWRKY141 | 3.02 | 3.59 | 4.41 | 11.78 | 16.75 | 15.86 | 4.03 | 2.01 | 3.59 ± 0.53 |
| 100795177 | Glyma.06G307700 | GmWRKY66 | 1.85 | 2.13 | 1.23 | 7.68 | 8.35 | 7.12 | 4.44 | 2.15 | 4.13 ± 0.22 |
| 102666898 | Glyma.03G042700 | GmWRKY21 | 2.34 | 2.68 | 1.48 | 8.34 | 11.75 | 11.64 | 4.88 | 2.29 | 4.53 ± 0.84 |
| 100127375 | Glyma.08G021900 | GmWRKY78 | 0.58 | 0.85 | 0.58 | 2.10 | 4.09 | 4.09 | 5.13 | 2.36 | 5.55 ± 1.19 |
| 100787050 | Glyma.05G127600 | GmWRKY44 | 0.29 | 0.36 | 0.26 | 1.82 | 1.36 | 1.82 | 5.47 | 2.45 | 5.58 ± 0.98 |
| 100170747 | Glyma.16G026400 | GmWRKY147 | 0.87 | 0.77 | 0.70 | 5.04 | 7.62 | 4.47 | 7.31 | 2.87 | 8.22 ± 1.14 |
| 100798500 | Glyma.04G061400 | GmWRKY31 | 0.12 | 0.20 | 0.15 | 1.74 | 1.39 | 1.01 | 8.89 | 3.15 | 8.47 ± 1.21 |
| 100790050 | Glyma.19G254800 | GmWRKY185 | 0.17 | 0.45 | 0.74 | 4.05 | 4.74 | 4.00 | 9.41 | 3.23 | 10.77 ± 1.49 |
| 100779533 | Glyma.18G092200 | GmWRKY168 | 0.27 | 0.77 | 0.38 | 4.28 | 5.13 | 4.97 | 10.13 | 3.34 | 12.36 ± 1.37 |
| 100816891 | Glyma.08G082400 | GmWRKY81 | 0.32 | 0.65 | 0.28 | 4.53 | 5.10 | 5.47 | 12.11 | 3.60 | 14.56 ± 2.56 |
| 100812027 | Glyma.06G125600 | GmWRKY56 | 0.21 | 0.48 | 0.09 | 3.03 | 3.24 | 3.82 | 12.94 | 3.69 | 15.21 ± 1.74 |
| 100127378 | Glyma.08G011300 | GmWRKY76 | 0.06 | 0.27 | 0 | 0.31 | 2.27 | 1.73 | 13.30 | 3.73 | 13.97 ± 1.55 |
| 100127373 | Glyma.06G061900 | GmWRKY53 | 0.02 | 0.21 | 0.22 | 2.05 | 2.66 | 1.68 | 13.97 | 3.80 | 15.20 ± 2.48 |
| 100776906 | Glyma.04G238300 | GmWRKY39 | 0.05 | 0.06 | 0.03 | 0.31 | 0.67 | 1.38 | 18.34 | 4.20 | 20.24 ± 3.0 |
| 102667019 | Glyma.01G222300 | GmWRKY6 | 0.05 | 0.11 | 0 | 0.36 | 2.24 | 1.54 | 26.11 | 4.71 | 27.55 ± 2.76 |
| 100798375 | Glyma.13G370100 | GmWRKY126 | 0.03 | 0.07 | 0.28 | 2.26 | 4.43 | 4.22 | 28.21 | 4.82 | 28.42 ± 2.45 |
| 732586 | Glyma.19G094100 | GmWRKY180 | 1.04 | 0.48 | 0.64 | 20.04 | 30.36 | 28.53 | 36.42 | 5.19 | 44.87 ± 5.64 |
Expression Patterns of GmWRKY142
To further characterize GmWRKY142, qRT-PCR was employed to analyze tissue samples and determine Cd-induced expression patterns. As shown in Figure 1A, GmWRKY142 expression was detected in all examined tissues, with higher expression levels in roots, pods, seeds and leaves than stems, apex and flowers. To examine the expression patterns of GmWRKY142 following Cd stress, 5-day-old soybean roots were exposed to different Cd concentrations for varying periods of time. After 2 h of treatment, GmWRKY142 expression was significantly up-regulated by 10–50 μM Cd, whereby the fold change increased with increasing concentration (Figure 1B). Cd-induced GmWRKY142 expression was also observed in a time-dependent manner, whereby treatment with 25 μM Cd resulted in the highest 5.6-fold expression to be reached at the 4th hour, followed by a decrease at the 6th hour (Figure 1C).
FIGURE 1.
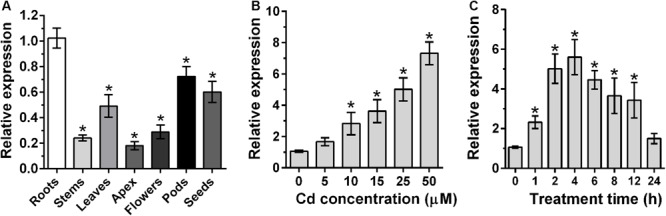
Analysis of expression patterns of GmWRKY142. (A) qRT-PCR analysis of the GmWRKY142 transcript in different tissues of the soybean variety Huaxia 7. mRNAs were isolated from roots, stems, leaves, apex, flowers, pods, and seeds. (B) Dose-dependent GmWRKY142 expression in roots. Roots were exposed to different Cd concentrations (0, 5, 10, 15, 25, and 50 μM) for 6 h. (C) For the time-course experiment, seedlings were exposed to 25 μM CdCl2 for 0, 1, 2, 4, 6, 8, 12, or 24 h. Samples were separately harvested for qRT-PCR analysis. Values are expressed as the means ± SD (n = 3). The experiment was performed with at least three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
Cloning and Characterization of GmWRKY142
The GmWRKY142 open reading frame (ORF) was isolated from the soybean variety Huaxia 7, based on putative sequence information available from the Phytozome database. The complete GmWRKY142 ORF (Accession number: MN639600) spanned 1,677 bp and encoded a 558 amino acid protein with an estimated molecular mass of 59.851 kDa and an isoelectric point of 6.81. Protein sequence alignment showed that GmWRKY142 shared homology with the corresponding genes of AtWRKY6 (51.7%), AtWRKY31 (51.0%), AtWRKY42 (55.8%), OsWRKY1 (48.8%), and OsWRKY43 (44.5%). In addition, GmWRKY142 and its homologous proteins contain a typical WRKY domain that includes the highly conserved amino acid sequence “WRKYGQK,” as well as a C2H2 zinc-finger motif (Figure 2A), indicating that GmWRKY142 is a member of the WRKY transcription factor family. Further analyses were performed to confirm the status of GmWRKY142 as a typical transcription factor. The subcellular localization of GmWRKY142 was investigated, whereby the full-length GmWRKY142 ORF without the stop codon was fused in-frame to the 5′ end of the green fluorescent protein (GFP), under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The GmWRKY142-GFP fusion protein was exclusively observed in the nucleus using confocal microscopy (Figure 2B). WRKY transcription factors are characterized by their common binding activity to the cis-element W-box (TTGACC/T) in promoters of downstream genes. A tandem DNA fragment consisting of two W-box motifs (TTGACT and TTGACC) was synthesized and used to determine the transcriptional activity of GmWRKY142. In the yeast one-hybrid (Y1H) assay, cotransformants carrying pGADT7-GmWRKY142 and the pAbAi-Wbox vectors could grow on SD/-Leu plates containing 150 ng/mL AbA. However, when the W-box sequences were mutated to TAAAAT or TAAAAT, the yeast cells could not grow, which is similar to the results of the blank vector (Figure 2C). Furthermore, a dual luciferase (LUC) assay involving N. benthamiana leaves was performed using the constructs shown in Figure 2D. Finally, the co-expression of Effecter-GmWRKY142 with Reporter-Wbox led to a higher LUC/REN ratio than that observed in the control and mutated reporter (Reporter-mWbox), indicating that GmWRKY142 may transcriptionally activate downstream target genes.
FIGURE 2.
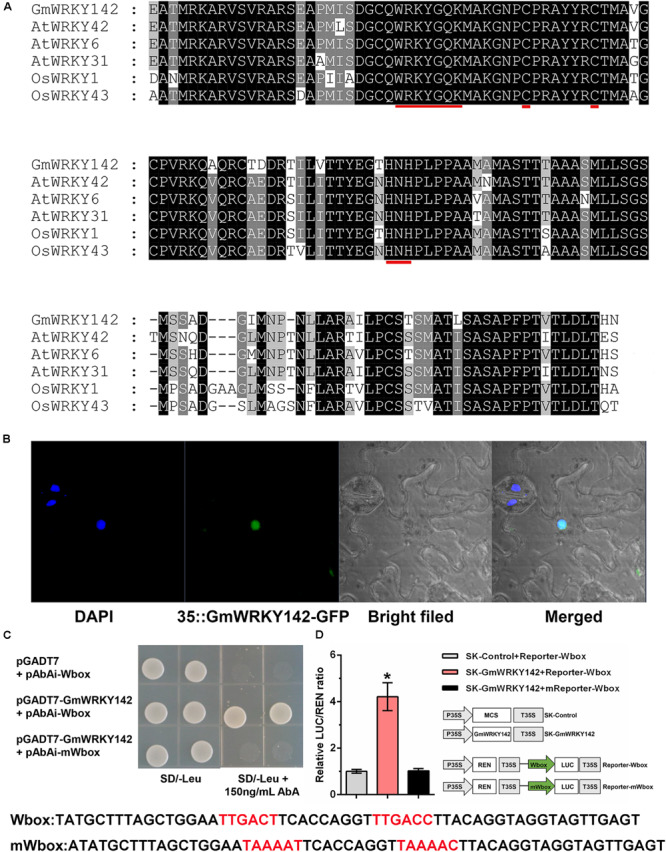
Sequence analysis, subcellular localization, and transcriptional activity assays of the GmWRKY142 protein. (A) Multiple alignments of the conserved WRKY domains between the WRKYs of soybean, rice and Arabidopsis. Identical amino acids are shown with a black background and analogous amino acids are shaded in gray. The WRKY motif and zinc-finger structures are denoted by red lines. (B) Subcellular localization of GmWRKY142 in Nicotiana benthamiana. DAPI was used to stain the nucleus. The overlapped images are shown on the right. (C) The growth phenotype of the cotransformant that harbored pGADT7-GmWRKY142 and bait vectors on a selective DO medium plate (SD/-Leu) with or without 150 ng mL–1 Aureobasidin A (AbA). (D) Schematic diagrams of the effector and reporter constructs used for transient luciferase (LUC) assays. Full-length CDS of GmWRKY142 was inserted into pGreen II 62-SK to produce an effector, while the original or mutated W-box elements were inserted into pGreen II 0800-LUC to generate reporters. MCS, multiple cloning site. P35S and T35S, the promoter and terminator of CaMV 35S, respectively. REN (Renilla luciferase) was used as an internal control for activity normalization. Transient expression assay of the promoter activity, shown as LUC/REN ratio, using tobacco leaves co-transformed with the effector and the reporters. LUC/REN ratio of the control co-transformed with the reporters and the empty effector vector (pGreen II 62-SK) was set as 1. Values are expressed as the means ± SD (n = 3). Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
Overexpression of GmWRKY142 in Transgenic A. thaliana and Soybean Hair Roots Confers Cd Tolerance
In order to determine the role of GmWRKY142 in Cd resistance, ectopic expression of GmWRKY142 was carried out in Arabidopsis. The seeds of three independent homozygous T3 transgenic lines exhibiting a higher gene expression, as well as WT plants were collected for functional gene analyses (Supplementary Figure S1). No differences in growth and development were observed between WT and GmWRKY142-OE plants under normal growth conditions. However, under Cd stress, GmWRKY142-OE plants displayed enhanced Cd tolerance compared with WT plants (Figure 3A). Leaf chlorosis of leaves and growth inhibition are two typical symptoms of Cd stress in plants (DalCorso et al., 2008). As shown in Figure 3A, the degree of chlorosis in leaves at 10 days of exposure to Cd stress was higher in WT than in GmWRKY142-OE plants. Further evaluations revealed that the plant height and fresh weights of the GmWRKY142-OE plants were significantly higher than those of WT (Figures 3B,C), and that Cd content was higher in WT compared to GmWRKY142-OE plants (Figures 3D,E).
FIGURE 3.
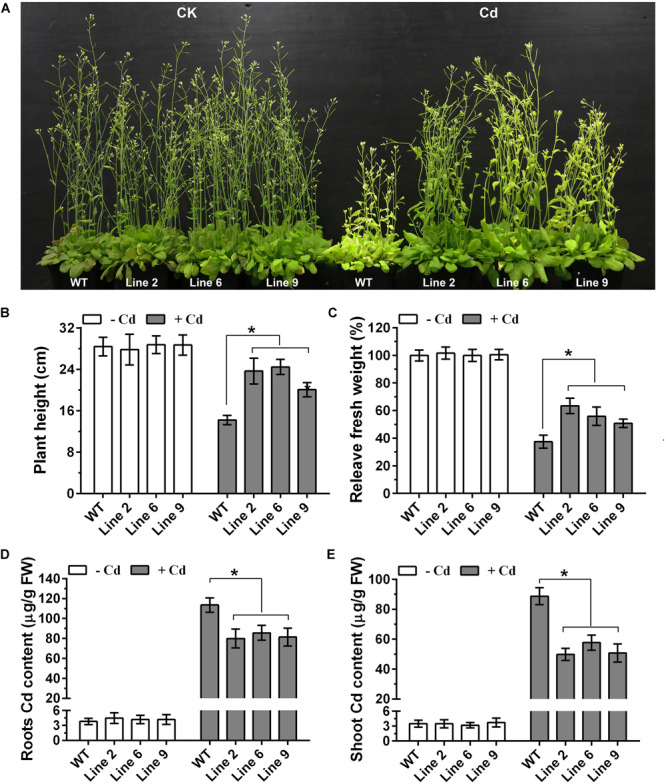
Overexpression of GmWRKY142 conferred enhanced Cd tolerance in transgenic Arabidopsis thaliana. (A) Phenotypes of A. thaliana transgenic lines (2, 6, and 9) and wild type (WT) under normal conditions or under Cd treatment conditions. (B–E) Plant height (B), relative fresh weight (C), root (D), and shoot (E) Cd content of the WT and transgenic lines. The experiment was performed with three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
To assess the effect of GmWRKY142 overexpression on Cd tolerance in soybean, transgenic hairy roots were produced using the A. rhizogenes-mediated transformation system. Analysis revealed that the transcripts for GmWRKY142 were up-regulated in GmWRKY142-OX hairy roots at an average fold of 5.63 (Supplementary Figure S2). WT and transgenic soybean hairy roots, each 2 cm long, were subsequently treated with 0 or 15 μM Cd for 5 days. As shown in Figure 4, no apparent difference was found between the GmWRKY142 transgenic and WT hairy roots in the absence of Cd. However, obvious differences were observed between transgenic and control hairy roots treated with 15 μM Cd. Relative root elongation was 45.18% in the WT, compared to 75.15% in transgenic GmWRKY142 hairy roots (Figure 4B). Moreover, Cd content in the transgenic GmWRKY142 hairy roots was significantly lower than in WT roots after Cd treatment, consistent with the phenotypes of the transgenic A. thaliana assay (Figure 3). These combined results suggest that GmWRKY142 overexpression in Arabidopsis and soybean hairy roots can enhance Cd tolerance.
FIGURE 4.
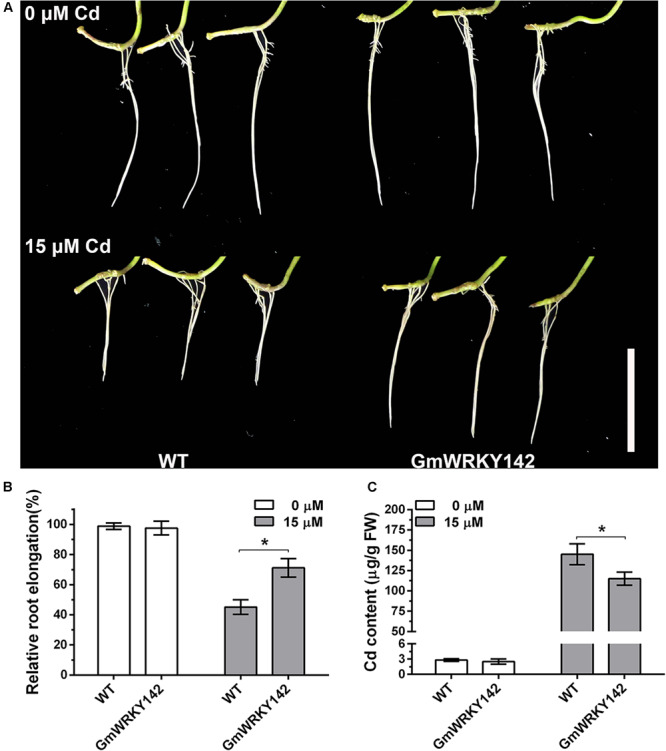
Overexpression of GmWRKY142 conferred enhanced Cd tolerance in transgenic soybean hairy roots. (A) Phenotypes of GmWRKY142 transgenic lines and wild type (WT) under normal conditions or under Cd treatment conditions. Relative root elongation (B) and Cd content (C) of the WT and transgenic lines were measured after 15 μM Cd treatment for 5 days. The experiment was performed with three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
GmWRKY142 Overexpression Leads to Extensive Transcriptional Reprogramming of Stress-Responsive Genes
RNA-seq was performed on an OX line (Line 6) and WT plants to determine how Cd resistance in Arabidopsis was mediated by GmWRKY142 and also to identify potential target genes that it may regulate. The difference in expression levels between two samples was determined as log2 Fold change >1.0 and false discovery rate <0.05. A total of 746 genes showed altered transcript levels (385 up-regulated and 239 down-regulated) in the transgenic line, compared with the WT (Figure 5A and Supplementary Table S4). Data obtained from RNA-seq were confirmed by analysis, using qRT-PCR, of the transcription of eight upregulated genes which were classified into either detoxification of Cd ion (GO:0071585) or response to Cd ion (GO:0046686) genes. As shown in Figure 5B, qRT-PCR analyses on expression patterns for all of the tested genes were highly consistent with the RNA-Seq, suggesting that DEG screening based on RNA-seq is reliable.
FIGURE 5.
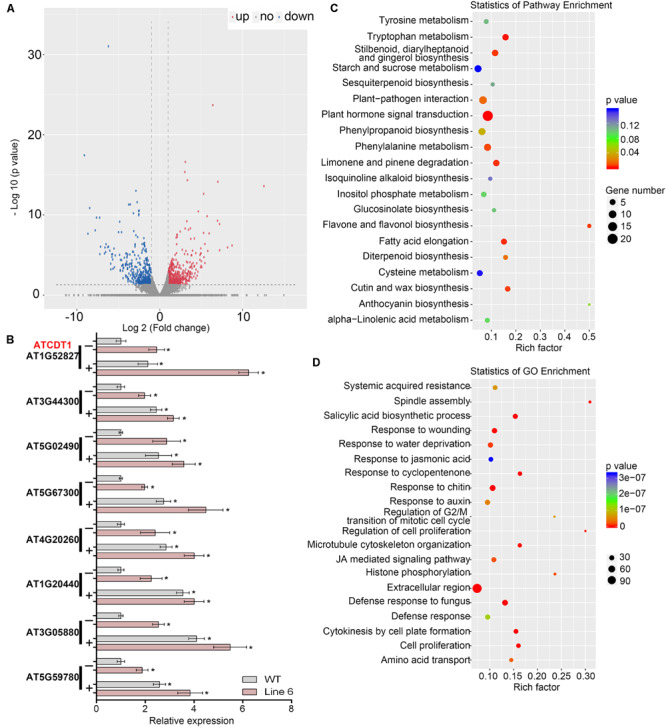
Overexpression of GmWRKY142 caused global transcriptional reprogramming in transgenic Arabidopsis thaliana. (A) Scatterplots of gene expression patterns in the transgenic line compared with the WT. Red represents up-regulated genes, while blue represents down-regulated ones. (B) Validation of the expression of eight selected Cd tolerance related genes by qRT-PCR. (C) KEGG pathways enriched among differentially expressed genes (DEGs). (D) GO analysis of DEGs. Three independent biological replicates were tested in qRT-PCR assay. Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
Analysis through the Kyoto Encyclopedia of Genes and Genomes (KEGG) suggested that 68 pathways were identified in DEGs (Supplementary Table S5). The top 11 most enriched pathways with p < 0.05 were plant hormone signal transduction, tryptophan metabolism, fatty acid elongation, limonene and pinene degradation, stilbenoid, diarylheptanoid and gingerol biosynthesis, flavone and flavonol biosynthesis, cutin, suberin, and wax biosynthesis, phenylalanine metabolism, plant-pathogen interaction, diterpenoid biosynthesis, and, finally, phenylpropanoid biosynthesis (Figure 5C). Gene ontology (GO) analysis showed that 237 GO were significantly enriched in the DEGs (Supplementary Table S6) with the top 20 most enriched GO shown in Figure 5D. Further analyses identified 255 up-regulated DEGs containing W-box in their promoters (Supplementary Table S7), which may be direct downstream genes. We noticed that a group of downstream genes that have been shown to play direct roles in stress tolerance were up-regulated in the transgenic line, including mental stress induced proteins, auxin-responsive proteins, cell wall structural proteins and an array of transcription factors that are known as positive regulators of stress response. It should be noted that 13 genes belonging to GO:0071585 (detoxification of Cd ion) and GO:0046686 (response to Cadmium ion) were identified, with AT1G52827 (Cadmium Tolerance 1, ATCDT1) being the only gene belonging to GO:0071585 (Supplementary Table S8). By homology search in the PANTHER (Protein Analysis Through Evolutionary Relationships) classification system6 using the PTHR35470 (PANTHER ID) of plant Cadmium Tolerance 1, two homologous genes in the soybean genome were identified on chromosomes 13 (Glyma.13G361300) and 15 (Glyma.15G012600), hereafter denoted as GmCDT1-1 and GmCDT1-2. Compared with expression levels in control hairy root (CK), the transcription levels of both GmCDT1-1 and GmCDT1-2 were significantly increased in the GmWRKY142-overexpressed hairy roots (Supplementary Figure S3). These results suggest that GmWKY142 activates ATCDT1, GmCDT1-1, and GmCDT1-2 expression in A. thaliana or soybean. Further work investigated whether ATCDT1 and its homologous genes GmCDT1-1 and GmCDT1-2 were direct target genes of GmWRKY142.
GmWRKY142 Directly Binds to and Activates the Promoter of ATCDT1, GmCDT1-1 and GmCDT1-2
Previous studies have demonstrated that ATCDT1 and its homologous genes (DcCDT1 and OsCDT1) confer Cd tolerance to yeast or A. thaliana (Song et al., 2004; Kuramata et al., 2009; Matsuda et al., 2009). The amino acid sequences of GmCDT1-1 and GmCDT1-2 peptides share a high level of identity with ATCDT1, DcCDT1, and OsCDT1, and all contain 10 conserved Cys residues clustered in their carboxy-distal regions (Figure 6A). Protein sequence alignment showed that GmCDT1-1 and GmCDT1-2 shared a high amino acid sequence similarity of up to 94.6%. Two soybean CDT1 genes shared homology with the corresponding genes of AtCDT1 (58%), DcCDT1 (64%), and OsCDT1 (67%). A phylogenetic tree constructed based on the GmCDT1-1 and GmCDT1-2 and using a total of 17 CDT1 genes from different plant species shows that they could be classified into two major groups (Figure 6B). From phylogenetic analysis, GmCDT1-1 and GmCDT1-2 are found to be closest to DcCDT1, BsCDT1, ZmCDT1, OsCDTs, HvCDTs, and MsCDT1, whereas AtCDT1 forms another clade with PsCDT1 and PtCDT1 (Figure 6B). In addition, GmCDT1-1 and GmCDT1-2 expression levels were induced by Cd (Figure 6C), suggesting that these two genes may play a role in Cd tolerance.
FIGURE 6.
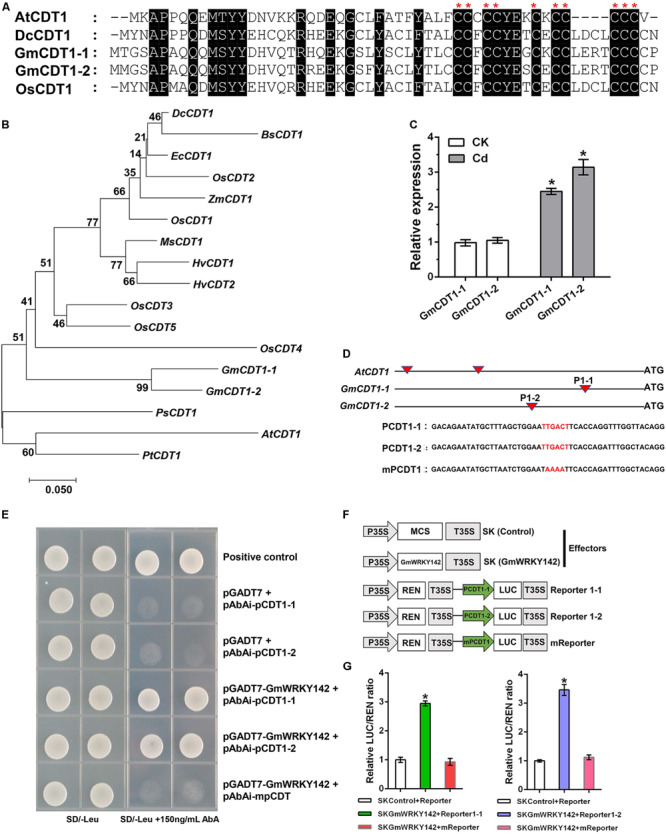
GmWRKY142 was bound to and activated the promoters of the GmCDT1-like genes. (A) Multiple alignments of the conserved CDT domains. Identical amino acids are shown with a black background. (B) Phylogenetic relationships between GmCDT1s and related proteins. Accession numbers are: AtCDT1, NM_202281; BsCDT1, ABL85053; EcCDT1, AB426477; HvCDT1, AK251605; HvCDT2, AK249080; MsCDT1, AB426478; OsCDT1, AK121052; OsCDT2, AK061597; OsCDT3, AK062450; OsCDT4, AK059641; OsCDT5, AK099514; PsCDT1, EF081525; PtCDT1, CU225257; ZmCDT1, AY103859. (C) GmCDT1-1 and GmCDT1-2 were induced by Cd. The Huaxia 7 roots cultured in 0 or 25 μM of CdCl2 for 6 h were sampled and qRT-PCR assay was performed. (D) Schematic diagrams of ATCDT1, GmCDT1-1, and GmCDT1-2 promoters and partial sequences containing W-box used in yeast one-hybrid assays. (E) Culture of yeast cells co-transformed with the prey and bait, as well as the positive control (pGADT7-Rec-p53+p53-AbAi) on selective medium with or without 150 ng mL–1 Aureobasidin A (AbA). (F) Schematic diagrams of the effector and reporter constructs used for transient luciferase (LUC) assays. Full-length CDS of GmWRKY142 was inserted into pGreen II 62-SK to produce an effector, while the original or mutated W-box elements were inserted into pGreen II 0800-LUC to generate reporters. MCS, multiple cloning site. P35S and T35S, the promoter and terminator of CaMV 35S, respectively. REN (Renilla luciferase) was used as an internal control for activity normalization. (G) Transient expression assay of the promoter activity, shown as LUC/REN ratio, using tobacco leaves co-transformed with the effector and the reporters. LUC/REN ratio of the control co-transformed with the reporters and the empty effector vector (pGreen II 62-SK) was set as 1. Values are expressed as the means ± SD (n = 3). The experiment was performed with at least three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
Most WRKY transcription factors regulate their target stress-related genes via binding to the W-box of promoters (Ülker and Somssich, 2004; Jiang et al., 2014; Sheng et al., 2019). Isolation of the promoter sequence spanning 2,000 bp upstream of the first ATG of ATCDT1, GmCDT1-1, and GmCDT1-2 identified 2, 1 and 1 W-boxes, respectively (Figure 6D). To test whether GmWRKY142 can directly bind to the identified W-box, yeast one-hybrid (Y1H) was performed using GmWRKY142 as prey, and promoter sequence fragments containing either original or mutated W-box elements as baits (Figure 6C and Supplementary Figure S4). Results showed that GmWRKY142 could interact with selected promoter sequence fragments that contained the original W-box. However, yeast cells co-transformed with the prey and mutated W-box bait or pGADT7 and original W-box elements did not exhibit normal growth (Figure 6E and Supplementary Figure S4), suggesting the ability of GmWRKY142 to bind to the W-box element in the ATCDT1, GmCDT1-1 and GmCDT1-2 promoters. To determine whether the GmWRKY142 transcription factor could activate GmCDT1-1 and GmCDT1-2 transcription, a dual luciferase (LUC) assay was performed in N. benthamiana leaves (Figure 6F). As shown in Figures 6G, the co-expression of Effecter-GmWRKY142 with Reporter1-1 and Reporter1-2 led to a higher LUC/REN ratio than that observed in the control and mutated reporter (mReporter), indicating that GmWRKY142 is a transcriptional activator of GmCDT1-1 and GmCDT1-2.
Overexpression of GmCDT1-1 and GmCDT1-2 in Transgenic A. thaliana Confer Cd Tolerance
The results described above strongly support that GmCDT1-1 and GmCDT1-2 are direct targets of GmWRKY142 and that they play a functional role in Cd tolerance. To explore the function of GmCDT1-1 and GmCDT1-2 during Cd stress, transgenic Arabidopsis and WT plants were subjected to treatment with CdCl2. The results showed that both GmCDT1-1 OX lines (OX 2 and 5) and GmCDT1-2 OX lines (OX D and H) exhibited a higher biomass accumulation than the WT (Figures 7A,B). The average fresh weight and plant height of OX lines were higher than those of the WT (Figures 7C,D). Analysis of the Cd content in the WT and OX plants under Cd stress revealed that higher Cd levels were detected in the root and shoot of WT plants compared to OX plants (Figures 7E,F). These results suggest that GmCDT1-1 and GmCDT1-2 overexpression in Arabidopsis can enhance Cd tolerance by limiting Cd uptake.
FIGURE 7.
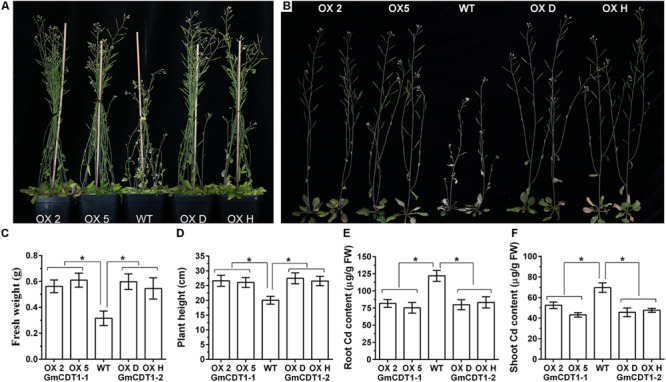
The overexpression Arabidopsis thaliana lines of GmCDT1-1 or GmCDT1-2 showed tolerance to Cd stress. (A,B) Phenotypes of A. thaliana transgenic lines and wild type (WT) after Cd treatment. (C–F) Fresh weight (C), plant height (D), root (E), and shoot (F) Cd content of the WT and transgenic lines. The experiment was performed with at least three independent biological replicates. Significant differences according to the one-way analysis of variance are denoted as follows: *p < 0.01.
Discussion
Heavy metal stresses such as high concentration of Cd, Cu and As are among the major environmental factors which not only limit the productivity and growth potential of plants, but also cause serious problems for human health (Gallego et al., 1996; der Maur et al., 1999; Matsuda et al., 2009). Fortunately, a number of strategies have been employed in plants to resist heavy metal stress, including uptake, accumulation, translocation, and detoxification of the metal. Various transcription factor-mediated metal stress sensing and coping strategies have been reported in plants (Liu et al., 2017; Zhu et al., 2018). The WRKY transcription factor family is one of the most important regulator families reported to be involved in a variety of heavy metal stresses. For example, several studies have revealed the differential transcript expression of WRKY transcription factors under Cd stress in different plant species (Yang et al., 2016; Hong et al., 2017; Cheng et al., 2018; Han et al., 2019; Sheng et al., 2019). An increasing number of WRKYs that were identified to confer Cd tolerance in plants also exemplify the importance of heavy metal tolerance in plants. To date, no report on the role of soybean WRKYs in Cd detoxification and tolerance has been published. In the present study, 29 GmWRKYs were detected as Cd-responsive DEGs using genome wide transcriptomic analysis in roots (Table 1). Of these, 26 GmWRKYs were up-regulated and 3 were down-regulated, suggesting that functional differentiation may exist. We further demonstrated that GmWRKY142 is involved in Cd resistance and provided several lines of evidence supporting this observation. Firstly, GmWRKY142 is a nucleic protein with transcriptional activation activity and its expression was rapidly induced by Cd in a dose-dependent manner (Figures 1, 2). Secondly, GmWRKY142 overexpression in Arabidopsis and soybean hair roots resulted in improved Cd resistance, indicating that GmWRKY142 is a positive regulator of Cd tolerance (Figures 3, 4). Thirdly, GmWRKY142 overexpression lead to the extensive transcriptional reprogramming of stress-responsive genes. We noticed that several downstream genes that have been shown to play direct roles in stress tolerance were prominently influenced by GmWRKY142 overexpression including crucial metabolic regulators, mental stress induced proteins, auxin-responsive proteins, cell wall structural proteins and an array of transcription factors. One of them, the salicylic acid biosynthetic
process (GO:0009697), which included 22 up-regulated and 9 down-regulated genes, had previously been found to be significantly associated with Cd tolerance in plants (Belkhadi et al., 2010; Chao et al., 2010; Szalai et al., 2013), and was enriched in the transgenic line (Figure 4 and Supplementary Table S9). Finally, the present study demonstrated that the GmWRKY142-AT/GmCDT1 cascade module confers Cd tolerance (Figures 5, 6). These findings suggest that GmWRKYs act as novel participants in the response of soybean to Cd stress, although further studies will be required to support this conclusion.
WRKYs can activate or repress the transcription of stress-related and co-regulated genes by directly recognizing and binding to the W-box element within the promoters of target genes (Eulgem et al., 2000; Ülker and Somssich, 2004; Han et al., 2019). In the present study, we demonstrated that overexpression of GmWRKY142 in transgenic A. thaliana or soybean hairy roots lead to enhanced tolerance to Cd and activated the expression of the ATCDT1 or GmCDT1-like genes (GmCDT1-1 and GmCDT1-2) encoding Cd-binding Cys-rich proteins (Figures 3, 5 and Supplementary Figure S3). The ATCDT1, GmCDT1-1, and GmCDT1-2 promoters were found to contain 2, 1, and 1 W-box elements, respectively (Figure 6C). We further demonstrated that GmWRKY142 activated the ATCDT1, GmCDT1-1, and GmCDT1-2 promoters through interactions with the W-box element using the yeast one-hybrid or dual luciferase (LUC) assays (Figures 6D,E and Supplementary Figure S4). So far, a plethora of target genes have been characterized for various WRKY members. However, only a limited number of WRKY TFs and their target genes were identified to regulate Cd tolerance in plants. AtWRKY13 is induced by Cd stress and directly activates the expression of the Cd2+ extrusion pump gene AtPDR8; therefore, AtWRKY13 overexpression in transgenic A. thaliana leads to an enhanced tolerance to Cd (Sheng et al., 2019). AtWRKY12 as a negative regulator directly targets GSH1 and indirectly represses PC synthesis-related genes expression to negatively regulate Cd accumulation and tolerance in Arabidopsis (Han et al., 2019). These two studies show that WRKY proteins can act as transcriptional activators or repressors to regulate Cd accumulation and tolerance. Herein, the characterization of ATCDT1 and GmCDT1-like genes as direct targets of GmWRKY142 provides valuable clues to better understand the WRKY regulatory mechanisms associated with Cd stress. However, RNA-seq results indicated that ATCDT1 and GmCDT1-like genes were not the only identified target genes (Figure 5), thereby highlighting the importance of confirming the identity of the other target genes to better understand the molecular mechanism of GmWRKY142 in Cd tolerance.
In the previous studies, three members of the CDT1 family, namely, DcCDT1, OsCDT1 and ATCDT1, from Digitaria ciliaris, Oryza sativa and A. thaliana were isolated and characterized for Cd tolerance (Song et al., 2004; Kuramata et al., 2009; Matsuda et al., 2009). In contrast to other Cys-rich peptides, CDT1 peptides are only present in angiosperms and gymnosperms, and are relatively small in size with 49 to 60 amino acid residues (Song et al., 2004). In the present study, two homologous CDT1s were identified in soybean and their expression induced by Cd stress (Figure 6B). Consistent with DcCDT1, OsCDT1, and ATCDT1, overexpression of GmCDT1-1 or GmCDT1-2 conferred Cd-tolerance to transgenic A. thaliana plants by lowering Cd accumulation in plants (Figure 7). A conserved sequence with 10 Cys-rich peptides, CC-(C/F)-CCYE-X-C-(K/E)-CC-(LDCL/LERT/delete)-CCC-(V/C), in their C-termini was identified in this study. Plants have evolved a set of versatile adaptive mechanisms to cope with Cd stress. One such means is by reducing the cellular accumulation of Cd by either enhancing Cd efflux or suppressing its uptake. The Cys-rich membrane proteins in different plant species have been found to play an important role in plant Cd resistance by chelating Cd at the cellular surface and limiting further entry of Cd into the cells (Song et al., 2004; Kuramata et al., 2009; Matsuda et al., 2009; Pirzadeh and Shahpiri, 2016; Niederwanger et al., 2017). Here, we have demonstrated that expression of GmCDT1-1 or GmCDT1-2 conferred Cd tolerance to Arabidopsis plants by reducing their cellular Cd contents. GmCDT1-1 and GmCDT1-2 are Cys-rich peptides, so it is highly likely that they can directly bind Cd. Based on our current findings, therefore, the most plausible mechanism we envisage for GmCDT1s and AtCDT1 function is that these proteins chelate Cd at the cellular surface and prevent further entry of Cd into the cells. These findings suggest that CDT1s play an important role in Cd tolerance.
In summary, 29 Cd-responsive WRKY genes were identified and confirmed in soybean roots, of which 26 were up-regulated and 3 were down-regulated. Among these GmWRKYs, the up-regulated GmWRKY142, in particular, directly activated expression of the ATCDT1 and GmCDT1-like genes, which resulted in reduced Cd accumulation, thereby conferring Cd tolerance (Figure 8). The identification of 29 Cd-responsive GmWRKYs as well as the GmWRKY142-CDT1 cascade module provides the first step in determining that GmWRKYs may be critical factors in Cd tolerance. More GmWRKYs-targeted cascade modules conferring Cd tolerance need to be further studied.
FIGURE 8.
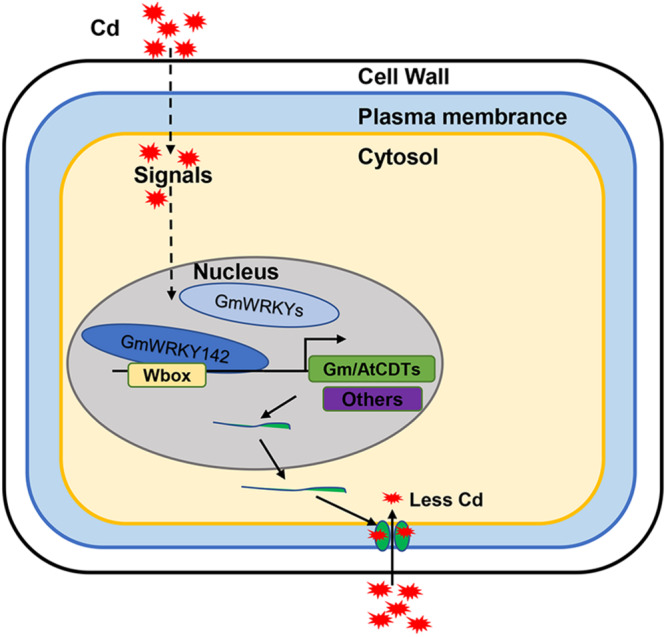
A working model for the role of GmWRKY142 in regulating Cd tolerance. Cd stress promotes GmWRKY142 expression and thereby directly activates the transcript of the GmCDT1-1 and GmCDT1-2 genes encoding Cys-rich peptides, which results in chelating Cd at the cellular surface and prevent further entry of Cd into the cells, and thus, conferring enhanced tolerance.
Data Availability Statement
The data sets supporting the results of this study are included in the article. The RNA-seq data have been deposited into the NCBI Short Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) under accession number PRJNA605520.
Author ContribUtions
ZC, PX, HW, and RL performed the experiments and data analyses. ZC and HN prepared the manuscript. HN planned, supervised, and financed this work. YC, TL, and QM reviewed and edited the manuscript. All authors have read and approved the final version of the manuscript to be published.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the Guangdong Subcenter of the National Center for Soybean Improvement for their daily help.
Funding. This work was supported by the National Key R&D Program of China (2018YFD0201006), the National Natural Science Foundation of China (31771816 and 31971965), and the Research Project of the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-b201804).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00724/full#supplementary-material
References
- Belkhadi A., Hediji H., Abbes Z., Nouairi I., Barhoumi Z., Zarrouk M., et al. (2010). Effects of exogenous salicylic acid pre-treatment on cadmium toxicity and leaf lipid content in Linum usitatissimum L. Ecotoxicol. Environ. Saf. 73 1004–1011. 10.1016/j.ecoenv.2010.03.009 [DOI] [PubMed] [Google Scholar]
- Cai S. Y., Zhang Y., Xu Y. P., Qi Z. Y., Li M. Q., Ahammed G. J., et al. (2017). HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal Res. 62:e12387. 10.1111/jpi.12387 [DOI] [PubMed] [Google Scholar]
- Chao Y.-Y., Chen C.-Y., Huang W.-D., Kao C. H. (2010). Salicylic acid-mediated hydrogen peroxide accumulation and protection against Cd toxicity in rice leaves. Plant Soil 329 327–337. [Google Scholar]
- Chen J., Yang L., Yan X., Liu Y., Wang R., Fan T., et al. (2016). Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiol. 171 707–719. 10.1104/pp.15.01882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. W., Jin J. F., Lou H. Q., Liu L., Kochian L. V., Yang J. L. (2018). LeSPL-CNR negatively regulates Cd acquisition through repressing nitrate reductase-mediated nitric oxide production in tomato. Planta 248 893–907. 10.1007/s00425-018-2949-z [DOI] [PubMed] [Google Scholar]
- Cheng D., Tan M., Yu H., Li L., Zhu D., Chen Y., et al. (2018). Comparative analysis of Cd-responsive maize and rice transcriptomes highlights Cd co-modulated orthologs. BMC Genomics 19:709. 10.1186/s12864-018-5109-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S., Aarts M. G., Thomine S., Verbruggen N. (2013). Plant science: the key to preventing slow cadmium poisoning. Trends Plant Sci. 18 92–99. 10.1016/j.tplants.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- DalCorso G., Farinati S., Maistri S., Furini A. (2008). How plants cope with cadmium: staking all on metabolism and gene expression. J. Integr. Plant Biol. 50 1268–1280. 10.1111/j.1744-7909.2008.00737.x [DOI] [PubMed] [Google Scholar]
- Dang F., Lin J., Chen Y., Li G. X., Guan D., Zheng S. J., et al. (2019). A feedback loop between CaWRKY41 and H2O2 coordinates the response to Ralstonia solanacearum and excess cadmium in pepper. J. Exp. Bot. 70 1581–1595. 10.1093/jxb/erz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P., Samantaray S., Rout G. (1997). Studies on cadmium toxicity in plants: a review. Environ. Pollut. 98 29–36. 10.1016/s0269-7491(97)00110-3 [DOI] [PubMed] [Google Scholar]
- der Maur A. A., Belser T., Elgar G., Georgiev O., Schaffner W. (1999). Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol. Chem. 380 175–185. 10.1515/BC.1999.026 [DOI] [PubMed] [Google Scholar]
- Dong J., Mao W., Zhang G., Wu F., Cai Y. (2007). Root excretion and plant tolerance to cadmium toxicity-a review. Plant Soil Environ. 53 193–200. [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 199–206. 10.1016/s1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10 366–371. 10.1016/j.pbi.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Gallego S. M., Benavides M. P., Tomaro M. L. (1996). Effect of heavy metal ion excess on sunflower leaves: evidence for involvement of oxidative stress. Plant Sci. 121 151–159. [Google Scholar]
- Gallego S. M., Pena L. B., Barcia R. A., Azpilicueta C. E., Iannone M. F., Rosales E. P., et al. (2012). Unravelling cadmium toxicity and tolerance in plants: insight into regulatory mechanisms. Environ. Exp. Bot. 83 33–46. 10.1016/j.jhazmat.2017.04.058 [DOI] [PubMed] [Google Scholar]
- Gill S. S., Tuteja N. (2011). Cadmium stress tolerance in crop plants: probing the role of sulfur. Plant Signal. Behav. 6 215–222. 10.4161/psb.6.2.14880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C., Buckley W., Bailey L., Selles F. (1998). Cadmium accumulation in crops. Can. J. Plant Sci. 78 1–17. [Google Scholar]
- Han Y., Fan T., Zhu X., Wu X., Ouyang J., Jiang L., et al. (2019). WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis. Plant Mol. Biol. 99 149–159. 10.1007/s11103-018-0809-7 [DOI] [PubMed] [Google Scholar]
- Hasan S. A., Fariduddin Q., Ali B., Hayat S., Ahmad A. (2009). Cadmium: toxicity and tolerance in plants. J. Environ. Biol. 30 165–174. [PubMed] [Google Scholar]
- Hoagland D. R., Arnon D. I. (1950). The water-culture method for growing plants without soil. Circular 347:32. [Google Scholar]
- Hong C., Cheng D., Zhang G., Zhu D., Chen Y., Tan M. (2017). The role of ZmWRKY4 in regulating maize antioxidant defense under cadmium stress. Biochem. Biophys. Res. Commun. 482 1504–1510. 10.1016/j.bbrc.2016.12.064 [DOI] [PubMed] [Google Scholar]
- Hu S., Yu Y., Chen Q., Mu G., Shen Z., Zheng L. (2017). OsMYB45 plays an important role in rice resistance to cadmium stress. Plant Sci. 264 1–8. 10.1016/j.plantsci.2017.08.002 [DOI] [PubMed] [Google Scholar]
- IBM Corp (2012). IBM SPSS Statistics for Windows, Version 21.0. Armonk, NY: IBM Corp. [Google Scholar]
- Ishiguro S., Nakamura K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and ß-amylase from sweet potato. Mol. Gen. Genet. 244 563–571. 10.1007/BF00282746 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid–and auxin-mediated signaling in jasmonic acid–induced leaf senescence. Plant Cell 26 230–245. 10.1105/tpc.113.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Pandey R., Siddiqi N. (2019). Oxidative stress biomarkers of cadmium toxicity in mammalian systems and their distinct ameliorative strategy. J. Appl. Biotechnol. Bioeng. 6 126–135. [Google Scholar]
- Kunihiro S., Saito T., Matsuda T., Inoue M., Kuramata M., Taguchi-Shiobara F., et al. (2013). Rice DEP1, encoding a highly cysteine-rich G protein γ subunit, confers cadmium tolerance on yeast cells and plants. J. Exp. Bot. 64 4517–4527. 10.1093/jxb/ert267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramata M., Masuya S., Takahashi Y., Kitagawa E., Inoue C., Ishikawa S., et al. (2009). Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant Cell Physiol. 50 106–117. 10.1093/pcp/pcn175 [DOI] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25 1966–1967. 10.1093/bioinformatics/btp336 [DOI] [PubMed] [Google Scholar]
- Lin T., Yang W., Lu W., Wang Y., Qi X. (2017). Transcription factors PvERF15 and PvMTF-1 form a cadmium stress transcriptional pathway. Plant Physiol. 173 1565–1573. 10.1104/pp.16.01729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. S., Feng S. J., Zhang B. Q., Wang M. Q., Cao H. W., Rono J. K., et al. (2019). OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 19:283. 10.1186/s12870-019-1899-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yu X., Feng Y., Zhang C., Wang C., Zeng J., et al. (2017). Physiological and transcriptome response to cadmium in cosmos (Cosmos bipinnatus Cav.) seedlings. Sci. Rep. 7:14691. 10.1038/s41598-017-14407-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Luo J.-S., Gu T., Yang Y., Zhang Z. (2019a). A non-secreted plant defensin AtPDF2.6 conferred cadmium tolerance via its chelation in Arabidopsis. Plant Mol. Biol. 100 561–569. 10.1007/s11103-019-00878-y [DOI] [PubMed] [Google Scholar]
- Luo J.-S., Yang Y., Gu T., Wu Z., Zhang Z. (2019b). The Arabidopsis defensin gene AtPDF2.5 mediates cadmium tolerance and accumulation. Plant Cell Environ. 42 2681–2695. 10.1111/pce.13592 [DOI] [PubMed] [Google Scholar]
- Margoshes M., Vallee B. L. (1957). A cadmium protein from equine kidney cortex. J. Am. Chem. Soc. 79 4813–4814. [Google Scholar]
- Matsuda T., Kuramata M., Takahashi Y., Kitagawa E., Youssefian S., Kusano T. (2009). A novel plant cysteine-rich peptide family conferring cadmium tolerance to yeast and plants. Plant Signal. Behav. 4 419–421. 10.4161/psb.4.5.8272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. F., Youssef R. M. (2016). Agrobacterium rhizogenes-based transformation of soybean roots to form composite plants. Bio Protoc. 6:e1708. [Google Scholar]
- Mekawy A. M. M., Assaha D. V. M., Ueda A. (2020). Constitutive overexpression of rice metallothionein-like gene OsMT-3a enhances growth and tolerance of Arabidopsis plants to a combination of various abiotic stresses. J. Plant Res. 133, 429–440. 10.1007/s10265-020-01187-y [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Moulis J.-M., Thévenod F. (2010). New Perspectives in Cadmium Toxicity: an Introduction. Berlin: Springer. [DOI] [PubMed] [Google Scholar]
- Nagajyoti P. C., Lee K. D., Sreekanth T. (2010). Heavy metals, occurrence and toxicity for plants: a review. Environ. Chem. Lett. 8 199–216. [Google Scholar]
- Niederwanger M., Dvorak M., Schnegg R., Pedrini-Martha V., Bacher K., Bidoli M., et al. (2017). Challenging the Metallothionein (MT) gene of Biomphalaria glabrata: unexpected response patterns due to cadmium exposure and temperature stress. Int. J. Mol. Sci. 18:1747. 10.3390/ijms18081747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okereafor U., Makhatha M., Mekuto L., Uche-Okereafor N., Sebola T., Mavumengwana V. (2020). Toxic metal implications on agricultural soils, plants, animals, aquatic life and human health. Int. J. Environ. Res. Public Health 17:2204. 10.3390/ijerph17072204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios Ò., Espart A., Espín J., Ding C., Thiele D. J., Atrian S., et al. (2014). Full characterization of the Cu-, Zn-, and Cd-binding properties of CnMT1 and CnMT2, two metallothioneins of the pathogenic fungus Cryptococcus neoformans acting as virulence factors. Metallomics 6 279–291. 10.1039/c3mt00266g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzadeh S., Shahpiri A. (2016). Functional characterization of a type 2 metallothionein isoform (OsMTI-2b) from rice. Int. J. Biol. Macromol. 88 491–496. 10.1016/j.ijbiomac.2016.04.021 [DOI] [PubMed] [Google Scholar]
- Rizwan M., Ali S., Adrees M., Ibrahim M., Tsang D. C., Zia-ur-Rehman M., et al. (2017). A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 182 90–105. 10.1016/j.chemosphere.2017.05.013 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Macdonald H., Huttly A. K., Lazarus C. M., Hooley R. (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 29 691–702. 10.1007/BF00041160 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Torres J. T., Parniske M., Wernert P., Hahlbrock K., Somssich I. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Yamaji N., Ma J. F. (2014). Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 65 6013–6021. 10.1093/jxb/eru340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanying H., Xiaoe Y., Zhenli H., Baligar V. C. (2017). Morphological and physiological responses of plants to cadmium toxicity: a review. Pedosphere 27 421–438. [Google Scholar]
- Sheng Y., Yan X., Huang Y., Han Y., Zhang C., Ren Y., et al. (2019). The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 42 891–903. 10.1111/pce.13457 [DOI] [PubMed] [Google Scholar]
- Smirnova I. V., Bittel D. C., Ravindra R., Jiang H., Andrews G. K. (2000). Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J. Biol. Chem. 275 9377–9384. 10.1074/jbc.275.13.9377 [DOI] [PubMed] [Google Scholar]
- Solis W. A., Childs N. L., Weedon M. N., He L., Nebert D. W., Dalton T. P. (2002). Retrovirally expressed metal response element-binding transcription factor-1 normalizes metallothionein-1 gene expression and protects cells against zinc, but not cadmium, toxicity. Toxicol. Appl. Pharmacol. 178 93–101. 10.1006/taap.2001.9319 [DOI] [PubMed] [Google Scholar]
- Song H., Wang P., Hou L., Zhao S., Zhao C., Xia H., et al. (2016). Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 7:9. 10.3389/fpls.2016.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.-Y., Martinoia E., Lee J., Kim D., Kim D.-Y., Vogt E., et al. (2004). A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 135 1027–1039. 10.1104/pp.103.037739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1 2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Szalai G., Krantev A., Yordanova R., Popova L. P., Janda T. (2013). Influence of salicylic acid on phytochelatin synthesis in Zea mays during Cd stress. Turk. J. Bot. 37 708–714. 10.1371/journal.pone.0160157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Mao B. G., Li Y. K., Lv Q. M., Zhang L. P., Chen C. Y., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7:14438. 10.1038/s41598-017-14832-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Charles T. M., Newton R. J. (2005). Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 59 603–617. 10.1007/s11103-005-0451-z [DOI] [PubMed] [Google Scholar]
- Ülker B., Somssich I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7 491–498. 10.1016/j.pbi.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Wu H., Chen C., Du J., Liu H., Cui Y., Zhang Y., et al. (2012). Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 158 790–800. 10.1104/pp.111.190983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Chen J., Yue X., Wei X., Zou J., Chen Y., et al. (2019). The zinc-regulated protein (ZIP) family genes and glutathione s-transferase (GST) family genes play roles in Cd resistance and accumulation of pak choi (Brassica campestris ssp. chinensis). Ecotoxicol. Environ. Saf. 183:109571. 10.1016/j.ecoenv.2019.109571 [DOI] [PubMed] [Google Scholar]
- Xu H., Watanabe K. A., Zhang L., Shen Q. J. (2016). WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 23 311–323. 10.1093/dnares/dsw025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Ge Y., Zhang W., Zhao Y., Yang G. (2018). The walnut JrVHAG1 gene is involved in cadmium stress response through ABA-signal pathway and MYB transcription regulation. BMC Plant Biol. 18:19. 10.1186/s12870-018-1231-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Wang C., Wang Y., Guo Y., Zhao Y., Yang C., et al. (2016). Overexpression of ThVHAc1 and its potential upstream regulator, ThWRKY7, improved plant tolerance of Cadmium stress. Sci. Rep. 6:18752. 10.1038/srep18752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang Y., Liu G., Yang C., Li C. (2011). Tamarix hispida metallothionein-like ThMT3, a reactive oxygen species scavenger, increases tolerance against Cd 2+, Zn 2+, Cu 2+, and NaCl in transgenic yeast. Mol. Biol. Rep. 38 1567–1574. 10.1007/s11033-010-0265-1 [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhou Y., Chi Y., Fan B., Chen Z. (2017). Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 7:17804. 10.1038/s41598-017-18235-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Cai Y., Yu D., Liang G. (2018). bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 60 691–702. 10.1111/jipb.12658 [DOI] [PubMed] [Google Scholar]
- Yi S. Y., Kim J.-H., Joung Y.-H., Lee S., Kim W.-T., Yu S. H., et al. (2004). The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol. 136 2862–2874. 10.1104/pp.104.042903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang N., Hu R., Xiang F. (2016). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus 5:920. 10.1186/s40064-016-2647-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wang R., Ju Q., Li W., Tran L.-S. P., Xu J. (2019). The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol. 180 529–542. 10.1104/pp.18.01380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. H., Zhou T., Tang T. J., Song H. X., Guan C. Y., Huang J. Y., et al. (2019). A multiomics approach reveals the pivotal role of subcellular reallocation in determining rapeseed resistance to cadmium toxicity. J. Exp. Bot. 70 5437–5455. 10.1093/jxb/erz295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F. J., Huang X. Y. (2018). Cadmium phytoremediation: call rice CAL1. Mol. Plant 11 640–642. 10.1016/j.molp.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Zheng S., Wang Q., Yuan Y., Sun W. (2020). Human health risk assessment of heavy metals in soil and food crops in the Pearl River Delta urban agglomeration of China. Food Chem. 316:126213. 10.1016/j.foodchem.2020.126213 [DOI] [PubMed] [Google Scholar]
- Zhu H., Ai H., Cao L., Sui R., Ye H., Du D., et al. (2018). Transcriptome analysis providing novel insights for Cd-resistant tall fescue responses to Cd stress. Ecotoxicol. Environ. Saf. 160 349–356. 10.1016/j.ecoenv.2018.05.066 [DOI] [PubMed] [Google Scholar]
- Zhu Y. L., Pilon-Smits E. A., Jouanin L., Terry N. (1999a). Overexpression of glutathione synthetase in Indian mustard enhances cadmium accumulation and tolerance. Plant Physiol. 119 73–80. 10.1104/pp.119.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. L., Pilon-Smits E. A., Tarun A. S., Weber S. U., Jouanin L., Terry N. (1999b). Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing γ-glutamylcysteine synthetase. Plant Physiol. 121 1169–1177. 10.1104/pp.121.4.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the results of this study are included in the article. The RNA-seq data have been deposited into the NCBI Short Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) under accession number PRJNA605520.


