Abstract
Cancer stemness represents one of the major mechanisms that predispose patients to tumor aggressiveness, metastasis, and treatment resistance. MicroRNA biogenesis is an important process controlling miRNA processing and maturation. Deregulation of miRNA biogenesis can lead to tumorigenesis and cancer stemness. DDX17 is a co-factor of the miRNA microprocessor. Misregulation of DDX17 can be associated with cancer stemness. K63-linked polyubiquitination of DDX17 presents a concerted mechanism of decreased synthesis of stemness-inhibiting miRNAs and increased transcriptional activation of stemness-related gene expression. K63-linked polyubiquitination of HAUSP serves as a scaffold to anchor HIF-1α, CBP, the mediator complex, and the super-elongation complex to enhance HIF-1α-induced gene transcription. Recent progress in RNA modifications shows that RNA N6-methyladenosine (m6A) modification is a crucial mechanism to regulate RNA levels. M6A modification of miRNAs can also be linked to tumorigenesis and cancer stemness. Overall, miRNA biogenesis and K63-linked polyubiquitination of DDX17 play an important role in the induction of cancer stemness. Delineation of the mechanisms and identification of suitable targets may provide new therapeutic options for treatment-resistant cancers.
Keywords: miRNA biogenesis, DDX17, Cancer stemness, HAUSP, K63-linked polyubiquitination, m6A
Tumorigenesis, cancer stemness, and hypoxia
The traditional concept of cancer stem cells (CSCs) is that these CSCs can self-renew through asymmetric cell division and the non-CSCs can differentiate into mature tumor cells [1]. However, recent results show that CSCs can derive from tumor initiating cells (TICs) or differentiated cells that receive niche signals, indicating the plasticity of adult cancer stem cells [2,3]. The stem cell niches that CSCs reside are different from the general tumor microenvironment [2]. These niches include cancer-associated fibroblasts, mesenchymal stem cells, inflammatory cells, etc [2]. Hypoxia also serves as a chemical microenvironmental factor and a crucial niche component [2]. The stem cell niches will maintain the main characteristics of CSCs, provide escape from immune surveillance, and promote metastatic capability [2]. Therefore, the abundance of CSCs will depend on the amplitude of niche signals that can promote either type of cells into CSCs [3]. However, as tumor progresses and becomes more aggressive, tumor cells acquire more mutations to become less dependent on niche signals [3]. Therefore, the degree of malignancy will depend on the degree of niche dependency [3]. One of the tumor metastatic mechanisms, epithelial–mesenchymal transition, can confer tumor cells with stem-like property and provide plasticity [4]. These stemness features of CSCs are associated with tumor aggressiveness, drug resistance, and metastasis in cancer patients [4]. Recent literature showed that cancer stemness features are correlated with immunotherapy response through machine learning algorithm [5]. All these results indicate the importance of the concept of “cancer stem cells” in the management of cancer patients [[1], [2], [3], [4], [5]].
Solid tumor hypoxia has been shown to increase tumor stemness [2,6]. Intratumoral hypoxia can program tumor cells into pluripotency through activation of several stemness-related genes [2,6]. Hypoxia as a microenvironmental factor and stem cell niche for solid tumors can serve as a good model to study the mechanisms of regulation of tumor stemness [2,6].
miRNA biogenesis
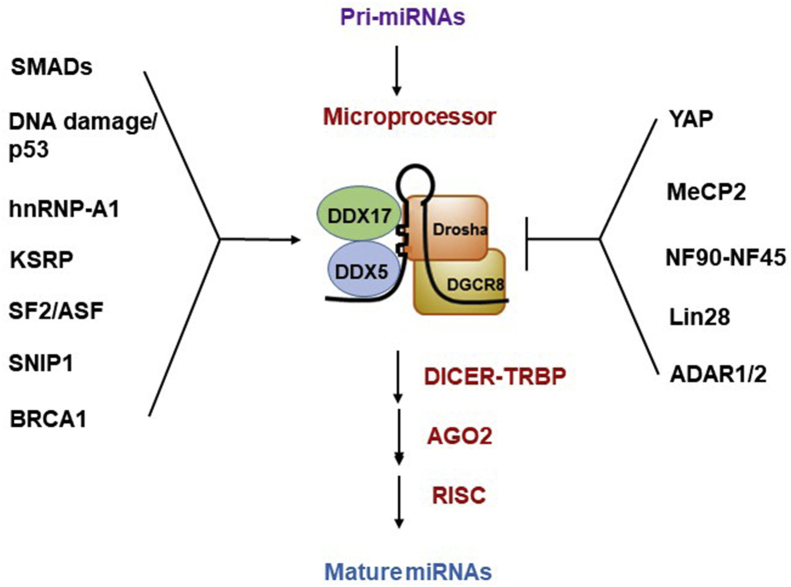
MiRNAs are ∼22 nucleotides in length and they play a regulatory role in various important biological processes, including development and cancer [[7], [8], [9]]. MiRNA biogenesis includes transcription of pri-miRNAs, processing by Drosha/DGCR8 (in the nucleus) and Dicer (in the cytoplasm), formation of RNA-induced silencing complex (RISC), and final processes of going through different modifications to become mature miRNAs [[7], [8], [9]]. The complex to process miRNAs is designated as the “microprocessor complex” [7,8]. The basic microprocessor complex includes Drosha and DGCR8 (a dsRNA-binding protein) [7]. The super microprocessor complex contains accessory proteins, including EWSR1, Fus, numerous heterogenous ribonucleoprotein (hnRNP) complex proteins, and the DEAD-box helicases, DDX5 and DDX17 [7,8]. MiRNAs are tightly controlled temporally and spatially through the regulation of miRNA biogenesis [9]. Specifically, DDX5 and DDX17 function as a hub for signaling transduction, including TGF-β signaling, DNA damage/p53, and cell density control/Hippo signaling [[10], [11], [12], [13]]. In addition, different co-factors may positively or negatively regulate miRNA biogenesis [10]. For positive regulation of miRNA processing, SMAD proteins are recruited by DDX5 to associate with the Drosha microprocessor complex and facilitate the processing of pri-miR-21 [11]. p53 under DNA damage interacts with DDX5 to facilitate miRNA processing [12]. Heterogenous ribonucleoprotein A1 (hnRNP-A1) and KH-type splicing regulatory protein (KSRP) recognize the terminal loop of certain pri-miRNAs to facilitate their cleavage by Drosha/DGCR8 [14,15]. Similar facilitation of miRNA cleavage is mediated by binding of mRNA splicing factor SRSF1 (SF2/ASF) to the stem region of specific pri-miRNAs [16]. Smad nuclear interacting protein 1 (SNIP1) directly binds to pri-miRNAs and associates with Drosha to enhance miRNA production [17]. BRCA1 associates with the super microprocessor complex to promote the processing of certain pri-miRNAs [18]. For negative regulation of miRNA processing, Methyl-CpG binding protein 2 (MeCP2) binds to DGCR8 to interfere with the assembly of Drosha/DGCR8 complex to inhibit miRNA processing [19]. NF90 and NF45 (components of the super microprocessor complex) inhibit Drosha-mediated cleavage of certain pri-miRNAs [20]. Lin-28, a RNA binding protein, specifically binds to the terminal loop of pri-let7 and inhibits its processing [21]. Finally, adenosine deaminase acting on RNA (ADARs), ADAR1 and ADAR2, convert adenosine to inosine in a subset of pri-miRNAs and these A-to-I converted pri-miRNAs suppress Drosha-mediated cleavage [22]. A detailed review of the above regulation of miRNA processing can be found in literature [10]. A summary of the co-factors regulating miRNA biogenesis is shown (Fig. 1).
Fig. 1.
Co-factors that regulate miRNA biogenesis. Positive and negative regulatory factors are summarized in the figure. The detailed mechanisms are described in the text.
Depending on their downstream targets, miRNAs may function as oncogenes or tumor suppressor genes [23]. However, impaired miRNA processing has been shown to enhance tumorigenesis [24]. MiRNAs can regulate both epithelial–mesenchymal transition (EMT) and cancer stem cells (CSCs) [25]. Deregulation of miRNA biogenesis has been involved in breast, ovarian, prostate, and colorectal cancers [26,27]. MiRNAs have been shown to promote cancer stemness and control pluripotency and reprogramming [28,29]. MiRNA deregulation has been delineated in cancer cells and the tumor microenvironment, contributing to drug resistance and metastasis [30,31]. Specifically, hypoxia represses DICER expression through activation of KDM6A/B to induce hypermethylation of the DICER promoter [32]. Hypoxia also represses DROSHA expression through activation of ETS1 and ELK to methylate the DROSHA promoter [33]. The combined repression of DICER and DROSHA causes downregulation of miR-200 family member, miR-Let-7a, miR-135a, miR-146a, and miR-30c to induce EMT, stem cell phenotypes, and the expression of prometastatic genes [32,33]. In addition, EGFR-induced phosphorylation of AGO2 under hypoxia decreases the interaction between AGO2 and DICER, resulting in decreased miRNA biogenesis activity [34]. Finally, hypoxia and miRNAs can induce metabolic reprogramming and cancer stemness and contribute to tumor aggressiveness [35,36]. The prominent example is miR-210 that is regulated by hypoxia to repress TCA cycle activity and produce lactate, an oncometabolite that promotes tumor initiating cells (TICs) [35,36].
K63-linked polyubiquitination, HectH9, and histone 3 lysine 56 acetylation (H3K56Ac)
K63-linked polyubiquitination is a well-established post-translational modification that plays a significant role in many important biological processes present in cells [37,38]. In contrast to K48-linked polyubiquitination of proteins that leads to protein degradation, different types of regulations, including signal transduction, DNA repair, protein localization, endocytosis, and protein–protein interaction, can be regulated by K63-linked polyubiquitination of different proteins [37,38]. One specific example is that K63-linked polyubiquitination of HAUSP has been shown to be a scaffold for HIF-1α-mediated gene transcription and dictate H3K56Ac on the promoters of HIF-1α target genes [39]. Therefore, K63-linked polyubiquitination of proteins presents multiple aspects of regulation of protein functions.
HectH9 (Huwe1, Mule, ARF-BP1) is one of the few E3 ligases that can trigger both K63- and K48-linked ubiquitination [40,41]. HectH9 is shown to be linked to tumorigenesis and poor prognosis [42,43]. HectH9 is also implicated in the maintenance and development of stem cells [[44], [45], [46]]. A detailed review of HectH9 functions has been presented [41].
Histone 3 lysine 56 acetylation (H3K56Ac) is a special histone mark that is closely associated with stemness property and pluripotency [[47], [48], [49]]. Specifically, H3K56Ac is linked to the core transcriptional network in human ES cells [47]. Stemness factor Oct4 interacts directly with H3K56Ac to promote the pluripotency of ES cells [48]. Finally, p53-regulated lncRNA lncPRESS1 interacts with SIRT6 to prevent SIRT6 chromatin localization and maintain high levels of H3K56Ac at the promoters of pluripotency genes [49]. Therefore, lncPRESS1 safeguards the pluripotency state of human ES cells by sequestering SIRT6 [49].
DDX17 and miRNA biogenesis
DEAD-box RNA-binding proteins (RBPs) play a critical role in miRNA biogenesis [50]. Among the DEAD-box RBPs, DDX17 is a well-characterized co-factor of the Drosha/DGCR8 Microprocessor that mediates recognition and processing of primary miRNAs (pri-miRNAs) to mature into miRNAs as mentioned above [[8], [9], [10]]. Further results show that nuclear Yes-associated protein 1 (YAP, a downstream target of the Hippo signaling pathway) sequesters DDX17 to dissociate DDX17 from the microprocessor complex and decrease the processing of a subset of pri-miRNAs under low cell density [13]. DDX17 also plays a different role in regulating miRNA biogenesis by up-regulating Ago2 levels post-transcriptionally [51]. DDX17 regulates the levels of certain miRNAs (e.g. miR-17) that lead to the stabilization of Ago2 [51]. Decrease in DDX17 reduces the levels of certain miRNAs that result in a decrease in unloaded Ago2 levels and cause its degradation [51]. Recent crystallography results show that DDX17 has a RMFQ motif that recognizes RNA [52]. DDX17 can enhance the miRNA processing by remodeling the 3’ flanking region of the pri-miRNA [52]. N-terminal domain of DDX17 has an intramolecular interaction with its DEAD box domain to regulate the ATPase activity [52]. This result demonstrates the molecular details of DDX17-mediated miRNA processing by structural analysis [52]. Overall, DDX17 plays an essential role in the regulation of miRNA biogenesis [[8], [9], [10],51,52].
The linkage between DDX17 in miRNA biogenesis and K63-linked polyubiquitination leading to H3K56Ac modification
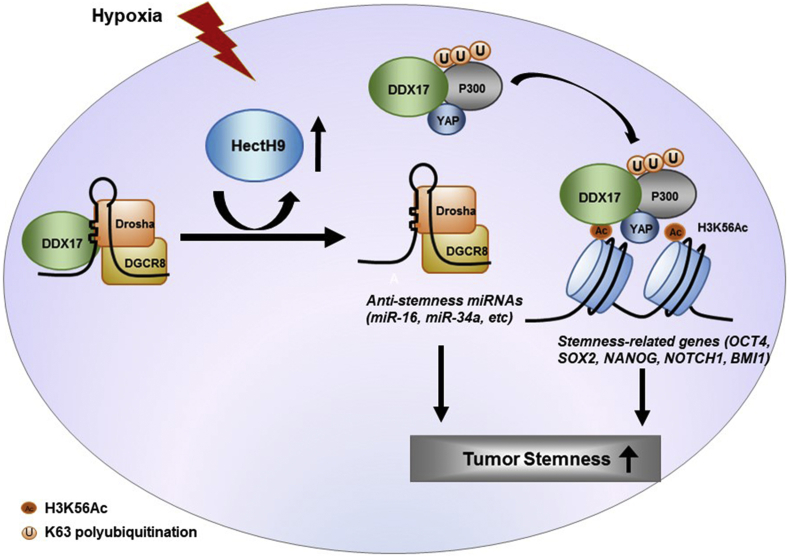
In addition to serving as a co-factor of the Microprocessor [[8], [9], [10]], DDX17 also functions as a transcriptional co-activator for various transcription factors [[53], [54], [55]], including nuclear hormone receptors, p53, MyoD, Smads, Sox2, etc [[53], [54], [55]]. DDX17 can also interact with a REST repressor complex to cause gene repression [56], in contrast to its role in serving as a co-activator to activate gene expression [[53], [54], [55]]. DDX17 can interact with a chromatin modifier (e.g. HDAC1) or go through post-translational modifications (e.g. sumoylation, acetylation) to exert its functions [53,55]. Under the scenario that DDX17 can serve as a co-activator of certain transcription factors (e.g. stemenss-related transcription factors) to regulate gene expression (e.g. to induce cancer stemness), hypoxia induces K63-linked polyubiquitintion of DDX17 and dephosphorylation of YAP to cause nuclear translocation of YAP that participates in the sequestration of the K63-polyuitinated DDX17 into nucleus [57]. As shown by co-immunoprecipitation assays, hypoxia increases the interaction of DDX17 with YAP and decreases its interaction with Drosha and DGCR8, eschewing its role toward the side of a co-factor for transcription factor [57]. In the meanwhile, the dissociation of DDX17 from Drosha and DGCR8 will decrease the biogenesis of certain miRNAs (e.g. anti-stemness miRNAs) [57].
After the sequestration of K63-polyubiquitinated DDX17 by the dephosphorylated YAP inside the nucleus, DDX17 interacts with a ubiquitin receptor, p300, to form a K63-polyubiquitinated DDX17/dephophorylated YAP/p300 complex [57]. Since p300 is a histone acetyltransferase, the formation of a K63-polyubiquitinated DDX17/dephophorylated YAP/p300 complex would induce H3K56Ac specifically as demonstrated by DDX17 knockdown experiments [57]. This K63-polyubiquitinated DDX17/dephophorylated YAP/p300 complex will interact with the transcription factors to further enhance the activation of downstream target genes (e.g. stemness-related transcription factors) [57]. From the published literature, this report is the first demonstration to provide the linkage between DDX17 in miRNA biogenesis and K63-linked polyubiquitintion and bring the mechanisms of miRNA biogenesis and K63-linked polyubiquitination together through DDX17 [57].
DDX17, tumorigenesis, and cancer stemness
DDX17 has been shown to regulate tumorigenesis, including colon, breast, and prostate cancers [58]. DDX17 can activate β-catenin or repress Beclin 1 expression to promote tumor progression in non-small cell lung cancer and glioma cells, respectively [59,60]. DDX17 can also promote hepatocellular carcinoma progression [61]. Interaction of DDX5/DDX17 with U3-, U8-, U13-snoRNAs also have consequences in cell proliferation and survival [62]. In human cancers, sequestration of DDX17 by YAP in the Hippo pathway has been reported to cause global downregulation of miRNAs that is linked to tumorigenesis [13,24].
DEAD-box RBPs have also been shown to regulate pluripotency and reprogramming [63]. Among these RBPs, DDX17 plays a role in pluripotency by interacting with the stemness factor, Sox2 [53,64]. As described in the previous sections, DDX17 can go through K63-polyubiquitination mediated by HectH9 under hypoxia to confer stem-like properties and tumor-initiating capabilities in tumor cells [57]. Dissociation of K63-polyubiquitinated DDX17 from the Drosha-DGCR8 microprocessor complex results in decreased biogenesis of anti-stemness miRNAs (e.g. miR-16, miR-34a, etc) [57]. These miRNAs will influence the levels of stemness factors such as BMI1, SOX2, and OCT4 [57]. As stated above, the formation of a K63-polyubiquitinated DDX17/dephophorylated YAP/p300 complex will induce H3K56 acetylation that deposits on the promoters of stemness-related genes (OCT4, SOX2, NANOG, NOTCH1, and BMI1) and activate their expression, consistent with the role of H3K56Ac in stemness property and pluripotency [[47], [48], [49],57]. This result is confirmed by qChIP assays showing the increased co-occupancy of DDX17, YAP, p300, and H3K56Ac on the promoters of OCT4, SOX2, NANOG, NOTCH1, and BMI1 genes and by reporter gene assays [57]. On the contrary, the DDX17 partner, DDX5, is not K63-polyubiquitinated under hypoxia due to lack of the lysine residue, further delineating the differential role between DDX17 and DDX5 in miRNA biogenesis [57]. The role of DDX17 in conferring tumorigenesis is confirmed by a decrease in tumor sphere formation activity (i.e. cancer stemenss) through knockdown of DDX17 or overexpressing a dominant negative DDX17K190R mutant [57]. Overexpression of the dominant negative DDX17K190R mutant also decreases in vitro colony formation activity and increases drug sensitivity, reflecting an inhibition of tumorigenesis [57]. Further survival analysis shows that high expression of HectH9 and six stemness-related-genes (BMI1, SOX2, OCT4, NANOG, NOTCH1, and NOTCH2) predicts poor survival in HNSCC and lung adenocarcinoma patients [57]. These results demonstrate that miRNA biogenesis and H3K56Ac modification can be coupled through K63-linked polyubiquitination of DDX17 to confer cancer stemness and promote tumorigenesis [57]. A summary of the above results is shown (Fig. 2).
Fig. 2.
Concerted regulation of miRNA biogenesis and transcriptional activation that mediates cancer stemness through K63-linked polyubiquitination of DDX17.
HAUSP (USP7), tumorigenesis, and stemness
Hypoxia has been demonstrated to induce stemness-related gene expression [2,6]. Recent results show that HAUSP (USP7), a USP type deubiquitinase, is a dequbiquitinase of HIF-1α [39]. HAUSP can undergo hypoxia-induced K63-linked polyubiquitination by HectH9 to enhances its ability to deubiquitinate HIF-1α and also serve as a scaffold of HIF-1α-induced gene transcription [39]. K63-polyubiquitinated HAUSP interacts with HIF-1α, CBP, the mediator complex, and the super-elongation complex to enhance HIF-1α-induced gene transcription [39]. In addition, K63-polyubiqutinated HAUSP exerts allosteric activation of CBP activity to restrict its acetyltransferase activity mostly on mediating H3K56Ac [39]. CBP is also identified as a new ubiquitin receptor to interact with K63-polyubiqutinated HAUSP and the H3K56Ac histone mark then is deposited on the promoters of HIF-1α target genes, including VEGF, Glut1, and Twist1 [39]. Therefore, K63-linked polyubiquitination of HAUSP presents a dual mechanism of increasing both the deubiquitinase activity and transcription complex scaffolding activity as well as converting a post-translational modification into a global histone modification mode [39]. This result also demonstrates the revised model of HIF-1α-induced gene transcription in which K63-polyubiquitinated HAUSP plays a crucial role [39]. HAUSP is also shown to maintain neural progenitor cells through stabilizing REST [65]. HAUSP is a direct target of Bach1 and Bach1 facilitates the deubiquitination and stabilization of stemness-related genes to regulate self-renewal of human embryonic stem cells [66]. Therefore, HAUSP also possesses stemness-promoting activity.
m6A, microRNA biogenesis, and cancer
N6-methyladenosine modification (m6A) is the most prevalent internal RNA modification that recently received great attention in the field of RNA modification [67,68]. Modifications of RNAs by m6A have been involved in different aspects of mRNA processing, including splicing, export, translation, and decay [69]. METTL3 and METTL14 are m6A writers that deposits m6As on RNAs [67,68]. Various m6A readers (YTHDC and YTHDF family proteins) and erasers (FTO, ALKBH5) are also identified [67,68]. M6A has been linked to different types of cancer, including AML, hepatoma, glioblastoma, breast cancer, lung cancer, cervical cancer, etc [[69], [70], [71]]. M6A modification of pri-miRNAs by METTL3 renders the recognition and processing by DGCR8 [72]. METTL3 enhances miRNA maturation through m6A deposition on the pri-miRNAs, indicating the important role of m6A modification in miRNA biogenesis [72]. Other results show that m6A has been discovered in miRNAs and m6A regulates the steady state levels of several miRNAs [73]. Another m6A writer, METTL14, interacts with DGCR8 to positively regulate miR-126 in a m6A-dependent manner [74]. METTL14 represses metastasis in hepatoma cells by promoting pri-miR-126 processing [74]. Increased miR-25-3p maturation can also be induced by cigarette smoke through m6A modification of miR-25-3p to promote pancreatic cancer progression [75]. DDX3 interacts with ALKBH5 to demethylate certain miRNAs and regulate cell growth and proliferation [76]. HNRNPA2/B1 is a mediator of m6A-dependent nuclear RNA processing events [77]. HNRNPA2/B1 can serve as a m6A reader to increase miRNA processing and contribute to endocrine resistance in breast cancer cells [78]. TAR RNA-binding protein 2 (TARBP2) can recruit METTL3/METTL14 complex to induce m6A modifications of its target transcripts, leading to intron retention and nuclear decay [79]. TARBP2 promotes lung cancer growth by downregulating ABCA3 and FOXN3 expression [79]. For the role of DDX17 in enhancing miRNA biogenesis through m6A modifications, it will be imperative to investigate the possible interaction between DDX17 and m6A-methylated pri-miRNAs, METTL3, METTL14, or other proteins modulating the process of m6A modifications to see whether DDX17 plays a role in regulating miRNA biogenesis through m6A modifications of pri-miRNAs and/or other regulatory proteins.
In contrast, m6A methylation can also be regulated by miRNAs and promote reprogramming to pluripotency [80]. It is reasonable that non-coding RNAs can regulate the members of m6A modification process [81,82]. Detailed reviews in this aspect can be found in the literature [81,82].
Clinical relevance and therapeutic strategies
Since DDX17 polyubiquitination plays a concerted role in conferring cancer stemness, inhibition of DDX17 polyubiquitination by inhibitors against HectH9 may ablate this process. HectH9 inhibitors such as BI8622 and BI8626 were identified through a high-throughput screening of small molecules [83]. Inhibition of HectH9 causes the accumulation of Miz1 and represses c-Myc target gene expression in colorectal cancer cells [83]. HectH9 inhibition also increases p53 levels, reduces c-Myc transcriptional activity to promote apoptosis in c-Myc-driven B cell lymphomas [84]. Overexpression of DDX17 and six stemness markers can serve as a prognostic marker [57]. MicroRNA biogenesis genes (e.g. DROSHA, TARBP2, XPO5, TNRC6A) can also serve as prognostic markers in different cancer types [27]. Therefore, it will be important to apply different targets or markers for therapy or diagnosis. Finally, cancer stemness relies on stem cell niches and targeting niches will be an alternative for the management of treatment resistance [2,3].
Conclusions
Complicated signaling pathways are demonstrated for the regulation of miRNA biogenesis. One of the focal points of this regulation is DDX5/DDX17 [10]. DDX5 is shown to interact with different proteins to receive regulation [11,12]. Similarly, DDX17 also interacts with other proteins to facilitate the regulation [13]. However, post-translational modification (i.e. K63-linked polyubiquitination) of DDX17 adds another layer of regulation to further fine tune the miRNA biogenesis process [57]. In addition, this post-translational modification can turn into transcriptional activation of certain stemness-related genes due to the inherent nature of DDX17 as a transcriptional co-activator [[53], [54], [55],57]. Therefore, future approaches to investigate post-translational modifications (e.g. phosphorylation, polyubiquitination, etc) of the protein components inside the microprocessor complex will point to new regulatory mechanisms.
Due to the contribution of deregulation of miRNA biogenesis to tumorigenesis and cancer stemness [24,26,27,[32], [33], [34]], these detailed mechanisms should provide new thinking in order to facilitate future pharmaceutical design of new drugs that can be used to treat aggressive cancers. Recent advances in the investigation of RNA m6A modification will present new research venues to further understand the process of miRNA biogenesis and the contribution of m6A modification to tumorigenesis [[67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81]]. As the concepts of “cancer stem cells” have been modified by incorporating “niche signals” [2,3], further approaches to target these niche components and signals will be one of the priorities in order to improve our understanding of tumor microenvironment and provide better management of treatment-resistant cancers.
Funding
This work was supported in part to K.J.W. by Ministry of Science and Technology Summit and Frontier grants (MOST 107-2745-B-039-001, MOST 108-2321-B-182A-005), Chang Gung Memorial Hospital (OMRPG3I0011, NMRPG3H0651, CORPG3J0231, NMRPG3J0671).
Conflicts of interest
There is no competing interest since there is only one author.
Acknowledgements
Due to the limitation of space, I apologize to the authors whose papers are not cited in the manuscript.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Pattabiraman D.R., Weinberg R.A. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plaks V., Kong N., Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batlle E., Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 4.Shibue T., Weinberg R.A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malta T.M., Sokolov A., Gentles A.J., Burzykowski T., Poisson L., Weinstein J.N. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu J., Zhang Z., Zhou W., Wang A.J., Heddleston J.M., Pinna C.M.A. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory R.I., Yan K.-P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 8.Newman M.A., Hammond S.M. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 10.Shen J., Hung M.C. Signaling-mediated regulation of microRNA processing. Cancer Res. 2015;75:783–791. doi: 10.1158/0008-5472.CAN-14-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 13.Mori M., Triboulet R., Mohseni M., Schlegelmilch K., Shrestha K., Camargo F.D. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guil S., Caceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 15.Trabucchi M., Briata P., Garcia-Mayoral M., Haase A.D., Filipowicz W., Ramos A. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H., Sun S., Tu K., Gao Y., Xie B., Krainer A.R. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai S., Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng T.L., Wang Z., Liao Q., Zhu Y., Zhou W.H., Xu W. MeCP2 suppresses nuclear MicroRNA processing and dendritic growth by regulating the DGCR8/drosha complex. Dev Cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto S., Aoki K., Higuchi T., Todaka H., Morisawa K., Tamaki N. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 22.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romano G., Veneziano D., Acunzo M., Croce C.M. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485–491. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 25.Ceppi P., Peter M.E. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene. 2014;33:269–278. doi: 10.1038/onc.2013.55. [DOI] [PubMed] [Google Scholar]
- 26.Khan S., Ayub H., Khan T., Wahid F. MicroRNA biogenesis, gene silencing mechanisms and role in breast, ovarian and prostate cancer. Biochimie. 2019;167:12–24. doi: 10.1016/j.biochi.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Vychytilova-Faltejskova P., Svobodova Kovarikova A., Grolich T., Prochazka V., Slaba K., Machackova T. MicroRNA biogenesis pathway genes are deregulated in colorectal cancer. Int J Mol Sci. 2019;20:4460. doi: 10.3390/ijms20184460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan A.Q., Ahmed E.I., Elareer N.R., Junejo K., Stenhoff M., Uddin S. Role of miRNA-regulated cancer stem cells in the pathogenesis of human malignancies. Cells. 2019;8:840. doi: 10.3390/cells8080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao J., Duan F.F., Wang Y. MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:95–103. doi: 10.1016/j.gde.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Rupaimoole R., Calin G.A., Lopez-Berestein G., Sood A.K. miRNA deregulation in cancer cells and the tumor microenvironment. Canc Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raza U., Zhang J.D., Sahin O. MicroRNAs: master regulators of drug resistance, stemness, and metastasis. J Mol Med. 2014;92:321–336. doi: 10.1007/s00109-014-1129-2. [DOI] [PubMed] [Google Scholar]
- 32.Rupaimoole R., Wu S.Y., Pradeep S., Ivan C., Pecot C.V., Gharpure K.M. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun. 2014;5:5202. doi: 10.1038/ncomms6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van den Beucken T., Koch E., Chu K., Rupaimoole R., Prickaerts P., Adriaens M. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen J., Xia W., Khotskaya Y.B., Huo L., Nakanishi K., Lim S.O. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullmann P., Nurmik M., Begaj R., Haan S., Letellier E. Hypoxia- and microRNA-induced metabolic reprogramming of tumor-initiating cells. Cells. 2019;8:528. doi: 10.3390/cells8060528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macharla L.W., Wanjiru C.M., Mureithl M.W., Pereira C.M., Ferrer V.P., Moura-Neto V. MicroRNAs, hypoxia and the stem-like state as contributors to cancer aggressiveness. Front Genet. 2019;10:125. doi: 10.3389/fgene.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komander D., Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 38.Silva G.M., Finley D., Vogel C. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol. 2015;22:116–123. doi: 10.1038/nsmb.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H.T., Kuo Y.C., Huang C.H., Hung J.J., Chen W.Y., Chou T.Y. K63-polyubiquitinated HAUSP deubiquitinates HIF-1α and dictates H3K56 acetylation promoting hypoxia-induced tumor progression. Nat Commun. 2016;7:13644. doi: 10.1038/ncomms13644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Kao S.H., Wu H.T., Wu K.J. Ubiquitination by Huwe1 in tumorigenesis and beyond. J Biomed Sci. 2018;25:67. doi: 10.1186/s12929-018-0470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Confalonieri S., Quarto M., Goisis G., Nuciforo P., Donzelli M., Jodice G. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene. 2009;28:2959–2968. doi: 10.1038/onc.2009.156. [DOI] [PubMed] [Google Scholar]
- 43.Yang D., Cheng D., Tu Q., Yang H., Sun B., Yan L. HUWE1 controls the development of non-small cell lung cancer through down-regulation of p53. Theranostics. 2018;8:3517–3529. doi: 10.7150/thno.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urbán N., van den Berg D.L., Forget A., Andersen J., Demmers J.A., Hunt C. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science. 2016;353:292–295. doi: 10.1126/science.aaf4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King B., Boccalatte F., Moran-Crusio K., Wolf E., Wang J., Kayembe C. The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells. Nat Immunol. 2016;17:1312–1321. doi: 10.1038/ni.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee H.J., Li C.F., Ruan D., He J., Montal E.D., Lorenz S. Non-proteolytic ubiquitination of Hexokinase 2 by HectH9 controls tumor metabolism and cancer stem cell expansion. Nat Commun. 2019;10:2625. doi: 10.1038/s41467-019-10374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie W., Song C., Young N.L., Sperling A.S., Xu F., Sridharan R. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan Y., Xue Y., Song C., Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc Natl Acad Sci USA. 2013;110:11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain A.K., Xi Y., McCarthy R., Allton K., Akdemir K.C., Patel L.R. LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol Cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linder P., Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 51.Connerty P., Bajan S., Remenyi J., Fuller-Pace F.V., Hutvagner G. The miRNA biogenesis factors, p72/DDX17 and KHSRP regulate the protein level of Ago2 in human cells. Biochim Biophys Acta. 2016;1859:1299–1305. doi: 10.1016/j.bbagrm.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Ngo T.D., Partin A.C., Nam Y. RNA specificity and autoregulation of DDX17, a modulator of MicroRNA biogenesis. Cell Rep. 2019;29:4024–4035. doi: 10.1016/j.celrep.2019.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuller-Pace F.V. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta. 2013;1829:756–763. doi: 10.1016/j.bbagrm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 54.Dardenne E., Polay Espinoza M., Fattet L., Germann S., Lambert M.-P., Neil H. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014;7:1900–1913. doi: 10.1016/j.celrep.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Giraud G., Terrone S., Bourgeois C.F. Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep. 2018;51:613–622. doi: 10.5483/BMBRep.2018.51.12.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lambert M.P., Terrone S., Giraud G., Benoit-Pilven C., Cluet D., Combaret V. The RNA helicase DDX17 controls the transcriptional activity of REST and the expression of proneural microRNAs in neuronal differentiation. Nucleic Acids Res. 2018;46:7686–7700. doi: 10.1093/nar/gky545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kao S.H., Cheng W.C., Wang Y.T., Yeh H.Y., Chen Y.J., Wu H.T. Regulation of miRNA biogenesis and histone modification by K63-polyubiquitinated DDX17 controls cancer stem-like features. Cancer Res. 2019;79:2549–2563. doi: 10.1158/0008-5472.CAN-18-2376. [DOI] [PubMed] [Google Scholar]
- 58.Janknecht R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17) Am J Transl Res. 2010;2:223–234. [PMC free article] [PubMed] [Google Scholar]
- 59.Li K., Mo C., Gong D., Chen Y., Huang Z., Li Y. DDX17 nucleocytoplasmic shuttling promotes acquired gefitinib resistance in non-small cell lung cancer cells via activation of β-catenin. Cancer Lett. 2017;400:194–202. doi: 10.1016/j.canlet.2017.02.029. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z., Tian H., Miao Y., Feng X., Li Y., Wang H. Upregulation of p72 enhances malignant Migration and invasion of glioma cells by repressing Beclin1 expression. Biochemistry (Mosc) 2016;81:574–582. doi: 10.1134/S0006297916060031. [DOI] [PubMed] [Google Scholar]
- 61.Xue Y., Jia X., Li C., Zhang K., Li L., Wu J. DDX17 promotes hepatocellular carcinoma progression via inhibiting Klf4 transcriptional activity. Cell Death Dis. 2019;10:814. doi: 10.1038/s41419-019-2044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ismael H., Altmeyer S., Stahl H. Regulation of the U3-, U8-, and U13snoRNA expression by the DEAD box proteins Ddx5/Ddx17 with consequences for cell proliferation and survival. Noncoding RNA. 2016;2:11. doi: 10.3390/ncrna2040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guallar D., Wang J. RNA-binding proteins in pluripotency, differentiation, and reprogramming. Front Biol. 2014;9:389–409. doi: 10.1007/s11515-014-1326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alqahtani H., Gopal K., Gupta N., Jung K., Alshareef A., Ye X. DDX17 (P72), a Sox2 binding partner, promotes stem-like features conferred by Sox2 in a small cell population in estrogen receptor-positive breast cancer. Cell Signal. 2016;28:42–50. doi: 10.1016/j.cellsig.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Huang Z., Wu Q., Guryanova O.A., Cheng L., Shou W., Rich J.N. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei X., Guo J., Li Q., Jia Q., Jing Q., Li Y. Bach1 regulates self-renewal and impedes mesendodermal differentiation of human embryonic stem cells. Sci Adv. 2019;5 doi: 10.1126/sciadv.aau7887. eaau7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nachtergaele S., He C. Chemical modifications in the life of an mRNA transcript. Annu Rev Genet. 2018;52:349–372. doi: 10.1146/annurev-genet-120417-031522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J., Harada B.T., He C. Regulation of gene expression by N6-methyladenosine in cancer. Trends Cell Biol. 2019;29:487–499. doi: 10.1016/j.tcb.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dai D., Wang H., Zhu L., Jin H., Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124. doi: 10.1038/s41419-017-0129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tuncel G., Kalkan R. Importance of m N6-methyladenosine (m6A) RNA modification in cancer. Med Oncol. 2019;36:71. doi: 10.1007/s12032-019-1260-6. [DOI] [PubMed] [Google Scholar]
- 71.He L., Li H., Wu A., Peng Y., Shu G., Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alarcon C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berulava T., Rahmann S., Rademacher K., Klein-Hitpass L., Horsthemke B. N6-adenosine methylation in MiRNAs. PloS One. 2015;10 doi: 10.1371/journal.pone.0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J., Bai R., Li M., Ye H., Wu C., Wang C. Excessive miR-25-3pmaturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:18. doi: 10.1038/s41467-019-09712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah A., Rashid F., Awan H.M., Hu S., Wang X., Chen L. The DEAD-Box RNA helicase DDX3 interacts with m(6)A RNA demethylase ALKBH5. Stem Cells Int. 2017;2017:8596135. doi: 10.1155/2017/8596135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klinge C.M., Piell K.M., Tooley C.S., Rouchka E.C. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep. 2019;9:9430. doi: 10.1038/s41598-019-45636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fish L., Navickas A., Culbertson B., Xu Y., Nguyen H.C.B., Zhang S. Nuclear TARBP2 drives oncogenic dysregulation of RNA splicing and decay. Mol Cell. 2019;75:967–981. doi: 10.1016/j.molcel.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen T., Hao Y.J., Zhang Y., Li M.M., Wang M., Han W. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 81.Fazi F., Fatica A. Interplay between N6-methyladenosine (m6A) and non-coding RNAs in cell development and cancer. Front Cell Dev Biol. 2019;7:116. doi: 10.3389/fcell.2019.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma S., Chen C., Ji X., Liu J., Zhou Q., Wang G. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peter S., Bultinck J., Myant K., Jaenicke L.A., Walz S., Müller J. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6:1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qi C.F., Kim Y.S., Xiang S., Abdullaev Z., Torrey T.A., Janz S. Characterization of ARF-BP1/HUWE1 interactions with CTCF, MYC, ARF and p53 in MYC-driven B cell neoplasms. Int J Mol Sci. 2012;13:6204–6219. doi: 10.3390/ijms13056204. [DOI] [PMC free article] [PubMed] [Google Scholar]