Coordination of the expression of genes associated with bacterial virulence and environmental adaptation is often dependent on quorum sensing (QS). The QS circuitry of the nonpathogenic bacterium Burkholderia thailandensis, widely used as a model system for the study of the human pathogen Burkholderia pseudomallei, is complex. We found that the LysR-type transcriptional regulator, ScmR, which is highly conserved and involved in the control of virulence/survival factors in the Burkholderia genus, is a global regulator mediating gene expression through the multiple QS systems coexisting in B. thailandensis, as well as QS independently. We conclude that ScmR represents a key QS modulatory network element, ensuring tight regulation of the transcription of QS-controlled genes, particularly those required for acclimatization to the environment.
KEYWORDS: N-acyl-l-homoserine lactone, quorum sensing, secondary metabolism, transcriptional regulation
ABSTRACT
The nonpathogenic soil saprophyte Burkholderia thailandensis is a member of the Burkholderia pseudomallei/B. thailandensis/B. mallei group, which also comprises the closely related human pathogens B. pseudomallei and Burkholderia mallei responsible for the melioidosis and glanders diseases, respectively. ScmR, a recently identified LysR-type transcriptional regulator in B. thailandensis, acts as a global transcriptional regulator throughout the stationary phase and modulates the production of a wide range of secondary metabolites, including N-acyl-l-homoserine lactones and 4-hydroxy-3-methyl-2-alkylquinolines and virulence in the Caenorhabditis elegans nematode worm host model, as well as several quorum sensing (QS)-dependent phenotypes. We have investigated the role of ScmR in B. thailandensis strain E264 during the exponential phase. We used RNA sequencing transcriptomic analyses to identify the ScmR regulon, which was compared to the QS-controlled regulon, showing a considerable overlap between the ScmR-regulated genes and those controlled by QS. We characterized several genes modulated by ScmR using quantitative reverse transcription-PCR or mini-CTX-lux transcriptional reporters, including the oxalate biosynthetic gene obc1 required for pH homeostasis, the orphan LuxR-type transcriptional regulator BtaR5-encoding gene, and the bsa (Burkholderia secretion apparatus) type III secretion system genes essential for both B. pseudomallei and B. mallei pathogenicity, as well as the scmR gene itself. We confirmed that the transcription of scmR is under QS control, presumably ensuring fine-tuned modulation of gene expression. Finally, we demonstrated that ScmR influences virulence using the fruit fly model host Drosophila melanogaster. We conclude that ScmR represents a central component of the B. thailandensis QS regulatory network.
IMPORTANCE Coordination of the expression of genes associated with bacterial virulence and environmental adaptation is often dependent on quorum sensing (QS). The QS circuitry of the nonpathogenic bacterium Burkholderia thailandensis, widely used as a model system for the study of the human pathogen Burkholderia pseudomallei, is complex. We found that the LysR-type transcriptional regulator, ScmR, which is highly conserved and involved in the control of virulence/survival factors in the Burkholderia genus, is a global regulator mediating gene expression through the multiple QS systems coexisting in B. thailandensis, as well as QS independently. We conclude that ScmR represents a key QS modulatory network element, ensuring tight regulation of the transcription of QS-controlled genes, particularly those required for acclimatization to the environment.
INTRODUCTION
Quorum sensing (QS) is a global regulatory mechanism of gene expression depending on bacterial density (1). Gram-negative bacteria often possess homologues of the LuxI/LuxR system initially characterized in the bioluminescent marine bacterium Vibrio fischeri (2). The signaling molecules N-acyl-l-homoserine lactones (AHLs), which are typically produced by LuxI-type synthases, accumulate in the environment as bacterial growth progresses until a threshold concentration is reached allowing bacteria to synchronize their activities and to function as multicellular communities. These AHLs activate LuxR-type transcriptional regulators that modulate the expression of QS target genes, which usually contain a lux box sequence in their promoter region (3).
Burkholderia thailandensis is a nonpathogenic soil saprophyte belonging to the Burkholderia pseudomallei/B. thailandensis/B. mallei group (Bptm group), which also comprises the closely related human pathogens Burkholderia pseudomallei and Burkholderia mallei responsible for melioidosis and glanders, respectively (4). B. thailandensis is considered the avirulent version of B. pseudomallei (5) and is thus commonly used as a surrogate model for the study of B. pseudomallei, which is considered a potential bioterrorism agent and whose manipulation is consequently restricted to biosafety level 3 (BSL3) laboratories (6). The members of the Bptm group carry multiple LuxI/LuxR QS systems that are associated with the biosynthesis of numerous AHL signaling molecules (4, 7–9). These QS systems are referred to as the BtaI1/BtaR1 (QS-1), BtaI2/BtaR2 (QS-2), and BtaI3/BtaR3 (QS-3) QS systems in B. thailandensis (10, 11). The QS-1 system is composed of the BtaR1 transcriptional regulator and the BtaI1 synthase, which synthesizes N-(octanoyl)-l-homoserine lactone (C8-HSL) (12, 13). The BtaR2 transcriptional regulator and the BtaI2 synthase that catalyzes the biosynthesis of both N-(3-hydroxy-decanoyl)-l-homoserine lactone (3OHC10-HSL) and N-(3-hydroxy-octanoyl)-l-homoserine lactone (3OHC8-HSL) constitute the QS-2 system (13, 14). The QS-3 system is composed of the BtaR3 transcriptional regulator and the BtaI3 synthase also responsible for 3OHC8-HSL production (12, 13). Furthermore, B. thailandensis, B. pseudomallei, and B. mallei, carry orphan luxR homologues, namely, btaR4 (malR) and btaR5 in B. thailandensis (15, 16).
QS is involved in the regulation of several virulence factors in B. pseudomallei and B. mallei and is essential to their full capacity to cause infections (7, 8, 17, 18). Other QS-controlled phenotypic traits among the Bptm group members have been reported, such as colony morphology, the development of biofilm, self-aggregation, motility, and pH homeostasis, as well as the production of secondary metabolites (9, 11, 12, 14, 16, 18–25).
A LysR-type transcriptional regulator (LTTR) involved in secondary metabolism regulation, hence designated ScmR, was recently identified in Bptm group members (26). LTTRs are part of a large family and display a well-conserved structure with an N-terminal DNA-binding helix-turn-helix motif and a C-terminal cofactor-binding domain (27). LTTRs are typically negatively autoregulated and frequently positively modulate expression of adjacent genes (27). Nevertheless, LTTRs were also described as global regulators acting positively or negatively (27). Mao et al. (26) demonstrated that ScmR constitutes a global regulator of gene expression in B. thailandensis and influences the production of a wide range of secondary metabolites, including AHLs and the putative 4-hydroxy-3-methyl-2-alkylquinoline (HMAQ) signaling molecules, virulence in the nematode worm model host Caenorhabditis elegans, and several QS-dependent phenotypes. In addition, expression of the scmR gene is under QS control (11, 26).
The central goal of the present study was to further characterize the molecular mechanism of action of the B. thailandensis E264 ScmR transcriptional regulator. We found that ScmR is a global regulator mediating gene expression through the QS-1, QS-2, and/or QS-3 systems, as well as independently of QS. Furthermore, we identified novel genes modulated by ScmR, including the oxalate biosynthetic gene obc1 that is essential for pH homeostasis in the Burkholderia genus, the orphan LuxR-type transcriptional regulator BtaR5-encoding gene, and the bsa (Burkholderia secretion apparatus) type III secretion system (T3SS) genes required for both B. pseudomallei and B. mallei pathogenicity. Moreover, we showed that scmR is negatively autoregulated, and we confirmed that its transcription is QS controlled, ensuring tight regulation of gene expression by ScmR in B. thailandensis. Finally, we demonstrated that ScmR represses virulence using the Drosophila melanogaster fruit fly host model. All in all, this study contributes to a better appreciation of the ScmR regulatory mechanism of the expression of genes in B. thailandensis and in particular those related to pathogenicity in B. pseudomallei.
RESULTS
The ScmR regulon comprises many QS-controlled genes.
ScmR was recently described as a global transcriptional regulator impacting gene expression during the stationary phase of bacterial growth in B. thailandensis (26). We used RNA sequencing (RNA-Seq) transcriptomic analyses to further characterize the regulon of the ScmR transcriptional regulator. We identified the ScmR-regulated genes by comparing the transcripts in the wild type and in the scmR mutant strains of B. thailandensis E264 throughout the logarithmic growth phase. We found that ScmR both positively and negatively influenced the expression of genes located on the two B. thailandensis E264 chromosomes (see Fig. S1A in the supplemental material). Using a 3-fold difference in transcription as a cutoff, we identified 907 genes that were positively modulated by ScmR and 397 genes that were negatively modulated by ScmR (Fig. S1A). These findings confirm that ScmR constitutes a global regulator of gene expression in B. thailandensis E264 (26). Our RNA-Seq analyses identified genes known to be under ScmR control or genes encoding functions known to be controlled by ScmR. Indeed, Mao et al. (26) recently demonstrated that ScmR stimulates the production of the putative HMAQ signaling molecules. RNA-Seq confirmed that expression of the hmqABCDEFG operon, which is required for HMAQ production (28), is activated by ScmR (Table S1). Furthermore, ScmR represses the biosynthesis of burkholdac, a hybrid polyketide/nonribosomal peptide and a potent inhibitor of some histone deacetylases (26). Consistently, expression of the bhc gene cluster, responsible for burkholdac biosynthesis (29), was increased in the scmR mutant compared to the wild-type strain (Table S1). Moreover, we observed that ATP synthesis and stress response genes were downregulated in the absence of ScmR (Table S1), as recently reported (26). Finally, RNA-Seq showed that transcription of the putative exopolysaccharide genes bceABCDEFGHIJ and bceNOPRSTU is affected by ScmR (Table S1). This is in agreement with the finding that ScmR influences colony morphology, as well as pellicle and biofilm formation of B. thailandensis E264 (26).
The ScmR transcriptional regulator was shown to influence the biosynthesis of C8-HSL, 3OHC10-HSL, and 3OHC8-HSL (26), the main AHLs produced by B. thailandensis E264 (10–12, 14). Therefore, we assumed that ScmR could intervene in the regulation of gene expression, inter alia, by impacting the QS-1, QS-2, and/or QS-3 systems of B. thailandensis E264. Indeed, Mao et al. (26) demonstrated that QS-dependent phenotypes, including colony morphology, as well as the development of biofilm, are influenced by ScmR, and we accordingly found several previously reported QS-controlled genes in the ScmR regulon (Table S1). Consequently, we also compared the transcripts in the wild-type strain of B. thailandensis E264 and in the AHL-defective ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant under the same growth conditions to characterize the genes specifically modulated by ScmR independently of its effect on QS. Our RNA-Seq analyses indicated that QS positively regulated expression of 1,088 genes and negatively modulated expression of 547 genes on the two chromosomes of B. thailandensis E264 (Fig. S1B). We identified genes known to be controlled by QS or genes encoding functions known to be under QS control. In B. thailandensis E264, QS stimulates contact-dependent growth inhibition (CDI) (11, 30), and we indeed observed that the transcription of the CDI genes was decreased in the absence of AHLs (Table S1). Furthermore, RNA-Seq indicated that the transcription of the bactobolin biosynthetic genes (14), as well as the obc1 gene expression, encoding the oxalate biosynthetic enzyme Obc1 that is essential to pH homeostasis (25), is activated by QS (Table S1), as previously reported (11). Moreover, RNA-Seq confirmed that the expression of both flagellar genes and methyl-accepting chemotaxis protein genes was upregulated in the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant in comparison to the wild-type strain (11) (Table S1), which is consistent with the observation that B. thailandensis E264 QS mutants are hypermotile (12).
Interestingly, we found a considerable overlap between the genes regulated by ScmR and those that are QS controlled (Table S2). We identified 681 genes that were activated by both ScmR and QS (Fig. S2A), whereas 310 genes were repressed by both ScmR and QS (Fig. S2B). Other patterns of coregulation were observed, including positive regulation by ScmR and negative regulation by QS (Fig. S2C), as well as negative regulation by ScmR and positive regulation by QS (Fig. S2D). While we identified 1,019 genes that were coregulated by both ScmR and QS, 901 genes appeared to be independently regulated by either ScmR or QS under the conditions of our experiments (Fig. S2). Altogether, these results support the idea that ScmR regulates the transcription of many genes through modulation of the QS-1, QS-2, and/or QS-3 systems in B. thailandensis E264. In addition, we found that ScmR affected the expression of genes encoding transcriptional factors, including the QS-controlled orphan transcriptional regulator BtaR5-encoding gene (Table S2). Thus, many genes could be modulated by ScmR indirectly through auxiliary regulators, as recently proposed (26).
ScmR modulates AHL biosynthesis but not the transcription of the AHL synthase-coding genes.
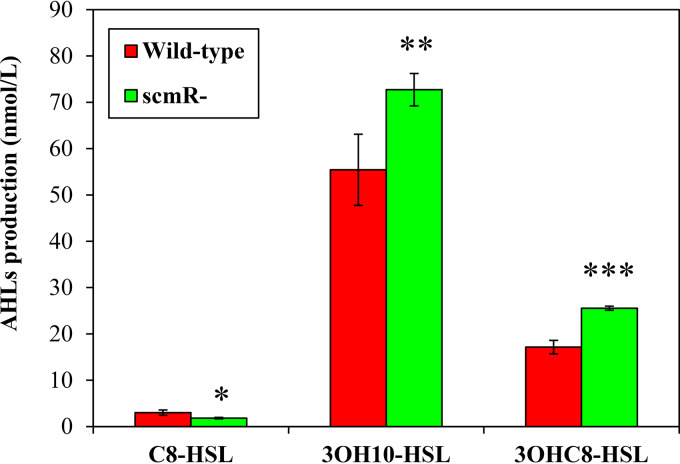
The influence of ScmR on C8-HSL, 3OHC10-HSL, and 3OHC8-HSL production was demonstrated throughout the stationary phase of growth (26), but its effect on QS during the logarithmic phase had not been investigated yet. To determine whether the biosynthesis of the AHLs synthesized by B. thailandensis E264 was impacted by ScmR in the exponential growth phase, we compared the concentrations of these AHLs in the wild-type strain of B. thailandensis E264 and in the scmR mutant. We confirmed that the levels of C8-HSL were decreased in the absence of ScmR (Fig. 1), as previously reported (26), suggesting that ScmR is an activator on the QS-1 system. In contrast to stationary-phase observations (26), however, we detected increasing concentrations of 3OHC10-HSL and 3OHC8-HSL in the scmR mutant versus the wild-type strain under our conditions (Fig. 1), indicating that the QS-2 and/or QS-3 system might be repressed by ScmR.
FIG 1.
ScmR influences the production of the main AHLs found in B. thailandensis E264. C8-HSL, 3OHC10-HSL, and 3OHC8-HSL biosynthesis was measured using LC-MS/MS in cultures of wild-type and scmR mutant strains of B. thailandensis E264 during the exponential growth phase (OD600, ∼4.0). The values are means ± SD (error bars) for three replicates. Values that are significantly different are indicated by asterisks (***, P < 0.001; **, P < 0.01; *, P < 0.05).
ScmR stimulates the production of C8-HSL, 3OHC10-HSL, and 3OHC8-HSL during the stationary phase, but neither the expression of the BtaI1-, BtaI2-, and BtaI3-encoding genes responsible for the production of these AHLs nor the transcription of the btaR1, btaR2, and btaR3 genes was downregulated in an scmR mutant in comparison with the wild-type strain (26). To gain insights into the ScmR-dependent modulation of C8-HSL, 3OHC10-HSL, and 3OHC8-HSL biosynthesis, we determined the expression profiles of the AHL synthase-coding genes btaI1, btaI2, and btaI3 throughout the bacterial growth phases in cultures of the scmR mutant versus the B. thailandensis E264 wild-type strain harboring a chromosomal btaI1-lux, btaI2-lux, or btaI3-lux transcriptional fusion. No discernible difference in expression from the btaI1, btaI2, and btaI3 promoters was found in the scmR mutant compared to the wild-type strain (Fig. S3). Accordingly, our RNA-Seq analyses indicated that ScmR had no impact on btaI1, btaI2, and btaI3 transcription (Table S1). The btaR1, btaR2, and btaR3 genes, encoding the BtaR1, BtaR2, and BtaR3 transcriptional regulators, respectively, were also not affected by ScmR (Table S1). Taken together, these data confirm that the effect of ScmR on C8-HSL, 3OHC10-HSL, and 3OHC8-HSL biosynthesis does not result from modulation of expression of the QS-1, QS-2, and/or QS-3 system genes.
ScmR contributes to pH homeostasis.
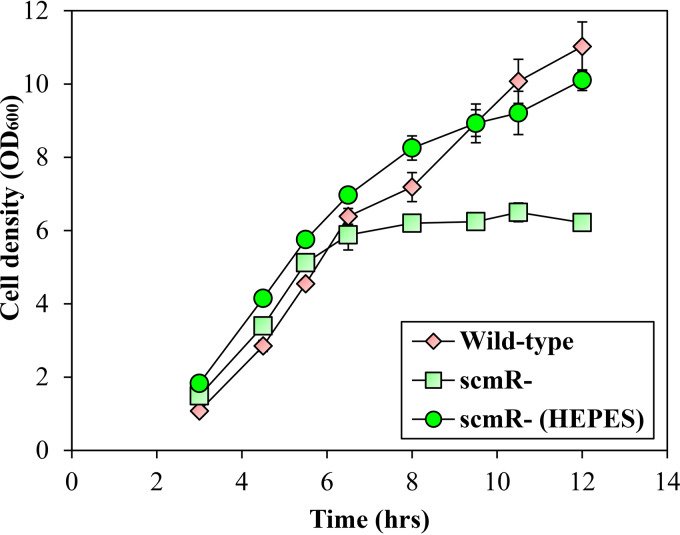
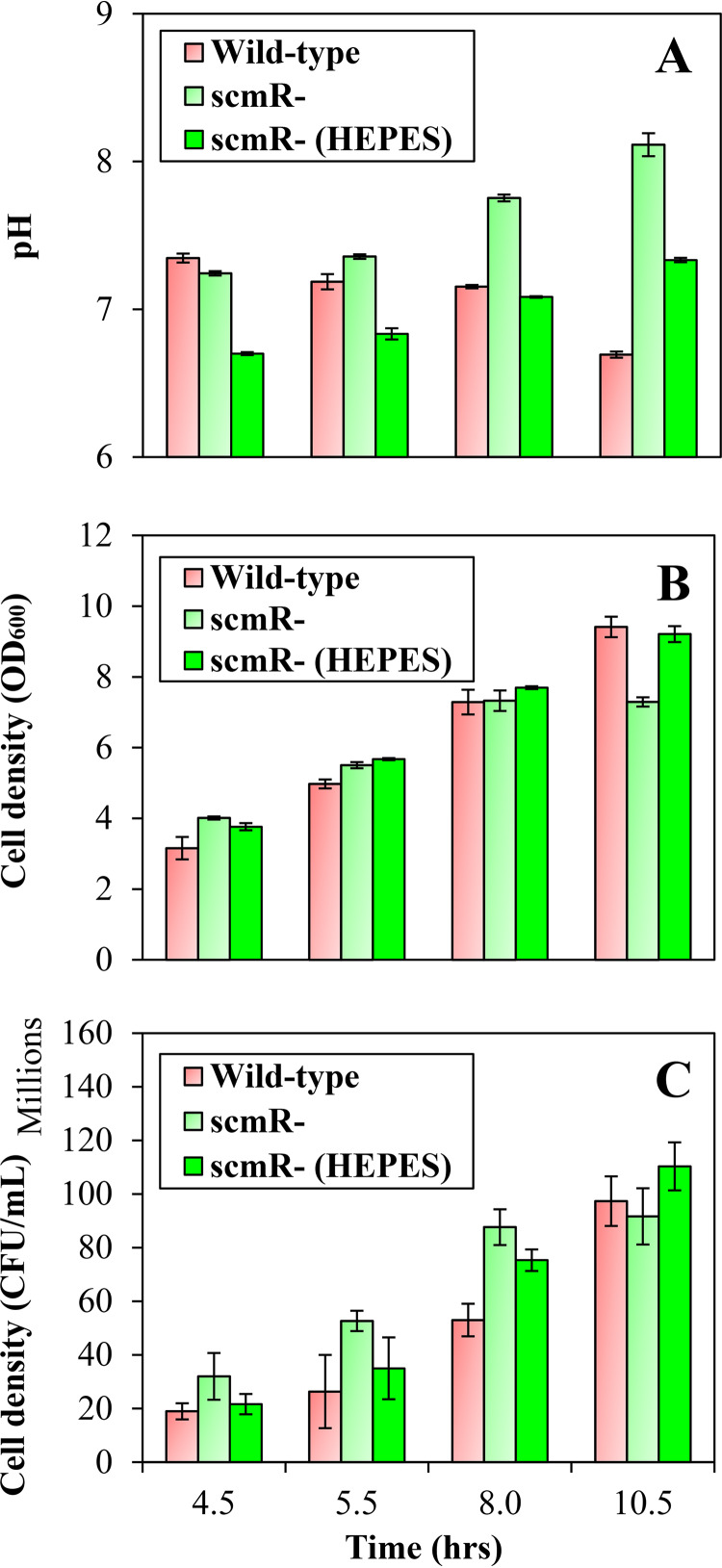
Interestingly, we noticed growth differences between the B. thailandensis E264 wild-type strain and the scmR mutant under the conditions of our experiments. Indeed, inactivation of the scmR gene resulted in reduced optical density at 600 nm (OD600) during the stationary phase (OD600, ∼8.0) but not during the exponential phase (OD600, ∼4.0) (Fig. 2). Since pH was reported to significantly affect the growth of B. thailandensis E264, B. pseudomallei Bp82, and Burkholderia glumae BGR1 (25, 31), we hypothesized that it could be involved in the scmR mutant phenotype. We analyzed the implication of ScmR in pH homeostasis by measuring the pH in cultures of the B. thailandensis E264 wild-type strain and the scmR mutant throughout the different stages of the bacterial growth. The pH in cultures of both the wild-type strain and the scmR mutant was ∼7.0 during the exponential phase (Fig. 3A). On the other hand, the pH in wild-type strain cultures decreased to between 7.0 and 6.5 throughout the stationary phase, whereas the pH in scmR mutant cultures increased to between 7.5 and 8.0, apparently correlating with the OD600 stabilization (Fig. 3A and B). To verify whether the reduction in OD600 could be caused by alkaline toxicity, we buffered cultures of the scmR mutant with 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.0) and observed that the effect on the OD600 was alleviated (Fig. 2; Fig. 3A and B), supporting the hypothesis that culture medium alkalization is the cause of the wild-type strain and the scmR mutant growth variations.
FIG 2.
Impact of scmR inactivation on bacterial growth. B. thailandensis E264 wild-type strain and scmR mutant strain growth curves are shown. Cultures were buffered with 100 mM HEPES. Water only was added to the controls. The error bars represent the SD of the averages for three replicates.
FIG 3.
ScmR influences pH homeostasis. (A) The pH was measured with a pH electrode and meter (Mettler-Toledo, Mississauga, Ontario, Canada) at various times during growth in cultures of the B. thailandensis E264 wild-type strain and the scmR mutant strain. Cultures were buffered with 100 mM HEPES. Water only was added to the controls. (B) The cell density was monitored by measuring turbidity, expressed in 600-nm absorption units (OD600). (C) CFU were determined by plate-counting methods. The error bars represent the SD of the averages for three replicates.
We recently demonstrated that the OD600 is sometimes not representative for growth measurement in B. thailandensis (32). B. thailandensis can synthesize polyhydroxyalkanoates (PHAs), important carbon storage compounds that accumulate intracellularly in form of inclusion bodies (PHA granules) under conditions of carbon source oversupply (33). Accumulation of PHA granules influences both the sizes and the shapes of cells and, consequently, the light scattering, making the OD600 values unreliable for growth evaluation in B. thailandensis (32). To verify that the wild-type strain and the scmR mutant growth variations are due to culture medium alkalization, we performed viable-cell counts. We noticed differences between OD measurements and CFU (Fig. 3B and C). Importantly, no change in viable-cell counts was detected between the B. thailandensis E264 wild-type strain and the scmR mutant strain cultures buffered or not buffered with HEPES (Fig. 3C), revealing that under our conditions pH variations resulting from the absence of ScmR do not actually affect bacterial growth. Considering that fewer PHAs are produced in the absence of ScmR (S. Martinez and E. Déziel, unpublished data), PHA biosynthesis is likely responsible for the OD600 differences between the wild-type strain and the scmR mutant strain of B. thailandensis E264.
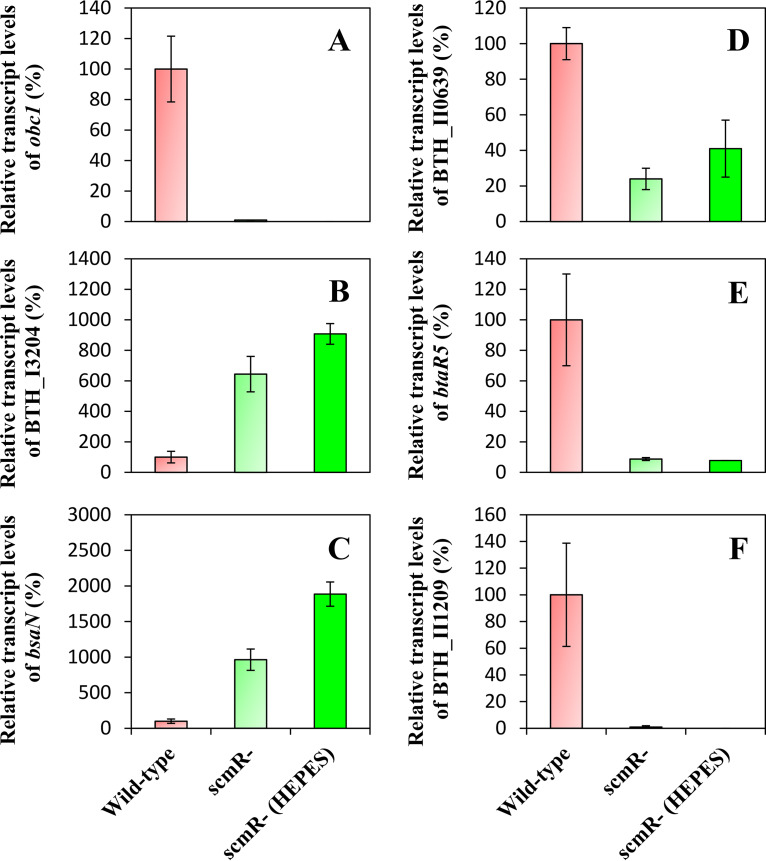
To further characterize the underlying regulatory mechanisms directing pH homeostasis through ScmR, we investigated the effect of ScmR on expression of the obc1 gene, encoding the oxalate biosynthetic enzyme Obc1, which influences the pH in several Burkholderia spp. (25, 31). Oxalic acid was indeed reported to be essential to neutralize alkalization in stationary-phase cultures of the wild-type strain of B. thailandensis E264, B. pseudomallei Bp82, and B. glumae BGR1 (25, 31). Both the expression of obc1 and oxalic acid production were shown to be QS controlled (11, 25). Our RNA-Seq analyses indicated that obc1 transcription was downregulated in the AHL-defective ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant in comparison to the wild-type strain (∼35-fold) (Table S2), confirming that QS activates the obc1 gene expression. Furthermore, we noticed a drastic reduction in expression of obc1 in the scmR mutant compared to the wild-type strain (∼72-fold) (Table S2), revealing that the transcription of obc1 is also strongly enhanced by ScmR. To ascertain the involvement of ScmR in the regulation of the obc1 gene, the expression of obc1 was assessed by quantitative reverse transcription-PCR (qRT-PCR) experiments in cultures of the B. thailandensis E264 wild-type strain and the scmR mutant buffered with HEPES or not buffered with HEPES during the logarithmic growth phase. We observed that the transcription of obc1 was completely abolished in the absence of ScmR (Fig. 4A), indicating that the expression of obc1 is tightly controlled by ScmR. Moreover, the finding that obc1 expression is stimulated by ScmR under neutral conditions confirms previous observations that alkaline stress does not induce obc1 transcription (11, 25). All in all, these findings suggest that ScmR intervenes in pH homeostasis by regulating oxalic acid biosynthesis.
FIG 4.
Expression of several ScmR-regulated genes in cultures of B. thailandensis E264 wild-type and scmR mutant strains buffered or not buffered with HEPES. The relative transcript levels of obc1 (A), BTH_I3204 (B), bsaN (C), BTH_II0639 (D), btaR5 (E), and BTH_II1209 (F) were assessed by qRT-PCR in cultures of B. thailandensis E264 wild-type and scmR mutant strains. Cultures were buffered with 100 mM HEPES. Water only was added to the controls. The results are presented as relative quantification of transcription of the gene compared to the wild-type strain, which was set at 100%. The error bars represent the SD of the averages for three replicates.
Considering the impact of the absence of ScmR on pH, we hypothesized that some of the regulatory effects observed in our RNA-Seq analyses could result from pH imbalance. Consequently, we measured the transcription of several genes located on the two B. thailandensis E264 chromosomes that were strongly upregulated or downregulated in the scmR mutant strain in comparison to the wild-type strain of B. thailandensis E264, namely, BTH_I3204, bsaN, BTH_II0639, btaR5, and BTH_II1209, encoding a lipoprotein, the T3SS transcriptional regulator BsaN, a lipase, the orphan LuxR-type transcriptional regulator BtaR5, and a hypothetical protein, respectively. Their expression was monitored by qRT-PCR during the exponential phase in cultures of the B. thailandensis E264 wild-type and the scmR mutant strains supplemented with HEPES or not supplemented with HEPES. According to our transcriptomic data, expression of the BTH_I3204 and bsaN genes is repressed by ScmR (∼26-fold and ∼17-fold, respectively), whereas expression of the BTH_II0639, btaR5, and BTH_II1209 genes is stimulated by ScmR (∼16-fold, ∼27-fold, and ∼274-fold, respectively) (Table S1). We observed that buffering scmR mutant cultures did not restore normal expression of any of these genes to wild-type levels (Fig. 4B to F), showing that the effects observed on these genes in the scmR mutant did not result from culture medium alkalization.
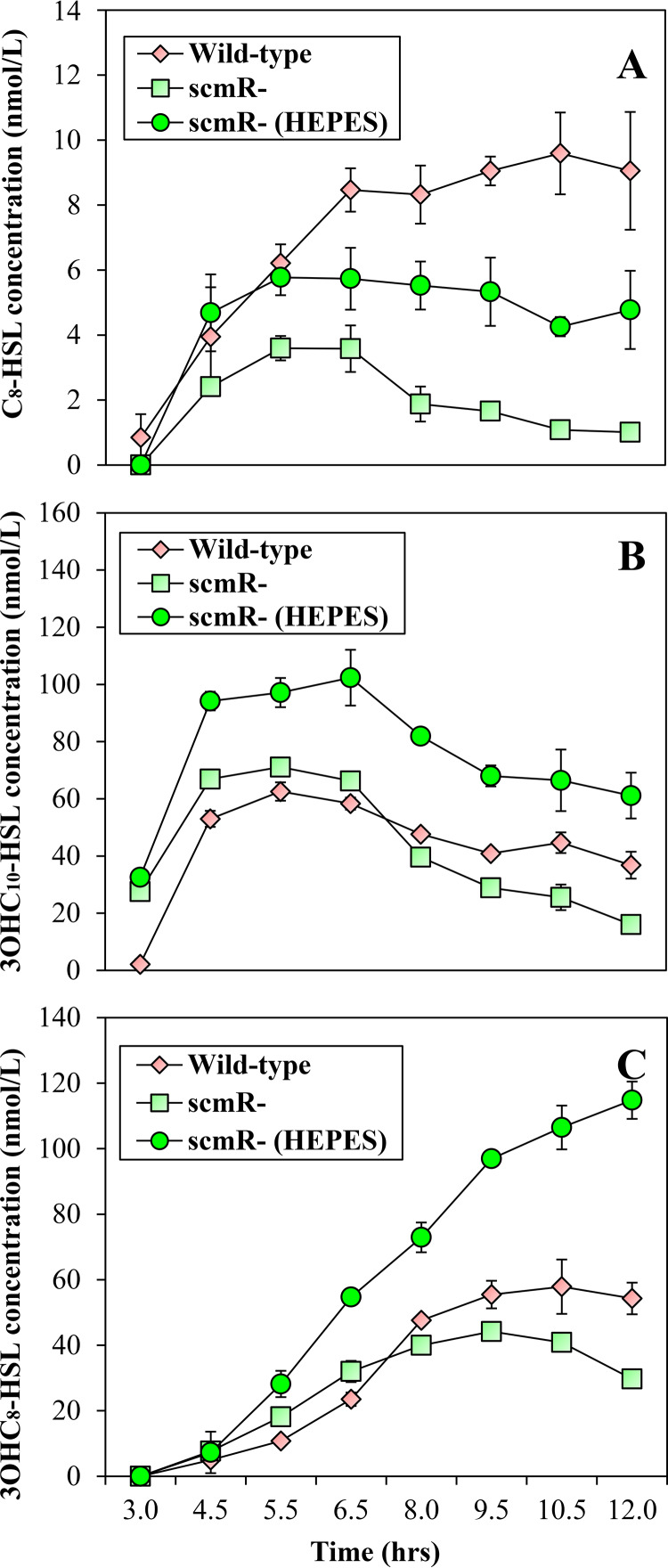
pH affects the integrity of AHL signaling molecules. AHLs are stable at neutral and acidic pH, whereas alkaline conditions cause AHL hydrolysis (34, 35). Therefore, we sought to determine whether ScmR could influence C8-HSL, 3OHC10-HSL, and 3OHC8-HSL stability by impacting pH homeostasis. The concentrations of C8-HSL, 3OHC10-HSL, and 3OHC8-HSL were monitored in the B. thailandensis E264 wild-type strain and the scmR mutant throughout the different stages of bacterial growth. We confirmed that the levels of C8-HSL were reduced in the scmR mutant compared to the wild-type strain in the early stages of the bacterial growth (Fig. 5A), whereas 3OHC10-HSL and 3OHC8-HSL concentrations were increased (Fig. 5B and C). As expected, the concentrations of all three AHLs were decreased in the scmR mutant compared to the wild-type strain in the late stages of bacterial growth (Fig. 5). We then examined the effect of pH buffering on AHLs levels in scmR mutant cultures. The production of all three AHLs was increased in buffered cultures of the scmR mutant (Fig. 5). Taken together, these observations indicate that the impact of ScmR on the QS-1, QS-2, and/or QS-3 systems might result, inter alia, from its influence on pH homeostasis.
FIG 5.
Effect of culture medium alkalization on the levels of AHLs in B. thailandensis E264. The concentrations of C8-HSL (A), 3OHC10-HSL (B), and 3OHC8-HSL (C) were assessed by LC-MS/MS throughout the bacterial growth phases in cultures of wild-type and scmR mutant strains of B. thailandensis E264. Cultures were buffered with 100 mM HEPES. Water only was added to the controls. The error bars represent the SD of the averages for three replicates.
QS regulation of the scmR gene.
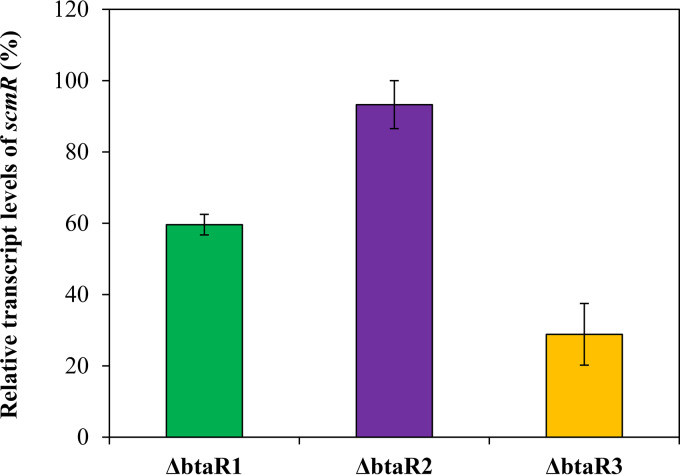
The transcription of the scmR gene is activated by QS (11, 26). It was established that all three AHLs stimulate scmR expression (11). Accordingly, our RNA-Seq analyses indicate that scmR transcription is diminished in the AHL-null ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant compared to the wild-type strain of B. thailandensis E264 (∼4-fold) (Table S1), confirming that scmR transcription is positively modulated by QS. However, the respective influence of the BtaR1, BtaR2, and BtaR3 transcriptional regulators on scmR expression was not investigated (11). To gain insights into the QS-dependent modulation of the scmR gene, we measured its transcription in the ΔbtaR1, ΔbtaR2, and ΔbtaR3 mutants versus the B. thailandensis E264 wild-type strain during the logarithmic growth phase. While no obvious change in scmR transcription was visible in the absence of the BtaR2 transcriptional regulator, the expression of scmR was decreased in both the ΔbtaR1 and the ΔbtaR3 mutant backgrounds (Fig. 6). Collectively, these observations indicate that the transcription of scmR is stimulated by the QS-1 and QS-3 systems, whereas the QS-2 system is not apparently involved in the modulation of scmR expression.
FIG 6.
The QS-1 and QS-3 systems activate the transcription of scmR. The relative transcript levels of scmR were assessed by qRT-PCR in cultures of wild-type and ΔbtaR1, ΔbtaR2, and ΔbtaR3 mutant strains of B. thailandensis E264. The results are presented as relative quantification of transcription of the gene compared to the wild-type strain, which was set at 100%. The error bars represent the SD of the averages for three replicates.
While a putative lux box sequence was found in the promoter region of the B. thailandensis E264 scmR gene (26), we do not know whether the BtaR1 and/or BtaR3 transcriptional regulators directly control its transcription. We found a putative lux box sequence in the promoter region of scmR homologues in both B. pseudomallei K96243 and B. mallei ATCC 23344 (Fig. S4). Accordingly, Klaus et al. (36) and Majerczyk et al. (19) demonstrated that the expression of scmR in B. pseudomallei Bp82 and B. mallei GB8, respectively, is stimulated by QS. Burkholderia cenocepacia J2315 also possesses an scmR homologue, which was shown to be QS controlled in B. cenocepacia K56-2 (37), but no putative lux box sequence was found in its promoter region (38). Altogether, these observations suggest that the QS-dependent regulation of the scmR gene is conserved among Burkholderia spp.
Since scmR is directly adjacent to its downstream gene, namely, ldhA, encoding a putative lactate dehydrogenase, on the genomes of B. thailandensis E264, B. pseudomallei K96243, B. mallei ATCC 2344, and B. cenocepacia J2315, and is transcribed in the same direction (Fig. S5A), we sought to determine whether they could be cotranscribed. The scmR gene is indeed predicted to be arranged in operon with ldhA (http://www.burkholderia.com/), and we observed that ldhA transcription is also activated by QS (Table S2). However, both our transcriptomic data (Fig. S5B) and RT-PCR experiments (Fig. S5C) indicated that scmR is not cotranscribed with the ldhA gene.
Interestingly, expression of ldhA was decreased in the scmR mutant compared to the wild-type strain (Table S2), highlighting that the ldhA gene is positively modulated by ScmR as well. Of note, the reduction in expression of ldhA was substantially greater in the scmR mutant (∼17-fold) than in the ΔbtaI1 ΔbtaI2 ΔbtaI3 mutant (∼3-fold) compared to the wild-type strain (Table S2), suggesting that QS activates ldhA transcription indirectly via positive regulation of the scmR gene.
Since LdhA was hypothesized to influence pH homeostasis in B. thailandensis E264 (26), we tested its involvement in the ScmR-dependent control of pH homeostasis by measuring the pH in cultures of the B. thailandensis E264 wild-type strain and the scmR and ldhA mutants during the stationary phase of growth. While the pH in both the wild-type strain and the ldhA mutant was between 6.5 and 7.0, the pH in cultures of the scmR mutant was approximately 9.0 (Fig. S6A), showing that LdhA does not affect pH in B. thailandensis E264 under the conditions of our experiments. Of note, inactivation of the ldhA gene was not associated with a change in OD600 (Fig. S6B and C). Altogether, these observations indicate that LdhA is not likely involved in the ScmR-dependent control of pH homeostasis in B. thailandensis E264.
scmR is negatively autoregulated.
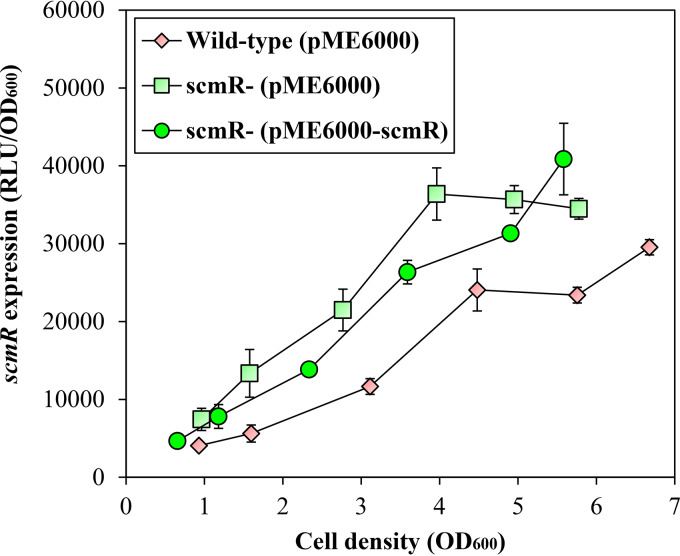
Since LTTRs are typically negatively autoregulated (27), we investigated the impact of ScmR on its own transcription. Considering that the use of an scmR mutant to perform our RNA-Seq analyses precludes clear assessment, we measured the expression of scmR in the B. thailandensis E264 wild-type strain and in the scmR mutant strain harboring a chromosomal scmR-lux transcriptional fusion. We observed an increase in scmR expression in the scmR mutant in comparison with the wild-type strain (Fig. 7), revealing that scmR is negatively autoregulated. However, this repression appears to be less straightforward than the QS activation, as shown by the partial in trans complementation with a plasmid-borne scmR (Fig. 7). We believe this incomplete restoration is explained by an effect of the putative ScmR ligand; while LTTRs are typically negatively autoregulated, this usually requires binding of their cognate ligand, whose availability is unknown and possibly imbalanced when ScmR is overexpressed.
FIG 7.
The scmR gene is negatively autoregulated. The luciferase activity of the chromosomal scmR-lux transcriptional fusion was monitored at various times during growth in cultures of the B. thailandensis E264 wild-type strain and the scmR mutant strain carrying either pME6000 or pME6000-scmR. The error bars represent the SD of the averages for three replicates. The luminescence is expressed in relative light units per optical density of the culture (RLU/OD600).
A heterologous host Escherichia coli expression reporter system was developed to examine the possibility of direct interaction of ScmR with the promoter region of the scmR gene. E. coli DH5α recombinant strains were generated containing the chromosomal scmR-lux transcriptional reporter and either pMLS7 or pMLS7-scmR for constitutive expression of the ScmR transcriptional regulator. We observed no repression of scmR transcription in the presence of ScmR (data not shown), supporting the idea that ScmR might not directly repress its own expression or that an additional unknown factor(s), which might be absent in the E. coli background, is required for scmR negative autoregulation. Indeed, LTTRs generally function in association with ligands to modulate the expression of genes (27).
ScmR represses virulence in the fruit fly model host D. melanogaster.
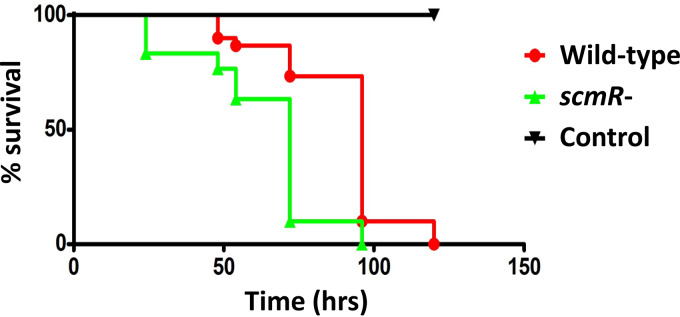
The cytotoxin malleilactone was reported to contribute to pathogenicity in both B. thailandensis E264 and B. pseudomallei Bp82 (15, 36). Interestingly, an scmR mutant of B. thailandensis E264, which is known to overproduce malleilactone, is hypervirulent toward the C. elegans nematode worm host model in comparison with the wild-type strain (26). Consistently, we found that our scmR mutant was significantly more virulent than the wild-type strain using the fruit fly model host D. melanogaster (P < 0.001) (Fig. 8). However, according to our transcriptomic data and in contrast to Mao et al. (26) observations, ScmR had no impact on the transcription of either malleilactone biosynthetic genes or the btaR4 gene, encoding the orphan LuxR-type transcriptional regulator BtaR4 (MalR), which activates the mal gene cluster responsible for malleilactone biosynthesis (15, 16), indicating that ScmR might not regulate malleilactone production under the conditions of our experiments (data not shown). Hence, the ScmR-dependent regulation of pathogenicity in B. thailandensis E264 is not exclusively mediated through control of the biosynthesis of malleilactone.
FIG 8.
Virulence of the wild-type strain and the scmR mutant strain of B. thailandensis E264 toward the fruit fly D. melanogaster. Thirty flies, which were distributed in three vials containing the fruit fly dry medium, as well as bacterial suspensions of either the B. thailandensis E264 wild-type strain or the scmR mutant, were used to carry out the infection tests of D. melanogaster. Significance between survival curves was assessed using the log rank (Mantel-Cox) test.
DISCUSSION
The function of the ScmR transcriptional regulator was recently addressed in B. thailandensis, revealing its importance in secondary metabolism regulation, as well as its involvement in the modulation of additional QS-controlled phenotypes (26). While Mao et al. (26) defined the ScmR regulon during the stationary phase, we established the impact of ScmR on the expression of genes during the logarithmic phase of growth. It must be emphasized that the growth stage is an important variable when investigating QS, and this is especially relevant for B. thailandensis, a bacterium for which we reported significant differences in QS regulation depending on the growth stage (10). We confirmed that ScmR is a global regulator of gene expression in B. thailandensis E264 (see Fig. S1A in the supplemental material). Mao et al. (26) reported that ScmR modulates the production of several secondary metabolites during the stationary phase, including the main AHL signaling molecules found in this bacterium, namely, C8-HSL, 3OHC10-HSL, and 3OHC8-HSL. On the other hand, our RNA-Seq analyses revealed that almost no genes encoding enzymes involved in the production of secondary metabolites were modulated by ScmR during the exponential phase (Table S1). Still, we demonstrated that AHL biosynthesis is affected by ScmR during the logarithmic growth phase as well (Fig. 1), which suggests that ScmR might actually control the transcription of many genes through its effect on the QS-1, QS-2, and/or QS-3 systems. This is also further supported by the finding that ScmR modulates QS-controlled phenotypic traits, such as colony morphology, as well as pellicle and biofilm formation (26). Consistently, we noticed a considerable overlap between the ScmR-regulated genes and those controlled by QS (Fig. S2). Furthermore, we confirmed that the scmR gene is regulated by QS (Fig. 6), showing that ScmR is deeply integrated into the QS modulatory network of B. thailandensis E264. We assume that the QS-dependent regulation of scmR transcription allows tightly controlled coordination of the expression of genes.
Interestingly, we found that the expression of many genes that encode transcriptional regulators, including the orphan transcriptional regulator BtaR5-encoding gene (Fig. 4E), is modulated either positively or negatively by ScmR (Table S1). Consequently, we propose that ScmR controls many genes through different and not mutually exclusive mechanisms, i.e., (i) regulation of AHL signaling molecule biosynthesis, (ii) direct binding of target genes, and (iii) indirect modulation of some genes via intermediate regulators. It will therefore be important to further investigate the molecular mechanism of action of ScmR in order to decipher between directly and indirectly ScmR-regulated genes. Moreover, the characterization of an ScmR-binding motif will contribute to the identification of promoters that are directly modulated versus those that are indirectly modulated.
The production of oxalic acid, which is required for pH homeostasis, is under QS control in several Burkholderia spp. (25, 31). Our RNA-Seq analyses confirmed the implication of AHLs in the regulation of expression of the oxalate biosynthetic gene obc1, and we showed that the transcription of obc1 is stringently modulated by ScmR as well (Fig. 4A). Furthermore, we noticed that the impact of ScmR on obc1 expression was more pronounced than the effect of AHLs (Table S2), suggesting that QS activates obc1 transcription indirectly via positive regulation of the scmR gene. Whether the ScmR-dependent control of the obc1 gene is direct or not remains to be determined. It is possible that the ScmR-mediated control of the homeostasis of pH is not exclusively dependent on regulation of oxalate production. We indeed observed that several genes involved in ATP synthesis are modulated by ScmR (Table S1), as formerly highlighted (26). In addition, our RNA-Seq analyses revealed that ScmR stimulates expression of the putative lactate dehydrogenase LdhA-encoding gene, which is directly adjacent to scmR, and transcribed in the same direction in several Burkholderia spp. (26, 39) (Fig. S5A). Because lactate dehydrogenase, by reducing pyruvate to lactate, was suggested to affect pH (26), ScmR could also intervene in pH homeostasis through the activation of ldhA transcription. Indeed, Silva et al. (39) demonstrated that the Burkholderia multivorans ATCC 17616 homologue, called LdhR, influences pH homeostasis by activating both expression of the ldhA gene and lactate production. Still, we noticed no difference in pH between cultures of the B. thailandensis E264 wild-type strain and the ldhA mutant (Fig. S6A), suggesting that LdhA is not involved in the ScmR-dependent control of pH homeostasis in B. thailandensis E264. Other discrepancies between the B. thailandensis E264 ScmR and the B. multivorans ATCC 17616 LdhR homologues were reported, such as their negative and positive effects, respectively, on the development of biofilm (39). These findings indicate that these two proteins could be functionally different. More experiments will therefore be necessary to further understand the precise underlying molecular mechanism of action of ScmR in the control of pH homeostasis in B. thailandensis.
The observation that medium alkalization influences the concentration of the main AHLs produced by B. thailandensis E264, namely, C8-HSL, 3OHC10-HSL, and 3OHC8-HSL, reveals that the impact of ScmR on the production of these AHLs might result, inter alia, from its influence on pH homeostasis (Fig. 5). This would then explain why no visible change in expression from the btaI1, btaI2, and btaI3 promoters was observed in the scmR mutant compared to the wild-type strain (Fig. S3). However, the ScmR-dependent regulation of the QS-1, QS-2, and/or QS-3 systems might be more complex. Indeed, while the pH is known to affect the integrity of AHLs (34, 35), we did not detect any hydrolyzed AHLs in the B. thailandensis E264 wild-type strain (by liquid chromatography coupled to tandem mass spectrometry [LC-MS/MS]) and the scmR mutant strain cultures buffered or not buffered with HEPES (data not shown). An explanation could be that ScmR, through its effect on pH, affects the activity of unknown regulatory elements involved in the modulation of AHLs biosynthesis, which are responsive to pH variations. Thus, the ScmR-dependent regulation of the multiple QS systems coexisting in B. thailandensis E264 will need further investigation.
In agreement with the fact that LTTRs are typically negatively autoregulated (27), we highlighted that ScmR represses its own expression (Fig. 7). Still, we observed no direct effect of the ScmR transcriptional regulator on the promoter region of the scmR gene when coexpressed in a heterologous host E. coli expression reporter system. An explanation could be that scmR-negative autoregulation requires additional modulatory elements, including molecular ligands. Indeed, ligands are recognized as being crucial for the function of LTTRs (27). These ligands frequently participate to a self-inducing loop in which a product or an intermediate product of a given metabolic/synthesis pathway that is commonly stimulated by an LTTR acts as the ligand required for transcriptional regulation (27). Therefore, it will be important to determine the putative ligand(s) of ScmR in order to uncover the precise regulatory mechanism underlying scmR-negative autoregulation.
While the pathogenicity of B. multivorans ATCC 17616 in the Galleria mellonella larvae model of infection is apparently not regulated by the ScmR homologue LdhR (39), the loss of ScmR in both B. thailandensis E264 and B. pseudomallei Bp82 results in hypervirulence toward the model host C. elegans (26, 36). In addition, Melanson et al. (40) demonstrated that the rice-pathogenic bacterium Burkholderia glumae 336 gr-1 homologue, called NtpR, is a negative regulator of toxoflavin production, which is considered the primary virulence factor of B. glumae, suggesting that similarly to the B. thailandensis E264 and B. pseudomallei Bp82 ScmR homologues, NtpR influences virulence. In B. thailandensis and B. pseudomallei, ScmR was proposed to modulate the infection process by repressing the biosynthesis of malleilactone (26, 36). We demonstrated that ScmR of B. thailandensis E264 contributes to pathogenicity using the D. melanogaster host model (Fig. 8). However, we observed no difference in the transcription of the mal gene cluster, which encodes the principal enzymes responsible for malleilactone biosynthesis (15, 36), between the B. thailandensis E264 wild-type and scmR mutant strains (data not shown), suggesting that ScmR is not necessarily involved the production of the cytotoxin malleilactone. Any negative effect of ScmR on malleilactone biosynthesis in vivo is currently unknown. Still, we do not exclude the possibility that ScmR reduces virulence by modulating expression of additional virulence/survival factors. For instance, we highlighted that the transcription of the bsa T3SS genes, which are crucial for the pathogenicity of both B. pseudomallei and B. mallei (41, 42), is repressed by ScmR (Table S1). The involvement of other potential virulence factors in the ScmR-mediated control of pathogenicity in B. thailandensis is under investigation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. Unless otherwise stated, all bacteria were cultured at 37°C in tryptic soy broth (TSB; BD Difco, Mississauga, Ontario, Canada), with shaking (240 rpm) in a TC-7 roller drum (NB, Canada), or on petri dishes containing TSB solidified with 1.5% agar. When required, antibiotics were used at the following concentrations: 200 μg/ml tetracycline (Tc), 100 μg/ml kanamycin (Km), and 100 μg/ml trimethoprim (Tp) for B. thailandensis E264, while Tc was used at 15 μg/ml for E. coli DH5α. All measurements of the OD600 were acquired using a Thermo Fisher Scientific NanoDrop ND-1000 spectrophotometer.
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| E. coli | ||
| χ7213 | thr-1 leuB6 fhuA21 lacY1 glnV44 recA1 ΔasdA4 Δ(zhf-2::Tn10) thi-1 RP4-2-Tc::Mu[λpir] | Lab collection |
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Lab collection |
| B. thailandensis | ||
| E264 | Wild type | 5 |
| ED1023 | E264 scmR::pUT-mini-Tn5-Km; Kmr | Our unpublished data |
| JBT112 | E264 ΔbtaI1 ΔbtaI2 ΔbtaI3 | 12 |
| JBT107 | E264 ΔbtaR1 | 12 |
| JBT108 | E264 ΔbtaR2 | 12 |
| JBT109 | E264 ΔbtaR3 | 12 |
| BT08944 | E264 ldhA::IslacZ-PrhaBo-Tp/FRT; Tpr | 45 |
| ED3330 | E264::btaI1-lux | 10 |
| ED3510 | E264 scmR::btaI1-lux | This study |
| ED3331 | E264::btaI2-lux | 10 |
| ED3511 | E264 scmR::btaI2-lux | This study |
| ED3332 | E264::btaI3-lux | 10 |
| ED3512 | E264 scmR::btaI3-lux | This study |
| ED3993 | E264::scmR-lux (pME6000) | This study |
| ED3994 | E264 scmR::scmR-lux (pME6000) | This study |
| ED3995 | E264 scmR::scmR-lux (pME6000-scmR) | This study |
Construction of plasmids.
All plasmids used in this study are described in Table 2. Amplification of the promoter region of scmR was performed from genomic DNA from B. thailandensis E264 using the appropriate primers (see Table S3 in the supplemental material). The amplified product was digested with the FastDigest restriction enzymes XhoI and BamHI (Thermo Fisher Scientific) and inserted by T4 DNA ligase (Bio Basic, Inc., Markham, Ontario, Canada) within the corresponding restriction sites in the mini-CTX-lux plasmid (43), generating the transcriptional reporter pSLG01. Amplification of scmR was accomplished from genomic DNA from B. thailandensis E264 using the primers shown in Table S3. The amplified product was digested with the restriction enzymes KpnI and HindIII before ligation within the corresponding restriction sites in the pME6000 plasmid (44), generating the constitutive expression vector pJPD03. All primers were from Alpha DNA (Montreal, Quebec, Canada).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| Mini-CTX-lux | Integration vector with promoterless luxCDABE; Tcr | 43 |
| pSLG02 | btaI1 promoter inserted in XhoI-BamHI restriction sites in mini-CTX-lux; Tcr | 10 |
| pSLG03 | btaI2 promoter inserted in XhoI-BamHI restriction sites in mini-CTX-lux; Tcr | 10 |
| pSLG04 | btaI3 promoter inserted in XhoI-BamHI restriction sites in mini-CTX-lux; Tcr | 10 |
| pSLG01 | scmR promoter inserted in XhoI-BamHI restriction sites in mini-CTX-lux; Tcr | This study |
| pME6000 | Broad-host-range cloning vector; Tcr | 44 |
| pJPD03 | scmR inserted in KpnI-HindIII restriction sites in pME6000; Tcr | This study |
| pFLPe4 | Site-specific excision vector; Cbr Kmr | 46 |
Construction of reporter strains.
Chromosomal integration of the mini-CTX-btaI1-lux, mini-CTX-btaI2-lux, mini-CTX-btaI3-lux, and mini-CTX-scmR-lux transcriptional reporters at the attB locus in B. thailandensis E264 strains was performed through conjugation with the auxotrophic E. coli χ7213, as described previously (13). Successful chromosomal insertion of the btaI1-lux, btaI2-lux, btaI3-lux, and scmR-lux plasmids was confirmed by PCR using appropriate primers. Excision of the tetracycline cassette in the chromosomal scmR-lux transcriptional fusion was carried out using the pFLPe4 plasmid as previously described (46).
Construction of recombinant strains.
The pME6000 and pME6000-scmR constitutive expression vectors were introduced in B. thailandensis E264 strains by electroporation. Briefly, bacterial cultures were grown to an OD600 of 1.0, pelleted by centrifugation, and washed several times with 1 ml of sterile water. The pellets were concentrated 100-fold in 100 μl of sterile water and electroporated using a 1-mm-gap disposable electroporation cuvette (Bio-Rad Laboratories, Mississauga, Ontario, Canada) at 1.8 kV with an Electroporator 2510 (Eppendorf Scientific, Inc., Westbury, NY). Cells were grown for 1 h in 1 ml of lysogeny broth (LB; Alpha Biosciences, Inc., Baltimore, MD) at 37°C and then plated on Tc selective media.
RNA isolation.
Total RNA of B. thailandensis E264 cultures at an OD600 of 4.0 was extracted with the PureZOL RNA isolation reagent (Bio-Rad Laboratories) and treated twice with a Turbo DNA-Free kit (Ambion Life Technologies, Inc., Burlington, Ontario, Canada) according to the manufacturer’s instructions. Extractions were done on two different bacterial cultures for RNA-Seq analysis and on three different bacterial cultures for qRT-PCR and RT-PCR experiments. Quality and purity controls were confirmed by agarose gel electrophoresis and UV spectrophotometric analysis, respectively. Quantification of total RNA was accomplished on a Corbett Life Science Rotor-Gene 6000 thermal cycler using the QuantiFluor RNA system (Promega, Madison, WI), according to the manufacturer’s protocol.
RNA-Seq library construction and sequencing.
The RNA-Seq library construction and sequencing using an Illumina HiSeq 2000 PE100 were performed by the McGill University and Génome Québec Innovation Centre (Montreal, Quebec, Canada). The RNA-Seq libraries were prepared using a TruSeq stranded mRNA sample preparation kit (Illumina, Inc., San Diego, CA) and a Ribo-Zero rRNA removal kit (Epicentre, Madison, WI).
RNA-Seq mapping and analyses.
All computations were made on the supercomputer Briarée from the Université de Montréal, managed by Calcul Québec and Compute Canada. Raw reads were filtered to remove low-quality reads using the FASTX toolkit by discarding any reads with more than 10% nucleotides with a PHRED score < 20. Reads were then aligned with the reference genome (GenBank accession numbers: chromosome 1 of strain E264, CP000086.1; chromosome 2 of strain E264, CP000085.1) using Bowtie (v2.2.3) with default parameters. Chromosome 1 and chromosome 2 sequence alignments were separately processed to allow expression analysis between the two chromosomes. SAMtools (v0.1.18) and BEDtools (v2.20.1) were used for the generation of sam and bam files, respectively. The GC content of B. thailandensis E264 genes was calculated using BEDtools (v2.20.1), prior to normalization. Normalization of the read count was done using the RPKM normalization function of the NOIseq package in R (47). To exclude features with low read counts, a low-count filter was applied using a CPM method with a CPM value of 1 and a cutoff of 100 for the coefficient of variation. Cutoff values of 3-fold were used to consider differential expression biologically significant.
LC-MS/MS quantification of AHLs.
The concentrations of AHLs were determined by LC-MS/MS from samples of B. thailandensis E264 cultures obtained at different time points during bacterial growth. The samples were prepared and analyzed as described previously (48). 5,6,7,8-Tetradeutero-4-hydroxy-2-heptylquinoline (HHQ-d4) was used as an internal standard. All experiments were performed in triplicate and conducted at least twice independently. For experiments with additions of HEPES, cultures were buffered or not buffered with 100 mM HEPES (Sigma-Aldrich Co., Oakville, Ontario, Canada) from a stock prepared in ultrapure water. Water only was added to the controls.
Measurement of the activities of btaI1-lux, btaI2-lux, btaI3-lux, and scmR-lux reporters.
Expression from the promoter regions of btaI1, btaI2, btaI3, or scmR was quantified by measuring the luminescence of B. thailandensis E264 cultures carrying the corresponding chromosomal mini-CTX-lux transcriptional reporter, as described previously (10). Overnight bacterial cultures were diluted in TSB to an initial OD600 of 0.1 and incubated as indicated above. The luminescence was regularly determined from culture samples using a multimode microplate reader (Cytation 3; BioTek Instruments, Inc., Winooski, VT) and expressed in relative light units per optical density unit of the culture (RLU/OD600). All experiments were performed with three biological replicates and repeated at least twice.
Quantitative reverse transcription-PCR and reverse transcription-PCR experiments.
cDNA synthesis was performed using iScript reverse transcription supermix (Bio-Rad Laboratories), and amplification was accomplished on a Corbett Life Science Rotor-Gene 6000 thermal cycler using the SsoAdvanced universal SYBR green supermix (Bio-Rad Laboratories), according to the manufacturer’s protocol. The reference gene was ndh (49). The ndh gene displayed stable expression under the different genetic contexts tested. All primers used for cDNA amplification are presented in Table S4. Differences in gene expression between B. thailandensis E264 strains were calculated using the 2−ΔΔCT formula (50). A threshold of 0.5 was chosen as significant. All experiments were performed in triplicate and conducted at least twice independently.
Infection of the D. melanogaster fruit fly model host.
The fruit flies were infected by feeding according to the previously described protocol (51). Briefly, 1 g of fruit fly dry medium was put into infection vials. Bacteria were harvested from LB-grown cultures adjusted to an OD600 of 4.0 by centrifugation at 10,000 × g for 5 min. The pellets were suspended in 0.02× phosphate-buffered saline (PBS) containing 1 mM CaCl2 and 1 mM MgCl2, as well as 500 μg/ml ampicillin (Ap) to avoid infection with nonspecific bacteria. Then, 2 ml of bacterial suspension was added to the dry food. Six- to seven-day-old male flies were anesthetized with CO2 and added to the vials in groups of 10. The control vials contained the PBS solution only. Fly survival was scored daily, and survival curves were processed using Prism 5 (GraphPad Software, Inc., San Diego, CA) to perform a statistical log rank (Mantel-Cox) test.
Data analysis.
Unless stated otherwise, data are reported as means ± standard deviations (SD). Statistical analyses were performed with the R software version 3.3.3 (http://www.R-project.org/) using one-way analysis of variance. Probability values of <0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Everett Peter Greenberg (Department of Microbiology, University of Washington School of Medicine, Seattle, WA) for providing the B. thailandensis E264 strains. We especially thank Sylvain Milot, François D’Heygere, Koyomi Ozaki, and Jean-Philippe Dumais for their technical help.
This study was supported by Canadian Institutes of Health Research operating grants MOP-97888 and MOP-142466 to E.D. E.D. holds the Canada Research Chair in Sociomicrobiology.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104:313–322. doi: 10.1128/JB.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuqua C, Greenberg EP. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 4.Majerczyk CD, Greenberg EP, Chandler JR. 2013. Quorum sensing in Burkholderia, p 40–57. In Vasil M, Darwin A (ed), Regulation of bacterial virulence. ASM Press, Washington, DC. [Google Scholar]
- 5.Brett PJ, DeShazer D, Woods DE. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 48(Pt 1):317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 6.Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. 2008. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect Immun 76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulrich RL, Deshazer D, Brueggemann EE, Hines HB, Oyston PC, Jeddeloh JA. 2004. Role of quorum sensing in the pathogenicity of Burkholderia pseudomallei. J Med Microbiol 53:1053–1064. doi: 10.1099/jmm.0.45661-0. [DOI] [PubMed] [Google Scholar]
- 8.Ulrich RL, Deshazer D, Hines HB, Jeddeloh JA. 2004. Quorum sensing: a transcriptional regulatory system involved in the pathogenicity of Burkholderia mallei. IAI 72:6589–6596. doi: 10.1128/IAI.72.11.6589-6596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulrich RL, Hines HB, Parthasarathy N, Jeddeloh JA. 2004. Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J Bacteriol 186:4350–4360. doi: 10.1128/JB.186.13.4350-4360.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Guillouzer S, Groleau MC, Déziel E. 2017. The complex quorum sensing circuitry of Burkholderia thailandensis is both hierarchically and homeostatically organized. mBio 8:e01861-17. doi: 10.1128/mBio.01861-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, Phattarasokul S, Bunt R, Greenberg EP. 2014. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol 196:1412–1424. doi: 10.1128/JB.01405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandler JR, Duerkop BA, Hinz A, West TE, Herman JP, Churchill ME, Skerrett SJ, Greenberg EP. 2009. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. J Bacteriol 191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Guillouzer S, Groleau MC, Déziel E. 2018. Two rsaM homologues encode central regulatory elements modulating quorum sensing in Burkholderia thailandensis. J Bacteriol 200. doi: 10.1128/JB.00727-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerkop BA, Varga J, Chandler JR, Peterson SB, Herman JP, Churchill ME, Parsek MR, Nierman WC, Greenberg EP. 2009. Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J Bacteriol 191:3909–3918. doi: 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biggins JB, Ternei MA, Brady SF. 2012. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J Am Chem Soc 134:13192–13195. doi: 10.1021/ja3052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truong TT, Seyedsayamdost M, Greenberg EP, Chandler JR. 2015. A Burkholderia thailandensis acyl-homoserine lactone-independent orphan LuxR homolog that activates production of the cytotoxin malleilactone. J Bacteriol 197:3456–3462. doi: 10.1128/JB.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valade E, Thibault FM, Gauthier YP, Palencia M, Popoff MY, Vidal DR. 2004. The PmlI-PmlR quorum-sensing system in Burkholderia pseudomallei plays a key role in virulence and modulates production of the MprA protease. J Bacteriol 186:2288–2294. doi: 10.1128/jb.186.8.2288-2294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, Xie C, Ong YM, Gan YH, Chua KL. 2005. The BpsIR quorum-sensing system of Burkholderia pseudomallei. J Bacteriol 187:785–790. doi: 10.1128/JB.187.2.785-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majerczyk CD, Brittnacher MJ, Jacobs MA, Armour CD, Radey MC, Bunt R, Hayden HS, Bydalek R, Greenberg EP. 2014. Cross-species comparison of the Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia mallei quorum-sensing regulons. J Bacteriol 196:3862–3871. doi: 10.1128/JB.01974-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng BS, Majerczyk CD, Passos da Silva D, Chandler JR, Greenberg EP, Parsek MR. 2016. Quorum sensing influences Burkholderia thailandensis biofilm development and matrix production. J Bacteriol 198:2643–2650. doi: 10.1128/JB.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulrich RL. 2004. Quorum quenching: enzymatic disruption of N-acylhomoserine lactone-mediated bacterial communication in Burkholderia thailandensis. Appl Environ Microbiol 70:6173–6180. doi: 10.1128/AEM.70.10.6173-6180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ooi WF, Ong C, Nandi T, Kreisberg JF, Chua HH, Sun G, Chen Y, Mueller C, Conejero L, Eshaghi M, Ang RM, Liu J, Sobral BW, Korbsrisate S, Gan YH, Titball RW, Bancroft GJ, Valade E, Tan P. 2013. The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet 9:e1003795. doi: 10.1371/journal.pgen.1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramli NS, Eng Guan C, Nathan S, Vadivelu J. 2012. The effect of environmental conditions on biofilm formation of Burkholderia pseudomallei clinical isolates. PLoS One 7:e44104. doi: 10.1371/journal.pone.0044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mongkolrob R, Taweechaisupapong S, Tungpradabkul S. 2015. Correlation between biofilm production, antibiotic susceptibility and exopolysaccharide composition in Burkholderia pseudomallei bpsI, ppk, and rpoS mutant strains. Microbiol Immunol 59:653–663. doi: 10.1111/1348-0421.12331. [DOI] [PubMed] [Google Scholar]
- 25.Goo E, Majerczyk CD, An JH, Chandler JR, Seo YS, Ham H, Lim JY, Kim H, Lee B, Jang MS, Greenberg EP, Hwang I. 2012. Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc Natl Acad Sci U S A 109:19775–19780. doi: 10.1073/pnas.1218092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao D, Bushin LB, Moon K, Wu Y, Seyedsayamdost MR. 2017. Discovery of scmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. Proc Natl Acad Sci U S A 114:E2920–E2928. doi: 10.1073/pnas.1619529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 28.Vial L, Lépine F, Milot S, Groleau M-C, Dekimpe V, Woods DE, Déziel E. 2008. Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J Bacteriol 190:5339–5352. doi: 10.1128/JB.00400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biggins JB, Gleber CD, Brady SF. 2011. Acyldepsipeptide HDAC inhibitor production induced in Burkholderia thailandensis. Org Lett 13:1536–1539. doi: 10.1021/ol200225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majerczyk C, Schneider E, Greenberg EP. 2016. Quorum sensing control of type VI secretion factors restricts the proliferation of quorum-sensing mutants. Elife 5:14712. doi: 10.7554/eLife.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goo E, An JH, Kang Y, Hwang I. 2015. Control of bacterial metabolism by quorum sensing. Trends Microbiol 23:567–576. doi: 10.1016/j.tim.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Martinez S, Déziel E. 2020. Changes in polyhydroxyalkanoate granule accumulation make optical density measurement an unreliable method for estimating bacterial growth in Burkholderia thailandensis. Can J Microbiol 66:256–262. doi: 10.1139/cjm-2019-0342. [DOI] [PubMed] [Google Scholar]
- 33.Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byers JT, Lucas C, Salmond GP, Welch M. 2002. Nonenzymatic turnover of an Erwinia carotovora quorum-sensing signaling molecule. J Bacteriol 184:1163–1171. doi: 10.1128/jb.184.4.1163-1171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Camara M, Smith H, Williams P. 2002. N-Acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun 70:5635–5646. doi: 10.1128/iai.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klaus JR, Deay J, Neuenswander B, Hursh W, Gao Z, Bouddhara T, Williams TD, Douglas J, Monize K, Martins P, Majerczyk C, Seyedsayamdost MR, Peterson BR, Rivera M, Chandler JR. 2018. Malleilactone is a Burkholderia pseudomallei virulence factor regulated by antibiotics and quorum sensing. J Bacteriol 200:e00008-18. doi: 10.1128/JB.00008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Grady EP, Viteri DF, Malott RJ, Sokol PA. 2009. Reciprocal regulation by the CepIR and CciIR quorum sensing systems in Burkholderia cenocepacia. BMC Genomics 10:441. doi: 10.1186/1471-2164-10-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambers CE, Lutter EI, Visser MB, Law PP, Sokol PA. 2006. Identification of potential CepR regulated genes using a cep box motif-based search of the Burkholderia cenocepacia genome. BMC Microbiol 6:104. doi: 10.1186/1471-2180-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva IN, Ramires MJ, Azevedo LA, Guerreiro AR, Tavares AC, Becker JD, Moreira LM. 2017. Regulator LdhR and d-lactate dehydrogenase LdhA of Burkholderia multivorans play roles in carbon overflow and in planktonic cellular aggregate formation. Appl Environ Microbiol 83:e01343-17. doi: 10.1128/AEM.01343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melanson RA, Barphagha I, Osti S, Lelis TP, Karki HS, Chen R, Shrestha BK, Ham JH. 2017. Identification of new regulatory genes involved in the pathogenic functions of the rice-pathogenic bacterium Burkholderia glumae. Microbiology 163:266–279. doi: 10.1099/mic.0.000419. [DOI] [PubMed] [Google Scholar]
- 41.Warawa J, Woods DE. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol Lett 242:101–108. doi: 10.1016/j.femsle.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 42.Ulrich RL, DeShazer D. 2004. Type III secretion: a virulence factor delivery system essential for the pathogenicity of Burkholderia mallei. Infect Immun 72:1150–1154. doi: 10.1128/iai.72.2.1150-1154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–950, 952. doi: 10.2144/00295bm04. [DOI] [PubMed] [Google Scholar]
- 44.Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, Défago G. 1998. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88:678–684. doi: 10.1094/PHYTO.1998.88.7.678. [DOI] [PubMed] [Google Scholar]
- 45.Gallagher LA, Ramage E, Patrapuvich R, Weiss E, Brittnacher M, Manoil C. 2013. Sequence-defined transposon mutant library of Burkholderia thailandensis. mBio 4:e00604-13. doi: 10.1128/mBio.00604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol 74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. 2011. Differential expression in RNA-Seq: a matter of depth. Genome Res 21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapalain A, Groleau MC, Le Guillouzer S, Miomandre A, Vial L, Milot S, Déziel E. 2017. Interplay between 4-hydroxy-3-methyl-2-alkylquinoline and N-acyl-homoserine lactone signaling in a Burkholderia cepacia complex clinical strain. Front Microbiol 8:1021. doi: 10.3389/fmicb.2017.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subsin B, Chambers CE, Visser MB, Sokol PA. 2007. Identification of genes regulated by the cepIR quorum-sensing system in Burkholderia cenocepacia by high-throughput screening of a random promoter library. J Bacteriol 189:968–979. doi: 10.1128/JB.01201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–ΔΔC(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Pilatova M, Dionne MS. 2012. Burkholderia thailandensis is virulent in Drosophila melanogaster. PLoS One 7:e49745. doi: 10.1371/journal.pone.0049745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.