The LysR-type transcriptional regulators are present in viruses, archaea, bacteria, and eukaryotic cells. Furthermore, these proteins are the most abundant transcriptional factors in bacteria. Here, we demonstrate that two LysR-type proteins are generated from the ltrR gene. These proteins are genetically induced by pH and repressed at the promoter and coding regions by the global regulators H-NS and Lrp. Thus, novel basic aspects of the complex genetic regulation of the LysR-type transcriptional regulators are described.
KEYWORDS: Salmonella Typhi, LysR-type family, UP element, LtrR, H-NS, Lrp
ABSTRACT
LtrR is a LysR-type regulator involved in the positive expression of ompR to promote ompC and ompF expression. This regulatory network is fundamental for the control of bacterial transformation and resistance to the bile salt sodium deoxycholate in Salmonella enterica serovar Typhi. In this work, the transcriptional regulation of ltrR was characterized, revealing that the use of alternative promoters results in two transcripts. The larger one, the ltrR2 mRNA, was repressed at promoter and coding regions by H-NS, whereas Lrp repressed its expression at the coding region. In the case of the second and shorter ltrR1 transcript, it was repressed only at the coding region by H-NS and Lrp. Remarkably, pH 7.5 is a positive signal involved in the transcriptional expression of both ltrR units. Translational fusions and Western blot experiments demonstrated that ltrR2 and ltrR1 mRNAs encode the LtrR2 and LtrR1 proteins. This study adds new data on the complex genetic and regulatory characteristics of one of the most predominant types of transcriptional factors in bacteria, the LysR-type transcriptional regulators.
IMPORTANCE The LysR-type transcriptional regulators are present in viruses, archaea, bacteria, and eukaryotic cells. Furthermore, these proteins are the most abundant transcriptional factors in bacteria. Here, we demonstrate that two LysR-type proteins are generated from the ltrR gene. These proteins are genetically induced by pH and repressed at the promoter and coding regions by the global regulators H-NS and Lrp. Thus, novel basic aspects of the complex genetic regulation of the LysR-type transcriptional regulators are described.
INTRODUCTION
Salmonella enterica serovar Typhi is the etiological agent of typhoid fever. This bacterium is able to survive in the presence of bile salts in the small intestine and gallbladder. An association between S. Typhi and gallbladder cancer has also been reported (1, 2). In contrast to Salmonella enterica serovar Typhimurium, where a considerable amount of information has been reported regarding bile resistance (3–14), genetic studies related to the survival of S. Typhi in the gallbladder and in the presence of bile are limited (10, 16). In this regard, it was shown previously that the genetic network ltrR-ompR-ompC has a fundamental role in the resistance of S. Typhi IMSS-1 to the bile salt sodium deoxycholate. The ltrR gene encodes a LysR-type regulator that is highly conserved in the Salmonella genus, since its presence has been detected in the genome sequences of S. Typhimurium (17), S. Agona (18), S. enterica subsp. arizonae (19), S. Choleraesuis (20), S. Dublin (21), and S. Enteritidis (22), among other Salmonella serovars, and interestingly, it is absent in Escherichia coli. Microarray-based studies suggest that the ltrR gene homologue STM0030 is involved in the pathogenicity of S. Typhimurium (23, 24).
LtrR is one of the 45 LysR-type regulators present in the Salmonella Typhi genome (25). It consists of 331 amino acids (aa), with an N-terminal DNA-binding helix-turn-helix (HTH) domain from residues 13 to 71 and a C-terminal domain comprising residues 149 to 289. Since LtrR indirectly regulates porin synthesis and is involved in bacterial transformation and bile resistance (26), we decided to better understand the transcriptional regulation of the ltrR gene. The results showed that this LysR-type gene encodes two proteins whose transcriptional expression is regulated, at their promoters and coding regions, by environmental signals and transcriptional global regulators in S. Typhi IMSS-1.
(This research was conducted by J. E. Rebollar-Flores in partial fulfillment of the requirements for a doctoral degree from the Posgrado en Ciencias Biológicas of the Universidad Nacional Autónoma de México.)
RESULTS
The ltrR gene contains two independent promoters, ltrRP2 and ltrRP1, located in its regulatory region and coding sequence, respectively.
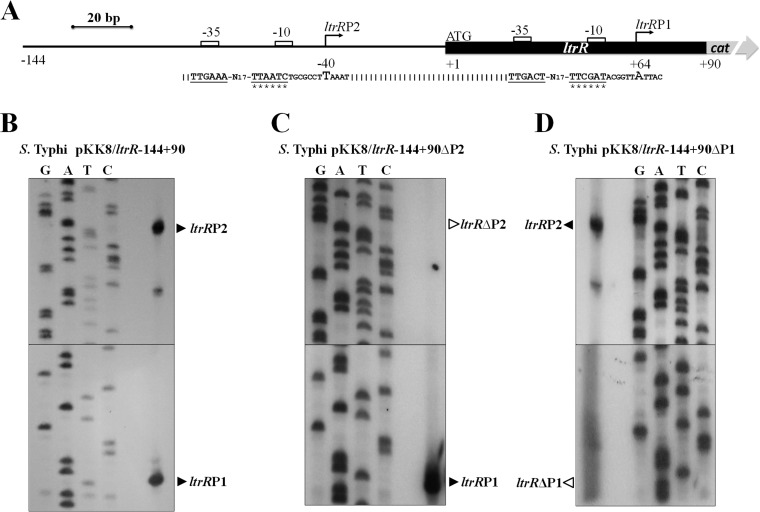
We previously demonstrated that ltrR was induced in N-minimal medium (N-MM) (26). To determine the promoter region that controls its expression, primer extension experiments were performed with total RNA obtained from S. Typhi harboring the pKK8/ltrR-144+90 plasmid. This construct was used since it harbors sequences upstream and downstream of the ltrR ATG translational start codon. The bacterial strain was grown in N-MM, and the primer extension results showed that ltrR contains two promoters. One of them, ltrRP2, includes a transcriptional start site localized 40 bp upstream of the ATG initiation codon. The second promoter, ltrRP1, contains a transcriptional initiation site 64 bp downstream of the ATG translational start site (Fig. 1A and B).
FIG 1.
ltrR transcriptional start sites. (A) Schematic diagram of the regulatory region of the ltrR gene. The bent arrows and bold letters represent the transcriptional starts sites; −10 and −35 boxes are shown as white rectangles, and the corresponding sequences are underlined; the nucleotide substitutions in the −10 box are indicated by asterisks; and ATG is the ltrR canonical translational initiation codon. (B) Primer extension of the ltrR regulatory region performed with total RNA of S. Typhi plus pKK8/ltrR-144+90. Two transcriptional initiation sites located upstream at nucleotide −40 and downstream at nucleotide +64, with respect to the initial canonical ATG codon, were identified, and the promoter regions were designated ltrRP2 and ltrRP1, respectively. (C) Primer extension using S. Typhi plus pKK8/ltrR-144+90ΔP2 containing a substitution of the ltrRP2 −10 box. (D) Primer extension using S. Typhi plus pKK8/ltrR-144+90ΔP1 containing a substitution of the ltrRP1 −10 box. Black triangles indicate the transcriptional start sites.
To confirm that these promoter regions control ltrR expression, specific substitutions of the −10 TATA box of each promoter (Fig. 1A) were obtained in the chromosome, and the mutations were cloned independently to generate plasmids pKK8/ltrR-144+90ΔP2 and pKK8/ltrR-144+90ΔP1. The plasmids were transformed into S. Typhi IMSS-1, and primer extension experiments were performed. The results obtained with the substitutions of the −10 box of the ltrRP2 promoter (pKK8/ltrR-144+90ΔP2) demonstrated the absence of the corresponding transcriptional initiation site and confirmed the presence of the ltrRP1 transcriptional start site (Fig. 1C). In the substitutions of the −10 box of the ltrRP1 promoter (pKK8/ltrR-144+90ΔP1), the presence of the ltrRP2 transcriptional start site and the absence of the ltrRP1 transcriptional initiation site were evident (Fig. 1D). Furthermore, transcriptional expression of the pKK8/ltrR-144+90 wild-type fusion showed transcriptional activity of 1,529 chloramphenicol acetyltransferase (CAT) units. However, in the fusions pKK8/ltrR-144+90ΔP2 and pKK8/ltrR-144+90ΔP1, the expression values were 1,007 and 659 CAT units, respectively, illustrating the contribution of each promoter to the expression of ltrR (Table 1).
TABLE 1.
Transcriptional activities of the ltrRP2 and ltrRP1 wild-type promoters and mutated derivativesa
| Plasmid | Mean CAT sp act (μmol min−1 mg−1) for S. Typhi IMSS-1 ± SD |
|---|---|
| pKK8/ltrR-144+90 | 1,529 ± 101 |
| pKK8/ltrR-144+90ΔP2 | 1,007 ± 6 |
| pKK8/ltrR-144+90ΔP1 | 659 ± 2 |
| pKK8/ltrRP2-471-1 | 521 ± 32 |
| pKK8/ltrRP1+7+90 | 1,109 ± 60 |
| pKK232-8 | 0 ± 0 |
The fusions were named according to the ltrR translational initiation site; the numbers in each fusion represent the distances upstream and downstream of the ATG start site, with the fusions pKK8/ltrR-144+90ΔP1 and pKK8/ltrR-144+90ΔP2 containing individual substitutions in the −10 box. pKK232-8 was introduced into the S. Typhi wild-type strain to determine basal activity. The right column represents the expression levels of the ltrR fusions evaluated in the wild-type S. Typhi IMSS-1 strain. The activities were determined at an OD595 of 0.6 in N-MM. The values are the means ± standard deviations from at least three independent experiments performed in duplicate (n ≥ 6).
To determine whether each promoter is expressed independently, a fusion containing only the ltrRP2 promoter (pKK8/ltrRP2-471-1) and a fusion containing only the ltrRP1 promoter (pKK8/ltrRP1+7+90) were constructed: their transcriptional activities rendered 521 and 1,109 CAT units, respectively (Table 1). Therefore, the presence of two ltrR promoters, ltrRP2 and ltrRP1, located upstream and downstream of the ltrR ATG initiation codon was determined by substitutions of the TATA boxes in the putative promoters fused to transcriptional gene fusions and assessment by primer extension experiments (Fig. 1 and Table 1).
H-NS and Lrp are involved in the negative regulation of ltrR2 and ltrR1.
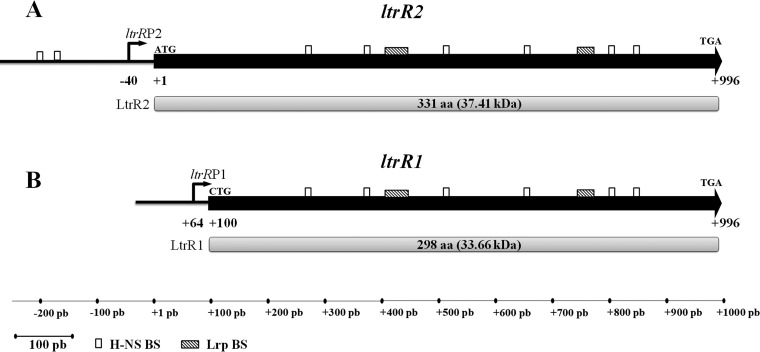
The LysR-type ltrR gene contains a transcriptional initiation site located 40 bp upstream of the ltrR ATG translational start site, which is followed by a complete ltrR open reading frame (ORF) of 996 bp. This large transcriptional unit was denoted ltrR2 (Fig. 2A). A second transcriptional start site was found within the ltrR coding region, 64 bp downstream of the canonical ltrR ATG translational start site, and the resulting 897-bp ORF initiates at nucleotide +100 (CTG-leucine-) and finishes at nucleotide +996 (TGA). This shorter transcriptional unit was denoted ltrR1 (Fig. 2B). Both ltrR2 and ltR1 encode independent putative proteins in the same ORF, denoted LtrR2 (331 aa) and LtrR1 (298 aa) (Fig. 2). To identify the regulatory sequences and genetic elements involved in the transcriptional control of ltrR2 and ltrR1, virtual footprinting was carried out using the RSAT matrix-scan program (http://embnet.ccg.unam.mx/rsat/matrix-scan_form.cgi) and the MAST program (http://meme-suite.org/tools/mast), selecting those sites with a higher probability and/or weight value. The results showed H-NS-binding motifs upstream of the ltrR2 transcriptional start site and within the ltrR2 coding region, whereas Lrp-binding motifs were observed only in the ltrR2 coding sequence (Fig. 2A). The same H-NS- and Lrp-binding sites located in the ltrR2 coding region were also present in the respective ltrR1 coding region (Fig. 2B).
FIG 2.
ltrR2 and ltrR1 genetic organization. Shown are schematic representations of ltrR2 and its corresponding LtrR2 protein of 331 aa (37.41 kDa) (A) and of ltrR1 and its corresponding LtrR1 protein of 298 aa (33.41 kDa) (B). The white rectangles show the putative H-NS-binding sites (BS) predicted by the RSAT matrix-scan program and MATS program (−207TTTATAAAAT−198, −175TTTATAAAAT−166, +266GCGATAAAAC+275, +369TCGATATATT+378, +508CAGATAAATT+517, +649TGAATAAATC+658, +798GCGTTATAAT+807, and +842TTCGTACATT+851). The following Lrp-binding sites (shaded rectangles) were also predicted: +403ACAGGGGAATACTCGCCAGCCTCCATGCTGACGCATGGCT+442 and +744GAAAAAAGCGATATGGTTGCGATTTTGCC+772. Bent arrows represent transcriptional start sites. Translational initiation and stop codons are indicated.
To determine the effect of these regulatory proteins on the transcriptional control of ltrR2 and ltrR1, fusions of different lengths were generated, and their expression was evaluated in wild-type S. Typhi IMSS-1, isogenic individual hns and lrp mutants, and an hns lrp doubly deficient strain grown in N-MM at an optical density at 595 nm (OD595) of 0.6.
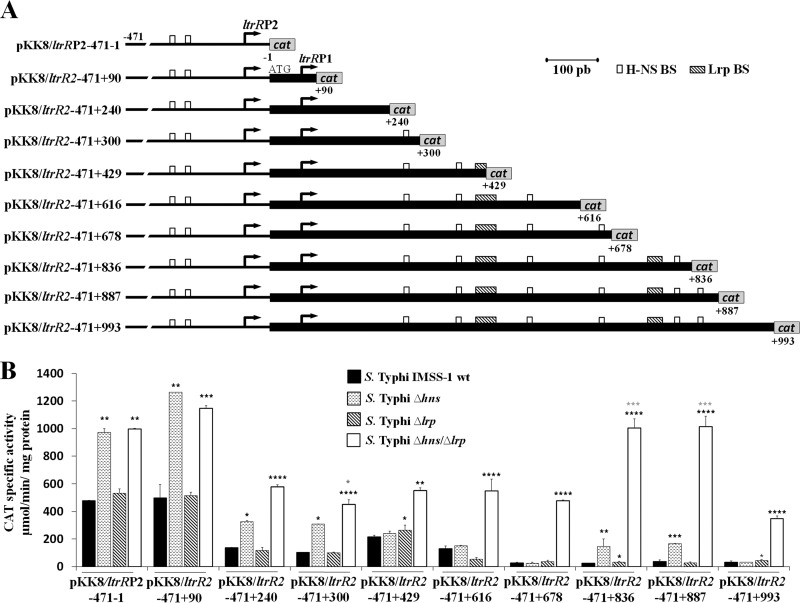
Initially, we evaluated a collection of fusions that contained the ltrR2 and ltrR1 regulatory regions as well as different lengths of the ltrR gene (Fig. 3A). In the S. Typhi IMSS-1 wild type, the activities of the fusions pKK8/ltrRP2-471-1 and pKK8/ltrR2-471+90 were higher than the activities of pKK8/ltrR2-471+240, pKK8/ltrR2-471+300, pKK8/ltrR2-471+429, pKK8/ltrR2-471+616, pKK8/ltrR2-471+678, pKK8/ltrR2-471+836, pKK8/ltrR2-471+887, and pKK8/ltrR2-471+993 (Fig. 3B). This indicated that from nucleotides +90 to +993, negative regulatory elements in cis modulate ltrR expression.
FIG 3.
Transcriptional profile of ltrR2. (A) Fusions were constructed in the pKK232-8 vector and labeled with respect to the ltrR2 ATG initiation codon. The coordinates are given. The white and shaded rectangles show the putative H-NS- and Lrp-binding sites, respectively. (B) Transcriptional profile of ltrR2 fusions evaluated in the wild-type S. Typhi IMSS-1, S. Typhi Δhns, S. Typhi Δlrp, and S. Typhi Δhns Δlrp strains in N-MM at an OD595 of 0.6. The values are the means ± standard deviations from at least three independent experiments performed in duplicate (n ≥ 6). Statistically different values are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) comparing wild-type (wt) S. Typhi IMSS-1 versus mutant strains expression levels (black asterisks) and S. Typhi Δhns Δlrp versus S. Typhi Δhns (gray asterisks).
Since the in silico analysis suggested the presence of H-NS-binding motifs upstream of the ltrR2 transcriptional start site as well as in its coding region (Fig. 2A), we determined whether H-NS specifically repressed the ltrR2 promoter. The pKK8/ltrRP2-471-1 fusion, containing only the ltrR2 promoter sequence, was analyzed individually in the hns mutant, rendering transcriptional values that were higher than those of the wild-type strain (Fig. 3B). Remarkably, two H-NS-binding sites (positions −207 to −198 and −175 to −166) are present in the ltrR2 promoter region according to the bioinformatic analysis. Thus, the in silico and CAT specific activity results showed that the ltrR2 promoter is repressed by H-NS.
H-NS also repressed the ltrR2 coding sequence, since the transcriptional expression levels of the fusions pKK8/ltrR2-471+90, pKK8/ltrR2-471+240, pKK8/ltrR2-471+300, pKK8/ltrR2-471+836, and pKK8/ltrR2-471+887 were higher in the hns-deficient strain than in the wild-type strain (Fig. 3B). Interestingly, the last three constructs contained in silico-predicted H-NS-binding sites located at +266GCGATAAAAC+275, +798GCGTTATAAT+807, and +842TTCGTACATT+851, respectively. Therefore, the in silico analysis supported the transcriptional results on the negative role of H-NS in ltrR2 expression.
Regarding the role of Lrp in the control of the ltrR2 promoter, analyses of a construct containing only this promoter, pKK8/ltrRP2-471-1, were performed in the wild-type and Δlrp strains. The expression results were similar, indicating that this protein alone does not regulate the ltrR2 promoter (Fig. 3B). Additionally, experiments were performed with the lrp single mutant and fusions harboring the ltrR2 coding region. The results showed that Lrp alone is not involved in ltrR2 repression at the coding region since the expression levels of all the fusions evaluated in this mutant were similar to those of the wild-type strain (Fig. 3B).
As mentioned above, the lrp mutant did not show a role in ltrR2 expression. To confirm these results, we decided to evaluate the activities of all ltrR2 fusions in an hns lrp double mutant. We found that H-NS is the only repressor of the ltrR2 promoter since the fusions pKK8/ltrRP2-471-1 and pKK8/ltrR2-471+90 displayed the same activity in the hns lrp double mutant as in the single Δhns strain. However, the fusions pKK8/ltrR2-471+240, pKK8/ltrR2-471+300, pKK8/ltrR2-471+429, pKK8/ltrR2-471+616, pKK8/ltrR2-471+678, pKK8/ltrR2-471+836, pKK8/ltrR2+471+887, and pKK8/ltrR2+471+993 showed higher transcriptional expression levels in S. Typhi Δhns Δlrp than in the single hns or lrp mutant strain or the wild-type strain (Fig. 3B). In spite of the null effect of the lrp single mutant on the expression of ltrR2, the activity values obtained with the hns lrp double mutant support the notion that H-NS and Lrp together repress the expression of the ltrR2 coding region (Fig. 3B).
The data mentioned above showed the negative roles of H-NS and Lrp in the expression of the ltrR2 coding region, and since this region is shared with ltrR1, we determined its transcriptional expression in a collection of fusions that contain only ltrR1 (Fig. 4A).
FIG 4.
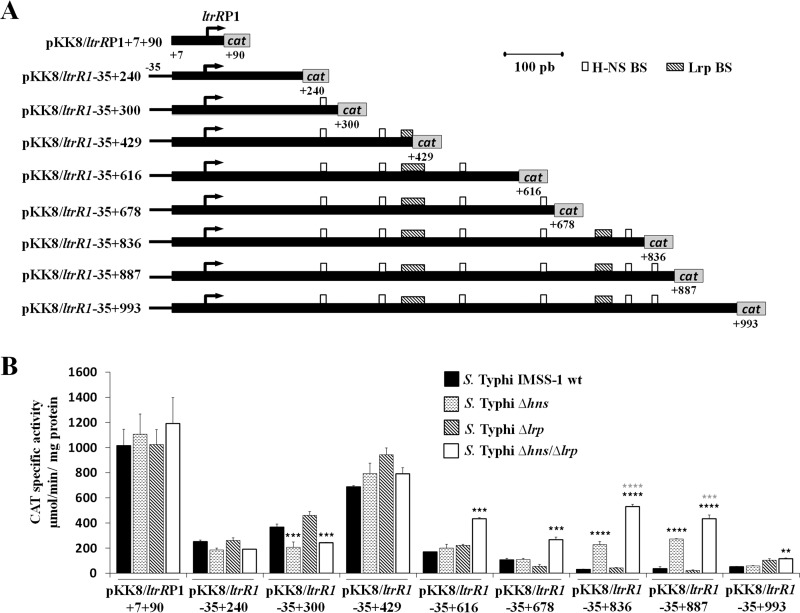
Transcriptional profile of ltrRP1. (A) The fusions were constructed in the pKK232-8 vector and labeled with regard to the ltrR ATG initiation codon. The coordinates are indicated. The white and shaded rectangles show the putative H-NS- and Lrp-binding sites, respectively. (B) Transcriptional profile of the fusions that contained only the ltrRP1 promoter and different regions of the ltrR1 coding sequence. Transcriptional expression (CAT units) was evaluated in the wild-type S. Typhi IMSS-1, S. Typhi Δhns, S. Typhi Δlrp, and S. Typhi Δhns Δlrp strains in N-MM at an OD595 of 0.6. The values are the means ± standard deviations from at least three independent experiments performed in duplicate (n ≥ 6). Statistically different values are indicated (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001) comparing wild-type S. Typhi IMSS-1 versus mutants strain expression levels (black asterisks) and S. Typhi Δhns Δlrp versus S. Typhi Δhns (gray asterisks).
The wild-type strain with a fusion that contained only the ltrR1 promoter (pKK8/ltrRP1+7+90) expressed 1,015 CAT units. The fusions containing the promoter and different lengths of ltrR1 (pKK8/ltrR1-35+240 and pKK8/ltrR1-35+300) had 251 and 364 CAT units, respectively. With pKK8/ltrR1-35+429, the value increased to 685 CAT units, and the values again decreased substantially in the fusions pKK8/ltrR1-35+616, pKK8/ltrR1-35+678, pKK8/ltrR1-35+836, pKK8/ltrR1-35+887, and pKK8/ltrR1-35+993 to 167, 105, 27, 35, and 49 CAT units, respectively. These results show that positive and negative genetic elements are involved in ltrR1 transcriptional expression (Fig. 4B).
Since ltrR1 is regulated by H-NS, we evaluated the role of this histone-like protein in ltrR1 expression, finding that the ltrR1 promoter is not repressed by H-NS since with a construct that contains only the ltrR1 promoter (pKK8/ltrRP1+7+90), the values for the wild-type strain and the hns mutant strain were similar (1,015 and 1,104 CAT units, respectively) (Fig. 4B). Nevertheless, H-NS regulates the ltrR1 coding region, since the activity values of the fusions pKK8/ltrR1-35+836 and pKK8/ltrR1-35+887, which include the ltrR1 coding region, were low for the wild-type strain (27 and 35 CAT units) compared with the high expression values for the hns mutant (224 and 372 CAT units) (Fig. 4B). Thus, H-NS represses the ltrR1 coding region only at nucleotides +678 to +887 with respect to the canonical ATG codon of ltrR. Interestingly, at bases +798 to +807 and +842 to +851, H-NS-binding sites were predicted. Thus, the in silico results together with the transcriptional expression of fusions encompassing these regions supported negative H-NS regulation of the ltrR1 coding region.
Regarding the role of Lrp in the regulation of the ltrR1 promoter, we evaluated the pKK8/ltrRP1+7+90 fusion in the Δlrp strain. The activity values obtained were similar to those of the wild-type strain (1,015 and 1,022 CAT units) (Fig. 4B). Therefore, Lrp does not regulate the ltrR1 promoter.
Moreover, the transcriptional expression of fusions containing the promoter and different lengths of the ltrR1 coding region (pKK8/ltrR1-35+240, pKK8/ltrR1-35+300, pKK8/ltrR1-35+429, pKK8/ltrR1-35+616, pKK8/ltrR1-35+678, pKK8/ltrR1-35+836, pKK8/ltrR1-35+887, and pKK8/ltrR1-35+993) was evaluated in the Δlrp strain. The specific CAT activities observed were similar in the wild-type strain and the lrp mutant (Fig. 4B), indicating that Lrp alone is not involved in the regulation of the ltrR1 coding region.
To confirm the data mentioned above, the expression of ltrR1 fusions was also evaluated in the hns lrp double mutant strain. The transcriptional values of the ltrR1 promoter (pKK8/ltrRP1+7+90) indicated that these two global regulatory proteins are not involved in ltrR1 regulation (Fig. 4B). However, the transcriptional expression of pKK8/ltrR1-35+616, pKK8/ltrR1-35+678, pKK8/ltrR1-35+836, and pKK8/ltrR1-35+887 increased to 432, 264, 529, and 432 CAT units in the hns lrp double mutant compared with the values observed for the wild type (167, 105, 27, and 35 CAT units, respectively) or for the individual hns (199, 107, 224, and 272 CAT units, respectively) and lrp (219, 51, 40, and 21 CAT units, respectively) mutant strains (Fig. 4B), supporting the notion that ltrR1 is repressed by both H-NS and Lrp at its coding region.
The data indicate that in the presence of H-NS, Lrp has a minor effect, although in its absence, Lrp represses the ltrR2 and ltrR1 coding regions (Fig. 3B and Fig. 4B), suggesting that Lrp compensates for the absence of H-NS.
In conclusion, all the data indicate that ltrR2 is repressed at its promoter and coding region by H-NS, whereas Lrp represses its expression only at the coding region. In the case of the second and shorter ltrR1 transcript, it was repressed only at the coding region by H-NS and Lrp.
H-NS and Lrp directly repress ltrR2 and ltrR1.
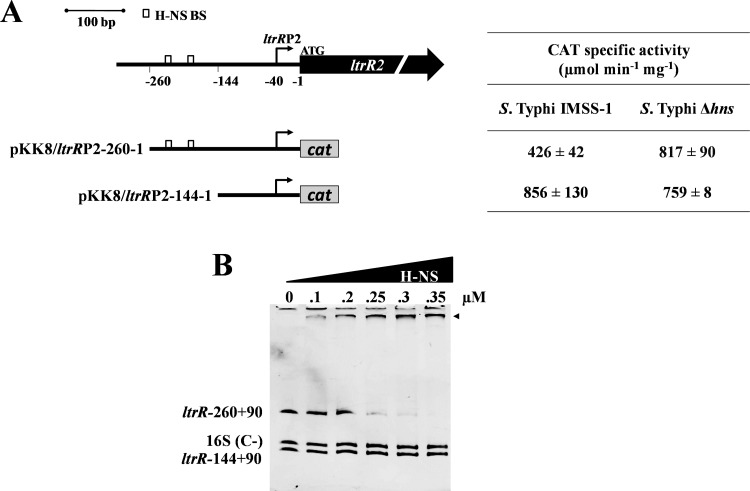
Since the in silico analysis suggested the presence of H-NS-binding motifs upstream and downstream of the ltrR2 transcriptional start site (Fig. 2A), and the transcriptional analysis also indicated a role for H-NS in regulating the ltrR2 promoter, we delimited the specific H-NS-binding sites located in the ltrR2 promoter. Thus, the fusions pKK8/ltrRP2-260-1, containing two putative H-NS-binding sites (residues −207 to −198 and −175 to −166), and pKK8/ltrRP2-144-1, lacking these H-NS motifs, were constructed, and their expression profiles were analyzed in the wild-type strain and the hns mutant (Fig. 5A). The results showed that H-NS represses the ltrR2 promoter at nucleotides −260 to −144 since the lack of this region (pKK8/ltrR2-144-1) showed a 2-fold increase in transcriptional activity in the wild-type strain (856 CAT units versus 426 CAT units when the region was present), and the values were similar to those observed in the hns mutant strain (759 CAT units) (Fig. 5A). Furthermore, this region is rich in A-T nucleotides, a characteristic of H-NS-binding sites. Considering these results, we determined whether H-NS directly repressed the ltrR2 promoter. A gel retardation experiment showed that H-NS binds at nucleotides −260 to +90 but does not bind to nucleotides −144 to +90, consistent with the notion that H-NS directly represses the ltrR2 promoter by interacting with nucleotides −260 to −144 (Fig. 5B).
FIG 5.
H-NS binds to ltrRP2. (A, left) Diagram of ltrRP2 regulatory region. White rectangles represent predicted in silico H-NS-binding sites (−207TTTATAAAAT−198 and −175TTTATAAAAT−166). Below the diagram of the ltrR2 regulatory region, transcriptional fusions are shown and were named according to the ltrR2 translational initiation site. (Right) Expression of the ltrR fusions evaluated in the wild-type S. Typhi IMSS-1 and S. Typhi Δhns strains. The activities were determined at an OD595 of 0.6 in N-MM. The values are the means ± standard deviations from three independent experiments performed in duplicate (n = 6). (B) EMSAs were performed with purified H-NS protein and the regions spanning residues −260 to +90 and −144 to +90 of the ltrR gene. A 262-bp fragment of the 16S rRNA of S. Typhi was used as a negative control [16S (C−)].
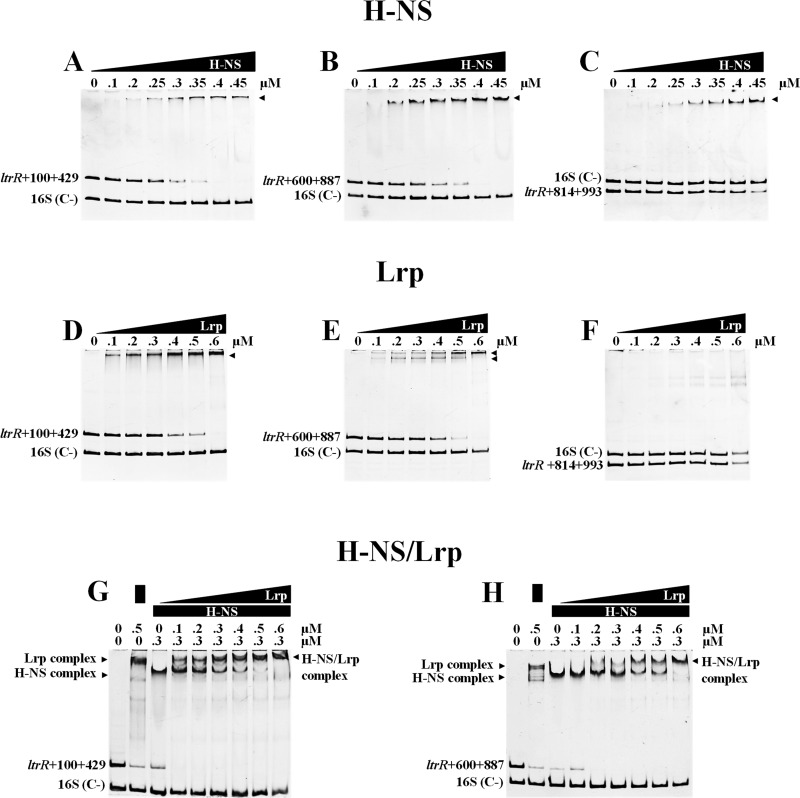
The transcriptional results showed that H-NS repressed the ltrR2 coding region at nucleotides +90 to +429 and +678 to +887 (Fig. 3B). Additionally, reports of chromatin immunoprecipitation on chip (ChIP-on-chip) analysis have shown that H-NS is able to bind to coding regions (27, 28). Thus, we evaluated whether H-NS could interact with fragments spanning residues +100 to +429, +600 to +887, and +814 to +993 of the ltrR2 coding region. Moreover, these fragments encompass H-NS-binding sites (+266GCGATAAAAC+275, +798GCGTTATAAT+807, and +842TTCGTACATT+851) predicted by virtual footprinting.
The gel shift experiment results showed that H-NS binds at nucleotides +100 to +429 (Fig. 6A), +600 to +887 (Fig. 6B), and +814 to +993, although this ltrR2 region showed less affinity for H-NS (Fig. 6C). These results support the notion that H-NS directly represses the ltrR2 coding region and are in agreement with the expression results showing that these ltrR2 regions are repressed by H-NS (Fig. 3B). Eight putative H-NS-binding sites were identified by bioinformatic analysis. However, transcriptional expression profiles together with electrophoretic mobility shift assay (EMSA) results support the notion that two H-NS motifs are located in the ltrR2 promoter region, at nucleotides −207 to −198 and −175 to −166, and that three are located in the ltrR2 coding region, at nucleotides +266 to +275, +798 to +807, and +842 to +851, which would also be involved in ltrR2 repression.
FIG 6.
H-NS and Lrp bind to ltrR2 and ltrR1. (A to C) EMSAs were performed with purified H-NS and different fragments of the ltrR coding region. (D to F) EMSAs with purified Lrp and different fragments of the ltrR coding region. (G and H) EMSAs with purified H-NS–Lrp proteins and different fragments of the ltrR coding region. The fragments were named according to the ltrR translational initiation site. As a negative control, we used a structural fragment (262 bp) of the 16S rRNA gene (STYr016) of S. Typhi [16S (C−)].
Furthermore, H-NS also binds and directly represses ltrR1, since nucleotides +600 to +887 and +814 to +993 are part of the ltrR1 coding region and include bases +678 to +887, which are implicated in ltrR1 repression, according to transcriptional fusions (Fig. 4B). It is relevant to mention that these fragments contained two putative H-NS-binding sites (residues +798 to +807 and +842 to +851) involved in ltrR2 and ltrR1 repression.
With respect to Lrp, EMSAs were also performed with the purified protein and the different fragments of the ltrR coding region. The results showed that this transcriptional factor binds at nucleotides +100 to +429 (Fig. 6D) and +600 to +887 (Fig. 6E) but not in the region at nucleotides +814 to +993 (Fig. 6F). These data and the transcriptional expression profiles from Fig. 3 showed that the region at nucleotides +100 to +429 contains an Lrp regulatory site implicated in ltrR2 repression. With respect to the fragment spanning nucleotides +600 to +887, it contains an Lrp-binding motif identified in silico (nucleotides +744 to +772). Thus, the EMSA data, transcriptional results (Fig. 3B and Fig. 4B), and bioinformatic analysis support the presence of Lrp regulatory regions involved in ltrR2 and ltrR1 negative regulation. These experiments indicate that Lrp represses activity only within the ltrR2 and ltrR1 coding regions but not at the upstream promoter region. Therefore, H-NS as well as Lrp are able to interact with the ltrR2 and ltrR1 coding regions, silencing their transcriptional expression.
Previously, we demonstrated that H-NS and Lrp together are able to interact with the CRISPR-Cas cse1 regulatory region from S. Typhi (29). To determine whether H-NS and Lrp interact simultaneously with the ltrR2 and ltrR1 coding regions, EMSAs using both proteins were performed. The experiments showed that the complexes formed by H-NS and ltrR fragments encompassing nucleotides +100 to +429 or +600 to +887, in the presence of increasing amounts of Lrp, displayed higher molecular weights than those obtained in the presence of only the H-NS or the Lrp protein (Fig. 6G and H), indicating that they are able to bind together to the ltrR2 and ltrR1 coding regions.
All these data support a model where H-NS and Lrp together directly modulate the expression of the independent transcriptional units ltrR2 and ltrR1 in S. Typhi IMSS-1.
Basic pH and an UP element are involved in the induction of ltrR2 and ltrR1 expression.
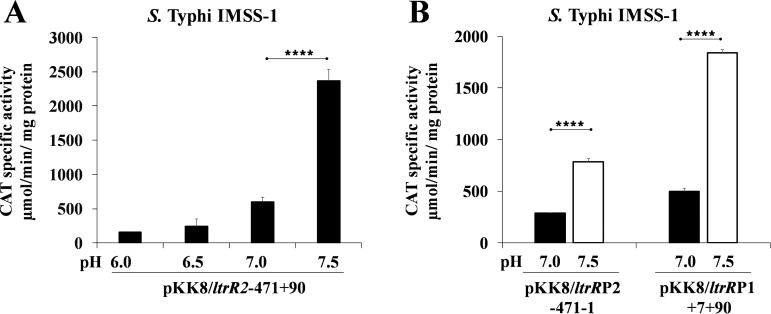
The results presented above demonstrated the presence of negative regulators involved in the expression of ltrR2 and ltrR1. Moreover, these transcriptional units were expressed in N-MM, and we previously demonstrated that ltrR was involved in resistance to the human bile salt sodium deoxycholate (26). To determine whether sodium deoxycholate or N-MM components, including pH, are responsible for ltrR2 and ltrR1 induction, we evaluated the influence of these factors on the expression of these transcriptional units. Initially, we evaluated the fusion pKK8/ltrR2-471+90, which contains both the ltrRP2 and ltrRP1 promoters. No effect was observed in the presence or absence of the human bile salt sodium deoxycholate or with any of the N-MM components, including KCl, (NH4)2SO4, K2SO4, KH2PO4, MgCl2, glycerol, and Casamino Acids. However, the transcriptional results at pH 6.0, 6.5, 7.0, and 7.5 showed that ltrR expression was upregulated by basic pH, with maximum values obtained at pH 7.5 (Fig. 7A). These experiments were also performed at pH 7.8 and 8.0. However, S. Typhi showed a deficient growth rate, and therefore, CAT activity was not measured at these pH values (data not shown).
FIG 7.
pH 7.5 is a positive signal for ltrR expression. Shown are transcriptional profiles of S. Typhi IMSS-1 harboring pKK8/ltrR2-471+90 grown in N-MM (Tris-HCl at 200 mM) at pH 6.0, 6.5, 7.0, and 7.5 (A) and the fusions pKK8/ltrRP2-471-1 and pKK8/ltrRP1+7+90 evaluated in N-MM (Tris-HCl at 200 mM) at pH 7.0 or 7.5 (B). Transcriptional activities were determined at an OD595 of 0.6. The values are the means ± standard deviations from two independent experiments performed in duplicate (n = 4). Statistically different values are indicated (****, P < 0.0001).
To determine whether the ltrRP2 or ltrRP1 promoter is upregulated by pH, independent fusions (pKK8/ltrRP2-471-1 and pKK8/ltrRP1+7+90) of each promoter were evaluated at pH 7.0 and pH 7.5 in the S. Typhi wild-type strain (Fig. 7B). The results showed that both promoters are induced by slightly alkaline pH, since higher activity values were obtained at pH 7.5 (Fig. 7B). Thus, pH 7.5 is involved in the positive control of both the ltrR2 and ltrR1 transcripts.
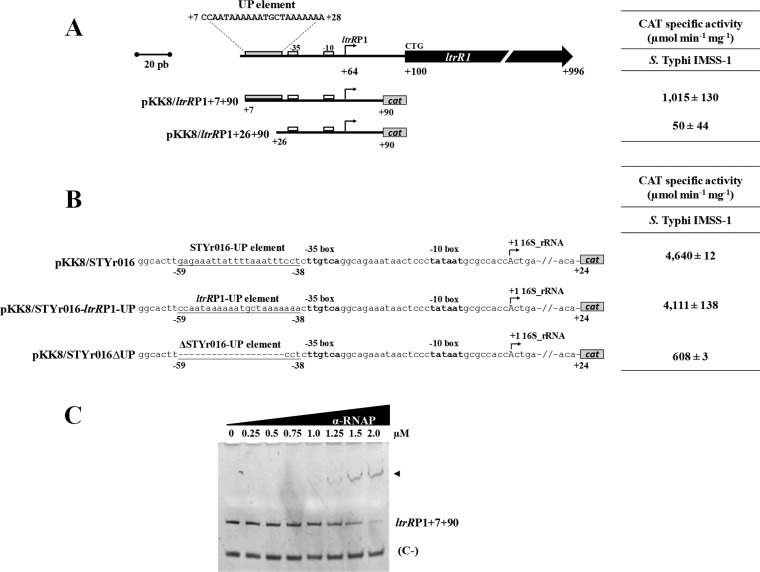
The data mentioned above showed that pH is involved in ltrR2 and ltrR1 expression. Since a characterization of the ltrRP2 promoter is described above, we decided to further identify additional elements involved in the regulation of ltrRP1. Therefore, the expression of the pKK8/ltrRP1+7+90 and pKK8/ltrRP1+26+90 constructs was analyzed in wild-type S. Typhi IMSS-1. The activity values obtained were 1,015 and 50 CAT units, respectively, indicating that from residues +7 to +26 with respect to the ltrR ATG codon, positive cis-regulatory elements are present for ltrRP1 (Fig. 8A).
FIG 8.
ltrR1 contains a functional UP element. (A, left) Schematic diagram of the regulatory region of ltrR1. The bent arrows represent the transcriptional start site, and the ltrRP1-UP element sequence and its localization (residues +7 to +28) are shown. Below the diagram, transcriptional fusions were named according to the canonical ltrR1 translational initiation site. (Right) Expression of the ltrR1 fusions evaluated in wild-type S. Typhi IMSS-1. The activities were determined at an OD595 of 0.6 in N-MM. The values are the means ± standard deviations from at least three independent experiments performed in duplicate (n = 6). (B, left) Transcriptional fusions of the STYr016 ribosomal promoter sequence of S. Typhi. The fusion pKK8/STYr016 contains an UP element as well as the −10 and −35 boxes fused to the cat reporter gene. In a second fusion, pKK8/STYr016-ltrRP1-UP, its native UP element was replaced by the ltrRP1-UP element. Finally, the fusion pKK8/STYr016ΔUP includes the −35 and −10 boxes without its native UP element. (Right) Expression of the fusions mentioned above evaluated upon growth in N-MM at an OD595 of 0.6. (C) EMSAs were performed with the purified RNA polymerase α-subunit and with ltrRP1+7+90 containing the native UP element. A 50-bp fragment of the cse1 promoter region of S. Typhi was used as a negative control (C−).
Remarkably, the region at residues +7 to +28 is rich in A-T nucleotides and resembles an UP element described for the rrnB 16S rRNA in E. coli (30) (Fig. 8A). Furthermore, this S. Typhi A-T region is located at the 5′ end of the −35 box of ltrRP1, a canonical characteristic of UP elements. To determine whether this region corresponded to an UP element, we cloned nucleotides +7 to +28 immediately after the −35 box of the 16S rRNA gene of S. Typhi (STYr016), obtaining the transcriptional fusion pKK8/STYr016-ltrRP1-UP. The expression results for this construct were similar (4,111 CAT units) to those of the fusion pKK8/STYr016 of the wild-type S. Typhi 16S rRNA promoter (4,640 CAT units) that contains its native UP element. Additionally, as expected, the lack of the native UP element in the 16S rRNA (pKK8/STYr016ΔUP) promoter showed a substantial decrease in transcriptional activity (608 CAT units) (Fig. 8B), indicating that nucleotides +7 to +28 from the ltrR2 ATG codon function as an UP element for the ltrRP1 promoter.
Previous studies demonstrated that UP elements interact with the α-subunit of the RNA polymerase (α-RNAP) (30). To evaluate whether the ltrRP1 UP element interacts with the α-RNAP, EMSAs were performed with the purified α-subunit and nucleotides +7 to +90. The experiments showed DNA-protein complexes using 1.25, 1.5, or 2.0 μM the α-RNAP (Fig. 8C). The amount of the interacting protein is in agreement with those used in previous studies (30–32). Therefore, all these results support the presence and functionality of an UP element for the ltrRP1 promoter.
ltrR encodes two proteins, LtrR2 and LtrR1.
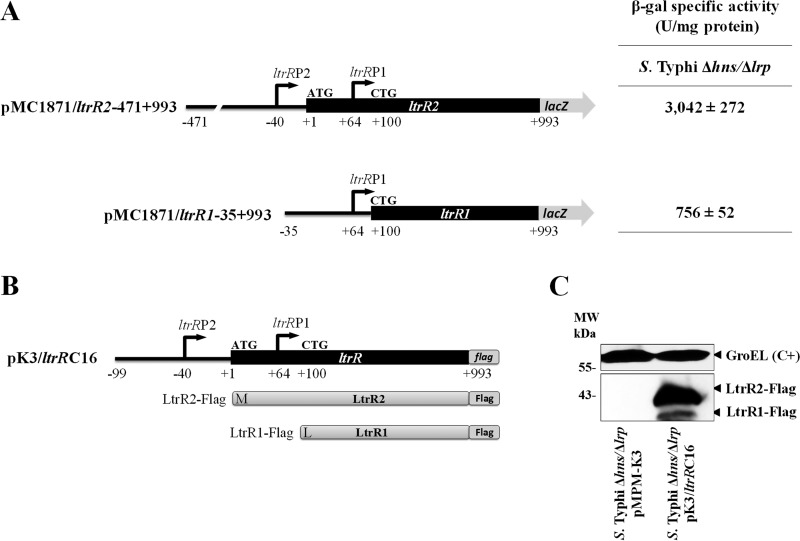
The results obtained in this work demonstrate that ltrR has two promoters. To evaluate whether they are able to promote the synthesis of two different proteins, translational lacZ fusions were obtained. The first fusion, pMC1871/ltrR2-471+993, included the ltrRP2 and ltrRP1 promoters as well as the ltrR gene without its TGA codon (1,464 bp). The second construct, pMC1871/ltrR1-35+993, included only the ltrRP1 promoter and the ltrR1 ORF without its TGA codon (894 bp) (Fig. 9A).
FIG 9.
The ltrR gene encodes two different proteins, LtrR1 and LtrR2. (A) Diagram of the translational lacZ fusions pMC1871/ltrR2-471+993 and pMC1871/ltrR1-35+993. The β-galactosidase (β-gal) specific activity evaluated in S. Typhi Δhns Δlrp is also shown. Activities were determined at an OD595 of 1.0 in N-MM. The values are the means ± standard deviations from two independent experiments performed in duplicate. (B) Scheme of ltrR-flag cloned into the pMPMK3 vector (pK3/ltrRC16). The fusion includes the ltrRP2 and ltrRP1 promoters, the structural ltrR2-ltrR1 sequence, and the Flag epitope. The LtrR2-Flag and LtrR1-Flag peptides are schematically shown (products of the pK3/ltrRC16 fusion). (C) Western blotting of overexpressed pK3/ltrRC16, generating LtrR2-Flag (40.15 kDa) and LtrR1-Flag (36.39 kDa) from S. Typhi Δhns Δlrp. S. Typhi Δhns Δlrp containing the empty pMPM-K3 vector was used as a negative control. These strains were cultivated in N-MM at an OD595 of 1.0. GroEL was used as a protein loading control (C+). Molecular weight (MW) markers are indicated.
Since the highest ltrR2 and ltrR1 transcriptional values were obtained with the hns lrp double mutant (Fig. 3B and Fig. 4B), we evaluated the ltrR2 and ltrR1 translational fusions in this genetic background. The β-galactosidase activity values of the fusions pMC1871/ltrR2-471+993 and pMC1871/ltrR1-35+993 were 3,042 and 756 units at an OD595 of 1.0, indicating that both the ltrRP2 and -P1 promoters and ltrRP1 alone are able to induce the expression of the ltrR2 and ltrR1 coding regions (Fig. 9A). All these data demonstrate that ltrRP2 and ltrRP1 are able to independently drive the expression of ltrR, which encodes two proteins.
The predicted masses of the LtrR2 and LtrR1 proteins are 37.41 and 33.66 kDa, respectively (Fig. 2). To determine the presence of the two LtrR proteins encoded by the same ORF, the ltrR gene containing the ltrRP2 and ltrRP1 promoters as well as its structural coding region of 993 bp encoding a Flag epitope at the C-terminal region was cloned into the pMPM-K3 vector, which contains an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Ptac promoter (named pK3/ltrRC16) (Fig. 9B). Western blot experiments in which this construct was overexpressed in the S. Typhi Δhns Δlrp mutant showed the presence of LtrR2-Flag with a molecular weight of 40.15 kDa and LtrR1-Flag with a molecular weight of 36.39 kDa (Fig. 9C), demonstrating that ltrR encodes two independent proteins that correspond to the previously predicted ORFs of LtrR2 and LtrR1 (Fig. 2).
Determination of the translational start sites of LtrR2 and LtrR1 proteins.
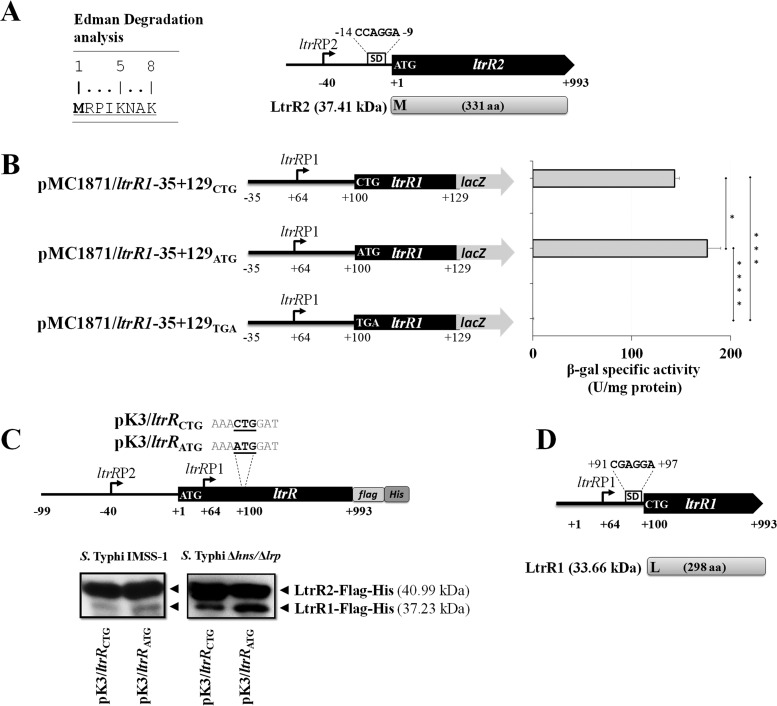
Since ltrR encodes two proteins, we determined the corresponding translational start sites. By Edman degradation of the purified LtrR2 protein, it was demonstrated that the ATG codon located 40 bp downstream of the ltrR2 transcriptional start site corresponds to the LtrR2 translational initiation codon. This codon was preceded by a putative CCAGGA Shine-Dalgarno sequence located 9 nucleotides upstream of the ATG codon (Fig. 10A).
FIG 10.
LtrR2 and LtrR1 translational start site determination. (A) Edman degradation analysis showing the N-terminal sequence (8 amino acids) of LtrR2. M (methionine) corresponds to the LtrR2 translational start codon. Shown is a schematic representation of ltrR2 showing its transcriptional start site ltrRP2; a putative Shine-Dalgarno sequence, CCAGGA, located 9 bp upstream of the LtrR2 translational start codon ATG; and the corresponding protein of 37.41 kDa. (B) Translational lacZ fusions pMC1871/ltrR1-35+129CTG, pMC1871/ltrR1-35+129ATG, and pMC1871/ltrR1-35+129TGA containing the CTG wild-type initiation codon as well as an ATG or a TGA substitution. The β-galactosidase specific activity was evaluated in the S. Typhi IMSS-1 strain at an OD595 of 1.0 in N-MM. The values are the means ± standard deviations from two independent experiments performed in duplicate (n = 4). Statistically different values are indicated (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001). (C) Western blotting was performed with 25 μg of purified proteins from S. Typhi IMSS-1 or S. Typhi Δhns Δlrp containing pK3/ltrRCTG or pK3/ltrRATG that expresses LtrR2-Flag-His (40.99 kDa), LtrR1CTG-Flag-His (37.23 kDa), or LtrR1ATG-Flag-His (37.24 kDa). (D) Genetic representation of the ltrR1 gene showing its transcriptional start site ltrRP1 (residue +64); a putative Shine-Dalgarno sequence, +91CGAGGA+97; and the corresponding LtrR1 protein of 33.66 kDa with its leucine translational start site (L).
In the case of LtrR1 translational initiation site determination, in silico analysis predicted a CTG codon located 100 bp downstream of the canonical ltrR2 ATG codon as the translational start site. Thus, fusions that lack the H-NS and Lrp negative regulatory regions were obtained and introduced into the wild-type strain. Such independent fusions, pMC1871/ltrR1-35+129CTG, pMC1871/ltrR1-35+129ATG, and pMC1871/ltrR1-35+129TGA, contained independently the ltrR1 promoter and a CTG, ATG, or TGA codon located 36 bp downstream of the ltrR1 transcriptional initiation site (Fig. 10B). The β-galactosidase specific activity results for these fusions showed null activity in the fusion pMC1871/ltrR1-35+129TGA containing a stop codon. However, the fusion pMC1871/ltrR1-35+129CTG containing the wild-type predicted CTG initiation codon as well as the fusion pMC1871/ltrR1-35+129ATG containing a canonical ATG codon rendered activity values of 143 and 176 β-galactosidase units, respectively (Fig. 10B). The data showed that a replacement of the wild-type predicted CTG initiation codon by an ATG codon presented similar transcriptional activity, supporting the notion that the CTG codon could be the LtrR1 translational start site.
Furthermore, plasmids pK3/ltrRCTG and pK3/ltrRATG were generated. The first construct contained the ltrRP2 and ltrRP1 promoters, the wild-type ltrR1 CTG initiation codon, the structural ltrR coding region of 993 bp, a Flag epitope at the C-terminal region, and a 6×His tag cloned into the pMPM-K3 vector, which contains an IPTG-inducible Ptac promoter. The second plasmid was similar, with only a replacement of the CTG codon by an ATG codon. These constructs were transformed independently into the S. Typhi wild-type strain, and Western blotting of the purified proteins from these strains showed that LtrR1 with its wild-type CTG initiation codon as well as LtrR1 with an ATG codon display polypeptides of similar masses of 37 kDa. Furthermore, purified proteins from S. Typhi Δhns Δlrp containing independently plasmids pK3/ltrRCTG and pK3/ltrRATG, showed an increased signal in LtrR1 containing the wild-type CTG codon or LtrR1 containing the ATG substitutions by Western blotting, supporting the proposed negative roles of H-NS and Lrp in LtrR1 expression (Fig. 10C).
Thus, by translational fusions and Western blot experiments, we suggest that the CTG codon located 100 bp downstream of the ltrR2 translational start site corresponds to the LtrR1 translational initiation codon. This start codon was preceded by a putative Shine-Dalgarno sequence, CGAGGA, 3 nucleotides upstream of the CTG codon (Fig. 10D).
DISCUSSION
The LysR-type proteins are the most abundant transcriptional regulators in bacteria, and their roles in metabolism, nitrogen fixation, virulence, quorum sensing, motility, biofilm formation, and other key biological processes have been reported (33). Therefore, many of these regulators could be considered essential components of bacterial cell maintenance. In S. Typhi, we previously described that the LysR-type protein LeuO is a global regulator involved in the transcriptional control of porin synthesis and detoxification and in the regulation of the CRISPR-Cas system (29, 34–36). Several other reports support the global regulatory role of LeuO in E. coli (37) and S. Typhimurium (38). Therefore, the study of the LysR-type regulators has and will continue to generate relevant knowledge on the biology of Enterobacteriaceae.
In an effort to know more about the function of this family of transcriptional regulators in S. Typhi, we previously described that the LysR-type protein LtrR binds to the regulatory region of ompR, and OmpR interacts with the ompC and ompF promoters to induce porin synthesis. Furthermore, biological functions in the control of bacterial transformation and in the growth of S. Typhi in the presence of one of the major bile salts found in the gut, sodium deoxycholate, have been assigned for the ltrR-ompR-ompC genetic network (26).
In the present work, we define some of the genetic elements involved in the transcriptional control of the ltrR gene. We identified two promoters, ltrRP2 localized upstream and ltrRP1 located downstream of the canonical ATG translational start site. We describe that the ltrRP2 promoter is positively regulated by pH and that H-NS is directly involved in its repression. Furthermore, both H-NS and Lrp negatively regulate the ltrR2 coding region. pH and an UP element are involved in the positive regulation of the ltrRP1 promoter, whereas H-NS and Lrp are simultaneously and directly involved in the negative regulation of ltrR1 at its coding region. Since pH is involved in ltrR2 and ltrR1 expression, we evaluated whether these genes are fundamental for the growth of S. Typhi at pH 7.5, finding that the strain deleted in both ORFs was able to grow like the wild-type strain at pH 7.5. Therefore, these genetic elements are induced transcriptionally by pH but are not essential for growth under these conditions. Interestingly, this pH is characteristic of the gallbladder or distal ileum (39–42), compartments that are invaded and colonized by S. Typhi. Thus, it is relevant to determine whether ltrR2 or ltrR1 has a role in the response to these human environments.
The involvement of H-NS and Lrp in gene regulation has been documented for spvR, and a negative role for these proteins has also been reported for the casA gene of the CRISPR-Cas system (29, 36, 43–45). Moreover, these nucleoid-associated proteins regulate the 16S rRNA genes, where it is postulated that H-NS and Lrp work together in a synergic fashion to repress the expression of 16S rRNA (46, 47). In agreement with the results of these studies, we found that H-NS and Lrp bind simultaneously to ltrR2 and ltrR1, and it is possible that a nucleosome-like complex formed between H-NS, Lrp, and the target DNA is needed to repress ltrR2 and ltrR1, as suggested for the 16S rRNA gene repression mediated by H-NS and Lrp (46, 47). Reports of ChIP-on-chip analysis showed that H-NS is able to bind to coding regions, and the results obtained in this work validate that notion (27, 28). The participation of global regulatory proteins such as H-NS and Lrp, which have been implicated in virulence, metabolism, and other biological processes (43, 48–52), suggests that ltrR2 and ltrR1 respond to a wide variety of environmental cues. Further studies of mutants affected in these genes are needed to evaluate their role in infection and virulence in S. Typhi, since LtrR is the master regulator of ompR (26), and this two-component regulator has been widely implicated in virulence.
In this study, we report that ltrR encodes two proteins, opening the possibility that other LysR-type genes also encode two proteins. In this respect, bioinformatic analysis showed that 17 LTTRs (LysR-type transcriptional regulators) in S. Typhi CT18 (STY0014, STY0048, STY0159, STY0341, STY0651, STY0730, STY1551, STY1386, STY1693, STY2278, STY2821, STY3037, STY3220, STY3749, STY3935, STY4468, and STY4867) also present two or more transcriptional initiation sites (see Fig. S1 in the supplemental material). Some of them present a transcriptional initiation site in the coding region. It would be interesting to evaluate whether each LtrR protein is able to regulate different regulons. Thus, it is possible that the LysR regulators modulate the expression of a considerably larger number of genes. In this respect, it is relevant to determine whether the LtrR1 protein, which is devoid of the HTH DNA-binding site, is able to regulate target genes since it is possible that it interacts with the carboxy termini of other LysR-type proteins and thus interacts with the genes that it regulates. A precedent for this suggestion exists, since QseD in enteropathogenic E. coli (EPEC) O157:H7 is a LysR-type protein devoid of the HTH and is able to promote the regulation of several genes (53). Alternatively, LtrR1 could function in the modulation of the LtrR2 protein or vice versa. From the literature, only virF and copA are able to encode two proteins from one gene (54, 55). Thus, ltrR is the third gene reported with this characteristic.
Finally, the study presented here is the first transcriptional characterization of a LysR-type gene that encodes two proteins. It provides new data on the complex genetic and regulatory characteristics of one of the most predominant types of transcriptional factors in bacteria, the LysR-type transcriptional regulators.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used are listed in Table S1 in the supplemental material. S. Typhi and E. coli strains were grown aerobically at 25°C or 37°C in LB (10 g tryptone, 5 g yeast extract, and 10 g NaCl per liter) or N-MM [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 100 mM Tris-HCl (pH 7.4), 200 μM MgCl2, 0.5% glycerol, and 0.1% Casamino Acids] (57). For pH experiments, 200 mM Tris-HCl was used to avoid pH changes during bacterial growth. When required, the following antibiotics were added: kanamycin (Km) at 30 μg ml−1, tetracycline (Tc) at 12 μg ml−1, and ampicillin (Ap) at 200 μg ml−1.
DNA manipulations.
Plasmid and genomic DNA isolation were carried out according to previously reported protocols (58). Primers for PCR amplifications were provided by the oligonucleotide synthesis facility at our institute (Table S2). Restriction enzymes, ligase, nucleotides, and polymerases were acquired from New England Biolabs or Thermo Fisher Scientific. For sequencing, double-stranded DNA was purified with the High Pure plasmid isolation kit (Roche) and sequenced with an automatic PerkinElmer/Applied Biosystems 377-18 system.
Construction of transcriptional and translational reporter fusions.
For the construction of transcriptional fusions, a collection of ltrR fragments of different lengths was generated. PCR products were doubly digested with BamHI-KpnI and ligated into pKK232-8, which contains the promoterless cat gene. For the construction of ltrR translational lacZ reporter fusions, blunted PCR fragments encompassing different lengths of the promoter regions as well as of the ltrR coding region were cloned into the SmaI site of the pMC1871 fusion vector (Pharmacia) (59) (Table S1). Fusions were sequenced to verify the correct DNA sequence of the PCR fragments.
CAT assay.
Chloramphenicol acetyltransferase (CAT) assays were done as follows (60). S. Typhi strains were grown in N-MM to an OD595 of 0.6. Cells were harvested, centrifuged, washed with 0.8 ml of TDTT buffer (50 mM Tris-HCl, 30 μM dl-dithiothreitol [DTT] [pH 7.8]), resuspended in 0.6 ml of TDTT buffer, and sonicated on ice for 9.9-s intervals with 9.9-s rest periods until the extract was clear. The homogenate was centrifuged, and the supernatant was used for activity measurement. For CAT assays, 5 μl of each extract was added in duplicate to a 96-well enzyme-linked immunosorbent assay (ELISA) plate, followed by the addition of 0.2 ml of a reaction mixture containing 1 mM DTNB [5,5′-dithiobis(2-nitrobenzoic acid)], 0.1 mM acetyl coenzyme A (acetyl-CoA), and 0.1 mM chloramphenicol in 0.1 M Tris-HCl (pH 7.8). The absorbance at 412 nm was measured every 5 s for 5 min using a Ceres 900 scanning autoreader and microplate workstation. The protein concentration of the cell extracts was obtained by using the bicinchoninic acid (BCA) protein assay reagent (Pierce). Protein values and the mean rate of product formation by CAT were used to determine CAT specific activity as micromoles per minute per milligram of protein.
Microplate protein and β-galactosidase determinations.
Protein concentrations were determined by the Lowry method (61), and β-galactosidase activity was adapted to a microtiter plate assay (62, 63). The results of translational fusions presented in the figures and tables are the means of data from three independent experiments performed in duplicate.
Construction of S. Typhi mutants.
Individual chromosomal hns and lrp deletions as well as an hns lrp double-deletion strain were obtained using the method of Datsenko and Wanner (64). Chromosomal substitutions in the −10 box of each ltrR promoter were also obtained. The mutations were PCR amplified and cloned into pKK232-8, generating plasmids pKK8/ltrR-144+90ΔP1 and pKK8/ltrR-144+90ΔP2.
RNA isolation and primer extension analysis.
S. Typhi strains were grown at 37°C in N-MM to an OD595 of 0.6. Bacterial cells (20 ml) were collected, and total RNA was isolated using a phenol-acid extraction method (34). The concentration of RNA was determined by measuring the absorbance at 260 nm. The integrity of RNA was determined by using a 1.5% agarose gel. For primer extension, 40 μg of total RNA was denatured at 95°C for 3 min and then slowly cooled to 45°C. The RNA was annealed with [γ-32P]ATP-labeled primers. Primers were extended with Maxima H Minus reverse transcriptase (Thermo Scientific) at 55°C for 30 min, and the extended products were then purified by 1-butanol precipitation and analyzed by electrophoresis in 8% polyacrylamide–8 M urea gels alongside sequencing ladders (63). Sequencing ladders were generated from a plasmid containing the regulatory regions of the ltrR gene.
Purification of H-NS, Lrp, and the LtrR2 and LtrR1 proteins.
Purification of H-NS was performed according to methods described previously by De la Cruz et al. (65). Briefly, the gene was cloned into the arabinose-inducible vector pBAD-Myc-His (Invitrogen) (Table S1). E. coli BL21(DE3) harboring the cloned gene was grown overnight in LB medium. One milliliter of this preculture was inoculated into 100 ml LB medium and allowed to grow to an OD600 of 0.4 before being induced for 4 h with l-arabinose (Sigma-Aldrich) at a final concentration of 1%. Samples were collected and centrifuged at 10,000 × g for 5 min at 4°C. The cell pellet was resuspended in binding buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0]), the cells were sonicated, and the suspension was centrifuged at 10,000 × g for 5 min at 4°C and kept at 4°C. A nickel affinity chromatography column (Ni-nitrilotriacetic acid [Ni-NTA]–agarose; Qiagen) was equilibrated with binding buffer. The solution containing the protein was loaded onto the column and washed with 10 volumes of urea buffer at pH 8.0 and urea buffer at pH 6.0. The bound protein was eluted with urea buffer (pH 4.5). Fractions containing H-NS–Myc–6×His were resolved by SDS-PAGE. The selected fractions were loaded into a Slyde-A-Lyzer 10K cassette (Pierce) and dialyzed at 4°C in a buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 20% glycerol, 500 mM NaCl, 0.1% Triton X-100, and 4 M urea. The same buffer containing 1 M and 0.2 M urea was sequentially used to complete dialysis. The purified protein was stored in buffer containing 30 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 20% glycerol, 500 mM NaCl, 0.1% Triton X-100, and 3 mM EDTA.
To purify Lrp, the corresponding gene was cloned into the arabinose-inducible vector pMPM-T6 (Invitrogen) (66). E. coli BL21(DE3) harboring the pMPM-T6-lrp vector was grown overnight in LB supplemented with Tc. One milliliter of this preculture was inoculated into 100 ml LB plus Tc at 37°C at 200 rpm and allowed to grow to an OD600 of 0.6 before being induced for 4 h with l-arabinose (Sigma-Aldrich) to a final concentration of 0.1%. The culture was then grown at 25°C with shaking at 100 rpm. Samples were collected and centrifuged at 10,000 × g for 5 min at 4°C. The cell pellet was resuspended in 30 ml of lysis buffer (0.3 M NaCl, 50 mM NaH2PO4, 10 mM imidazole, 1 mg ml−1 lysozyme, 0.4% sarcosyl [pH 7.4]) and disrupted by using a French press. The suspension was centrifuged at 10,000 × g for 5 min at 4°C to collect cell debris, and the supernatant was loaded onto a 1.5-ml Ni-nitrilotriacetic acid–agarose chromatographic column (Qiagen) equilibrated with wash buffer (0.3 M NaCl, 50 mM NaH2PO4, 10 mM imidazole [pH 7.4]). The column was washed with 10 volumes of the same buffer containing increasing amounts of imidazole (10, 20, 30, and 50 mM). Lrp was eluted with 10 ml of elution buffer (0.3 M NaCl, 50 mM NaH2PO4, 500 mM imidazole [pH 7.4]), and fractions were resolved by SDS-PAGE.
In order to purify the LtrR2 and LtrR1 proteins, the ltrR wild-type gene of 993 bp, containing the Flag and His tag epitopes at its C-terminal end, was cloned into the pMPM-K3 vector, generating plasmid pK3/ltrRCTG. A similar construct, containing a replacement of CTG by an ATG codon at nucleotide +100 of the ltrR gene, was obtained and designated pK3/ltrRATG (Table S1). These constructs were transformed into either S. Typhi IMSS-1 or S. Typhi Δhns Δlrp, which was grown overnight in LB medium supplemented with Km (30 μg ml−1), and 10 ml of this preculture was inoculated into Erlenmeyer flasks to a final volume of 1.1 liters of LB supplemented with Km (30 μg ml−1) and 0.2% glucose. The cultures were grown at 37°C at 200 rpm to an OD595 of 1.4. Samples were collected and centrifuged at 10,000 × g for 5 min at 4°C. The cell pellet was resuspended in 30 ml of lysis buffer containing 0.3 M NaCl and 50 mM NaH2PO4 (pH 8) plus 1 mg ml−1 lysozyme, 1% sarcosyl, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were disrupted by using a French press. The suspension was centrifuged at 10,000 × g for 10 min at 4°C to collect cell debris, and the supernatant was incubated for 60 min at 4°C with 3 ml of Ni-nitrilotriacetic acid–agarose (Qiagen). The total volume was loaded into a Ni-NTA chromatographic column setup. The column was washed with 15 volumes of lysis buffer containing 0.3 M NaCl and 50 mM NaH2PO4 (pH 8) with increasing amounts of imidazole (10, 25, and 50 mM). The proteins were eluted with 3 ml of elution buffer (0.3 M NaCl, 50 mM NaH2PO4, 500 mM imidazole [pH 8]). Proteins were concentrated in Amicon Ultra centrifugal filters (Merck KGaA, Darmstadt, Germany). The amount of purified proteins was determined by a Bradford assay (67), using bovine serum albumin (BSA) as the protein standard, and resolved by SDS-PAGE.
Amino acid sequence determination by Edman degradation.
The N terminus of LtrR2 was determined using Edman degradation with a PPSQ-31A protein sequencer (Shimadzu Scientific Biotech) at the Laboratorio Universitario de Proteómica (LUP), Instituto de Biotecnología-UNAM.
Electrophoretic mobility shift assays.
Nonradioactive electrophoretic mobility shift assays (EMSAs) were performed according to a previously described protocol (29). The DNA fragments were amplified by PCR using the primers shown in Table S3. A total of 35 ng of each DNA probe was mixed with increasing concentrations of purified H-NS protein in the presence of 10× H-NS-binding buffer (400 mM HEPES, 80 mM MgCl2, 500 mM KCl, 10 mM DTT, 0.5% Nonidet P-40, and 1 mg ml−1 BSA) or with increasing concentrations of Lrp incubated with 5× Lrp-binding buffer (100 mM Tris [pH 8.0], 2 mM EDTA, 250 mM NaCl, 5 mM MgCl2, 62.5% [vol/vol] glycerol, 500 mg ml−1 BSA, and 0.5 mM DTT). A total of 100 ng of DNA probes was mixed with increasing concentrations of α-RNAP and 5× α-RNAP-binding buffer (100 mM Tris [pH 7.9], 100 mM NaCl, 5 mM EDTA, 50% glycerol) (29). The mixtures were incubated for 20 min at room temperature and then separated by electrophoresis in 6% native polyacrylamide gels in 0.5× Tris-borate-EDTA buffer. The DNA bands were visualized by ethidium bromide staining.
Cooperative EMSAs were performed with 35 ng of the DNA probe and the Lrp and H-NS proteins in the presence of 10× H-NS buffer. The mixtures were incubated for 20 min at room temperature and then separated by electrophoresis in 5% native polyacrylamide gels in 0.5× Tris-borate-EDTA buffer.
Western blot experiments.
For the Western blot experiments in Fig. 9, S. Typhi Δhns Δlrp containing pK3/ltrRC16 was grown in N-MM (50 ml) to an OD595 of 1.0, a total-extract sample was collected and boiled, and the proteins were separated by electrophoresis on a 12% sodium dodecyl sulfate-polyacrylamide gel. Western blotting of purified proteins was performed using cultures of the S. Typhi wild-type or Δhns Δlrp strain containing pK3/ltrRCTG or pK3/ltrRATG from 1.1 liters of LB supplemented with Km (30 μg ml−1) and 0.2% glucose to an OD595 of 1.4. Proteins were transferred to nitrocellulose membranes using the Trans-Blot SD system (Bio-Rad). Membranes were blocked with 10% nonfat milk and incubated with anti-Flag M2 monoclonal antibody (Sigma) at a dilution of 1:2,500 or with anti-GroEL polyclonal antibody (StressGen) at a 1:50,000 dilution. Next, anti-mouse antibody (Pierce) or anti-rabbit antibody (Pierce), at a dilution of 1:10,000, was used as a secondary antibody, respectively. The blotted membranes were incubated with Western Lightning chemiluminescence reagent plus (PerkinElmer) and visualized with Carestream X-OMAT LS films.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Fernández-Mora, F. J. Santana, M. Caro, P. Gaytan, E. Bustos, S. Becerra, J. Yañez, L. Perezgasga, A. Vazquez, F. Zamudio-Zuñiga, M. M. Banda, C. C. Paredes-Amaya, A. Medrano-López, and M. I. Isidro-Coxca for technical help, J. L. Puente for allowing us to develop part of this work in his laboratory, and, finally, I. B. Olivar-Casique, J. Miranda, D. Georgellis, and M. Dunn for stimulating discussions and critical reading.
This work was supported by grants from the Dirección General de Asuntos del Personal Académico, DGAPA/UNAMIN203618 (to I.H.-L.), and a predoctoral fellowship (grant 413132) to J.E.R.-F. from the Consejo Nacional de Ciencia y Tecnología.
We declare that no competing interests exist.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kumar S, Kumar S, Kumar S. 2006. Infection as a risk factor for gallbladder cancer. J Surg Oncol 93:633–639. doi: 10.1002/jso.20530. [DOI] [PubMed] [Google Scholar]
- 2.Tewari M, Mishra RR, Shukla HS. 2010. Salmonella Typhi and gallbladder cancer: report from an endemic region. Hepatobiliary Pancreat Dis Int 9:524–530. [PubMed] [Google Scholar]
- 3.Lacroix FJ, Cloeckaert A, Grepinet O, Pinault C, Popoff MY, Waxin H, Pardon P. 1996. Salmonella Typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol Lett 135:161–167. doi: 10.1111/j.1574-6968.1996.tb07983.x. [DOI] [PubMed] [Google Scholar]
- 4.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. 2007. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar Typhimurium. J Bacteriol 189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prieto AI, Hernández SB, Cota I, Pucciarelli MG, Orlov Y, Ramos-Morales F, García-del Portillo F, Casadesús J. 2009. Roles of the outer membrane protein AsmA of Salmonella enterica in the control of marRAB expression and invasion of epithelial cells. J Bacteriol 191:3615–3622. doi: 10.1128/JB.01592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prieto AI, Jakomin M, Segura I, Pucciarelli MG, Ramos-Morales F, García-Del Portillo F, Casadesús J. 2007. The GATC-binding protein SeqA is required for bile resistance and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 189:8496–8502. doi: 10.1128/JB.01156-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prieto AI, Ramos-Morales F, Casadesús J. 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787–1794. doi: 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prieto AI, Ramos-Morales F, Casadesús J. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575–584. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prouty AM, Van Velkinburgh JC, Gunn JS. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J Bacteriol 184:1270–1276. doi: 10.1128/jb.184.5.1270-1276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Velkinburgh JC, Gunn JS. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect Immun 67:1614–1622. doi: 10.1128/IAI.67.4.1614-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos-Morales F, Prieto AI, Beuzón CR, Holden DW, Casadesús J. 2003. Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J Bacteriol 185:5328–5332. doi: 10.1128/jb.185.17.5328-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López-Garrido J, Cheng N, García-Quintanilla F, García-del Portillo F, Casadesús J. 2010. Identification of the Salmonella enterica damX gene product, an inner membrane protein involved in bile resistance. J Bacteriol 192:893–895. doi: 10.1128/JB.01220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 14.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 18.McCusker MP, Hokamp K, Buckley JF, Wall PG, Martins M, Fanning S. 2014. Complete genome sequence of Salmonella enterica serovar Agona pulsed-field type SAGOXB.0066, cause of a 2008 pan-European outbreak. Genome Announc 2:e01219-13. doi: 10.1128/genomeA.01219-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang CX, Zhu SL, Wang XY, Feng Y, Li B, Li YG, Johnston RN, Liu GR, Zhou J, Liu SL. 2015. Complete genome sequence of Salmonella enterica subspecies arizonae str. RKS2983. Stand Genomic Sci 10:30. doi: 10.1186/s40793-015-0015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson EJ, Limaye B, Inamdar H, Datta A, Manjari KS, Pullinger GD, Thomson NR, Joshi RR, Watson M, Stevens MP. 2011. Genome sequences of Salmonella enterica serovar Typhimurium, Choleraesuis, Dublin, and Gallinarum strains of well-defined virulence in food-producing animals. J Bacteriol 193:3162–3163. doi: 10.1128/JB.00394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harhay DM, Smith TPL, Harhay GP, Loneragan GH, Webb HE, Bugarel M, Haley BJ, Kim SW, Van Kessel JAS. 2018. Complete closed genome sequences of three Salmonella enterica subsp. enterica serovar Dublin strains isolated from cattle at harvest. Microbiol Resour Announc 7:e01334-18. doi: 10.1128/MRA.01334-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogunremi D, Devenish J, Amoako K, Kelly H, Dupras AA, Belanger S, Wang LR. 2014. High resolution assembly and characterization of genomes of Canadian isolates of Salmonella Enteritidis. BMC Genomics 15:713. doi: 10.1186/1471-2164-15-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 24.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog 2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O’Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 26.Villarreal JM, Becerra-Lobato N, Rebollar-Flores JE, Medina-Aparicio L, Carbajal-Gómez E, Zavala-García ML, Vázquez A, Gutiérrez-Ríos RM, Olvera L, Encarnación S, Martínez-Batallar AG, Calva E, Hernández-Lucas I. 2014. The Salmonella enterica serovar Typhi ltrR-ompR-ompC-ompF genes are involved in resistance to the bile salt sodium deoxycholate and in bacterial transformation. Mol Microbiol 92:1005–1024. doi: 10.1111/mmi.12610. [DOI] [PubMed] [Google Scholar]
- 27.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 29.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, Calva E, Hernández-Lucas I. 2011. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol 193:2396–2407. doi: 10.1128/JB.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, Gourse RL. 1999. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev 13:2134–2147. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross W, Ernst A, Gourse RL. 2001. Fine structure of E. coli RNA polymerase-promoter interactions: alpha subunit binding to the UP element minor groove. Genes Dev 15:491–506. doi: 10.1101/gad.870001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 33.Maddocks SE, Oyston PCF. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 34.Gallego-Hernández AL, Hernández-Lucas I, De la Cruz MA, Olvera L, Morett E, Medina-Aparicio L, Ramírez-Trujillo JA, Vázquez A, Fernández-Mora M, Calva E. 2012. Transcriptional regulation of the assT-dsbL-dsbI gene cluster in Salmonella enterica serovar Typhi IMSS-1 depends on LeuO, H-NS, and specific growth conditions. J Bacteriol 194:2254–2264. doi: 10.1128/JB.06164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernández-Lucas I, Gallego-Hernández AL, Encarnación S, Fernández-Mora M, Martínez-Batallar AG, Salgado H, Oropeza R, Calva E. 2008. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J Bacteriol 190:1658–1670. doi: 10.1128/JB.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medina-Aparicio L, Rebollar-Flores JE, Beltrán-Luviano AA, Vázquez A, Gutiérrez-Ríos RM, Olvera L, Calva E, Hernández-Lucas I. 2017. CRISPR-Cas system presents multiple transcriptional units including antisense RNAs that are expressed in minimal medium and upregulated by pH in Salmonella enterica serovar Typhi. Microbiology 163:253–265. doi: 10.1099/mic.0.000414. [DOI] [PubMed] [Google Scholar]
- 37.Shimada T, Bridier A, Briandet R, Ishihama A. 2011. Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol Microbiol 82:378–397. doi: 10.1111/j.1365-2958.2011.07818.x. [DOI] [PubMed] [Google Scholar]
- 38.Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesús J, Dorman CJ. 2012. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol 85:1072–1089. doi: 10.1111/j.1365-2958.2012.08162.x. [DOI] [PubMed] [Google Scholar]
- 39.Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ, Hardcastle JD. 1988. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fallingborg J. 1999. Intraluminal pH of the human gastrointestinal tract. Dan Med Bull 46:183–196. [PubMed] [Google Scholar]
- 41.Matton APM, de Vries Y, Burlage LC, van Rijn R, Fujiyoshi M, de Meijer VE, de Boer MT, de Kleine RHJ, Verkade HJ, Gouw ASH, Lisman T, Porte RJ. 2019. Biliary bicarbonate, pH, and glucose are suitable biomarkers of biliary viability during ex situ normothermic machine perfusion of human donor livers. Transplantation 103:1405–1413. doi: 10.1097/TP.0000000000002500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molinero N, Ruiz L, Milani C, Gutiérrez-Díaz I, Sánchez B, Mangifesta M, Segura J, Cambero I, Campelo AB, García-Bernardo CM, Cabrera A, Rodríguez JI, González S, Rodríguez JM, Ventura M, Delgado S, Margolles A. 2019. The human gallbladder microbiome is related to the physiological state and the biliary metabolic profile. Microbiome 7:100. doi: 10.1186/s40168-019-0712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall DG, Sheehan BJ, Dorman CJ. 1999. A role for the leucine-responsive regulatory protein and integration host factor in the regulation of the Salmonella plasmid virulence (spv) locus in Salmonella typhimurium. Mol Microbiol 34:134–145. doi: 10.1046/j.1365-2958.1999.01587.x. [DOI] [PubMed] [Google Scholar]
- 44.O’Byrne CP, Dorman CJ. 1994. Transcription of the Salmonella typhimurium spv virulence locus is regulated negatively by the nucleoid-associated protein H-NS. FEMS Microbiol Lett 121:99–105. doi: 10.1111/j.1574-6968.1994.tb07082.x. [DOI] [PubMed] [Google Scholar]
- 45.Robbe-Saule V, Schaeffer F, Kowarz L, Norel F. 1997. Relationships between H-NS, sigma S, SpvR and growth phase in the control of spvR, the regulatory gene of the Salmonella plasmid virulence operon. Mol Gen Genet 256:333–347. doi: 10.1007/s004380050577. [DOI] [PubMed] [Google Scholar]
- 46.Pul U, Wurm R, Lux B, Meltzer M, Menzel A, Wagner R. 2005. LRP and H-NS—cooperative partners for transcription regulation at Escherichia coli rRNA promoters. Mol Microbiol 58:864–876. doi: 10.1111/j.1365-2958.2005.04873.x. [DOI] [PubMed] [Google Scholar]
- 47.Pul U, Wurm R, Wagner R. 2007. The role of LRP and H-NS in transcription regulation: involvement of synergism, allostery and macromolecular crowding. J Mol Biol 366:900–915. doi: 10.1016/j.jmb.2006.11.067. [DOI] [PubMed] [Google Scholar]
- 48.Calvo JM, Matthews RG. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev 58:466–490. doi: 10.1128/MMBR.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellison DW, Miller VL. 2006. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J Bacteriol 188:5101–5112. doi: 10.1128/JB.00862-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison JA, Pickard D, Higgins CF, Khan A, Chatfield SN, Ali T, Dorman CJ, Hormaeche CE, Dougan G. 1994. Role of hns in the virulence phenotype of pathogenic salmonellae. Mol Microbiol 13:133–140. doi: 10.1111/j.1365-2958.1994.tb00408.x. [DOI] [PubMed] [Google Scholar]
- 51.Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol 53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 52.Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B. 2004. The virF promoter in Shigella: more than just a curved DNA stretch. Mol Microbiol 51:523–537. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 53.Habdas BJ, Smart J, Kaper JB, Sperandio V. 2010. The LysR-type transcriptional regulator QseD alters type three secretion in enterohemorrhagic Escherichia coli and motility in K-12 Escherichia coli. J Bacteriol 192:3699–3712. doi: 10.1128/JB.00382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Martino ML, Romilly C, Wagner EGH, Colonna B, Prosseda G. 2016. One gene and two proteins: a leaderless mRNA supports the translation of a shorter form of the Shigella VirF regulator. mBio 7:e01860-16. doi: 10.1128/mBio.01860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drees SL, Klinkert B, Helling S, Beyer DF, Marcus K, Narberhaus F, Lubben M. 2017. One gene, two proteins: coordinated production of a copper chaperone by differential transcript formation and translational frameshifting in Escherichia coli. Mol Microbiol 106:635–645. doi: 10.1111/mmi.13841. [DOI] [PubMed] [Google Scholar]
- 56.Reference deleted.
- 57.Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol 31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 59.Shapira SK, Chou J, Richaud FV, Casadaban MJ. 1983. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene 25:71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- 60.Martinez-Laguna Y, Calva E, Puente JL. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol Microbiol 33:153–166. doi: 10.1046/j.1365-2958.1999.01460.x. [DOI] [PubMed] [Google Scholar]
- 61.Lowry OH, Rosebrough NJ, Farr NL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]
- 62.Martinez-Flores I, Cano R, Bustamante VH, Calva E, Puente JL. 1999. The ompB operon partially determines differential expression of OmpC in Salmonella typhi and Escherichia coli. J Bacteriol 181:556–562. doi: 10.1128/JB.181.2.556-562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oropeza R, Sampieri CL, Puente JL, Calva E. 1999. Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella typhi: a novel regulatory mechanism that involves OmpR. Mol Microbiol 32:243–252. doi: 10.1046/j.1365-2958.1999.01329.x. [DOI] [PubMed] [Google Scholar]
- 64.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De la Cruz MÁ, Fernández-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vázquez A, Calva E. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol 66:727–743. doi: 10.1111/j.1365-2958.2007.05958.x. [DOI] [PubMed] [Google Scholar]
- 66.Mayer MP. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41–46. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- 67.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.