Abstract
The COVID‐19 epidemic was not the first coronavirus epidemic of this century and represents one of the increasing number of zoonoses from wildlife to impact global health. SARS CoV‐2, the virus causing the COVID‐19 epidemic is distinct from, but closely resembles SARS CoV‐1, which was responsible for the severe acute respiratory syndrome (SARS) outbreak in 2002. SARS CoV‐1 and 2 share almost 80% of genetic sequences and use the same host cell receptor to initiate viral infection. However, SARS predominantly affected individuals in close contact with infected animals and health care workers. In contrast, CoV‐2 exhibits robust person to person spread, most likely by means of asymptomatic carriers, which has resulted in greater spread of disease, overall morbidity and mortality, despite its lesser virulence. We review recent coronavirus‐related epidemics and distinguish clinical and molecular features of CoV‐2, the causative agent for COVID‐19, and review the current status of vaccine trials.
Keywords: coronavirus, COVID‐19, otolaryngology, head and neck surgery, review
In early December 2019, Li Wenliang, a physician from Wuhan, a large metropolitan area in China's Hubei province, reported in a group chat that he noticed a series of patients showing signs of a severe acute respiratory syndrome or SARS‐like illness which was subsequently reported to the WHO Country Office in China on December 31, 2019. On January 12, Chinese scientists published the genome of the virus, and the World Health Organization (WHO) asked a team in Berlin to use that information to develop a diagnostic test to identify active infection, which was developed and shared 4 days later. On January 30, 2020, the outbreak was declared by the WHO a Public Health Emergency of International Concern (PHEIC). The first case of the disease due to local person to person spread in the United States was confirmed in mid‐February 2020. On March 11, WHO declared COVID‐19 a pandemic.
1. LINKS TO PRIOR CORONAVIRUSES CAUSING SARS
Because of the genome's homology with the coronavirus that caused the SARS outbreak (SARS‐CoV‐1) in China in 2002 to 2003, this virus was renamed SARS‐CoV‐2. Coronaviruses are known to be the causative agent for the “common cold,” accounting for up to 30% of upper respiratory tract infections in adults. Coronaviruses, like other RNA viruses, mutate frequently and evolve in vast animal reservoirs. The overwhelming majority of coronaviruses pose no threat to humans, but recombination events, natural selection and genetic drift permit particularly virulent coronaviruses to jump to human hosts and to subsequently acquire the capacity for efficient person to person spread. 1
For reasons that are not well understood, zoonoses from wildlife has been increasing over the last half‐century, and represent the most significant, growing threat to global health of all emerging infectious diseases. 1 Geographic hotspots, or maps reflecting zoonotic infectious disease risk have been identified in South American, Africa and South Asia 2 (Figure 1). Both SARS‐CoV‐1 and 2 arose in one of these “hot‐spots.” Future outbreaks are believed to be all but inevitable.1, 3
FIGURE 1.
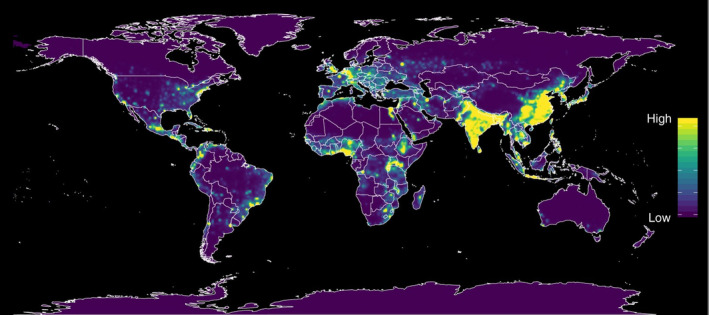
Heat maps of predicted relative risk distribution of zoonotic emerging disease threats. The CoV‐2 coronavirus arose in one of these “hot‐spots” 2 [Color figure can be viewed at wileyonlinelibrary.com]
2. A NEW EPIDEMIC?
The SARS‐CoV‐1 epidemic in 2002 to 2003 was the first coronavirus pandemic in modern times, which spread to two dozen countries with approximately 8000 cases and 800 deaths before it was contained. 4 In 2012, another outbreak, referred to as MERS for the Middle Eastern Respiratory Syndrome, and also caused by a coronavirus, resulted in over 1000 infections and 400 deaths through 2015. Since SARS and MERS coronavirus exhibited reduced person to person spread, the global impact of each was ultimately limited. Health care settings were the most frequent sites of person to person disease transmission.5, 6 Health care workers and those in close contact with infected individuals were at greatest risk of contracting and succumbing to disease, while the general public was relatively spared during these outbreaks. In particular, otolaryngologists were at greater risk of infection due to shedding of virus from nasal and pharyngeal mucosa. 7 The implementation of infection control methods, aggressive contact tracing and isolation limited the spread of disease in 2003 and 2012. Antiviral treatments and vaccines were never developed.
SARS CoV‐2, the virus which causes COVID19, is similar to, but not identical with the SARS‐CoV‐1 virus, which caused the SARS epidemic originating in China in 2002 to 2003. Full‐length genome sequences obtained from infected Chinese patients in 2019 showed 79.6% sequence homology to SARS‐CoV‐1. 8 SARS‐CoV‐2 is more closely related to two bat‐derived severe acute respiratory syndrome (SARS)‐like coronaviruses, bat‐SL‐CoVZC45 and bat‐SL‐CoVZXC21, collected in 2018 in Zhoushan, eastern China(with 88% identity), and a strain isolated from pangolins (99% identity) than to SARS‐CoV‐1, suggesting SARS‐CoV‐2 should be considered a new human‐infecting coronavirus, rather than re‐emergence of SARS‐CoV‐1. 9 Transmission is believed to have occurred from a bat‐CoV to an intermediate host after which the virus jumped to human hosts at live animal markets. 10 Suggestions that SARS CoV‐2 was constructed in a laboratory have been widely discredited.11, 12
The infectivity of SARS CoV‐2, relative to CoV‐1, is likely accounted for by relatively high rates of asymptomatic carriers which promote the spread of infection to susceptible populations, 13 rather than greater stability of virus in aerosolized droplets or on surfaces. 14 Despite variations and limitations in testing, CoV‐2 appears to be less virulent than CoV‐1. 15
3. SARS‐COV2: CONSERVED SEQUENCES AND GENETIC DRIFT
While the viral genome continues to evolve since entering the human population, sequences critical to human transmission will likely remain preserved. All coronaviruses exhibit a spike on its surface, which gives it the appearance of a crown (ie, Corona in Latin) on electron micrographs, mediates binding to host cell receptors, determines viral host range and tissue tropism, and induces host immune response. SARS‐CoV2, like SARS‐CoV‐1, gain entry into human cells via the angiotensin 1 converting enzyme 2 (ACE2) receptor, expressed on human lung and other tissues. 8 This receptor mediates the effects of angiotensin, which affects vasoconstriction and hypertension in the lungs, heart, kidneys and intestines. An identical ACE receptor is also found in some animals; this permit cross‐species infection. 10
The spike or S protein, expressed on the surface of coronaviruses plays an important role in the first step of viral infection by binding to the host ACE‐2 receptor, and initiating virus‐host membrane fusion. The amino acid sequences of the protein, particularly its receptor‐binding domains (RBDs) are highly conserved, since they are necessary to propagate the infection in person to person spread. The amino acid sequence of the SARS‐CoV‐2 spike (S) protein, particularly its RBD, serves as an antigen for the immune system, which could be used in the development of serologic tests and vaccines. So‐called “escape mutants,” viruses with amino acid alterations just flanking but not disrupting the RBD could potentially compromise durable immunity and the effectiveness of vaccines against SARS‐CoVs. 16 The clinical significance of escape mutations in coronaviruses is uncertain. In general, immune responses to coronavirus infections confer prolonged immunity. Neutralizing antibodies to SARS CoV‐1, that prevent viral infection by preventing binding of the S protein to the host cell receptor, have been detected in patients with SARS almost 20 years after recovering from disease suggesting that patients retain protection over long periods after mounting an immune response. 17
While the genetic sequences critical to transmission are preserved, noncritical viral sequences have undergone genetic drift. Compared to CoV‐2 sequences isolated in January in China, strains from other parts of the world exhibited genetic sequences which distinguish European from Asian SARS CoV‐2 coronavirus strains. 18 Differences between viral strains has permitted evaluations of the effectiveness of public health interventions. For example, analyses of different strains showed that predominance of COVID‐19 cases in the New York City area resulted largely from infected patients who flew in from Europe, rather than Asia. Consequently, barring US entry to recent visitors to China on January 31st, and from European countries on March 11th was largely ineffective in preventing the COVID outbreak in the United States. 19
4. SEROLOGICAL TESTING AND VACCINE DEVELOPMENT
The identification of viral epitopes that stimulate the production of antibodies that serve as markers of an immune response to CoV‐2 infection are necessary for the development of serologic testing and vaccines. Currently numerous serologic tests using antibodies against specified antigens are commercially available, but most have not been adequately validated. At best, tests reflect an immune response without necessarily indicating host immunity. 20 The availability of conserved epitopes also allows the structure‐based design of a SARS‐CoV‐2 vaccine. However, not all antibodies prevent viral infection, despite targeting virus specific epitopes. For example, antibodies raised against SARS CoV‐1 cross‐react with the CoV‐2 viral epitopes, but do not prevent CoV‐2 binding to ACE‐1 receptors; these antibodies may provide some clinical protection or attenuate the disease without actually preventing infection.21, 22 Neutralizing antibodies that prevent viral attachment to host cell receptors protect against viral infection. However, subtle amino acid alterations may provide a “back door” for viral escape mutants 16 . A cocktail of viral epitopes to stimulate a robust neutralizing immunologic response has been recommended to address this phenomenon.23, 28, 29, 30, 31
Currently, the need for a safe and effective vaccine against coronaviruses is generally appreciated. Numerous human vaccine trials for are under way targeting SARS CoV‐2. Historically, support for vaccine development after the SARS epidemic was short‐lived. Vaccine development for SARS included a phase 1 clinical trial of a SARS‐CoV‐1 vaccine which showed that the vaccine was safe, and effectively stimulated the production of neutralizing antibodies. 24 Unfortunately, when the SARS epidemic ended, vaccine development was abandoned. 25 The impact of the COVID‐19 epidemic suggests that interest in the development of a vaccine will be more enduring.
Nevertheless, potential impediments to vaccine development, noted during the SARS, remain potentially problematic. For example, concerns about vaccine‐mediated immune enhancement were raised during the vaccine development for SARS. Anecdotal reports of individuals developing more severe disease after exposure in individuals vaccinated against SARS CoV‐1 raised concerns about vaccine safety and efficacy. 26 Animal models demonstrated an analogous effect. Prior immunization with severe acute respiratory syndrome (SARS)‐associated coronavirus (SARS‐Cove) nucleocapsid protein caused severe pneumonia in mice infected with SARS‐CoV. 27 Demonstrations of safety and efficacy of vaccines against CoV‐2 are crucial, but take time. In the meantime, other forms of prevention (social distancing, protective device use) will limit transmission rates, and ultimately morbidity and mortality.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Lango MN. How did we get here? Short history of COVID‐19 and other coronavirus‐related epidemics. Head & Neck. 2020;42:1535–1538. 10.1002/hed.26275
REFERENCES
- 1. Jones KE, Patel NG, Levy MA, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990‐993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen T, Murray KA, Zambrana‐Torrelio C, et al. Global hotspots and correlates of emerging zoonotic diseases. Nat Commun. 2017;8(1):1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parrish CR, Holmes EC, Morens DM, et al. Cross‐species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72(3):457‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan JF, Li KS, To KK , Cheng VC, Chen H, Yuen KY. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV‐EMC) the beginning of another SARS‐like pandemic? J Infect. 2012;65(6):477‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peeri NC, Shrestha N, Rahman MS, et al. The SARS, MERS and novel coronavirus (COVID‐19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2020. 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alexander AJ, Tan AK, Evans GA, Allen J. Infection control for the otolaryngologist in the era of severe acute respiratory syndrome. J Otolaryngol. 2003;32(5):281‐287. [DOI] [PubMed] [Google Scholar]
- 8. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li JY, You Z, Wang Q, et al. The epidemic of 2019‐novel‐coronavirus (2019‐nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect. 2020;22(2):80‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hao P, Zhong W, Song S, Fan S, Li X. Is SARS‐CoV‐2 originated from laboratory? A rebuttal to the claim of formation via laboratory recombination. Emerg Microbes Infect. 2020;9(1):545‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS‐CoV‐2. Nat Med. 2020;26(4):450‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID‐19. Jama. 2020;323:1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N Engl J Med. 2020;382(16):1564‐1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang S, Cao P, Du P, et al. Early estimation of the case fatality rate of COVID‐19 in mainland China: a data‐driven analysis. Ann Transl Med. 2020;8(4):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rockx B, Corti D, Donaldson E, et al. Structural basis for potent cross‐neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol. 2008;82(7):3220‐3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Petherick A. Developing antibody tests for SARS‐CoV‐2. Lancet. 2020;395(10230):1101‐1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stefanelli P, Faggioni G, Lo Presti A, et al. Whole genome and phylogenetic analysis of two SARS‐CoV‐2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020;25(13):2000305. 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zimmer C. Most New York coronavirus cases came from Europe, genomes show. New York Times. April 8, 2020, 2020.
- 20. Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic testing for severe acute respiratory syndrome‐related coronavirus‐2:a narrative review. Ann Intern Med. 2020. 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor‐binding domains of SARS‐CoV‐2 and SARS‐CoV. Science. 2020;368:630‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coughlin MM, Babcook J, Prabhakar BS. Human monoclonal antibodies to SARS‐coronavirus inhibit infection by different mechanisms. Virology. 2009;394(1):39‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin JT, Zhang JS, Su N, et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir Ther. 2007;12(7):1107‐1113. [PubMed] [Google Scholar]
- 25. Lurie N, Saville M, Hatchett R, Halton J. Developing Covid‐19 vaccines at pandemic speed. N Engl J Med. 2020. 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 26. Tirado SM, Yoon KJ. Antibody‐dependent enhancement of virus infection and disease. Viral Immunol. 2003;16(1):69‐86. [DOI] [PubMed] [Google Scholar]
- 27. Yasui F, Kai C, Kitabatake M, et al. Prior immunization with severe acute respiratory syndrome (SARS)‐associated coronavirus (SARS‐CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS‐CoV. J Immunol. 2008;181(9):6337‐6348. [DOI] [PubMed] [Google Scholar]
- 28. Mitsuki YY, Ohnishi K, Takagi H, et al. A single amino acid substitution in the S1 and S2 Spike protein domains determines the neutralization escape phenotype of SARS‐CoV. Microbes Infect. 2008;10(8):908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rockx B, Donaldson E, Frieman M, et al. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. J Infect Dis. 2010;201(6):946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. ter Meulen J, van den Brink EN, Poon LL, et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coughlin MM, Prabhakar BS. Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential. Rev Med Virol. 2012;22(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]


