Abstract
Recently, Coronavirus Disease 2019 (COVID‐19) pandemic is the most significant global health crisis. In this study, we conducted a meta‐analysis to find the association between liver injuries and the severity of COVID‐19 disease. Online databases, including PubMed, Web of Science, Scopus, and Science direct, were searched to detect relevant publications up to 16 April 2020. Depending on the heterogeneity between studies, a fixed‐ or random‐effects model was applied to pool data. Publication bias Egger's test was also performed. Meta‐analysis of 20 retrospective studies (3428 patients), identified that patients with a severe manifestation of COVID‐19 exhibited significantly higher levels of alanine aminotransferase, aspartate aminotransferase, and bilirubin values with prolonged prothrombin time. Furthermore, lower albumin level was associated with a severe presentation of COVID‐19. Liver dysfunction was associated with a severe outcome of COVID‐19 disease. Close monitoring of the occurrence of liver dysfunction is beneficial in early warning of unfavorable outcomes.
Keywords: COVID‐19, liver function, meta‐analysis, outcome, SARS‐CoV‐2
Highlights
Liver dysfunction was associated with a severe outcome of COVID‐19
Sever cases of COVID‐19 patients have high serum levels of ALT, AST and bilirubin with prolonged prothrombin time.
Low serum albumin is associated with a severe presentation of COVID‐19
Close monitoring of the occurrence of liver dysfunction is beneficial in early warning of unfavorable COVID‐19 outcomes.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- AKI
acute kidney injury
- ALT
alanine aminotransferase
- ARDS
acute respiratory distress syndrome
- AST
aspartate aminotransferase
- COVID‐19
Coronavirus Disease 2019
- PT
prothrombin time
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- SMD
the standardized mean difference
- TSA
trial sequential analysis
1. INTRODUCTION
In December 2019, a novel virus known as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) was identified as a causative pathogen for a cluster of pneumonia initially detected in Wuhan City, China. 1 As of 3 May 2020, the World Health Organization has reported Worldwide 3 267 184 confirmed cases and 229 971 deaths. The United States has reported 1 067 127 confirmed cases and 57 406 deaths. 2
Coronavirus Disease 2019 (COVID‐19) is typically characterized by the symptoms of viral pneumonia, such as fever, fatigue, dry cough, anosmia, and headache, which may evolve to respiratory failure. 3 , 4 The pathogen, however, displays a wide range of severity causing difficulty in determining infection outcome. COVID‐19 may cause hepatic, intestinal, and respiratory diseases, and lead to respiratory distress syndrome, organ failure, and even death in severe cases. 5 , 6
Currently, studies about the relationship between underlying mechanisms of COVID‐19 and liver dysfunction are limited. COVID‐19 uses the angiotensin‐converting enzyme 2 (ACE2) as the binding site to enter the host cell in the lungs, kidneys, and heart. 7 Chai et al 8 found that both liver cells and bile duct cells express ACE2. However, the ACE2 expression of bile duct cells is much higher than that of liver cells. 9 These findings suggest that liver injury in patients with COVID‐19 may be the result of damage to bile duct cells. Various studies have reported the laboratory findings and the clinical characteristics associated with different degrees of liver dysfunction in patients with COVID‐19 disease. 10 , 15
However, to date, there is still limited research regarding the concomitant association between the COVID‐19 and the hepatobiliary system. Therefore, by meta‐analyzing data in the observational studies available so far, our study aimed to assess liver dysfunction among patients infected with SARS‐CoV‐2 to investigate the potential relationship between acute liver injury and COVID‐19.
2. METHODS
2.1. Literature search strategy
A comprehensive literature review of all qualifying studies was conducted to identify the association of COVID‐19 with acute liver injury based on Preferred Reporting Items for Systematic Reviews and Meta‐Analyses (PRISMA) guidelines. 16 Two authors (RE, MY) independently screened the following medical electronic database: PubMed, Web of Science, Scopus, and Science direct for relevant data published up to 16 April 2020, using a combination of the following keywords and medical subjects headings (MeSHs): (“COVID‐19” OR “SARS‐CoV‐2” OR “severe acute respiratory syndrome coronavirus 2” OR “coronavirus SARS‐CoV‐2” OR “2019‐nCoV” OR “Wuhan coronavirus” OR “Wuhan pneumonia”) AND (“Liver” OR “Acute Liver injury” OR “Liver enzymes” Chronic Liver”) AND (“outcome” OR “survival” OR “mortality” OR “complications” Or “infection”). The reference list of previous studies and systematic reviews were also searched for identifying eligible studies. The identified records were screened for the inclusion criteria specified for the present systematic review and meta‐analysis.
2.2. Eligibility criteria
We applied the following criteria to all extracted studies: (a) Types of studies: observational, retrospective cohort, prospective case‐control, or clinical trials reporting laboratory features of COVID‐19 patients, including alanine aminotransferase (ALT), aspartate aminotransferase (AST), bilirubin, albumin, and prothrombin time (PT); (b) Subjects: diagnosed patients with COVID‐19, and (c) Severity: mild cases of COVID‐19 disease with patients that do not require extraordinary measures to manage the diseases and severe cases of COVID‐19 infection who developed COVID‐related complications such as acute respiratory distress syndrome (ARDS) and respiratory failure, or expired. Exclusion criteria were as follows: (a) duplicate data (b) case reports, series, abstract‐only articles, conference article and comment, editorials and expert opinions (c) studies with insufficient outcome data, and (d) preprints (articles in the peer‐review stage).
2.3. Data extraction
Data extraction was conducted by four authors (MY, GZ, AA, and AF). The process included using a two‐step approach: first, we screened titles and abstracts for eligibility according to the study objective, and second, we screened the full‐text article of relevant abstracts.
2.4. Quality assessment
The Newcastle‐Ottawa Scale was used for assessing the quality of eligible manuscripts. Publication bias was assessed with the Newcastle‐Ottawa Quality Assessment Scale cohort studies. 17
2.5. Pairwise comparison and heterogeneity assessment
The pooled estimates were extracted using RevMan version 5.3. Descriptive summary statistics in the form of mean, standard deviation, and range for continuous parametric measures were tabulated. Pairwise comparison between mild and severe COVID‐19 patients was performed. Overall pooled odds ratio (OR) or standardized mean difference (SMD) with 95% confidence intervals (CI) were estimated for categorical and quantitative variables, respectively. A Fixed‐effects model was employed unless significant heterogeneity was detected. In this case, the Random‐effects model has applied. 18 Heterogeneity was considered significant if the I2 value exceeds 50%, or its P value was less than .1.
Subgroup analyses by the location of the patients, publication date, sample size, and quality score were performed. Sensitivity analysis was carried out by removing one study each time, to reflect its effect size on the overall OR.
Publication bias was assessed via Begg's funnel plot and Egger's linear regression approach using Comprehensive Meta‐analysis software. 19 An asymmetric funnel‐shape or a P value less than .1 indicated significant bias. 20
2.6. Meta‐regression analysis
Meta‐regression analysis was employed using OpenMeta Analyst software, taking into consideration the following study characteristics; sample size, mean age of patients, percentage of males, city of the hospital, publication date, and quality score.
2.7. Trial sequential analysis
To evaluate the reliability of statistical appraisal of this meta‐analysis study, we used trial sequential analysis (TSA) software (version 0.9.5.10 beta) by merging several available sample sizes of applicable studies with the threshold of statistical influence to reduce the unintentional miscalculations and improve the strength of anticipations. We used two‐side trials and type I error with a calculated power of 5% and 80%. If the cumulative Z‐curve crosses the monitoring boundaries, no additional trials would be required. On the contrary, if the Z‐curve did not accomplish the boundary levels, the necessary threshold requires additional records to achieve a prominent significance.
3. RESULTS
3.1. Characteristics of the included studies
Following the removal of duplicates (n = 1870), our database search identified 2582 unique citations, of which 186 full‐text articles were assessed. A total of 20 eligible retrospective cohort studies, including 3428 positively confirmed COVID‐19 patients, were enrolled in the current meta‐analysis. The workflow of the process of study selection is demonstrated in Figure 1. All articles were published during the period between 30 January and 16 April 2020. Most of them were from Wuhan city (13), three from Zhejiang, one from Guangdong, one from Hubei, one from Guangdong, and one from Anhui. As depicted in Table 1, the sample size of studies ranged from 21 to 651 cohorts. The mean age of patients was 53.8 years, and 57.8% were men. In the included studies, the severe disease was detected in 36.2% of patients and the average survival rate was 72.18%. All studies except for three scored more than 5 on the scale. Two studies scored a three, and one study scored a two.
Figure 1.
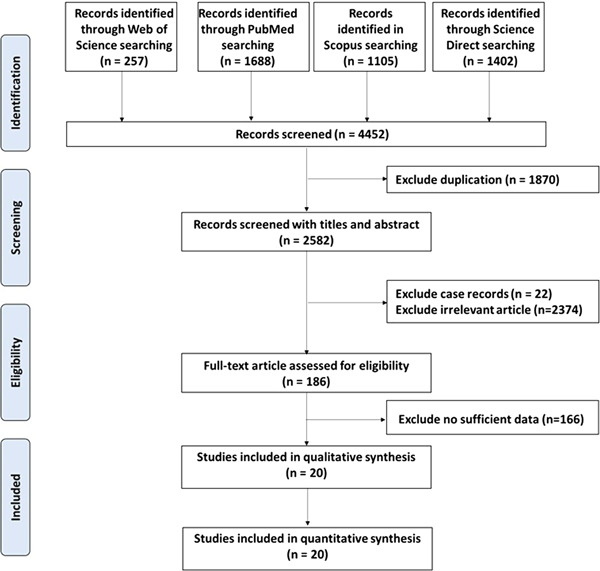
The workflow of the selection process
Table 1.
Characteristics of the included studies
| Author | Date of publication | Year | Ref | Journal name | Study | Country | City | Sample Size | Mean age | Male/female | Severe cases (%) | Survival rate (%) | Quality score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Huang 4 | 30 Jan | 2020 | [4] | The Lancet | Retro | China | Wuhun | 41 | 49 | 30/11 | 31.7 | 85.37 | 7 |
| Wang 21 | 7 Feb | 2020 | [21] | JAMA | Retro | China | Wuhun | 138 | 58.5 | 75/63 | 26.1 | NA | 5 |
| Yang 22 | 21 Feb | 2020 | [22] | The Lancet | Retro | China | Wuhun | 52 | 58.25 | 35/17 | 61.5 | 38.46 | 6 |
| Liu 23 | 28 Feb | 2020 | [23] | Chin Med J (Engl) | Retro | China | Wuhun | 78 | 51.5 | 39/39 | 14.1 | NA | 5 |
| Ruan 24 | 3 Mar | 2020 | [24] | Intensive Care Med | Retro | China | Wuhun | 150 | 58.5 | 102/48 | 45.3 | 54.67 | 5 |
| Zhou 25 | 11 Mar | 2020 | [25] | The Lancet | Retro | China | Wuhun | 191 | 60.5 | 119/72 | 28.3 | 71.73 | 8 |
| Gao 11 | 13 Mar | 2020 | [11] | J Med Virol | Retro | China | Anhui | 43 | 44 | 26/17 | 34.9 | NA | 5 |
| Wu 14 | 13 Mar | 2020 | [14] | JAMA Intern Med | Retro | China | Wuhun | 201 | 53.25 | 128/73 | 41.8 | 78.11 | 5 |
| Zhang 15 | 15 Mar | 2020 | [15] | Int J Infect Dis | Retro | China | Zhejiang | 645 | 40.77 | 328/317 | 88.8 | NA | 7 |
| Mo 10 | 16 Mar | 2020 | [10] | Clin Infect Dis | Retro | China | Wuhun | 155 | 53.5 | 86/69 | 54.8 | NA | 5 |
| Wang 26 | 16 Mar | 2020 | [26] | Clin Infect Dis | Retro | China | Wuhun | 69 | 53.75 | 32/37 | 20.3 | 92.75 | 8 |
| Chen 12 | 17 Mar | 2020 | [12] | BMJ | Retro | China | Wuhun | 274 | 59.5 | 171/103 | 41.2 | 58.76 | 8 |
| Qian 27 | 17 Mar | 2020 | [27] | QJM | Retro | China | Zhejiang | 91 | 57.5 | 37/54 | 9.9 | NA | 3 |
| Qu 28 | 17 Mar | 2020 | [28] | J Med Virol | Retro | China | Guangdong | 30 | 54.72 | NA | 10.0 | NA | 2 |
| Deng 29 | 20 Mar | 2020 | [29] | Chin Med J (Engl) | Retro | China | Wuhun | 225 | 54.5 | 124/101 | 48.4 | 51.56 | 7 |
| Wan 30 | 21 Mar | 2020 | [30] | J Med Virol | Retro | China | Chongqing | 135 | 50 | 72/63 | 29.6 | 99.26 | 8 |
| Jin 31 | 24 Mar | 2020 | [31] | Gut | Retro | China | Zhejiang | 651 | 45.61 | 331/320 | 11.4 | NA | 8 |
| Chen 13 | 27 Mar | 2020 | [13] | J Clin Invest | Retro | China | Wuhun | 21 | 56.5 | 17/4 | 52.4 | 80.95 | 6 |
| Pan 32 | 14 Apr | 2020 | [32] | Am J Gastroenterol | Retro | China | Hubei | 204 | 52.9 | 107/97 | 50.5 | 82.35 | 7 |
| Zhou 33 | 16 Apr | 2020 | [33] | J Infect | Retro | China | Wuhun | 34 | 65 | 17/17 | 23.5 | NA | 3 |
Abbreviations: Ref, reference number; Retro, retrospective.
3.2. Pooled analysis of laboratory findings
Table 2 summarizes pairwise comparison, heterogeneity analysis, and publication bias of the meta‐analysis. Patients who had severe presentations of COVID disease had higher levels of AST (SMD = 0.36; 95% CI = 0.27; 0.44; P < .001), ALT (SMD = 0.44; 95% CI = 0.35, 0.52; P < .001), bilirubin (SMD = 0.40; 95% CI = 0.31, 0.50; P < .001), and PT (SMD = 0.69; 95% CI = 0.57, 0.81; P < .001). In contrast, lower albumin level was associated with severe presentation (SMD = −0.68; 95% CI = −0.7, −0.58; P < .001) (Figure S1). Apart from ALT data, significant heterogeneity was detected in laboratory results. Subgroup analysis by the origin of the hospital, publication date, sample size, and quality score of the studies failed to resolve the obvious heterogeneity.
Table 2.
Summarizing results of pooled estimates of liver function tests and clinical parameters of chronic liver patients
| Characteristics | Number studies | Sample size | Test of association | Effect size | Heterogeneity | Publication bias | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Mild | Severe | Method | Model | Estimate | 95% CI | P value | I 2 | P value | P (Egger's) | ||
| Laboratory tests | ||||||||||||
| ALT | 19 | 3376 | 1953 | 1423 | SMD, IV | Fixed | 0.35 | 0.27, 0.43 | .073 | 34.13% | <.001 | 0.279 |
| AST | 19 | 3376 | 1953 | 1423 | SMD, IV | Random | 0.44 | 0.17, 0.70 | <.001 | 88.80% | .001 | 0.940 |
| Bilirubin | 11 | 2512 | 1365 | 1147 | SMD, IV | Random | 0.41 | 0.20, 0.62 | <.001 | 75.02% | <.001 | 0.773 |
| Albumin | 11 | 2605 | 1434 | 1171 | SMD, IV | Random | −0.84 | −1.20, −0.48 | <.001 | 91.4% | <.001 | 0.204 |
| PT | 10 | 1300 | 799 | 501 | SMD, IV | Random | 0.62 | 0.32, 0.91 | <.001 | 81.34% | <.001 | 0.512 |
| Comorbidities | ||||||||||||
| Hypertension | 13 | 2141 | 1024 | 1117 | OR, M‐H | Fixed | 2.36 | 1.86, 3.01 | .30 | 14.01% | <.001 | 0.976 |
| Chronic kidney dis | 7 | 1675 | 690 | 985 | OR, M‐H | Fixed | 7.28 | 3.25, 16.26 | .54 | 0.00% | <.001 | 0.279 |
| Diabetes | 14 | 2193 | 1044 | 1149 | OR, M‐H | Fixed | 2.72 | 2.05, 3.60 | .05 | 41.58% | <.001 | 0.453 |
| Cardiovascular dis | 12 | 2327 | 1086 | 1241 | OR, M‐H | Random | 5.11 | 2.03, 12.83 | <.001 | 77.27% | <.001 | 0.061 |
| Chronic liver dis | 9 | 1659 | 685 | 974 | OR, M‐H | Fixed | 1.17 | 0.66, 2.06 | .87 | 0.00% | .58 | 0.824 |
| Malignancy | 12 | 2132 | 990 | 1142 | OR, M‐H | Fixed | 2.20 | 1.28, 3.77 | .90 | 0.00% | .004 | 0.890 |
| Cerebrovascular dis | 5 | 769 | 435 | 334 | OR, M‐H | Fixed | 5.73 | 2.52, 13.04 | .20 | 32.59% | <.001 | 0.041 |
| Treatment | ||||||||||||
| Antiviral | 10 | 1685 | 1002 | 683 | OR, M‐H | Random | 0.70 | 0.42, 1.16 | .16 | 62.06 | <.001 | 0.52 |
| Antibiotics | 7 | 1991 | 1387 | 604 | OR, M‐H | Random | 2.13 | 0.86, 5.29 | .10 | 81.93 | <.001 | 0.64 |
| Glucocorticoids | 13 | 2981 | 1651 | 1330 | OR, M‐H | Random | 3.17 | 2.03, 4.97 | <.001 | 73.41 | <.001 | 0.49 |
| Immunoglobulins | 6 | 1101 | 605 | 496 | OR, M‐H | Random | 2.75 | 1.09, 6.94 | .032 | 89.10 | <.001 | 0.32 |
| Outcomes | ||||||||||||
| ARDS | 9 | 2204 | 1230 | 974 | OR, M‐H | Random | 18.84 | 5.39, 65.87 | <.001 | 89.58% | <.001 | 0.106 |
| AKI | 6 | 1300 | 516 | 784 | OR, M‐H | Random | 7.20 | 1.38, 37.74 | .003 | 71.59% | <.001 | 0.511 |
| Sepsis | 5 | 1259 | 488 | 771 | OR, M‐H | Random | 21.19 | 4.21, 106.73 | .085 | 50.99% | <.001 | 0.680 |
| Acute liver injury | 2 | 1296 | 649 | 647 | OR, M‐H | Fixed | 1.93 | 1.11, 3.34 | .60 | 0.00% | .001 | NA |
| Myocardial injury | 3 | 464 | 334 | 130 | OR, M‐H | Random | 11.19 | 0.44, 285.9 | <.001 | 90.44% | <.001 | 0.408 |
| Mortality | 11 | 1563 | 922 | 641 | OR, M‐H | Random | 55.22 | 12.62, 241.66 | <.001 | 90.82% | <.001 | 0.282 |
Abbreviations: AKI, acute kidney injury; ALT, alanine transaminase; ARDS, acute respiratory distress syndrome; AST, aspartate aminotransferase; CI, confidence interval; I2, the ratio of true heterogeneity to total observed variation; IV, inverse variance; M‐H, Mantel‐Haenszel; OR, odds ratio; PT, prothrombin time; SMD, standardized mean difference.
3.3. Pooled analysis of comorbidities
The analysis showed that patients with hypertension (OR = 2.37; 95% CI = 1.86‐3.01; P < .001), chronic kidney disease (OR = 7.28; 95% CI = 3.26‐16.26; P < .001), and diabetes (OR = 2.72; 95% CI = 2.06‐3.61; P < .001) were nearly twofold more risk to develop severe presentation of COVID‐19. Patients with underlying cardiovascular disease or cerebrovascular disease were five‐times more liable to develop severe phenotype (OR = 5.11; 95% CI = 2.04‐12.83; P < .0001 and OR = 5.73; 95% CI = 2.52‐13.04; P < .0001, respectively). Cancer patients also exhibited severe manifestations of the disease (OR = 2.20; 95% CI = 1.28‐3.78; P = .004) (Figure S2). Apart of cardiovascular disease, homogeneity between studies was detected.
3.4. Pooled analysis of treatment
A total of 17 studies reported treatment to be administered to COVID‐19 patients. On comparison between the two groups, severe patients were nearly three times more likely to receive steroids (OR = 3.17; 95% CI = 3.02‐4.97; P < .001) and immunoglobulins (OR = 2.75; 95% CI = 1.09‐6.94; P = .032). Sensitivity analysis revealed that the studies of Wang 21 and Zhang 15 contributed in the significant heterogeneity observed in treatment results (Table 2).
3.5. Pooled analysis of COVID‐19 outcomes
Our analysis confirmed that patients with severe COVID‐19 disease had higher odds of developing ARDS (OR = 18.84; 95% CI = 5.39‐65.87; P < .0001) and sepsis (OR = 21.19; 95% CI = 4.21‐106.7; P < .001). Similarly, acute liver injury (OR = 1.93; 95% CI = 1.12‐3.34; P = .001) and acute kidney injury (OR = 7.2; 95% CI = 1.38‐37.74; P < .001) were more prevalent among patient with severe disease. Moreover, our analysis revealed that mortality was more likely to occur among patients with severe COVID‐19 patients (OR = 55.22; 95% CI = 12.62‐241.66; P < .001) (Figure S3). Considerable heterogeneity was observed for the outcomes. Meta‐regression analysis for study characteristics showed higher odds of mortality in articles involving Wuhun hospitals (coefficient = 4.30; 95% CI = 3.07‐5.54; P < .001) (Table S1).
3.6. Publication bias
The funnel plot of laboratory and clinical parameters is shown in Figure S4. Egger's test showed no publication bias for all variables (P > .1) except for two; cardiovascular and cerebrovascular diseases (P = .061 and .041) (Table 2).
3.7. Trial sequential analysis
We applied TSA on mortality rate available among all eligible articles of COVID‐19 patients with a mild and severe exhibition and indicated that the cumulative Z‐curve transverses the monitoring boundaries before reaching the required sample size and achieving considerable significant and so no further studies are necessary (Figure 2).
Figure 2.
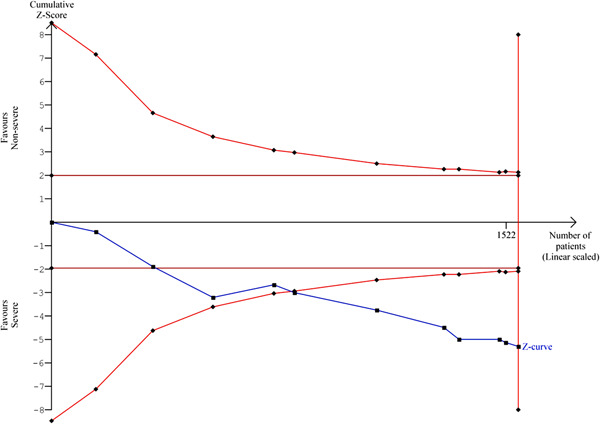
Trial sequential analysis for mortality
4. DISCUSSION
Our meta‐analysis including 3428 subjects from 20 retrospective studies explored the potential relationship between liver injury and the severity of COVID‐19 disease. We found that liver dysfunction seemed to be higher in patients with severe outcomes from COVID‐19 infection.
Our results were in agreement with a previous study review. 36 Previously, liver injury has been reported as an important risk factor for severe outcome and death in SARS and Middle East Respiratory Syndrome. 35 , 37 , 39
Patients in our study who had severe presentations of COVID‐19 disease had higher levels of AST, ALT, bilirubin, and lower albumin levels. Our results are consistent with recent studies on COVID‐19 disease that showed that the incidence of liver injury ranged from 58% to 78%, mainly indicated by elevated AST, ALT, and total bilirubin levels accompanied by slightly decreased albumin levels. 40 , 41 In a recent study, Guan et al 42 documented that higher serum levels of AST were observed in nearly 18% of patients with nonsevere COVID‐19 disease and approximately 56% of patients with severe COVID‐19 infection. Moreover, in that study, higher serum levels of ALT were also observed in nearly 20% of patients with nonsevere COVID‐19 presentation, and approximately 28% of patients with severe COVID manifestation. 42 Similar findings in Huang et al 4 were also observed, where patients with severe COVID‐19 features had an increased incidence of liver injury.
Postmortem liver biopsies specimens were observed in deceased COVID‐19 patients. The findings showed mild lobular and portal activity along with microvascular stenosis, indicating the injury could have been caused by either COVID‐19 disease or drug‐induced liver injury. 3 Similar to the treatment of SARS, steroids, antivirals, and antibiotics are widely used for the treatment of COVID‐19. 34 , 43 , 44 These drugs are all potential causes of liver injury during COVID‐19 treatment but have not yet been evident. 22 A recent study reported that the liver injury observed in COVID‐19 patients might be caused by lopinavir, which is used as an antiviral for the treatment of SARS‐CoV‐2 infection. 45 It is worth noting that the specific underlying causes of liver injury and elevated levels of liver enzymes in COVID‐19 patients are still limited. However, collectively the proposed mechanisms might include “hyperactivated immune responses and cytokine storm‐related systemic inflammation, psychological stress, drug toxicity, and progression of pre‐existing liver diseases” as detailed by Li and Fan. 46
Further studies are needed to investigate the mechanisms of liver dysfunction in COVID‐19 disease as a direct outcome of infection and the possible effects that treatment has on the liver.
Limitations of our study include the following; First, all the studies included in this meta‐analysis used a case‐control or cohort design, which are susceptible to recall and selection biases. Second, we could not distinguish if the liver dysfunction in COVID‐19 patients was an acute liver injury or exacerbated chronic liver disease. Last, the enrolled studies focused on Chinese patients, which restricted a more precise estimation of liver dysfunction in the context of other races.
5. CONCLUSIONS
In this meta‐analysis, we comprehensively analyzed liver dysfunction in accordance with the severity of clinical outcomes in COVID‐19 patients. Liver dysfunction was associated with severe COVID‐19 infection. Patients presented with abnormal liver function tests are at higher risk of severe clinical outcomes. Close monitoring of the presence of liver dysfunction may be beneficial as an early indicator of worse outcomes. This may serve to better prepare the treatment of patients.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
MY, ASA, RE: study design; MY, ASA, MO, GZ, AE, AS: study identification and data extraction; MH, RE, EAT: statistical analysis; MH, RE, EAT, MSF: data interpretation; MY, ASA, RE, MO, GZ, AF, EAT, MSF: original draft preparation. All authors revised and approved the final version of the manuscript.
Supporting information
Supplementary Information
Youssef M, Hussein M, Attia AS, et al. COVID‐19 and liver dysfunction: A systematic review and meta‐analysis of retrospective studies. J Med Virol. 2020;92:1825–1833. 10.1002/jmv.26055
Contributor Information
Mohammad H Hussein, Email: mhussein1@tulane.edu.
Abdallah S Attia, Email: aattia@tulane.edu.
Rami M Elshazli, Email: Relshazly@horus.edu.eg.
Mahmoud Omar, Email: Mahmoud.omar@waimg.org.
Ghassan Zora, Email: gzora@tulane.edu.
Ashraf S Farhoud, Email: afarhoud@tulane.edu.
Ahmad Elnahla, Email: aelnahla@tulane.edu.
Eman A Toraih, Email: etoraih@tulane.edu.
Manal S Fawzy, Email: manal_mohamed@med.suez.edu.eg.
Emad Kandil, Email: ekandil@tulane.edu.
REFERENCES
- 1. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507‐513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization (WHO) . Coronavirus disease 2019 (COVID‐19) Situation Report—84. World Heal Organ. 2020;323:1122. 10.1001/jama.2020.2633 [DOI] [Google Scholar]
- 3. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420‐422. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peiris J, Lai S, Poon L, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319‐1325. 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727‐733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoffmann M, Kleine‐Weber H, Krueger N, Mueller MA, Drosten C, Poehlmann S. The novel coronavirus 2019 (2019‐nCoV) uses the SARS‐coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. 2020. 10.1101/2020.01.31.929042 [DOI] [Google Scholar]
- 8. Chai X, Hu L, Zhang Y, et al. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019‐nCoV Infection. bioRxiv. 2020. 10.1101/2020.02.03.931766 [DOI] [Google Scholar]
- 9. Banales JM, Huebert RC, Karlsen T, Strazzabosco M, LaRusso NF, Gores GJ. Cholangiocyte pathobiology. Nat Rev Gastroenterol Hepatol. 2019;16:269‐281. 10.1038/s41575-019-0125-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mo P, Xing Y, Xiao Y, et al. Clinical characteristics of refractory COVID‐19 pneumonia in Wuhan, China. Clin Infect Dis. 2020. 10.1093/cid/ciaa270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID‐19. J Med Virol. 2020:jmv.25770. 10.1002/jmv.25770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020:m1091. 10.1136/bmj.m1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen G, Wu D, Guo W, et al. Clinical and immunologic features in severe and moderate Coronavirus Disease 2019. J Clin Invest. 2020;130:2620‐2629. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with Coronavirus Disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang X, Cai H, Hu J, et al. Epidemiological, clinical characteristics of cases of SARS‐CoV‐2 infection with abnormal imaging findings. Int J Infect Dis. 2020;94:81‐87. 10.1016/j.ijid.2020.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stang A. Critical evaluation of the Newcastle‐Ottawa scale for the assessment of the quality of nonrandomized studies in meta‐analyses. Eur J Epidemiol. 2010;25:603‐605. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 18. DerSimonian R, Laird N. Meta‐analysis in clinical trials. Control Clin Trials. 1986;7(3):177‐188. [DOI] [PubMed] [Google Scholar]
- 19. Pierce CA, Software Review: Borenstein, M., Hedges, L. V., Higgins, J. P. T., & Rothstein, H. R. (2007). Comprehensive Meta‐Analysis (Version 2.2.027) [Computer software]. Englewood, NJ: Biostat. Organizational Research Methods, 2008. 11(1): p. 188‐191. 10.1177/1094428106296641 [DOI]
- 20. Elshazli RM, Toraih EA, Elgaml A, Kandil E, Fawzy MS. Genetic polymorphisms of TP53 (rs1042522) and MDM2 (rs2279744) and colorectal cancer risk: an updated meta‐analysis based on 59 case‐control studies. Gene. 2020;734:144391. [DOI] [PubMed] [Google Scholar]
- 21. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA—J Am Med Assoc. 2020;323:1061. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med. 2020;8:475‐481. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu W, Tao ZW, Wang L, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J (Engl). 2020;133:1032‐1038. 10.1097/CM9.0000000000000775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID‐19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846‐848. 10.1007/s00134-020-05991-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054‐1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with Coronavirus Disease 2019 in Wuhan, China. Clin Infect Dis. 2020;ciaa272. 10.1093/cid/ciaa272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qian GQ, Yang NB, Ding F, et al. Epidemiologic and clinical characteristics of 91 hospitalized patients with COVID‐19 in Zhejiang, China: a retrospective, multi‐centre case series. QJM. 2020;113:474–481. 10.1093/qjmed/hcaa089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qu R, Ling Y, Zhang Y, et al. Platelet‐to‐lymphocyte ratio is associated with prognosis in patients with coronavirus disease‐19. J Med Virol. 2020:jmv.25767. 10.1002/jmv.25767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng Y, Liu W, Liu K, et al. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 (COVID‐19) in Wuhan, China: a retrospective study. China Med J (Engl). 2020:1. 10.1097/CM9.0000000000000824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wan S, Xiang Y, Fang W, et al. Clinical features and treatment of COVID‐19 patients in northeast Chongqing. J Med Virol. 2020;92:797–806. 10.1002/jmv.25783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus‐infected disease 2019 (COVID‐19) with gastrointestinal symptoms. Gut. 2020;69:1002‐1009. 10.1136/gutjnl-2020-320926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pan L, Mu M, Yang P, et al. Clinical characteristics of COVID‐19 patients with digestive symptoms in Hubei, China: a Descriptive, cross‐sectional, multicenter study. Am J Gastroenterol. 2020;115:766‐773. 10.14309/ajg.0000000000000620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou B, She J, Wang Y, Ma X. The clinical characteristics of myocardial injury in severe and very severe patients with 2019 novel coronavirus disease. J Infect. 2020;81:147–178. 10.1016/j.jinf.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu L, Liu J, Lu M, Yang D, Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int. 2020;40:998‐1004. 10.1111/liv.14435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saad M, Omrani AS, Baig K, et al. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single‐center experience in Saudi Arabia. Int J Infect Dis. 2014;29:301‐306. 10.1016/j.ijid.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Assiri A, Al‐Tawfiq JA, Al‐Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13:752‐761. 10.1016/S1473-3099(13)70204-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Al‐Hameed F, Wahla AS, Siddiqui S, et al. Characteristics and outcomes of middle east respiratory syndrome coronavirus patients admitted to an intensive care unit in Jeddah, Saudi Arabia. J Intensive Care Med. 2014;31:344‐348. 10.1177/0885066615579858 [DOI] [PubMed] [Google Scholar]
- 38. Chang HL, Chen KT, Lai SK, et al. Hematological and biochemical factors predicting SARS fatality in Taiwan. J Formos Med Assoc. 2006;105:439‐450. 10.1016/S0929-6646(09)60183-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Y, Zhou H, Yang R, Xu Y, Feng X, Gong P. Clinical characteristics of 36 non‐survivors with COVID‐19 in Wuhan, China. medRxiv. 2020. 10.1101/2020.02.27.20029009 [DOI] [Google Scholar]
- 40. Zhang B, Zhou X, Qiu Y, et al. Clinical characteristics of 82 death cases from COVID‐19. PLoS ONE. 2020;15:e0235458. 10.1371/journal.pone.0235458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708‐1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stebbing J, Phelan A, Griffin I, et al. COVID‐19: combining antiviral and anti‐inflammatory treatments. Lancet Infect Dis. 2020;20(4):400‐402. 10.1016/S1473-3099(20)30132-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Little P. Non‐steroidal anti‐inflammatory drugs and covid‐19. BMJ. 2020;368:m1185. 10.1136/bmj.m1185 [DOI] [PubMed] [Google Scholar]
- 44. Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Götte M. The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295:4773‐4779. 10.1074/jbc.AC120.013056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fan Z, Chen L, Li J, et al. Clinical features of COVID‐19‐related liver damage. SSRN Electron J. 2020. 10.2139/ssrn.3546077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li J, Fan JG. Characteristics and Mechanism of Liver Injury in 2019 Coronavirus Disease. J Clin Transl Hepatol. 2020;8(1):13‐17. 10.14218/JCTH.2020.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


