Abstract
In January 2020, we identified two severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)‐infected patients in a familial cluster with one person coming from Wuhan, China. The complete genome sequences of two SARS‐CoV‐2 strains isolated from these patients were identical and 99.98% similar to strains isolated in Wuhan. This is genetically suggestive of human‐to‐human transmission of SARS‐CoV‐2 and indicates Wuhan as the most plausible origin of the early outbreak in Vietnam. The younger patient had a mild upper respiratory illness and a brief viral shedding, whereas the elderly with multi‐morbidity had pneumonia, prolonged viral shedding, and residual lung damage. The evidence of nonsynonymous substitutions in the ORF1ab region of the viral sequence warrants further studies.
Keywords: complete genome sequencing, coronavirus, COVID‐19, respiratory infections, SARS‐CoV‐2, Vero
Highlights
Transmission of SARS‐CoV‐2 is a global public health and clinical concern.
This report describes clinical features, virus isolation, and complete genome sequences from the first two SARS‐CoV‐2 infections in Vietnam.
Epidemiological and phylogenetic analysis suggested evidence of human‐to‐human transmission of SARS‐CoV‐2. Comparison of SARS‐CoV‐2 strains isolated from these two patients with those from Wuhan showed high similarities.
Nonsynonymous substitutions existed in the ORF1ab region of the viral sequence.
Compared with mild clinical and virological manifestations in the younger patient, the elderly suffered from pneumonia, prolonged viral shedding, and residual lung damage.
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the etiologic agent of the ongoing coronavirus disease 2019 (Covid‐19) pandemic. Globally, it has affected approximately 5.4 million people and caused over 345 400 deaths as of 25 May 2020. 1 In Vietnam, between 22 January and 25 May 2020, there have been 326 laboratory‐confirmed infections of SARS‐CoV‐2 with no deaths. Despite an increase of SARS‐CoV‐2 infections in the past 4 months, clinical features of patients with Covid‐19 were poorly described in this country. 2 , 3 , 4 Infections were initially detected among overseas travelers to Vietnam and their close contacts. The recent occurrence of SARS‐CoV‐2 infections among health care workers and the general public is an evidence of potential community transmission of SARS‐CoV‐2 and the subsequent expansion of this outbreak in Vietnam. In such a context, virus isolation and an understanding of genomic characterization through complete genome sequencing approach are important for optimal design and assessment of diagnostic tests and the development of vaccines for Covid‐19.
In this report, we describe clinical features, virus isolation, and complete genome sequences from the first two SARS‐CoV‐2 infections in Vietnam.
2. MATERIALS AND METHODS
2.1. Patients
In response to the outbreak of the novel coronavirus first reported in December 2019, on 21 January 2020, Vietnam's Ministry of Health (MoH) published its interim guidelines on the surveillance, prevention, and control of Covid‐19, which mandates all health facilities to report any suspected and confirmed Covid‐19 cases to the MoH within 24 hours. A suspected case was defined as a person with an acute respiratory tract illness, who reported that he/she had fever and cough (with and without difficulty in breathing) and that within 14 days before the onset of disease, he/she had returned or came from areas where SARS‐CoV‐2 was spreading or had exposure to a laboratory‐confirmed case of Covid‐19. Laboratory confirmation required detection of SARS‐CoV‐2 in nasopharyngeal (NP) and oropharyngeal (OP) swab specimens by real‐time reverse transcription polymerase chain reaction (RT‐PCR) assays. Apart from the respiratory specimens, blood specimens were also collected from suspected cases during acute and recovery phases of illness for serological assessment of SARS‐CoV‐2 antibodies.
We identified a familial cluster of two patients with SARS‐CoV‐2 infection at a referral hospital in Ho Chi Minh City in southern Vietnam through the notifiable communicable disease surveillance system. Demographic, epidemiological, and clinical data were collected through interviews with the patients and their relatives, and a review of medical records. NP and OP swab specimens of the patients were collected in a single tube containing 3 mL virus transport media and tested for SARS‐CoV‐2 by real‐time RT‐PCR assays described previously. 5 The limits of detection of the assays for the E and RdRp genes were reported as low as 3.9 and 3.6 copies per reaction, respectively. We performed both the E and RdRp gene assays for the confirmation of the SARS‐CoV‐2 infection and the RdRp gene assay for treatment follow‐up. The E or RdRp gene assay was considered positive if the cycle threshold (Ct) value was less than 40. Written informed consent was obtained from the two patients.
2.2. Isolation of virus
After being confirmed positive for SARS‐CoV‐2, NP and OP swab specimens, which were taken from the two patients on admission and during the treatment follow‐up period, were stored at −70°C at the Pasteur Institute of Ho Chi Minh City until used. After thawing the specimens, virus isolation was performed using the Vero E6 cell line. In brief, Vero cells were inoculated with 400 µL of filtered specimens in 25‐cm2 flasks, each containing 5 mL of maintenance media. To increase the chance of virus isolation, we used two maintenance media: Dulbecco's modified Eagle's media (DMEM) supplemented with 2% fetal bovine serum (FBS) and DMEM supplemented with 0.2% bovine serum albumin (BSA) and 16 μg/mL tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK)‐trypsin. 6 We monitored the flasks for the formulation of cytopathic effect (CPE) at 48, 60, and 72 hours postinoculation. Once the CPE was observed under the microscope, cell culture supernatants were harvested, divided into aliquots, tested for the presence of SARS‐CoV‐2 by real‐time RT‐PCR assays, and stored at −70°C until virus titration and sequencing were performed. We performed virus titration on Vero E6 cells by using the standard plaque assay. 7 The virus titer was summarized as plaque‐forming units (PFU) per milliliter.
2.3. Viral genome sequencing and variant analysis
We synthesized complementary DNA and used random primers for amplification. Thereafter, we performed next‐generation sequencing analysis with complete genome sequences of isolates generated by the MiSeq platform (Illumina, San Diego, CA). 8 , 9 The consensus genome sequences were generated and mapped to the reference genome of SARS‐CoV‐2 (MN975262.1) using CLC Genomics Workbench version 10.1.1. We then aligned our complete genome sequences with other related coronavirus sequences archived from GenBank/GISAID using Mafft software version 7.452. Estimation of the best fitting substitution model (GTR+G+I) and inference of the phylogenetic tree were conducted by a maximum likelihood approach using MEGA‐X software version 10.0.5 with 1000 bootstrap replicates. Viral sequences were classified as the method of Forster et al. 10
Furthermore, the sequences were also analyzed to identify nucleotide variants, which were then translated into amino acid variants and compared with the Wuhan strain (MT019529). These variations were characterized as synonymous and non‐synonymous amino acid substitutions. The complete genome sequences were deposited to GenBank under accession numbers MT192772 and MT192773.
3. RESULTS
3.1. Clinical and epidemiological features
Patients’ travel history and epidemiological and clinical features obtained shortly after hospitalization have been reported elsewhere. 11 Briefly, the index case (Patient 1) was a 65‐year‐old Chinese man who traveled from Wuhan to Hanoi by airplane on 13 January 2020 when Covid‐19 was still spreading in his hometown (Wuchang district, Wuhan, China). His pre‐existing medical conditions included hypertension, type 2 diabetes, coronary heart disease for which a stent had been implanted, and lung cancer. He had no history of exposure to the wet‐market in Wuhan or any laboratory‐confirmed infection of SARS‐CoV‐2.
On admission, he complained of fever and fatigue for about 6 days. On 22 January 2020, his NP and OP swab specimens were collected and tested positive on real‐time RT‐PCR (Ct values 31.08 and 35.22 for the E and RdRp genes, respectively). He was diagnosed with pneumonia on January 22 (day 1 of hospitalization), with the presence of infiltrates on chest radiography. Laboratory abnormalities included severe lymphopenia of 0.4 × 109 cells/L (reference range, 0.8‐4.4 × 109 cells/L), moderate thrombocytopenia of 95 × 109 cells/L (reference range, 200‐400 × 109 cells/L), elevated C‐reactive protein level of 45.6 mg/L (reference level, <6 mg/L), and elevated glucose level of 128 mg/dL (reference range, 70‐110 mg/dL) (Table S1). By the first week of hospitalization, the thrombocytopenia resolved, whereas lymphopenia prevailed in the patient. There were signs of disease progression, including cough with sputum, crackles over the left lung, hypoxemia since day 3 of hospitalization, and several pulmonary infiltrates on chest radiographs. The patient was given oseltamivir, several broad‐spectrum antibacterials (including ceftriaxone, cefoperazone‐sulbactam, levofloxacin, linezolid, and imipenem‐cilastatin) drugs for hypertension and diabetes, and supplementary therapies. During treatment, he was also requested to use chlorhexidine mouthwashes two to six times a day; however, the compliance was poor due to dyspnea. Oxygen supplementation was started since day 4 because of an occurrence of respiratory failure. Fever resolved on day 4 and clinical improvement was observed from day 5. Viral RNA was detected in respiratory specimens for up to 18 days postadmission, with a Ct range of 26.89‐32.87 (Figure 1A). At the time of hospital discharge on February 12 (day 22), residual lung abnormalities were still observed on chest x‐rays. On the contrary, his wife, who accompanied him on the trip to Vietnam, had no signs and symptoms of Covid‐19 during the 2‐week quarantine period and her NP and OP swab specimens collected on February 9 (18 days after the last contact with the patient) tested negative for SARS‐CoV‐2.
Figure 1.
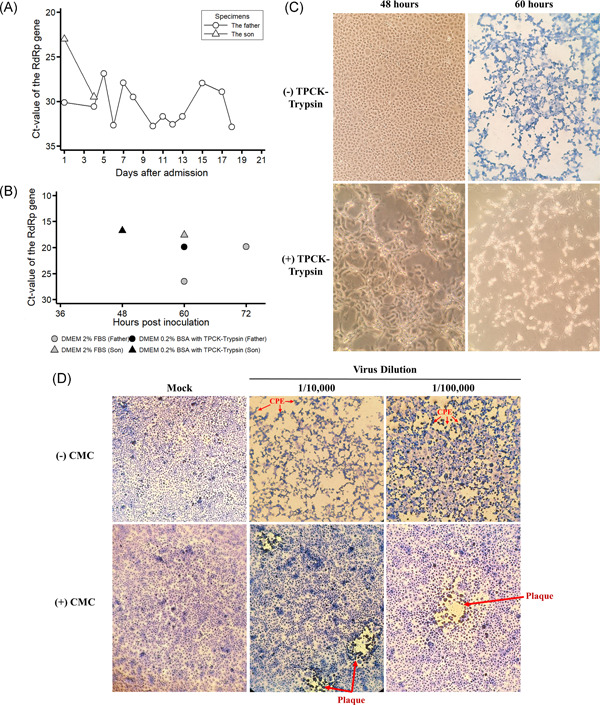
Viral shedding, isolation, and titration of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) from the first two infections identified in Vietnam. A, Results of real‐time reversetranscriptasepolymerasechainreaction (RT‐PCR) of nasopharyngeal and oropharyngeal swab specimens taken from the father (Patient 1) and son (Patient 2). Additionally, a sputum specimen was taken from Patient 1 on day 17 of hospitalization. B, Threshold cycle (Ct) values of the RdRp gene assay obtained from real‐time RT‐PCR assay of the supernatant collected from infected Vero E6 cells. The cut‐off value of Ct for a positive assay was less than 40. C, SARS‐CoV‐2‐infected Vero E6 cells by swab specimens of Patient 2 taken on admission in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% fetal bovine serum (top panels) and DMEM supplemented with 0.2% bovine serum albumin (BSA) and 16 μg/mL tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK)‐trypsin (bottom panels). D, Titration of the stock virus dilution in DMEM supplemented with 2% fetal bovine serum (FBS) with and without carboxymethyl cellulose (CMC). Red arrows indicate the presence of cytopathic effect (CPE) or plaque under the microscope. SARS‐CoV‐2. severe acute respiratory syndrome coronavirus 2
Another Covid‐19 case was his 27‐year‐old son (Patient 2), who had been in Vietnam for the past 4 months, and was apparently healthy. His NP and OP swab specimens had Ct values of 22.43 and 26.77 for the E and RdRp genes, respectively. Except for an increased C‐reactive protein level of 13.9 mg/L and lymphopenia of 0.55 × 109 cells/L, laboratory parameters and chest radiographs showed normal results. Treatment involved oseltamivir and supplementary therapies, including the daily use of chlorhexidine mouthwashes. Fever resolution was noted on day 3 of hospitalization. The last NP and OP swab specimens that tested positive for SARS‐CoV were collected on hospital day four postadmission (Figure 1A) and the first negative test was obtained on day 6. The patient was discharged from the hospital on February 4 (day 14).
3.2. Isolation of virus
SARS‐CoV‐2 was successfully isolated from NP and OP swab specimens collected from Patient 1 day five after hospitalization and from Patient 2 on admission. In flasks containing DMEM supplemented with 2% FBS, the CPEs were observed in the Vero cells at 60 hours postinoculation of Patient 2's specimens (Figure 1C, the top right panel). Cell supernatants showed the Ct value for the E and RdRp genes as low as 15.26 and 17.57 on real‐time RT‐PCR assays, respectively (Figure 1B), attesting to active viral infection. Despite the detection of SARS‐CoV‐2 in cell supernatants by real‐time RT‐PCR at 60 hours after inoculation of Patient 1′s specimen (the Ct value for the E and RdRp genes of 22.27 and 26.51), the CPEs were observed at 72 hours postinoculation with a lower Ct value of 17.64 for the E gene and 19.88 for the RdRp gene by real‐time RT‐PCR. In flasks containing DMEM supplemented with 0.2% BSA and 16 μg/mL TPCK‐trypsin, infected Vero cells became loose sooner at 48 hours postinoculation of Patient 2's specimen (Figure 1C, the bottom left panel), whereas it took 60 hours inoculation in the case of Patient 1's specimen, with a laboratory‐confirmed SARS‐CoV‐2 infection in cell supernatants (Figure 1B).
The lowest virus dilution for a visible plaque in a single field of view under the microscope was 10−5 dilution of the stock virus from Patient 2 (Figure 1D), which corresponds to the viral titer of 9.5 × 106 PFU/mL. The viral titer of the stock virus from Patient 1 was 3.3 × 102 PFU/mL.
3.3. Viral genome sequencing and variant analysis
The lengths of genome sequences of viral strains isolated from the two patients were 29 891 and 29 890 bp with no gaps and high coverage (664× and 1897×, respectively). Phylogenetic analyses revealed that the strains isolated in Vietnam belonged to Betacoronavirus B type and shared 99.98% sequence similarity at the nucleotide level with the strain isolated from patients in Wuhan (MT019529). It had more than 96.11% and 90.56% similarity with SARS‐CoV isolated from bat (MN996532) and pangolin (EPIISL410721) (Figure 2), respectively. The full genome sequence from Patient 2 was identical to that of Patient 1, genetically indicative of human‐to‐human transmission.
Figure 2.
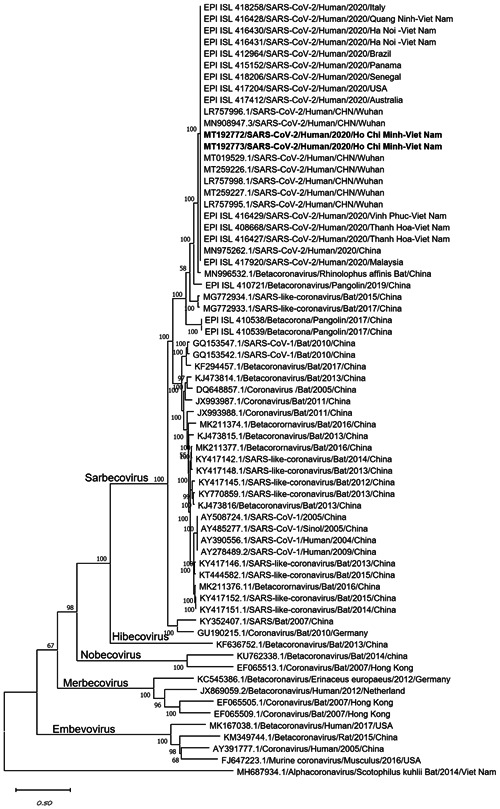
Phylogenetic tree of the complete genomes of severe acute respiratory syndrome coronavirus 2 and representative viruses of the genus β‐coronavirus. Texts highlighted in blue indicate the SARS‐CoV‐2 strains isolated in this study (MT192772 from Patient 2 and MT192773 from Patient 1). Complete genome sequences were aligned with other related coronavirus sequences archived from GenBank/GISAID using Mafft software and constructed a phylogenetic tree using the maximum likelihood method with 1000 bootstrap replicates in MEGA‐X using the general time‐reversible model. The bootstrap values were indicated on branches. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Compared to the reference sequence obtained from Wuhan (MT019529), four mutations were identified in the ORF1ab coding sequence. Three of the four mutations were classified as nonsynonymous mutations, including G8388A (serine to asparagine), A8987T (isoleucine to phenylalanine), and C10232T (arginine to cysteine). A synonymous mutation G3778A was also detected (Table S2).
4. DISCUSSION
This is a comprehensive report of clinical, epidemiological, and virological features of the first SARS‐CoV‐2 infections in Vietnam. We found that the SARS‐CoV‐2 strains isolated in Vietnam share a high similarity with the virus that originated in Wuhan, China. Considering Wuhan as the origin of the Covid‐19 first patients in Vietnam, our data demonstrate that the causative strains of the early Vietnam outbreak and the Wuhan outbreak were the same. This also implies that international air travel would have acted as the key pathway for the importation of Covid‐19 cases to Vietnam and, subsequently, triggered chains of transmission and spreading outbreak. Indeed, for all countries, border screening is needed. However, it is thought to be of limited effectiveness in detecting cases at borders, 12 due to the median incubation period of 5.2 days for SARS‐CoV‐2, 13 which is much longer than the duration of most international flights. To detect missing cases during border screening activities, implementation of appropriate strategies such as quarantine and testing for all overseas travelers may be useful to reduce the risk of community transmission of SARS‐CoV‐2. 14
Genome sequences of the virus strains isolated from both patients were found to be identical and this also indicates human‐to‐human transmission of SARS‐CoV‐2 (from the symptomatic infected Patient 1 to Patient 2). 11 Symptomatic as well as asymptomatic transmission of SARS‐CoV‐2 has been reported. 15 , 16
Despite an infection with the same strain of SARS‐CoV‐2, clinical manifestations between the two patients were distinct. The development of pneumonia, a prolonged viral shedding, and residual lung damage after hospital discharge were observed in the 65‐year‐old patient (Patient 1). These manifestations are consistent with other studies. 17 , 18 , 19 A declined immune function associated with aging and several underlying health conditions likely contribute to severe illness in older people. 20 , 21 Conversely, the 27‐year‐old patient (Patient 2) showed a mild upper respiratory tract infection with a high viral load. These data highlight the importance of contact tracing attempts to find, isolate, and test all contacts of a laboratory‐confirmed case for preventing community transmission. 22 In this young patient with high compliance chlorhexidine mouthwash, viral RNA was detected for a relatively shorter duration of illness (day 8, postonset of illness) compared to older patients, such as his 65‐year‐old father (day 24, postonset of illness). Such longer viremia among older patients have also been reported in Thailand (median age 61 years, median day 14, post‐onset of illness). 23 Of note, the small size of this report limited our ability to generalize clinical findings to the wider population.
There are concerns about the rapid evolution of SARS‐CoV‐2. 24 These are partly reinforced by our results showing nucleotide mutations in the ORF1ab region of the viral genome. The virological significance of the change, however, is unknown. Given the widespread transmission of SARS‐CoV‐2, additional sequences of new strains isolated are required to understand how the virus has spread and evolved. The broad availability of complete genome sequences of the virus isolated allows local and international agencies to adapt and validate diagnostic test kits for SARS‐CoV‐2 in the context of community transmission in different settings worldwide.
In conclusion, these data demonstrate the homogeneity of the SARS‐CoV‐2 isolated from Vietnam to the strains isolated from Wuhan and reconfirm its human‐to‐human transmission, and also highlight the need for compressive clinical and virological data of Covid‐19 patients in Vietnam to improve clinical practice and public health measures.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
QDP and MHD interpreted the data and wrote the first draft of the manuscript. HQL, SNN, and TANV obtained clinical specimens and data and contributed to the interpretation of clinical findings. LCQ and TVN collected and interpreted epidemiological data. LKH, HTN, QHN, TPH, MHD, NHPV, HTTP, AHN, NTN, TNTN, LTN, and TMC performed cell culture, sequencing, and contributed to the interpretation of virological findings. LTP, TVN, and TTN assisted in the interpretation of findings and overviewed the study. All authors have reviewed the manuscript, contributed to the critical revision of the manuscript, and approved the final version.
Supporting information
Supporting information
ACKNOWLEDGMENTS
This work would not be completed without the involvement of the patients and colleagues at Cho Ray Hospital and at the Pasteur Institute of Ho Chi Minh City, Vietnam. The authors thank Dr. Tu Ngoc Le at the Pasteur Institute of Ho Chi Minh City for medical record reviews. The authors acknowledge the authors, the originating, and submitting laboratories of the GISAID sequences. The authors thank Dr. Shyam Prakash Dumre at the Nagasaki University, Nagasaki, Japan for English language editing and proof‐reading before submission. The views in this publication do not necessarily represent the position of the Vietnamese Government. The Cho Ray Hospital and the Pasteur Institute of Ho Chi Minh City are affiliated with Vietnam's Ministry of Health. This study was supported by Vietnam's Ministry of Science and Technology [32/20‐ĐTĐL.CN‐CNN].
Phan LT, Nguyen TV, Huynh LKT, et al. Clinical features, isolation, and complete genome sequence of severe acute respiratory syndrome coronavirus 2 from the first two patients in Vietnam. J Med Virol. 2020;92:2209–2215. 10.1002/jmv.26075
Lan T. Phan and Thuong V. Nguyen contributed equally to this article and act as joint first authors.
REFERENCES
- 1. Johns Hopkins University Medicine. Coronavirus Resource Center . https://coronavirus.jhu.edu/. Accessed on May 25, 2020.
- 2. Than HM, Nong VM, Nguyen CT, Thi Tran NH, Do CD, Pham TN. Management of mild cases of COVID‐19 in low‐resource countries: An experience in Vietnam. J Microbiol Immunol Infect. 2020;S1684‐1182(20):30106‐30107. 10.1016/j.jmii.2020.1004.1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le TQM, Takemura T, Moi ML, et al. Severe acute respiratory syndrome coronavirus 2 shedding by travelers, Vietnam, 2020. Emerg Infect Dis. 2020;26(7), 10.3201/eid2607.200591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Cuong L, Giang HTN, Linh LK, et al. The first Vietnamese case of COVID‐19 acquired from China. Lancet Infect Dis. 2020;20(4):408‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baer A, Kehn‐Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J Vis Exp. 2014;93:e52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li B, Si H‐R, Zhu Y, et al. Discovery of bat coronaviruses through surveillance and probe capture‐based next‐generation sequencing. mSphere. 2020;5(1):e00807‐e00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS‐CoV‐2 genomes. Proc Natl Acad Sci USA. 2020;117(17):9241‐9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Phan LT, Nguyen TV, Luong QC, et al. Importation and human‐to‐human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382(9):872‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quilty BJ, Clifford S, Flasche S, Eggo RM. Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019‐nCoV). Euro Surveill. 2020;25(5):2000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferretti L, Wymant C, Kendall M, et al. Quantifying SARS‐CoV‐2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368(6491):eabb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gostic K, Gomez AC, Mummah RO, Kucharski AJ, Lloyd‐Smith JO. Estimated effectiveness of symptom and risk screening to prevent the spread of COVID‐19. eLife. 2020;9:e55570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan JF‐W, Yuan S, Kok K‐H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li C, Ji F, Wang L, et al. Asymptomatic and human‐to‐human transmission of SARS‐CoV‐2 in a 2‐family cluster, Xuzhou, China. Emerg Infect Dis. 2020;26(7), 10.3201/eid2607.200718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469. 10.1038/s41586-41020-42196-x [DOI] [PubMed] [Google Scholar]
- 18. Wang Y, Dong C, Hu Y, et al. Temporal changes of CT findings in 90 patients with COVID‐19 pneumonia: A longitudinal study. Radiology. 2020:200843. 10.1148/radiol.2020200843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nikolich‐Zugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol. 2018;19(1):10‐19. [DOI] [PubMed] [Google Scholar]
- 21. Marquez EJ, Chung CH, Marches R, et al. Sexual‐dimorphism in human immune system aging. Nat Commun. 2020;11(1):751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hellewell J, Abbott S, Gimma A, et al. Feasibility of controlling COVID‐19 outbreaks by isolation of cases and contacts. Lancet Glob Health. 2020;8(4):e488‐e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pongpirul WA, Mott JA, Woodring JV, et al. Clinical characteristics of patients hospitalized with coronavirus disease, Thailand. Emerg Infect Dis. 2020;26(7), 10.3201/eid2607.200598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS‐CoV‐2. Nat Med. 2020;26(4):450‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


