Abstract
Piwi-interacting RNAs (piRNAs) are a novel type of small noncoding RNAs, which are 26–30 nt in length and bind to Piwi proteins. These short RNAs were originally discovered in germline cells and are considered as key regulators for germline maintenance. A growing body of evidence has now extended our views into piRNA biological significance showing that they can also regulate gene expression in somatic cells through transposon silencing, epigenetic programming, DNA rearrangements, mRNA turnover, and translational control. Mounting studies have revealed that the dysregulation of piRNAs may cause epigenetic changes and contribute to diverse diseases. This review illustrates piRNA biogenesis, mechanisms behind piRNA-mediated gene regulation, and changes of piRNAs in different diseases, especially in cancers.
Keywords: piRNA, Piwi, disease, cancer
Graphical Abstract
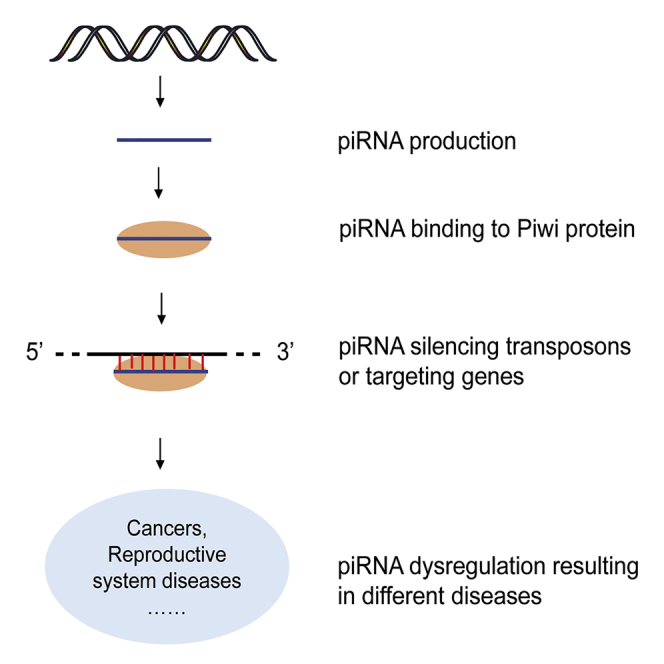
Shu and colleagues demonstrate the biogenesis, functions, changes, and probable roles of piRNAs in diseases of different systems, especially in cancers. piRNAs are promising novel biomarkers for early diagnosis and therapeutic targets for precise medicine.
Main Text
As reported, more than three-quarters of the human genome is capable of being transcribed into RNAs, but only 2% ultimately encodes proteins.1, 2, 3 Those RNAs that do not encode proteins are called noncoding RNAs (ncRNAs). With the establishment and development of high-throughput genome sequencing technology, it has been widely recognized that ncRNAs are of great importance in cellular activities and individual health. Basically, ncRNAs can be divided, based on the length, into long ncRNAs (>200 nt) and small ncRNAs (≤200 nt).4 Furthermore, small ncRNAs can be classified into several subclasses such as microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), small cytoplasmic RNAs (scRNAs), transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs).5,6 piRNAs were first isolated and purified from rat testes in four independent laboratories in 2006.7, 8, 9, 10 piRNAs are 26–30 nt in length, with 2′-O-methylation at their 3′ end.11 Actually, piRNAs promote gene silencing processes via interacting with Piwi proteins.7,12,13 There are four isoforms of Piwi proteins in human beings, i.e., PIWIL1 (Hiwi), PIWIL2 (Hili), PIWIL3, and PIWIL4 (Hiwi2); three in rodents, i.e., PIWIL1 (Miwi), PIWIL2 (Mili), and PIWIL4 (Miwi2); and three in Drosophila, i.e., PIWI, Aub, and Ago3.14,15 The initially identified functions of piRNAs included transposon silencing and gene integrity maintained in germline cells. To date, piRNAs have been detected both in the germline cells and somatic cells in many animals, such as nematodes, insects, fish, and mammals.16 It is apparent that piRNAs not only serve as transposon silencers but they also play crucial roles in regulating gene expression.17, 18, 19, 20 Aberrant piRNA expressions have been detected in diverse diseases, especially in neoplasms and reproductive system diseases. piRNAs are promising novel biomarkers for early diagnosis and therapeutic targets for precise medicine. This review explains the biogenesis, functions, changes, and probable roles of piRNAs in diseases of different systems.
Biogenesis of piRNAs
Generation of Precursors
A large proportion of piRNA precursors are produced from genetic regions named piRNA clusters, which can be divided into uni-strand or dual-strand clusters. Uni-strand clusters give rise to precursors mapping only to one strand, whereas dual-strand clusters produce precursors mapping to both genomic strands. Additionally, some piRNA precursors can be generated from the 3′ UTR of protein-coding genes, or from individual transposons.21,22
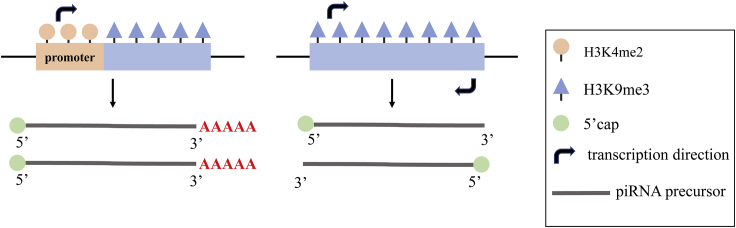
The transcription of uni-strand clusters is similar to the canonical mRNA transcription. Uni-strand clusters harbor the transcription-associated histone 3 lysine 4 demethylation (H3K4me2) mark at promoters. Additionally, piRNA precursors are 5′ methyl-guanosine capped and 3′ terminated.23,24 On the contrary, dual-strand clusters lack clear signatures of RNA polymerase II (RNA Pol II) promoters, such as H3K4me3 and peaks of RNA Pol II, and non-polyadenylated piRNA precursors are produced (Figure 1).23,25,26
Figure 1.
Difference between Uni-Strand and Dual-Strand Cluster Transcription
Uni-strand clusters (left) have distinct promoter regions, featured by H3K4me2, and generate 5′-methyl-guanosine-capped and 3′-terminated precursors from one direction. Dual-strand clusters (right) have no distinct promoter regions and generate non-polyadenylated piRNA precursors from two directions.
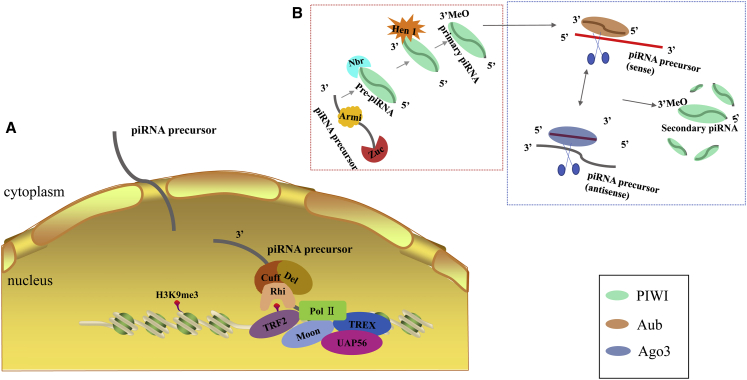
Dual-strand clusters are transcribed into piRNA precursors with the involvement of several proteins, including RNA Pol II, Rhino-Cutoff-Deadlock complex (RDC complex), Moonshiner, TATA-box binding protein-related factor (TRF2), and three prime repair exonuclease (TREX). First, RNA Pol II is recruited onto piRNA clusters. Then, the transcription is facilitated by the RDC complex, which includes three subunits: Rhino (Rhi), Deadlock (Del), and Cutoff (Cuff). Rhi, the paralog of heterochromatin protein HP1, binds to the histone 3 lysine 9 trimethylation (H3K9m3) mark. Although the H3K9me3 mark is usually known to inhibit transcription in heterochromatin, it is essential in the process of piRNA precursor biogenesis. Meanwhile, Rhi inhibits inaccurate splicing between dual-strand cluster transcripts and represses the use of canonical cleavage and polyadenylation sequence motifs.27 Cuff prevents premature termination and protects precursors from degradation.23,25 Moonshiner, the paralog of the germline-specific transcription initiation factor IIA subunit 1, also facilitates the initiation of transcription and represses the H3K9me3 mark by interacting with the RDC complex and TRF2.28 TREX prevents R-loop formation, the hybrids between nascent RNA and DNA.29, 30, 31 The nuclear DEAD box RNA helicase 56-kDa U2AF-associated protein (UAP56) suppresses the splicing of dual-strand cluster precursors (Figure 2A).27,32
Figure 2.
The Procedure of piRNA Production
(A) Biogenesis of piRNAs precursors from dual-strand clusters. piRNA precursors are produced with the involvement of RNA polymerase II, the Rhino (Rhi)-Cutoff (Cuff)-Deadlock (Del) complex, Moonshiner (Moon), TATA-box binding protein (TBP)-related factor 2 (TRF2), three prime repair exonuclease (TREX), and 56-kDa U2AF-associated protein (UAP56). Rhi binds to the H3K9me3 mark and inhibits transcript splicing, as well as represses the use of canonical cleavage and polyadenylation sequence motifs. Cuff prevents premature termination and protects RNA from degradation. Moonshiner promotes initiation of transcription and represses the H3K9me3 mark by interacting with the RDC complex and TRF2. TREX prevents the formation of R-loops. UAP56 suppresses the splicing of dual-strand cluster. (B) Biogenesis of mature piRNAs. The red dotted box represents the primary biogenesis of piRNAs. First, secondary structures of piRNA precursors are resolved by the RNA helicase Armitage (Armi). After despiralization, precursors are processed into pre-piRNAs with a 5′ monophosphate by the endonuclease Zucchini (Zuc). Then, pre-piRNAs are loaded onto the PIWI and trimmed by the 3′ to 5′ exonuclease Nibbler (Nbr). Concomitantly, the newly formed 3′ terminal ends are methylated at the 2′ oxygen by the small RNA 2′-O-methyltransferase Hen1. This procedure is called primary biogenesis. The dark blue dotted box represents the secondary biogenesis of piRNAs (the ping-pong cycle). Aub binds to antisense-strand piRNAs and then cleaves sense piRNA precursors, giving rise to sense piRNAs loaded into Ago3. In contrast, Ago3 binds to piRNAs from sense clusters. The Ago3/piRNA complex cleaves antisense piRNA precursors and produces antisense piRNAs that are loaded into Aub. The round of cleavage and trimming repeats and produces a number of piRNAs in a short time.
Uni-strand piRNA cluster transcriptions have been observed in many metazoans, such as sponges, hydras, silk worms, fruit flies, zebrafish, mice, and humans, which appear to be evolutionarily conserved.7, 8, 9,24,33, 34, 35, 36, 37, 38 Although dual-strand piRNA cluster transcriptions have been reported in Drosophila, whether they occur in other organisms needs more study.24
Generation of Mature piRNAs
Once the transcription is finished, piRNA precursors are transported out of the nucleus (Figure 2B). First, the secondary structures are resolved by the RNA helicase Armitage (Armi). After despiralization, piRNA precursors are processed into pre-piRNAs with a 5′ monophosphate through the cleavage of the mitochondria-associated endonuclease Zucchini (Zuc). Then, pre-piRNAs are loaded onto PIWI proteins and are trimmed at the 3′ ends by a 3′ to 5′ exonuclease, Nibbler (Nbr). Concomitantly, the newly formed 3′ terminal ends are methylated at the 2′ oxygen by the small RNA 2′-O-methyltransferase Hen1.33,39, 40, 41, 42, 43, 44, 45, 46, 47 It has been suggested that 2′-O-methylation may enhance piRNA stability.41,45, 46, 47 This procedure is called primary biogenesis of piRNAs, and piRNAs produced in this way are named primary piRNAs.
Primed by primary piRNAs, the generation of piRNAs is amplified with the involvement of Ago3 and Aub proteins. Aub binds to antisense-strand piRNAs and then cleaves sense piRNA precursors, giving rise to sense piRNAs bound by Ago3. In contrast, Ago3 binds to sense-strand piRNAs and cleaves antisense piRNA precursors, producing antisense piRNAs that load onto Aub.48,49 The round of cleavage repeats and produces a number of piRNAs in a short time. The way in which large amounts of piRNAs are produced is called secondary biogenesis of piRNAs, as well as the ping-pong cycle. These piRNAs are then bound by PIWI proteins and transported back to the nucleus to silence target genes.
The exact procedure of piRNA biogenesis remains elusive, and most of our understanding comes from Drosophila germline cells. A similar pathway has been observed in mice just with the involvement of different Piwi proteins. Meanwhile, there are clues indicating that this procedure in humans may be similar to Drosophila.50,51
Functions of piRNAs
It was initially reported that piRNAs defended against transposon mobilization in fly germline cells, which was later verified in organisms ranging from hydras to humans.15,33,40,52, 53, 54, 55, 56, 57 Transposons, known as jumping genes, are similar to endogenous viruses. They threaten the stability of genes as they “copy and paste” their own DNA in the host genome to reproduce themselves, which causes a series of consequences. Exon insertions hinder the coding sequence, and the insertion into introns may change splicing patterns, which may lead to the production of novel and potentially deleterious fusion proteins.58 If transcripts of transposons are inserted into promoter or enhancer regions, gene transcriptions can also be transformed. Meanwhile, insertions into 5′ UTRs or 3′ UTRs can affect post-transcriptional gene regulation. Transposon insertions also lead to DNA nicks and double-strand breaks; moreover, errors in the repair of these lesions can lead to recombination between transposon repeats, triggering chromosomal duplication, deletion, translocation, and inversion.59
At the transcriptional level, piRNAs and Piwi proteins directly modify chromatin structure and histone proteins in the nucleus via the regulation of DNA methyltransferase (DNMT). DNMT methylates CpG islands in promoter regions, which suppresses the initiation of transcription. Piwi proteins guide DNMT to bind with transposable elements or target genes. As a consequence, transposable elements or target genes are silenced. However, little is known about the exact mechanisms by which piRNAs and Piwi proteins regulate DNMT protein expression. piRNAs and Piwi proteins also interact with histone methylation machinery and regulate the methylation of histone lysine residues (H3K and H4K). They recruit histone methyltransferases onto target transcripts to repress transcription.60,61 Currently, the relationships between histone modification and piRNAs remain unresolved.
At the post-transcriptional level, piRNAs and Piwi proteins induce mRNA degradation via mRNA decay machinery. The major way to decay mRNA and impede translation is to shorten the poly(A) tail of mRNAs, which is called deadenylation.51
Changes in Diseases
As piRNAs are involved in gene regulation, there has been a budding interest in defining their roles in human diseases. A number of studies have revealed that the dysregulation of piRNAs can both promote or suppress the occurrence and development of various diseases, especially cancers.62
Cancers
Lung Cancer
Lung cancer ranks first with highest incidence and mortality among all cancers in both sexes worldwide.63 Unlike miRNAs and circRNAs, which have been largely studied in lung cancer pathogenesis, there have been limited studies demonstrating the relationship between piRNAs and lung cancer.64,65 However, current studies can extend our insights into the occurrence and development of lung cancer and can provide new therapeutic strategies. A recent study compared piRNA expression profiles between eight non-small-cell lung cancer (NSCLC) tissue samples and three human bronchial epithelial cell samples.66 piR-L-163 in NSCLC cells, the top downregulated one, inhibited cell migration and invasion by maintaining the function of phosphorylated ezrin-radixin-moesin (p-ERM), which connects transmembrane proteins, such as ERM-binding phosphoprotein 50 (EBP50) and filamentous actin (F-actin).66 In another study, piR-55490 was found to suppress lung cancer cell proliferation by binding to the 3′ UTR of mammalian target of rapamycin (mTOR) mRNA.67 It was identified that upregulated piR-651 promoted cell proliferation, migration, and invasion, as well as suppressed cell death in NSCLC A549 and HCC827 cell lines, but its mechanism needs further study.68 Moreover, piR-57125 was significantly downregulated in metastatic lung adenocarcinoma cells compared with primary cancer cells.69 Other piRNAs, such as piR-34871, piR-35127, piR-46545, piR-52200, and piR-57125, were also determined to be downregulated in lung cancer cells, and they comprehensively promoted the proliferation of lung cancer cell lines.70 These studies suggest that aberrant piRNA expressions are involved in the tumorigenesis, invasion, and metastasis of lung cancer, which may provide potential functional targets and novel biomarkers for precise medicine.
Gastric Cancer
Gastric cancer remains an important cancer, making it the fifth most frequently diagnosed cancer and the third leading cause of cancer death worldwide,63 while it is the second most commonly diagnosed cancer in males and third in females, and it is the second leading cause of cancer death in both men and women in China.71 It was identified that piR-823 significantly downregulated in gastric cancer tissues. Cell proliferation was inhibited with piR-823 mimics, and a xenograft nude mouse model verified that piR-823 suppressed tumor growth.72 Moreover, patients with gastric cancer had low expression of piR-651 and piR-823 in peripheral blood. piR-32105, piR-58099, and piR-59056 were reported to increase in gastric cancer tissues.73 It is well known that the early stage of gastric cancer can be cured by surgery or endoscopic treatment, but the prognosis of advanced gastric cancer is unsatisfied. Immunotherapy, similar to anti-programmed cell death 1/programmed cell death ligand 1 (PD-1/PD-L1) therapy, is a promising therapy in cancer, while patients with advanced gastric cancer are not very sensitive to immunotherapy alone or combined with chemotherapy in first-line therapy. More studies should be performed to identify whether piRNAs are related to immune escape, and whether those piRNAs, similar to piR-823, which can be detected in peripheral blood, could be promising biomarkers for predicting immunotherapy outcomes and for diagnosis in gastric cancer.
Liver Cancer
Liver cancer is predicted to be the sixth most commonly diagnosed cancer and the fourth leading cause of cancer death worldwide. Rates of both incidence and mortality are 2- to 3-fold higher among men in most world regions.63 Additionally, liver cancer is the third most commonly diagnosed cancer in men and the third leading cause of cancer-related deaths among both men and women in China.71 The formation of liver cancer can be divided into several steps: cirrhotic nodules (CNs), low-grade dysplastic nodules (LGDNs), high-grade dysplastic nodules (HGDNs), early hepatocellular carcinoma (eHCC), and progressed HCC (pHCC).74 Each stage was characterized by several piRNAs. For example, piR_LLi_24894 was only expressed in CNs; piR-LLi-30552 and hsa-piR-020498 were mainly expressed in HGDNs, eHCCs, and pHCCs; and hsa-piR-013306 was accumulated only in HCC. Downregulated piRNAs in early HCC-targeted genes such as tumor necrosis factor (TNF) receptor, phosphatidylinositol 3-kinase (PI3K)/AKT (serine/threonine kinase), WNT/β-catenin, growth arrest and DNA damage-inducible protein 45 (GADD45), adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK), high mobility group box 1 (HMGB1), and phosphatase and tensin homolog (PTEN), which control cell cycle regulation, telomerase activity, protein ubiquitination, DNA methylation, and apoptosis.74 Furthermore, piR-823 promoted the production of α-smooth muscle actin (α-SMA), and collagen type I alpha 1 (COL1a), as well as components of extracellular matrix, which finally resulted in cirrhosis.75 piR-Hep1 may have a role in suppressing cell death by activating the signal transducer and activator of transcription 3 (Stat3)/Bcl-xL signaling pathway.76 Actually, most HCCs develop from cirrhosis for which hepatitis B virus infection and alcohol abuse are the most common causes. It is important to explore potential relationships between piRNAs and these two kinds of cirrhosis, which are valuable for the prevention of cirrhosis transforming into HCC.
Colorectal Cancer
Colorectal cancer (CRC) is among the five most commonly diagnosed cancers, and the incidence rate increases year by year.71 piR-1245 was predominantly overexpressed in CRC tissues and regulated the expression of vital genes that control cellular processes such as cell death and survival, DNA replication and repair, and cell-cell communication.77 These genes included activating transcription factor 3 (ATF3), BTG anti-proliferation factor 3 (BTG1), dual specificity phosphatase 1 (DUSP1), Fas cell surface death receptor (FAS), nuclear factor κB (NF-κB) inhibitor α (NFκBIα), uridine phosphorylase 1 (UPP1), sestrin 2 (SESN2), and tumor protein p53 inducible nuclear protein 1 (TP53INP1).77 Consistently, the downregulation of piR-1245 induced cell apoptosis, while the overexpression of piR-1245 promoted cell proliferation.77 It was found that upregulated piR-54265 promoted CRC cell proliferation and invasiveness through the STAT3 pathway, which resulted in the increase of metastasis-causing molecules, such as matrix metallopeptidase (MMP)2 and MMP9. Moreover, piR-54265 hindered chemotherapy, and patients with higher serum piR-54265 levels presented a significantly poorer response to chemotherapy.78 piR-823 was reported to activate the expression of heat shock transcription factor 1 (HSF1) by promoting its phosphorylation at Ser326, and promote colorectal carcinogenesis.79 piR-5937, piR-001311, piR-004153, piR-017723, piR-017724, piR-020365, piR-28876, piR-32105, piR-58099, and piR-59056 were also downregulated in CRC patients.80, 81, 82 Patients with different primary sites of CRCs had different clinical manifestations, therapeutic responses, prognosis, and other characteristics. As many piRNAs are involved in the development of CRC, whether piRNAs present divergent expressions in left colon cancer, right colon cancer, or rectal cancer and whether they are associated with prognosis are worthy of further study.
Clear Cell Renal Cell Carcinoma
There are several types of kidney cancers, and most cases are clear cell renal cell cancers (ccRCCs).83 piR-34536 and piR-51810 were downregulated in ccRCC tumor tissues, and their low expression was related to shorter disease-free survival (DFS) and overall survival (OS). piR-823 was significantly decreased in tumor tissues, and a low level was associated with a longer DFS.84,85 Additionally, piR-32051, piR-39894, and piR-43607 were detected to be significantly upregulated in embryonic renal cancer and all ccRCC cell lines, accompanied by a shorter cancer-specific survival and a higher chance of tumor metastasis.86 Immune checkpoint inhibitors are effective in the treatment of advanced kidney cancer, yet there appear to be no studies about piRNAs and kidney immunotherapy.87 The relationship between piRNAs and immunotherapy can be further explored.
Bladder Cancer
It was reported that the mortality of bladder cancer ranked ninth in China in 2016, and the overall incidence of bladder cancer is increasing in men.88 Up to date, there have been very few studies illustrating roles of piRNAs in bladder cancer, to which more attention could be paid. A total of 197 piRNAs were differentially expressed in bladder cancer cells compared with normal cells, among which piR-60152 was the most downregulated.89 It was inferred that piR-60152 suppressed bladder cancer EJ cell apoptosis by targeting TNFSF4, which is a binding partner for the TNF superfamily member, OX40. However, it was not observed that piR-60152 promoted cell cycle progression, cell migration, and invasion.89
Breast Cancer
Breast cancer is the most common cancer diagnosed among women.90 Breast cancers can be classified into molecular subtypes according to different expressions of cellular markers, such as estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor-2 (HER2), Ki-67, and basal cytokeratin (CK). The six subtypes are luminal A, luminal B, luminal-HER2, HER2-enriched, basal-like, and TNP (triple-negative phenotype)-nonbasal.91 It was found that piR-36712 may suppress breast cancer cell proliferation by inhibiting selenoprotein W1 (SEPW1) expression through a retroprocessed pseudogene of SEPW1, SEPW1P. SEPW1 inhibited the expression of p53 and p21 by inducing mRNA degradation, and the knockdown of SEPW1 resulted in G1 cell cycle arrest. A lower piR-36712 level was related to a shorter progression-free survival (PFS) and a higher risk of axillary lymph node metastasis.92 Moreover, piR-36712 appeared to enhance the effect of chemotherapeutic agents.93 piR-021285 promoted invasion through methylating the 5′ UTR first exon of the pro-invasive Rho GTPase activating protein 11A (ARHGAP11A) gene.94 In another study, upregulated piR36026 promoted cancer cell proliferation by inhibiting serpin family A member 1 (SERPINA1) and lecithin retinol acyltransferase (LRAT) gene expression, which are known for their tumor-suppressing function.95 It was found that piR-932 was significantly overexpressed in breast cancer cells, which led to low latexin (a type of tumor-suppressing protein) expression.96 High expression of piR-20365, piR-20485, and piR-4987 was correlated with metastasis.97 In addition, piR-23662, piR-26526, piR-26527, piR-26528, piR-30293, piR-34377, piR-34736, piR-35407, piR-36318, and piR-36249 were significantly downregulated, while piR-651, piR-1282, piR-21131, piR-23672, piR-31106, piR-32745, and piR-36743 were upregulated in breast cancer cells.98,99 The specific mechanisms behind those changes are still unknown. Although many piRNAs were dysregulated in breast cancers, these studies were not histologically classified. Many questions remain unknown, such as, whether there is any difference between piRNA expressions in different luminal subtypes; whether aberrant piRNA expressions are associated with the phenotypic impact of genomic alterations; and whether piRNAs influence responses to treatment in different subtypes of breast cancer.
Ovarian Cancer
Ovarian cancer accounts for 2.5% of all female malignancies, but 5% of female cancer deaths.100 In endometrioid ovarian cancer (ENOCa) samples, the upregulation of piR-52207 repressed four target genes, nudix (nucleoside diphosphate linked moiety X)-type motif 4 (NUDT4), methionine synthetase (MTR), eukaryotic translation initiation factor 2 subunit 3 (EIF2S3), and M-phase phosphoprotein 8 (MPHOSPH8), which are involved in 3-phosphoinositide biosynthesis, D-myo-inositol-5-phosphate biosynthesis, folate transformation, vascular endothelial growth factor (VEGF), and EIF2 signaling pathways, respectively.101 In serous ovarian cancer (SOCa) samples, piR-52207 repressed the expression of actin-related protein 10 (ACTR10) and pleckstrin homology domain-containing family A member 5 (PLEKHA5), which are associated with the PI3K/Akt signaling pathway and dynactin’s pointed-end complex biogenesis, respectively.101 Furthermore, piR-33733 was identified to target the lipoic acid synthase (LISA) gene, which is involved in the synthesis of lipoic acid, a well-known antioxidant that induces apoptosis.101 It is suggested that mutations of breast cancer susceptibility genes (BRCA1 and BRCA2) account for almost 40% of ovarian cancer cases in women with a family history of the disease.102 It is worth of learning whether piRNAs are related to BRCA mutation. If piRNAs do associate with BRCA mutations, they can serve as assistant factors for genomic evaluation and indicators for anti-BRCA drug resistance.
Neurological Cancer
Neurogliomas are the most common primary malignant brain tumors in adults. Neuroglioma can be divided into four grades (I–IV) based on malignant behavior.103 piR-598 was identified as a tumor suppressor because its target genes were related to cell death, such as BAX, which regulates p53-mediated cell death.104 It was found that piR-DQ593109 regulated blood-tumor barrier (BTB) permeability through the MEG3/miR-330-5p/RUNX3 axis.105 Additionally, piR-39980 promoted tumor growth and facilitated drug resistance, yet piR-8041 suppressed glioma growth.28,106 Neuroblastoma is a complex and therapeutically challenging malignant cancer in children. Seven piRNAs (piR-32287, piR32512, piR-36095, piR-38581, piR-52205, piR-52206, and piR-57816) were enriched in neuroblastoma cells, and their target genes included BRCA1, eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2), histone deacetylase 2 (HDAC2), and peroxisome proliferator activated receptor alpha (PPARA), which are demonstrated to promote tumorigenesis.107 Currently, gliomas are treated with surgery and lack effective drugs, as drugs can hardly penetrate the blood-brain barrier. If more piRNAs are detected in cerebral spinal fluid or embed with nanoparticle biointerfacing by platelet membrane cloaking, such as piR-DQ593109, which could increase the permeability of the blood-brain barrier, the drug treatment to neurological cancer will be more efficient.
Multiple Myeloma
Multiple myeloma (MM) is a commonly diagnosed hematological neoplasm and is a progressive and mostly incurable disease.108 MM cells are characterized by permeating into end organs and growing into tumors with the help of endothelial cells (ECs), an important type of cells in tumor microenvironments. Increased piR-823 promoted EC proliferation by repressing the caspase-3/Bax signaling pathway, and it enhanced angiogenesis by inducing the secretion of VEGF and interleukin (IL)-6.109 Additionally, piR-823 induced intercellular adhesion molecule 1 (ICAM-1) and C-X-C motif chemokine receptor 4 (CXCR4) secretion to promote EC invasion.109 In another study, piRNA-823 induced DNA methylation via DNMTs (DNMT3A and DNMT3B) and suppressed a methylation-silenced tumor suppressor, p16INK4A.110 Meanwhile, piRNA-4987 and piRNA-Hep1 were overexpressed in MM cells.111 As piRNAs could result in the development of MM through DNA methylation, and histone-lysine N-methyltransferase EZH2 inhibitors have been applied for MM treatment, whether piRNAs interact with EZH2 in MM pathogenesis is worth further study.
Head and Neck Squamous Cell Carcinoma
Head and neck squamous cell carcinoma (HNSCC) can occur in the oral cavity, oropharynx, hypopharynx, larynx, and nasopharynx. It is well known that virus infection and tobacco and alcohol consumption increase the risk of development of HNSCC.112 In HPV16/18-associated HNSCC tissues, the loss of the piRNA FR140858 could potentially promote the expression of minichromosomal maintenance complex component 7 (Mcm7), which controls the initiation and elongation steps of eukaryotic DNA replication.113 In smoking-related HNSCC tissues, four upregulated piRNAs (NONHSAT015828, NONHSAT081250, NONHSAT113708, and NONHSAT123636) are connected with advanced tumor stage and metastasis, and the downregulated NONHSAT067200 was related to longer patient survival.114 Similarly, in alcohol-related HNSCC tissues, piR-34946, piR-38034, piR-43219, and piR-58510 were upregulated, and piR-70732 was downregulated.115 More efforts should be made so that deeper knowledge between piRNAs and HNSCC can be identified and more therapeutic opportunities will be available.
Osteosarcoma
Osteosarcoma is the most common primary solid tumor of bone in childhood and adolescence and mainly occurs in the second decade of life.116 Current therapeutic methods include surgical resection of all cancer tissues and systemic chemotherapy. The 5-year event-free survival rate of patients with localized osteosarcoma is approximately 70%, while that of patients with metastatic or recurrent osteosarcoma is less than 20%.117 Few studies showed that piRNAs are associated with osteosarcoma. piR-39980 was reported to promote the apoptosis of fibrosarcoma cells and suppress cell proliferation by inhibiting ribonucleoside-diphosphate reductase subunit M2 expression, an enzyme indispensable in DNA synthesis and repair.118 Because most sarcomas are not sensitive to chemotherapy, it is far-reaching to explore how piRNAs weaken the efficacy of systemic chemotherapy to sarcomas.
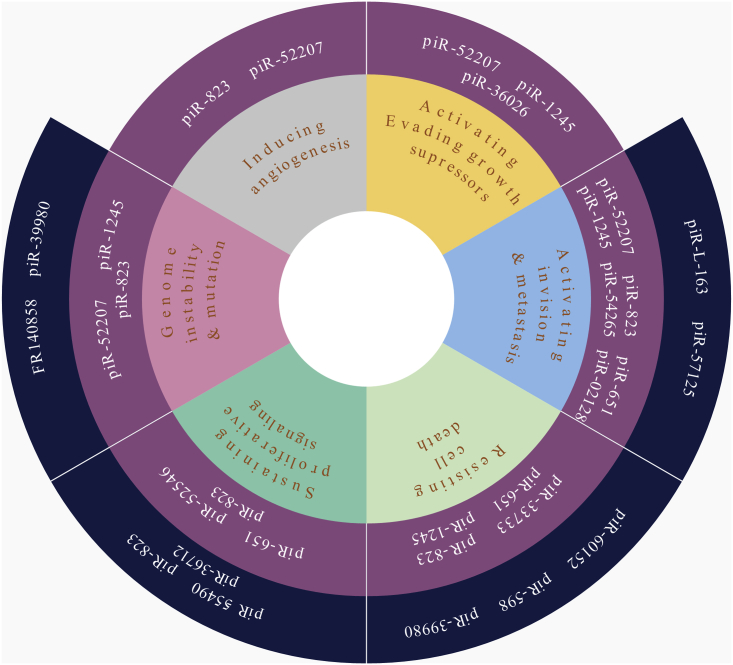
Overall, intensive studies have indicated that aberrant piRNA expressions may lead to tumorigenesis. It has been proposed that cancer cells present certain hallmarks, such as sustaining proliferative signaling, evading growth suppressors, inducing angiogenesis, activating invasion and metastasis, and others.119 Those aberrant piRNAs may influence the acquisition of hallmark capacities (Figure 3). For example, piR-L-163 inhibited cell migration and invasion by maintaining the function of p-ERM and F-actin;66 piR-823 enhanced angiogenesis by inducing the secretion of VEGF and IL-6.109 piRNAs also are strongly correlated with tumor cell malignant phenotype, clinical stage, as well as clinical response, and they can be a predictive factor for prognosis. Most piRNAs are unique to individual cancer types, while several piRNAs are common to many cancer types, such as piR-823. piR-823 suppressed the development of gastric cancer and kidney cancer, yet it promoted the development of liver cancer, MM, and CRCs. Specific piRNAs in particular cancers can serve as particular biomarkers, and those common ones can be complementary to other tests. However, specific mechanisms by which piRNAs lead to tumorigenesis remain unknown, such as whether piRNAs influence those targets transcriptionally or post-transcriptionally. Moreover, the relationship between cancer sensitivity to chemoradiotherapy and piRNAs is also worth further discussion. In recent years, immunotherapy has been effective in NSCLC, malignant melanoma, and kidney cancer, whereas it has had almost no effect in gastric, colorectal, and breast cancer. Is there any difference in piRNA expression profiles between immunotherapy-sensitive and -insensitive cancers? More studies are needed to decipher underlying molecular functions and deepen our understanding of therapeutic methods of piRNAs in cancer.
Figure 3.
piRNAs Related with Cancer Hallmarks
piRNAs in inner circle promote normal cells acquiring these cancer hallmark capabilities, and others in outer circle prevent normal cells from obtaining these cancer hallmarks.
Reproductive System Diseases
An evaluation published by World Health Organization in 2004 reported that one in every four couples in developing countries had been found to be affected by infertility. The report published at the end of 2012 showed that the overall burden of infertility remained similar.120 Infertility patients are under great psychological pressure and can finally result in mental illness. Additionally, infertility will lead to low birth rate and aggravate the aging of society. Premature ovarian insufficiency, polycystic ovary syndrome, endometriosis, uterine fibroids, and endometrial polyps are the main causes of female infertility. Testicular and post-testicular deficiencies may lead to male infertility.120 As a highly regulated developmental process, mammalian spermatogenesis consists of three steps in chronological order: mitosis, meiosis, and differentiation. This process results in a haploid gamete for sexual reproduction. Aberrant piRNA expression in testicular cells may cause spermatogenic failure. Five piRNAs (DQ589977, DQ591415, DQ598918, DQ601291, and DQ601609) were found to be downregulated in spermatogenic failure samples, and they targeted PIWI2 and tudor domain-containing protein 1 (TDRD1) gene promoters. Their expression was negatively correlated with the methylation degree of the PIWI2 and TDRD1 gene promoters.121 Other proposed target genes induced IL-16, kallikrein 1 (KLK1), G protein-coupled receptor 156 (GPR156), histone cluster 1 H2aa (HIST1H2AA), RAB24 (a member of the RAS oncogene family), sphingomyelin phosphodiesterase 3 (SMPD3), and dolichyl-phosphate mannosyltransferase polypeptide 1 (DPM1).121 It was observed that the rs508485 variation in the 3′ UTR of the HIWI2 gene led to idiopathic nonobstructive azoospermia. The mutation also affected mRNA stability or altered the binding affinity of regulatory miRNAs.122,123 hsa-piR-26399 was also expressed differently between sera of subfertile and control men.124
piRNAs were originally found in mouse testis cells and were confirmed to be key players in germ cell maintenance in mouse and Drosophila models. Because piRNAs act as versatile regulators of gene expression and epigenetic events during spermatogenesis, they may provide a potent tool for the diagnosis of infertility. It will be useful to establish cell and stage-specific expression profiles of piRNAs during mammalian spermatogenesis using microarray or real-time RT-PCR. Furthermore, piRNAs could be promising targets of novel drugs for treatment.
Other Diseases
Retinal Diseases
Oxidative stress is the most relevant biochemical cause of retinitis pigmentosa, and retinal pigment epithelium cells are very sensitive to oxidative stress.125 piRNAs, such as piR-41220 and piR-46956, were globally downregulated in retinal pigment epithelium cells treated with oxidized low-density lipoprotein (oxLDL). Specially, they defended against oxidative stress by targeting genes linked with visual function.126
Cardiovascular Diseases
piRNA expressions were profiled in cardiac progenitor cells (CPCs), such as cardiofibroblasts (CFs), cardiosphere-derived cells (CDCs), and cardiospheres.127 Upregulated piRNAs (such as DQ591926, DQ593270, and DQ593595) targeted transposons throughout the human genome, including LINE retrotransposons, of which LINE-1 was most targeted.127 Additionally, it was found that piRNAs stimulated cell viability through the AKT signaling pathway in the cardiac system.128 In another study, many piRNAs were downregulated in heart failure patients, among which hsa-piR-020009 and hsa-piR-006426 were the top two.129 piR-2106027 was increased significantly in myocardial infarction (MI) patients, except in troponin-I-negative MI patients.130
Nervous System Diseases
It was found that approximately 100 piRNAs were dysregulated in Alzheimer’s disease (AD) patients, and most AD-associated piRNAs were correlated with genome-wide single-nucleotide polymorphisms (SNPs).131 Roy et al.132 identified that three piRNAs (piR-38240, piR-34393, and piR-40666) that participated in maintaining hemostasis in the nervous system. Their overexpression may lead to cell apoptosis and neurodegeneration and affect the trafficking pathway regulating Aβ levels. Their target genes were cytochrome C somatic (CYCS), karyopherin subunit α6 (KPNA6), and Ras-related protein Rab-11A (RAB11A), which induce apoptosis and maintain cellular homeostasis via oxidative stress-induced modulation of the Nrf2 (nuclear factor E2-related factor 2)-dependent antioxidant response and recycled endosomes and vesicular trafficking. piRNAs or piRNA-like molecules derived from the LINE and SINE genes were significantly downregulated and failed to silence LINE and SINE transposons in patients with Parkinson’s disease (PD).133
So far, there have been few studies on other human diseases. The analysis of piRNA roles in these non-neoplastic diseases is far less thorough than that of tumors. Because it is difficult to obtain tissue samples, most studies are performed on model animal tissues. Actually, several studies suggested that piRNAs can be detected in body fluid. If relationships between piRNAs in body fluid and these diseases can be summarized, there will be more options for clinical diagnosis and treatment.
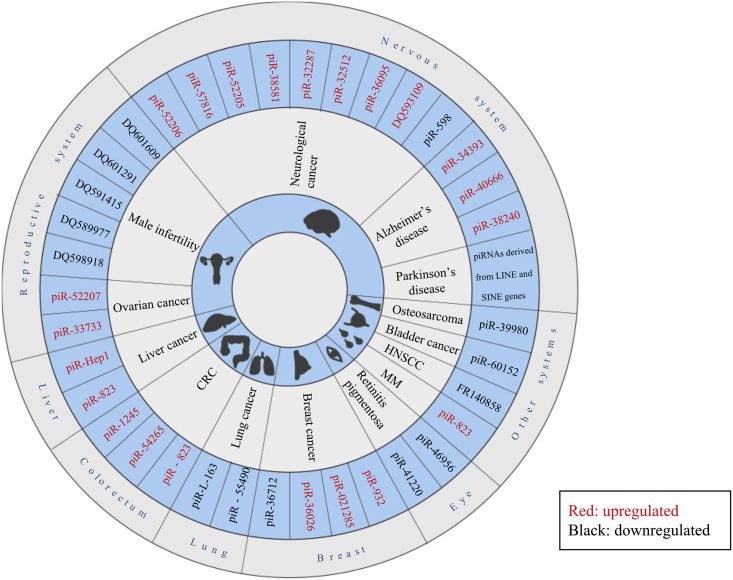
Although many piRNAs are dysregulated in cancers and other diseases, most mechanisms behind these changes are elusive. Those piRNAs with potential mechanisms are listed in Table 1 and are shown in Figure 4.
Table 1.
piRNAs in Different Diseases and Potential Mechanisms Behind Them
| Disease | piRNA | Changes | Mechanism | Role | Reference |
|---|---|---|---|---|---|
| Lung cancer | piR-L-163 | downregulated | bind to 3′ UTR of mTOR mRNA | suppressor | 66 |
| piR-55490 | maintain the function of phospho-ezrin-radaxin-moesin (p-ERM) | 67 | |||
| Liver cancer | piR-823 | upregulated | increase the production of α-SMA and COL1a1 | promotor | 75 |
| piR-Hep1 | activate the Stat3/Bcl-xL signaling pathway | 76 | |||
| Colorectal cancer | piR-1245 | upregulated | regulate the expression of ATF3, BTG1, DUSP1, FAS, NFKBIA, UPP1, SESN2, TP53INP1, and MDX1 | promotor | 77 |
| piR-54265 | regulate the STAT3 signaling pathway | 78 | |||
| piR-823 | activate the expression of HSF1 | 79 | |||
| Bladder cancer | piR-60152 | downregulated | regulate the expression of TNFSF4 | suppressor | 89 |
| Breast cancer | piR-021285 | upregulated | methylate 5′ UTR/first exon of the proinvasive ARHGAP11A gene | promotor | 94 |
| piR-36026 | suppress the expression of SERPINA1 and LRAT | 95 | |||
| piR-932 | suppress the expression of Latexin | 96 | |||
| piR-36712 | downregulated | suppress the expression of SEPW1 | suppressor | 92 | |
| Ovarian cancer | piR-52207 | upregulated | regulate the expression of NUDT4, MTR, EIF2S3, MPHOSPH8, ACTR10, and PLEKHA5 | promotor | 101 |
| piR-33733 | regulate the expression of LIAS | promotor | 101 | ||
| Neurological cancer | piR-32287 | upregulated | regulate the expression of four transfactors: BRCA1, EIF2AK2, HDAC2, and PPARA | promotor | 107 |
| piR-32512 piR-36095 piR-38581 | |||||
| piR-52205 | |||||
| piR-52206 | |||||
| piR-57816 | |||||
| DQ593109 | regulate the blood-tumor barrier (BTB) permeability | promotor | 106 | ||
| piR-598 | downregulated | regulate the expression of genes related with cell death, such as BAX | suppressor | 105 | |
| MM | piR-823 | upregulated | suppress the caspase-3/Bax signaling pathway | promotor | 109,110 |
| induce the secretion of VEGF, IL-6, ICAM-1, and CXCR4 secretion | |||||
| induce DNA methylation | |||||
| suppress the methylation of p16INK4A | |||||
| HNSCC | FR140858 | downregulated | regulate the expression of the minichromosomal maintenance complex component 7 (MCM7) | suppressor | 113 |
| Osteosarcoma | piR-39980 | downregulated | suppress the expression ribonucleoside-diphosphate reductase subunit M2 | suppressor | 118 |
| Male infertility | DQ589977 | downregulated | enhance the methylation level of several genes’ promoters, including PIWI2 and TDRD1, IL16, KLK1, GPR156, HIST1H2AA, SMPD3, and DPM1 | suppressor | 121 |
| DQ591415 | |||||
| DQ598918 | |||||
| DQ601291 DQ601609 | |||||
| Retinitis pigmentosa | piR-41220 | downregulated | regulate the expression of genes vital in the pathway involved in visual function | suppressor | 126 |
| piR-46956 | |||||
| Alzheimer’s disease | piR-34393 | upregulated | suppress the expression of CYCS, KPNA6, and RAB11A | promotor | 132 |
| piR-38240 | |||||
| piR-40666 | |||||
| Parkinson’s disease | piRNAs derived from LINE and SINE genes | downregulated | silence LINE and SINE transposons | suppressor | 133 |
Figure 4.
Aberrant piRNAs with Potential Mechanisms in Different Diseases
Various piRNAs are dysregulated in human diseases. piRNAs in red are upregulated and those in black are downregulated in different diseases.
Conclusion
Owing to the emergence of next-generation sequencing technologies, the expression of piRNAs can be readily observed. The production of piRNAs is complicated, and many vital proteins are involved. piRNAs regulate gene expression in a transcriptional or post-transcriptional manner and closely influence human health. A number of studies have reported dysregulated expressions of piRNAs in samples of different diseases and proposed their potential mechanisms. Most piRNAs are confined to particular disease models, while some are likely incidental, such as piR-651 and piR-823, which can be observed in different cancers. They could be either suppressors or promotors in different cancers because of the heterogeneity of cancers. Actually, the exact mechanism by which piRNAs regulate human health are still unresolved. Many questions remain unknown, such as whether the aberrant expression of piRNAs is the real cause of these diseases or just a byproduct of other molecular activities. What are the ideal thresholds for distinguishing healthy individuals from patients? Such ideas still face challenges, which will need to be addressed before piRNA-related therapies can transition from bench to bed. Further studies and multicenter clinical trials should be performed in the future to fully understand the basic biological mechanisms of piRNAs and their disruption.
Author Contributions
X.W., Y.P., and F.Y. drafted the manuscript. J.Z., M.X., F.Y., T.U., P.M., and W.L. discussed and revised the manuscript. Y.S., P.M., and W.L. designed the research and drafted the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 81802381, 81772475, and 81672896); the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801); and projects supported by Nanjing Medical University (NMUC2018005B and JX102GSP201727) and the National Key Research and Development Program: The Key Technology of Palliative Care and Nursing for Cancer Patients (2017YFC1309201). We also thank American Journal Experts (http://www.aje.com) for providing professional English polish.
Contributor Information
Pei Ma, Email: mapei@njmu.edu.cn.
Wei Li, Email: real.lw@163.com.
Yongqian Shu, Email: shuyongqian@csco.org.cn.
References
- 1.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 3.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Leung Y.Y., Kuksa P.P., Amlie-Wolf A., Valladares O., Ungar L.H., Kannan S., Gregory B.D., Wang L.S. DASHR: database of small human noncoding RNAs. Nucleic Acids Res. 2016;44(D1):D216–D222. doi: 10.1093/nar/gkv1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farazi T.A., Juranek S.A., Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 7.Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 8.Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 9.Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 10.Grivna S.T., Beyret E., Wang Z., Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan P., Damaraju S. The challenges and opportunities in the clinical application of noncoding RNAs: the road map for miRNAs and piRNAs in cancer diagnostics and prognostics. Int. J. Genomics. 2018;2018:5848046. doi: 10.1155/2018/5848046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox D.N., Chao A., Baker J., Chang L., Qiao D., Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klenov M.S., Sokolova O.A., Yakushev E.Y., Stolyarenko A.D., Mikhaleva E.A., Lavrov S.A., Gvozdev V.A. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalczykiewicz D., Pawlak P., Lechniak D., Wrzesinski J. Altered expression of porcine Piwi genes and piRNA during development. PLoS ONE. 2012;7:e43816. doi: 10.1371/journal.pone.0043816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 16.Grimson A., Srivastava M., Fahey B., Woodcroft B.J., Chiang H.R., King N., Degnan B.M., Rokhsar D.S., Bartel D.P. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dharap A., Nakka V.P., Vemuganti R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–1109. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizzo F., Hashim A., Marchese G., Ravo M., Tarallo R., Nassa G., Giurato G., Rinaldi A., Cordella A., Persico M. Timed regulation of P-element-induced wimpy testis-interacting RNA expression during rat liver regeneration. Hepatology. 2014;60:798–806. doi: 10.1002/hep.27267. [DOI] [PubMed] [Google Scholar]
- 19.Yan Z., Hu H.Y., Jiang X., Maierhofer V., Neb E., He L., Hu Y., Hu H., Li N., Chen W., Khaitovich P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L., Zhang J., Li A., Liu Z., He Z., Yuan X., Tuo S. Prediction of cancer-associated piRNA-mRNA and piRNA-lncRNA interactions by integrated analysis of expression and sequence data. Tsinghua Sci. Technol. 2018;23:115–125. [Google Scholar]
- 21.Robine N., Lau N.C., Balla S., Jin Z., Okamura K., Kuramochi-Miyagawa S., Blower M.D., Lai E.C. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr. Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 23.Mohn F., Sienski G., Handler D., Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Lim R.S., Kai T. A piece of the pi(e): the diverse roles of animal piRNAs and their PIWI partners. Semin. Cell Dev. Biol. 2015;47-48:17–31. doi: 10.1016/j.semcdb.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y.A., Stuwe E., Luo Y., Ninova M., Le Thomas A., Rozhavskaya E., Li S., Vempati S., Laver J.D., Patel D.J. Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol. Cell. 2016;63:97–109. doi: 10.1016/j.molcel.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Thomas A., Rogers A.K., Webster A., Marinov G.K., Liao S.E., Perkins E.M., Hur J.K., Aravin A.A., Tóth K.F. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Wang J., Schultz N., Zhang F., Parhad S.S., Tu S., Vreven T., Zamore P.D., Weng Z., Theurkauf W.E. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen P.R., Tirian L., Vunjak M., Brennecke J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature. 2017;549:54–59. doi: 10.1038/nature23482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strässer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A.G., Aguilera A., Struhl K., Reed R., Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 30.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 31.Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F., Wang J., Xu J., Zhang Z., Koppetsch B.S., Schultz N., Vreven T., Meignin C., Davis I., Zamore P.D. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 34.Murchison E.P., Kheradpour P., Sachidanandam R., Smith C., Hodges E., Xuan Z., Kellis M., Grützner F., Stark A., Hannon G.J. Conservation of small RNA pathways in platypus. Genome Res. 2008;18:995–1004. doi: 10.1101/gr.073056.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau N.C., Ohsumi T., Borowsky M., Kingston R.E., Blower M.D. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 2009;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedländer M.R., Adamidi C., Han T., Lebedeva S., Isenbarger T.A., Hirst M., Marra M., Nusbaum C., Lee W.L., Jenkin J.C. High-resolution profiling and discovery of planarian small RNAs. Proc. Natl. Acad. Sci. USA. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim R.S., Anand A., Nishimiya-Fujisawa C., Kobayashi S., Kai T. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev. Biol. 2014;386:237–251. doi: 10.1016/j.ydbio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Krishna S., Nair A., Cheedipudi S., Poduval D., Dhawan J., Palakodeti D., Ghanekar Y. Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata. Nucleic Acids Res. 2013;41:599–616. doi: 10.1093/nar/gks1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawaoka S., Izumi N., Katsuma S., Tomari Y. 3′ End formation of PIWI-interacting RNAs in vitro. Mol. Cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 40.Juliano C.E., Reich A., Liu N., Götzfried J., Zhong M., Uman S., Reenan R.A., Wessel G.M., Steele R.E., Lin H. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:337–342. doi: 10.1073/pnas.1320965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horwich M.D., Li C., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 42.Kirino Y., Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 44.Saito K., Sakaguchi Y., Suzuki T., Suzuki T., Siomi H., Siomi M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamminga L.M., Luteijn M.J., den Broeder M.J., Redl S., Kaaij L.J.T., Roovers E.F., Ladurner P., Berezikov E., Ketting R.F. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29:3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montgomery T.A., Rim Y.S., Zhang C., Dowen R.H., Phillips C.M., Fischer S.E.J., Ruvkun G. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 2012;8:e1002616. doi: 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Billi A.C., Alessi A.F., Khivansara V., Han T., Freeberg M., Mitani S., Kim J.K. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 2012;8:e1002617. doi: 10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiol J., Spinelli P., Laussmann M.A., Homolka D., Yang Z., Cora E., Couté Y., Conn S., Kadlec J., Sachidanandam R. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157:1698–1711. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z., Xu J., Koppetsch B.S., Wang J., Tipping C., Ma S.M., Weng Z., Theurkauf W.E., Zamore P.D. Heterotypic piRNA Ping-Pong requires Qin, a protein with both E3 ligase and tudor domains. Mol. Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams Z., Morozov P., Mihailovic A., Lin C., Puvvula P.K., Juranek S., Rosenwaks Z., Tuschl T. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 2015;13:854–863. doi: 10.1016/j.celrep.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 51.Rouget C., Papin C., Boureux A., Meunier A.C., Franco B., Robine N., Lai E.C., Pelisson A., Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 53.Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 54.Aravin A.A., Sachidanandam R., Bourc’his D., Schaefer C., Pezic D., Toth K.F., Bestor T., Hannon G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roovers E.F., Rosenkranz D., Mahdipour M., Han C.T., He N., Chuva de Sousa Lopes S.M., van der Westerlaken L.A., Zischler H., Butter F., Roelen B.A., Ketting R.F. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015;10:2069–2082. doi: 10.1016/j.celrep.2015.02.062. [DOI] [PubMed] [Google Scholar]
- 56.Praher D., Zimmermann B., Genikhovich G., Columbus-Shenkar Y., Modepalli V., Aharoni R., Moran Y., Technau U. Characterization of the piRNA pathway during development of the sea anemone Nematostella vectensis. RNA Biol. 2017;14:1727–1741. doi: 10.1080/15476286.2017.1349048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gainetdinov I., Skvortsova Y., Kondratieva S., Funikov S., Azhikina T. Two modes of targeting transposable elements by piRNA pathway in human testis. RNA. 2017;23:1614–1625. doi: 10.1261/rna.060939.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ayarpadikannan S., Kim H.S. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 2014;12:98–104. doi: 10.5808/GI.2014.12.3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hedges D.J., Deininger P.L. Inviting instability: gransposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat. Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugimoto K., Kage H., Aki N., Sano A., Kitagawa H., Nagase T., Yatomi Y., Ohishi N., Takai D. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem. Biophys. Res. Commun. 2007;359:497–502. doi: 10.1016/j.bbrc.2007.05.136. [DOI] [PubMed] [Google Scholar]
- 61.Lu Y., Zhang K., Li C., Yao Y., Tao D., Liu Y., Zhang S., Ma Y. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS ONE. 2012;7:e30999. doi: 10.1371/journal.pone.0030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y., Zhang J., Li A., Zhang Y., Li Y., Yuan X., He Z., Liu Z., Tuo S. Identification of PIWI-interacting RNA modules by weighted correlation network analysis. Cluster Comput. 2019;22:707–717. [Google Scholar]
- 63.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 64.Iqbal M.A., Arora S., Prakasam G., Calin G.A., Syed M.A. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol. Aspects Med. 2019;70:3–20. doi: 10.1016/j.mam.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Di X., Jin X., Li R., Zhao M., Wang K. circRNAs and lung cancer: biomarkers and master regulators. Life Sci. 2019;220:177–185. doi: 10.1016/j.lfs.2019.01.055. [DOI] [PubMed] [Google Scholar]
- 66.Mei Y., Wang Y., Kumari P., Shetty A.C., Clark D., Gable T., MacKerell A.D., Ma M.Z., Weber D.J., Yang A.J. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat. Commun. 2015;6:7316. doi: 10.1038/ncomms8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng L., Song L., Liu C., Lv X., Li X., Jie J., Zhao D., Li D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2016;37:2749–2756. doi: 10.1007/s13277-015-4056-0. [DOI] [PubMed] [Google Scholar]
- 68.Zhang S.J., Yao J., Shen B.Z., Li G.B., Kong S.S., Bi D.D., Pan S.H., Cheng B.L. Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer. Oncol. Lett. 2018;15:940–946. doi: 10.3892/ol.2017.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daugaard I., Venø M.T., Yan Y., Kjeldsen T.E., Lamy P., Hager H., Kjems J., Hansen L.L. Small RNA sequencing reveals metastasis-related microRNAs in lung adenocarcinoma. Oncotarget. 2017;8:27047–27061. doi: 10.18632/oncotarget.15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reeves M.E., Firek M., Jliedi A., Amaar Y.G. Identification and characterization of RASSF1C piRNA target genes in lung cancer cells. Oncotarget. 2017;8:34268–34282. doi: 10.18632/oncotarget.15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 72.Cheng J., Deng H., Xiao B., Zhou H., Zhou F., Shen Z., Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 73.Cui L., Lou Y., Zhang X., Zhou H., Deng H., Song H., Yu X., Xiao B., Wang W., Guo J. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin. Biochem. 2011;44:1050–1057. doi: 10.1016/j.clinbiochem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 74.Rizzo F., Rinaldi A., Marchese G., Coviello E., Sellitto A., Cordella A., Giurato G., Nassa G., Ravo M., Tarallo R. Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget. 2016;7:54650–54661. doi: 10.18632/oncotarget.10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang X., Xie X., Wang X., Wang Y., Jiang X., Jiang H. The combination of piR-823 and eukaryotic initiation factor 3 B (EIF3B) activates hepatic stellate cells via upregulating TGF-β1 in liver fibrogenesis. Med. Sci. Monit. 2018;24:9151–9165. doi: 10.12659/MSM.914222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Law P.T., Qin H., Ching A.K., Lai K.P., Co N.N., He M., Lung R.W., Chan A.W., Chan T.F., Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J. Hepatol. 2013;58:1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 77.Weng W., Liu N., Toiyama Y., Kusunoki M., Nagasaka T., Fujiwara T., Wei Q., Qin H., Lin H., Ma Y., Goel A. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol. Cancer. 2018;17:16. doi: 10.1186/s12943-018-0767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mai D., Ding P., Tan L., Zhang J., Pan Z., Bai R., Li C., Li M., Zhou Y., Tan W. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8:5213–5230. doi: 10.7150/thno.28001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin J., Jiang X.Y., Qi W., Ji C.G., Xie X.L., Zhang D.X., Cui Z.J., Wang C.K., Bai Y., Wang J., Jiang H.Q. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017;108:1746–1756. doi: 10.1111/cas.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qu A., Wang W., Yang Y., Zhang X., Dong Y., Zheng G., Wu Q., Zou M., Du L., Wang Y., Wang C. A serum piRNA signature as promising non-invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag. Res. 2019;11:3703–3720. doi: 10.2147/CMAR.S193266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vychytilova-Faltejskova P., Stitkovcova K., Radova L., Sachlova M., Kosarova Z., Slaba K., Kala Z., Svoboda M., Kiss I., Vyzula R. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol. Biomarkers Prev. 2018;27:1019–1028. doi: 10.1158/1055-9965.EPI-18-0318. [DOI] [PubMed] [Google Scholar]
- 82.Ng K.W., Anderson C., Marshall E.A., Minatel B.C., Enfield K.S., Saprunoff H.L., Lam W.L., Martinez V.D. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol. Cancer. 2016;15:5. doi: 10.1186/s12943-016-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wallen E.M., Pruthi R.S., Joyce G.F., Wise M., Urologic Diseases in America Project Kidney cancer. J. Urol. 2007;177:2006–2018, discussion 2018–2019. doi: 10.1016/j.juro.2007.01.126. [DOI] [PubMed] [Google Scholar]
- 84.Iliev R., Fedorko M., Machackova T., Mlcochova H., Svoboda M., Pacik D., Dolezel J., Stanik M., Slaby O. Expression levels of PIWI-interacting RNA, piR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer Res. 2016;36:6419–6423. doi: 10.21873/anticanres.11239. [DOI] [PubMed] [Google Scholar]
- 85.Zhao C., Tolkach Y., Schmidt D., Toma M., Muders M.H., Kristiansen G., Müller S.C., Ellinger J. Mitochondrial PIWI-interacting RNAs are novel biomarkers for clear cell renal cell carcinoma. World J. Urol. 2019;37:1639–1647. doi: 10.1007/s00345-018-2575-1. [DOI] [PubMed] [Google Scholar]
- 86.Li Y., Wu X., Gao H., Jin J.M., Li A.X., Kim Y.S., Pal S.K., Nelson R.A., Lau C.M., Guo C. Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol. Med. 2015;21:381–388. doi: 10.2119/molmed.2014.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Drake C.G., Lipson E.J., Brahmer J.R. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat. Rev. Clin. Oncol. 2014;11:24–37. doi: 10.1038/nrclinonc.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X., Jiang J., Yu C., Wang Y., Sun Y., Tang J., Chen T., Bi Y., Liu Y., Zhang Z.J. Secular trends in incidence and mortality of bladder cancer in China, 1990–2017: a joinpoint and age-period-cohort analysis. Cancer Epidemiol. 2019;61:95–103. doi: 10.1016/j.canep.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 89.Chu H., Hui G., Yuan L., Shi D., Wang Y., Du M., Zhong D., Ma L., Tong N., Qin C. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356(2 Pt B):561–567. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 90.Ahmad A. Breast cancer statistics: recent trends. Adv. Exp. Med. Biol. 2019;1152:1–7. doi: 10.1007/978-3-030-20301-6_1. [DOI] [PubMed] [Google Scholar]
- 91.Voduc K.D., Cheang M.C., Tyldesley S., Gelmon K., Nielsen T.O., Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 2010;28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 92.Hawkes W.C., Printsev I., Alkan Z. Selenoprotein W depletion induces a p53- and p21-dependent delay in cell cycle progression in RWPE-1 prostate epithelial cells. J. Cell. Biochem. 2012;113:61–69. doi: 10.1002/jcb.23328. [DOI] [PubMed] [Google Scholar]
- 93.Tan L., Mai D., Zhang B., Jiang X., Zhang J., Bai R., Ye Y., Li M., Pan L., Su J. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer. 2019;18:9. doi: 10.1186/s12943-019-0940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu A., Jacobs D.I., Hoffman A.E., Zheng T., Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36:1094–1102. doi: 10.1093/carcin/bgv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee Y.J., Moon S.U., Park M.G., Jung W.Y., Park Y.K., Song S.K., Ryu J.G., Lee Y.S., Heo H.J., Gu H.N. Multiplex bioimaging of piRNA molecular pathway-regulated theragnostic effects in a single breast cancer cell using a piRNA molecular beacon. Biomaterials. 2016;101:143–155. doi: 10.1016/j.biomaterials.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 96.Zhang H., Ren Y., Xu H., Pang D., Duan C., Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg. Oncol. 2013;22:217–223. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 97.Huang G., Hu H., Xue X., Shen S., Gao E., Guo G., Shen X., Zhang X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin. Transl. Oncol. 2013;15:563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 98.Hashim A., Rizzo F., Marchese G., Ravo M., Tarallo R., Nassa G., Giurato G., Santamaria G., Cordella A., Cantarella C., Weisz A. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget. 2014;5:9901–9910. doi: 10.18632/oncotarget.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koduru S.V., Tiwari A.K., Leberfinger A., Hazard S.W., Kawasawa Y.I., Mahajan M., Ravnic D.J. A comprehensive NGS data analysis of differentially regulated miRNAs, piRNAs, lncRNAs and sn/snoRNAs in triple negative breast cancer. J. Cancer. 2017;8:578–596. doi: 10.7150/jca.17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Torre L.A., Trabert B., DeSantis C.E., Miller K.D., Samimi G., Runowicz C.D., Gaudet M.M., Jemal A., Siegel R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh G., Roy J., Rout P., Mallick B. Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers. PLoS ONE. 2018;13:e0190485. doi: 10.1371/journal.pone.0190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alsop K., Fereday S., Meldrum C., deFazio A., Emmanuel C., George J., Dobrovic A., Birrer M.J., Webb P.M., Stewart C. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012;30:2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ostrom Q.T., Bauchet L., Davis F.G., Deltour I., Fisher J.L., Langer C.E., Pekmezci M., Schwartzbaum J.A., Turner M.C., Walsh K.M. The epidemiology of glioma in adults: a “state of the science” review. Neuro-oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jacobs D.I., Qin Q., Lerro M.C., Fu A., Dubrow R., Claus E.B., DeWan A.T., Wang G., Lin H., Zhu Y. PIWI-interacting RNAs in gliomagenesis: evidence from post-GWAS and functional analyses. Cancer Epidemiol. Biomarkers Prev. 2016;25:1073–1080. doi: 10.1158/1055-9965.EPI-16-0047. [DOI] [PubMed] [Google Scholar]
- 105.Shen S., Yu H., Liu X., Liu Y., Zheng J., Wang P., Gong W., Chen J., Zhao L., Xue Y. PIWIL1/piRNA-DQ593109 regulates the permeability of the blood-tumor barrier via the MEG3/miR-330-5p/RUNX3 axis. Mol. Ther. Nucleic Acids. 2018;10:412–425. doi: 10.1016/j.omtn.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jacobs D.I., Qin Q., Fu A., Chen Z., Zhou J., Zhu Y. piRNA-8041 is downregulated in human glioblastoma and suppresses tumor growth in vitro and in vivo. Oncotarget. 2018;9:37616–37626. doi: 10.18632/oncotarget.26331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roy J., Mallick B. Investigating piwi-interacting RNA regulome in human neuroblastoma. Genes Chromosomes Cancer. 2018;57:339–349. doi: 10.1002/gcc.22535. [DOI] [PubMed] [Google Scholar]
- 108.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 109.Li B., Hong J., Hong M., Wang Y., Yu T., Zang S., Wu Q. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene. 2019;38:5227–5238. doi: 10.1038/s41388-019-0788-4. [DOI] [PubMed] [Google Scholar]
- 110.Ai L., Mu S., Sun C., Fan F., Yan H., Qin Y., Cui G., Wang Y., Guo T., Mei H. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol. Cancer. 2019;18:88. doi: 10.1186/s12943-019-1011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan H., Wu Q.L., Sun C.Y., Ai L.S., Deng J., Zhang L., Chen L., Chu Z.B., Tang B., Wang K. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196–206. doi: 10.1038/leu.2014.135. [DOI] [PubMed] [Google Scholar]
- 112.Marur S., Forastiere A.A. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2016;91:386–396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 113.Firmino N., Martinez V.D., Rowbotham D.A., Enfield K.S.S., Bennewith K.L., Lam W.L. HPV status is associated with altered PIWI-interacting RNA expression pattern in head and neck cancer. Oral Oncol. 2016;55:43–48. doi: 10.1016/j.oraloncology.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Krishnan A.R., Korrapati A., Zou A.E., Qu Y., Wang X.Q., Califano J.A., Wang-Rodriguez J., Lippman S.M., Hovell M.F., Ongkeko W.M. Smoking status regulates a novel panel of PIWI-interacting RNAs in head and neck squamous cell carcinoma. Oral Oncol. 2017;65:68–75. doi: 10.1016/j.oraloncology.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saad M.A., Ku J., Kuo S.Z., Li P.X., Zheng H., Yu M.A., Wang-Rodriguez J., Ongkeko W.M. Identification and characterization of dysregulated P-element induced wimpy testis-interacting RNAs in head and neck squamous cell carcinoma. Oncol. Lett. 2019;17:2615–2622. doi: 10.3892/ol.2019.9913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ta H.T., Dass C.R., Choong P.F., Dunstan D.E. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev. 2009;28:247–263. doi: 10.1007/s10555-009-9186-7. [DOI] [PubMed] [Google Scholar]
- 117.Harrison D.J., Geller D.S., Gill J.D., Lewis V.O., Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther. 2018;18:39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 118.Das B., Roy J., Jain N., Mallick B. Tumor suppressive activity of PIWI-interacting RNA in human fibrosarcoma mediated through repression of RRM2. Mol. Carcinog. 2019;58:344–357. doi: 10.1002/mc.22932. [DOI] [PubMed] [Google Scholar]
- 119.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 120.Vander Borght M., Wyns C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018;62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 121.Heyn H., Ferreira H.J., Bassas L., Bonache S., Sayols S., Sandoval J., Esteller M., Larriba S. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS ONE. 2012;7:e47892. doi: 10.1371/journal.pone.0047892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kamaliyan Z., Pouriamanesh S., Soosanabadi M., Gholami M., Mirfakhraie R. Investigation of piwi-interacting RNA pathway genes role in idiopathic non-obstructive azoospermia. Sci. Rep. 2018;8:142. doi: 10.1038/s41598-017-17518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cao C., Wen Y., Wang X., Fang N., Yuan S., Huang X. Testicular piRNA profile comparison between successful and unsuccessful micro-TESE retrieval in NOA patients. J. Assist. Reprod. Genet. 2018;35:801–808. doi: 10.1007/s10815-018-1134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kumar K., Trzybulska D., Tsatsanis C., Giwercman A., Almstrup K. Identification of circulating small non-coding RNAs in relation to male subfertility and reproductive hormones. Mol. Cell. Endocrinol. 2019;492:110443. doi: 10.1016/j.mce.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 125.Inana G., Murat C., An W., Yao X., Harris I.R., Cao J. RPE phagocytic function declines in age-related macular degeneration and is rescued by human umbilical tissue derived cells. J. Transl. Med. 2018;16:63. doi: 10.1186/s12967-018-1434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Donato L., Scimone C., Rinaldi C., D’Angelo R., Sidoti A. Retraction. Sci. Rep. 2019;9:14012. doi: 10.1038/s41598-019-50646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vella S., Gallo A., Lo Nigro A., Galvagno D., Raffa G.M., Pilato M., Conaldi P.G. PIWI-interacting RNA (piRNA) signatures in human cardiac progenitor cells. Int. J. Biochem. Cell Biol. 2016;76:1–11. doi: 10.1016/j.biocel.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 128.Rajan K.S., Velmurugan G., Pandi G., Ramasamy S. miRNA and piRNA mediated Akt pathway in heart: antisense expands to survive. Int. J. Biochem. Cell Biol. 2014;55:153–156. doi: 10.1016/j.biocel.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 129.Yang J., Xue F.T., Li Y.Y., Liu W., Zhang S. Exosomal piRNA sequencing reveals differences between heart failure and healthy patients. Eur. Rev. Med. Pharmacol. Sci. 2018;22:7952–7961. doi: 10.26355/eurrev_201811_16423. [DOI] [PubMed] [Google Scholar]
- 130.Rajan K.S., Velmurugan G., Gopal P., Ramprasath T., Babu D.D., Krithika S., Jenifer Y.C., Freddy A., William G., Kalpana K., Ramasamy S. Abundant and altered expression of PIWI-interacting RNAs during cardiac hypertrophy. Heart Lung Circ. 2016;25:1013–1020. doi: 10.1016/j.hlc.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 131.Qiu W., Guo X., Lin X., Yang Q., Zhang W., Zhang Y., Zuo L., Zhu Y., Li C.R., Ma C., Luo X. Transcriptome-wide piRNA profiling in human brains of Alzheimer’s disease. Neurobiol. Aging. 2017;57:170–177. doi: 10.1016/j.neurobiolaging.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roy J., Sarkar A., Parida S., Ghosh Z., Mallick B. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s disease and their probable role in pathogenesis. Mol. Biosyst. 2017;13:565–576. doi: 10.1039/c6mb00699j. [DOI] [PubMed] [Google Scholar]
- 133.Schulze M., Sommer A., Plötz S., Farrell M., Winner B., Grosch J., Winkler J., Riemenschneider M.J. Sporadic Parkinson’s disease derived neuronal cells show disease-specific mRNA and small RNA signatures with abundant deregulation of piRNAs. Acta Neuropathol. Commun. 2018;6:58. doi: 10.1186/s40478-018-0561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]