Abstract
Background
Cervical cancer as one of the most common malignant tumors lead to bad prognosis among women. Some researches already focus on the carcinogenesis and pathogenesis of cervical cancer, but it is still necessary to identify more key genes and pathways.
Methods
Differentially expressed genes were identified by GEO2R from the gene expression omnibus (GEO) website, then gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyzed by DAVID. Meanwhile, protein–protein interaction network was constructed by STRING, and both key genes and modules were found in visualizing network through Cytoscape. Besides, GEPIA did the differential expression of key genes and survival analysis. Finally, the expression of genes related to prognosis was further explored by UNLCAN, oncomine, and the human protein atlas.
Results
Totally 57 differentially expressed genes were founded, not only enriched in G1/S transition of mitotic cell cycle, mitotic nuclear division, and cell division but also participated in cytokine–cytokine receptor interaction, toll‐like receptor signaling pathway, and amoebiasis. Additionally, 12 hub genes and 3 key modules were screened in the Cytoscape visualization network. Further survival analysis showed that TYMS (OMIM accession number 188350), MCM2 (OMIM accession number 116945), HELLS (OMIM accession number 603946), TOP2A (OMIM accession number 126430), and CXCL8 (OMIM accession number 146930) were associated with the prognosis of cervical cancer.
Conclusion
This study aim to better understand the characteristics of some genes and signaling pathways about cervical cancer by bioinformatics, and could provide further research ideas to find new mechanism, more prognostic factors, and potential therapeutic targets for cervical cancer.
Keywords: bioinformatics analysis, cervical cancer, diagnosis and prognosis, differentially expressed genes
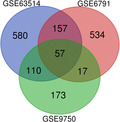
Venn diagram. Identification of the differentially expressed genes in the GSE63514, GSE6791, and GSE9750 gene expression profile datasets. The three datasets showed an overlap of 57 genes. DEGs, Differentially expressed genes.
1. INTRODUCTION
The morbidity of cervical cancer is the fourth malignant disease of women, and the mortality of cervical cancer is also the fourth in the women's malignancies. Moreover, the incidence of young female of cervical cancer increased in recent decades. Statistics showed that the number of deaths worldwide in 2018 was 311,000 (Bray et al., 2018). The total payments on cervix uteri and corpus uteri cancer treatments in China were estimated to be 11.5 billion RMB in 2015 (Li et al., 2017). With the popularization of cervical cancer screening and the application of HPV vaccine, most patients are diagnosed and treated at early stage. However, the effect of treatment about advanced or recurrent cervical cancer is not significant, and targeted treatment for specific gene mutations may be a new trend. With the application and development of high‐throughput sequencing, gene chip in research of tumor genomics, it is significant to find the mechanism, prognostic factors, and potential therapeutic targets of cervical cancer. Some studies have reported that mutations of FOXP3 (Cezar‐Dos‐Santos et al., 2019), PIK3CA (Razia et al., 2019), XRCC4 (Gupta, Kushwah, Singh, & Banerjee, 2019), FBXW7 (Liu et al., 2019), and some copy number variations of KLF10 and PSG (Marrero‐Rodriguez et al., 2018) are related to the carcinogenesis and pathogenesis of cervical cancer, but they were not complete so that to be further studied. This study aim to better understand the characteristics of some genes and signaling pathways that associate with cervical cancer through bioinformatics.
2. MATERIALS AND METHODS
2.1. Microarray data
We downloaded three gene expression profiles (GSE63514 (den Boon et al., 2015), GSE6791 (Pyeon et al., 2007), and GSE9750 (Scotto et al., 2008)) from the gene expression omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) (Edgar, Domrachev, & Lash, 2002), which is one of the most comprehensive public gene expression data resources available today. GSE63514 consists of 28 tumor samples and 24 normal cervical samples, and GSE6791 includes 20 tumor samples and 8 normal cervical samples. The gene chip platform of these two datasets is GPL570 Affymetrix Human Genome U133 Plus 2.0 Array. Moreover, GSE9750 analyzed 33 tumor samples and 24 normal cervical samples. The gene chip platform of GSE9750 is the GPL96 Affymetrix Human Genome U133A Array.
2.2. Identification of DEGs
We identified the DEGs, differentially expressed genes (DEGs) between cervical cancer and normal tissues by GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/). GEO2R is a method which could obtain the DEGs between two gene expression profiles in the GEO database by using Bioconductor's GEOquery and limma R software packages (Barrett et al., 2013). It was set to screen the DEGs by adjusted p value <.01 and logFC (fold change) >2, and we used the Venn diagram to find the overlapped DEGs.
2.3. GO and KEGG enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) (version 6.8) (Huang da, Sherman, & Lempicki, 2009) is a public online bioinformatic database which helps to identify the most significant enriched functional genes and biological pathways. To further analyze the DEGs, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed by using the DAVID online tool. GO analysis was used to annotate biological process (BP), cytological component (CC), and molecular function (MF) of genes (Ashburner et al., 2000; The Gene Ontology Consortium, 2019), and KEGG enrichment analysis was used to understand the relevant signaling pathways (Kanehisa & Sato, 2019). p value <.05 was considered to be statistically significant.
2.4. PPI network and key module analysis
We constructed the protein–protein interaction network (PPI) of DEGs by using Search Tool for the Retrieval of Interacting Genes (STRING; http://string‐db.org/) (version 11.0) (Szklarczyk et al., 2019) database based on the confidence scores. What's more, we further visualized the PPI by Cytoscape (version 3.7.0) (Shannon et al., 2003). And the Molecular Complex Detection (MCODE) plugin in Cytoscape was used to filter the key modules in the network with degree cutoff = 2, node score cutoff = 0.2, k‐core = 2, and max. depth = 100.
2.5. Key genes screening and analysis
The genes with degree ≥10 in the network were identified as key genes, and the co‐expressed genes were analyzed by cBioPortal (http://www.cbioportal.org) (Cerami et al., 2012; Gao et al., 2013) online platform. Gene Expression Profiling Interactive Analysis (GEPAI; http://gepia.cancer‐pku.cn) (Tang et al., 2017) is an interactive web application for gene expression analysis. We visualized the expression of key genes in cervical cancer tissues and normal tissues by box plots in GEPIA, and the overall survival analysis of key genes was also performed. Expression profiles of CXCL8 in human cancers were analyzed by UALCAN (http://ualcan.path.uab.edu/index.html) (Chandrashekar et al., 2017). Oncomine (https://www.oncomine.org) was used to analyze CXCL8 overexpression in multiple datasets comparing cervical cancer with normal tissues. The protein expression of CXCL8 in cervical cancer and normal tissues was analyzed by the human protein atlas (HAP; https://www.proteinatlas.org) (version 19) (Uhlen et al., 2017).
3. RESULTS
3.1. Identification of DEGs
The DEGs of GSE63514, GSE6791, and GSE9750 were obtained by using the GEO2R online analysis tool and we identified the intersection of the three profiles by Venn diagram (Figure 1). Total 57 differential genes were detected, of which 26 were upregulated and 31 were downregulated (Table 1).
FIGURE 1.
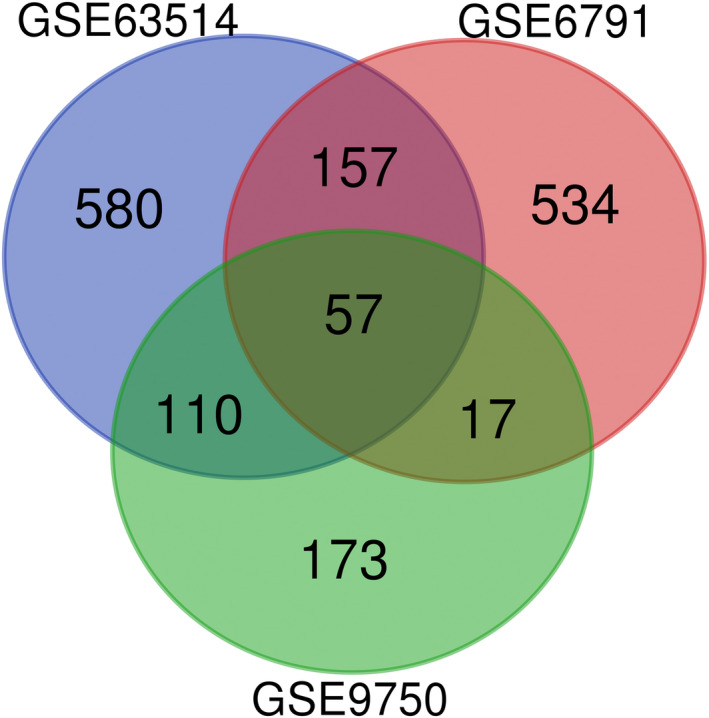
Venn diagram. Identification of the differentially expressed genes in the GSE63514, GSE6791, and GSE9750 gene expression profile datasets. The three datasets showed an overlap of 57 genes. DEGs, Differentially expressed genes
TABLE 1.
Name of 57 differentially expressed genes
| Differentially expressed genes | |||||||
|---|---|---|---|---|---|---|---|
| Upregulated genes | Downregulated genes | ||||||
| STAT1 | NUP62CL | TOP2A | HOXC6 | DSG1 | ENDOU | KRT4 | ALOX12 |
| AURKA | ISG15 | DTL | MMP12 | CWH43 | UPK1A | CLIC3 | PPP1R3C |
| NEK2 | CXCL8 | CDKN2A | SPP1 | GREB1 | BBOX1 | KLK11 | ESR1 |
| TYMS | GINS1 | FANCI | CEP55 | CXCL14 | GLTP | THSD4 | PDGFD |
| MCM2 | ECT2 | ENO2 | CDK1 | CRCT1 | EDN3 | EREG | SPINK5 |
| IFI44L | FN1 | HELLS | SYCP2 | KRT13 | HOPX | IVL | SOSTDC1 |
| INHBA | WDHD1 | CRNN | CRISP3 | KPT1 | MAL | ||
| DSC2 | KRT2 | IL1R2 | |||||
3.2. GO enrichment and KEGG pathway enrichment
In the GO analysis, the screened DEGs mainly participate in the biological process (BP) of G1/S transition of mitotic cell cycle, mitotic nuclear division, cell division, cell–cell signaling, and epithelial cell differentiation. As for the molecular function (MF), DEGs mainly involved in the protein binding, structural molecule activity, and growth factor activity. The cell composition (CC) of DEGs includes cytoplasm, extracellular exosome, nucleoplasm, extracellular space, and extracellular region. In the KEGG pathway analysis found that DEGs significantly enriched in the cytokine receptor interaction, toll‐like receptor signaling pathway, and amoebiasis (Table 2, Figure 2).
TABLE 2.
GO and KEGG pathway enrichment analysis of DEGs
| Term | Description | Gene count | p‐value |
|---|---|---|---|
| Biological processes | |||
| GO:0000082 | G1/S transition of mitotic cell | 5 | 3.21E‐04 |
| GO:0007067 | Mitotic nuclear division | 5 | 8.24E‐03 |
| GO:0051301 | Cell division | 5 | 2.59E‐02 |
| GO:0007267 | Cell–cell signaling | 4 | 4.83E‐02 |
| GO:0030855 | Epithelial cell differentiation | 4 | 1.48E‐03 |
| Cell component | |||
| GO:0005737 | Cytoplasm | 26 | 7.75E‐03 |
| GO:0070062 | Extracellular exosome | 22 | 4.64E‐05 |
| GO:0005654 | Nucleoplasm | 15 | 3.90E‐02 |
| GO:0005615 | Extracellular space | 14 | 1.74E‐04 |
| GO:0005576 | Extracellular region | 14 | 9.73E‐04 |
| Molecular function | |||
| GO:0005515 | Protein binding | 38 | 3.88E‐03 |
| GO:0005198 | Structural molecule activity | 5 | 6.99E‐03 |
| GO:0008083 | Growth factor activity | 4 | 1.36E‐02 |
| KEGG pathway | |||
| hsa04060 | Cytokine–cytokine receptor interaction | 4 | 4.08E‐02 |
| hsa04620 | Toll‐like receptor signaling pathway | 3 | 4.45E‐02 |
| hsa05146 | Amoebiasis | 3 | 4.45E‐02 |
Abbreviations: DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
FIGURE 2.
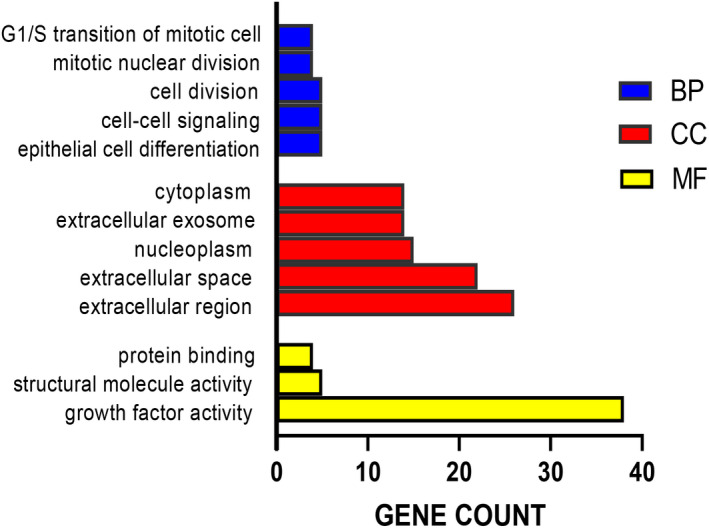
Gene ontology enrichment analysis. BP, Biological Process; CC, Cellular Components; MF, Molecular Function
3.3. PPI network construction and analysis
PPI network of 57 DEGs was constructed in STRING (Figure 3), the network visualized by Cytoscape including 42 nodes and 137 edges. The red nodes represent 23 upregulated genes, whereas the blue nodes represent 19 downregulated genes (Figure 4a). MOCD plug‐in screened out three key modules, two of which were composed by upregulated genes and one by downregulated genes (Figure 4b‐d). The degree ≥10 was used as the screening criteria for the identification of hub genes, and 12 key genes were found to compose the key modules; their names and functions are shown in Table 3. We used the cBioportal tool to explore the networks of these hub genes and their coexpressed genes (Figure 4e).
FIGURE 3.
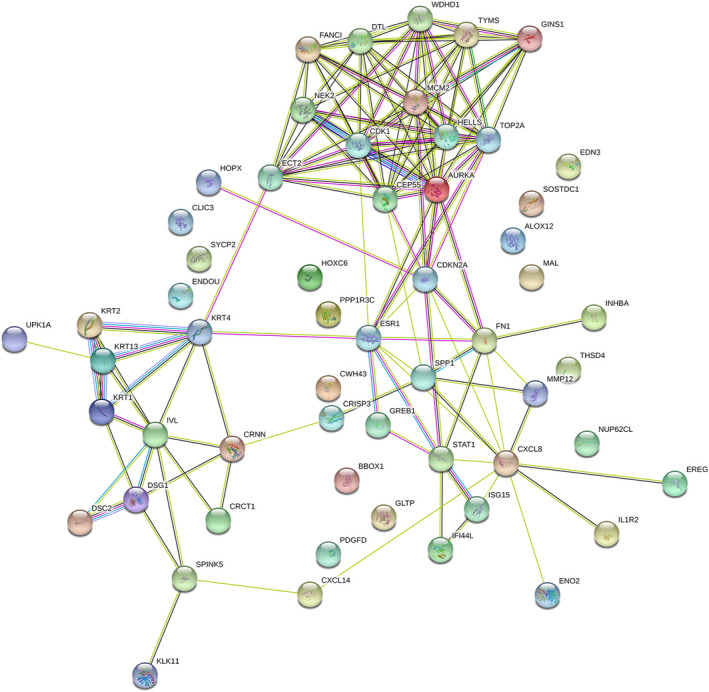
PPI network of 57 DEGs was constructed in STRING. DEGs, Differentially expressed genes
FIGURE 4.
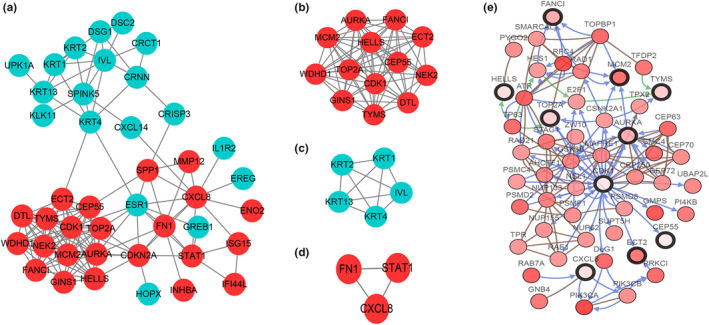
PPI network, most significant module of DEGs and Interaction network of the hub genes. (a) PPI network of DEGs was constructed in Cytoscape, red nodes represents upregulated genes, and blue nodes represents downregulated genes. (b–d) There modules were obtained from PPI network using MCODE plug‐in in Cytoscape. (e) Hub genes and their coexpression genes were analyzed by cBioPortal. Nodes with bold black outline mean hub genes. Nodes with thin black outline the coexpression genes. DEGs, differentially expressed genes; MCODE, molecular complex detection; PPI, protein–protein interaction
TABLE 3.
Functional roles of 12 key genes with degree ≥10
| Gene symbol | Full name | Function |
|---|---|---|
| ECT2 | Epithelial Cell Transforming 2 | ECT2 encodes guanine nucleotide exchange factor, and drives the proliferation and invasion of breast cancer, lung cancer, and gastric cancer. |
| WDHD1 | WD Repeat And HMG‐Box DNA Binding Protein 1 | WDHD1 is the replication initiation factor and involved in the mechanisms of the cancer promotion of E7 genes by evading mitotic checkpoints and inducing polyploid formation |
| TYMS | Thymidylate Synthetase | TYMS encodes thymidine synthase (TS), an important rate‐limiting enzyme that catalyzes the synthesis of pyrimidines, and also a target of fluorouracil medicines. |
| FANCI | FA Complementation Group I | FANCI and FANCD form heterodimers, and participate in DNA repairing and maintain the stability of chromosomes |
| CXCL8 | C‐X‐C Motif Chemokine Ligand 8 | CXCL8 is a chemotactic factor and induces inflammatory responses, neovascularization, and regulates immune responses |
| MCM2 | Minichromosome Maintenance Complex Component 2 | Participate in the initiation and elongation of DNA replication, form a cocktail of antibodies with TOP2A for cervical lesions |
| HELLS | Helicase, Lymphoid Specific | This gene encodes a lymphoid‐specific helicase. Participates in DNA replication, transcription and repair, and plays a role in cell proliferation |
| DTL | Denticleless E3 Ubiquitin Protein Ligase Homolog | DTL is E3 ubiquitin ligase. When DNA is damaged, DTL ubiquitin degrades relevant enzymes to prevent DNA duplication and maintain the stability of genome |
| CEP55 | Centrosomal Protein 55 | CEP55 is a member of the coiled‐coil protein family. Its main function is anchoring the microtubule polymerization‐related proteins, involving in spindle formation, then regulating cell proliferation. |
| CDK1 | Cyclin‐Dependent Kinase 1 | Mitotic serine/threonine kinase that contributes to the regulation of cell cycle progression |
| AURKA | Aurora kinase A | AURKA promotes tumor cell proliferation and invasion, which is related to the resistance of chemotherapy and radiosensitivity |
| TOP2A | DNA Topoisomerase II Alpha | TOP2A can control and alter the topologic states of DNA and can be used to identify advanced cervical lesions |
3.4. Key gene analysis
GEPIA expression analysis showed that these key genes were high expressing in cervical cancer, whereas low expressing in normal cancer tissues, which was consistent with GEO analysis (Figure 5). Moreover, overall survival analysis of GEPIA indicated that high expression of TYMS, MCM2, HELLS, and TOP2A in cervical cancer was correlated with better prognosis. On the other hand, high expression of CXCL8 in cervical cancer tumor tissues is associated with poor prognosis of patients (Figure 6). CXCL8 expression in various tissues from UCLCAN database showed that CXCL8 upregulated in multiple tumor types including Colorectal cancer, cervical cancer, Esophageal cancer, Head and neck cancer, and so on (Figure 7a). The DEGs between cervical cancer and normal tissues from four datasets analyzed by Oncomine showed that CXCL8 had high expression level (Figure 7b). Clinical immunohistochemistry samples of patients with cervical cancer were inquired through HPA database. Compared with normal tissues, CXCL8 protein was highly expressed in patients with cervical cancer (Figure 7c).
FIGURE 5.
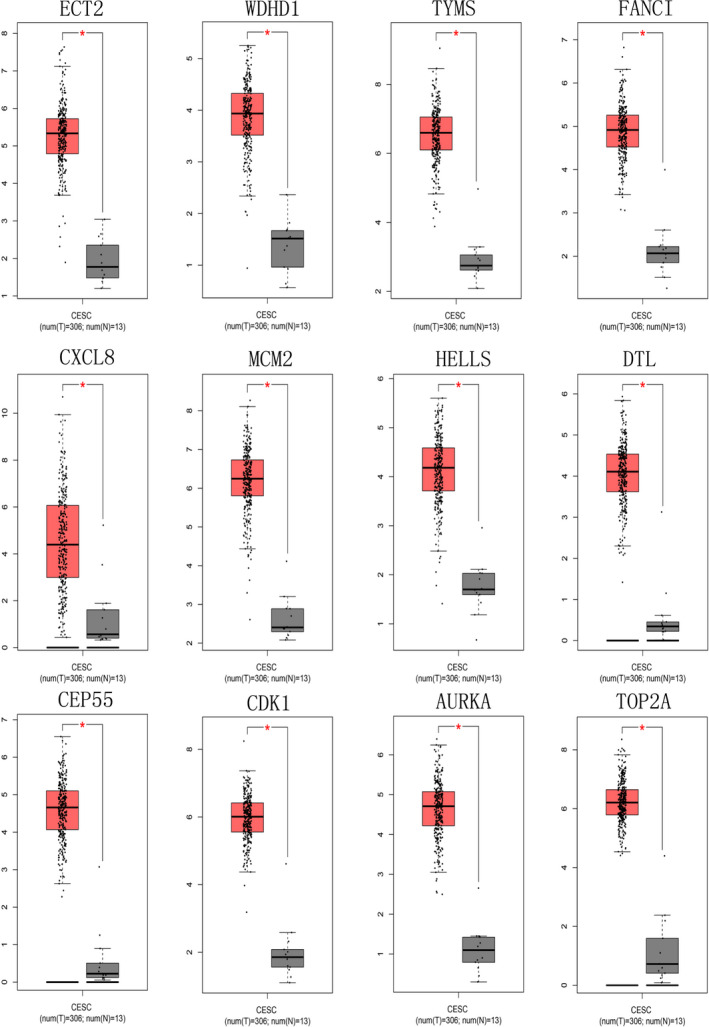
Expression boxplots of key genes using GEPIA website. ECT2, WDHD1, TYMS, FANCI, CXCL8, MCM2, HELLS, DTL, CEP55, CDK1, AURKA, and TOP2A were significantly upregulated in cervical cancer compared with normal tissues (p < .01). GEPIA, Gene Expression Profiling Interactive Analysis; ECT2: NC_000003.12; WDHD1: NC_000014.9; TYMS: NC_000018.10; FANCI: NC_000015.10; CXCL8: NC_00 0004.12; MCM2: NC_000003.12; HELLS: NC_000010.11; DTL:NC_000001.11; CEP55: NC_000010.11; CDK1: NC_000010.11; AURKA:NC_000020.11
FIGURE 6.
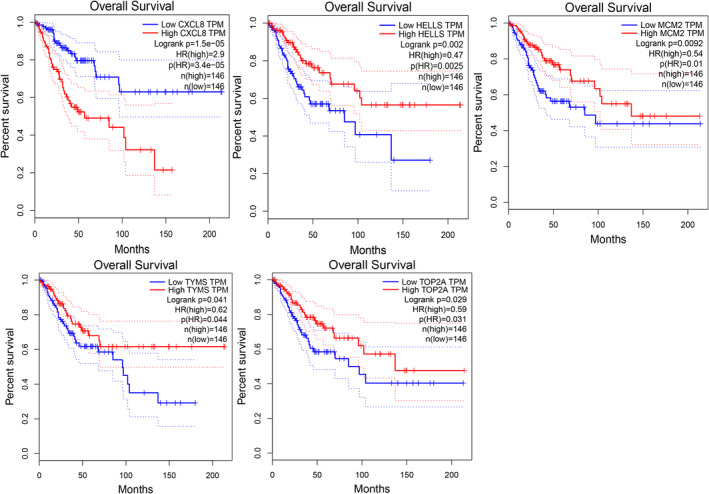
Survival analysis. Expression level of CXCL8, HELLS, MCM2, TYMS, and TOP2A was significantly related to the overall survival of patients with cervical squamous cancer (p < .05)
FIGURE 7.
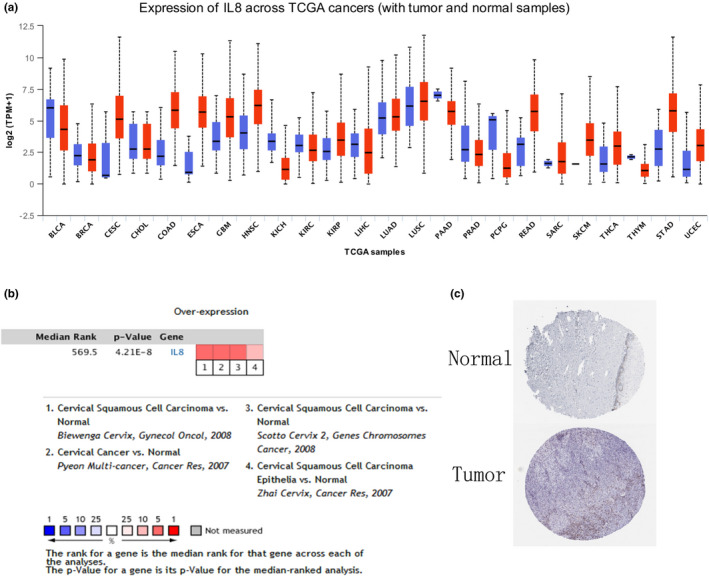
Expression profiles in human cancers, analysis comparison, and protein expression of CXCL8. (a) Expression profiles in human cancers of CXCL8 were analyzed by UALCAN. (b) The expression of the CXCL8 was compared during the four datasets. Normal tissues as control group. Values above the average were deemed to be overexpressed hub genes (red). (c) Immunohistochemistry results from human protein atlas database showed CXCL8 protein was upregulated
4. DISCUSSION
As we all know, the morbidity and mortality of cervical cancer are high in female. Although the chemoradiotherapy and surgery might cure early‐stage patients, many patients suffered from the recurrence and metastasis. And it is still unsatisfied of the effect of treatment to recurrence. Previous studies reported that some molecules and pathways involved in the HPV‐related and the non–HPV‐related cervical cancer. However, the molecule pathway is still needed to be explored.
Among 12 key genes we screened, CXCL8, with degree of 11 involved in all KEGG pathways and correlated with the prognosis of cervical cancer, was thought to be more relevant to cervical cancer. CXCL8, also known as interleukin 8 (IL‐8), is a pleiotropic chemokine. CXCL8 binds to its chemokine receptor CXCR1/2, and induces inflammatory responses, neovascularization, and regulates immune responses. Multiple studies have shown that CXCL8/ cxcr1/2 axis was highly expressed in lung cancer, malignant melanoma, breast cancer, colorectal cancer, osteosarcoma, pancreatic cancer, and other cancers. It plays an important role in regulating tumor growth, invasion, angiogenesis, and resistance through antiapoptosis, cell signal transduction, and immune evasion. CXCL8 could regulate VEGFR, matrix metalloprotein (MMP), and other mediators through autocrine and paracrine pathways to induce tumor angiogenesis (Kumar, Cherukumilli, Mahmoudpour, Brand, & Bandapalli, 2018). And, CXCL8 involves in the mechanisms of resistance to the apoptosis, thus induces tumorogenesis (Xiao et al., 2015). In addition, Epithelial mesenchymal transformation (EMT) modifies the tumor microenvironments resulting in tumor cells endowed with stem cell characteristics, immune escape, proliferation, and migration, and CXCL8 is conducive to the formation of EMT (Geng et al., 2019; Visciano et al., 2015). Multiple studies suggested that CXCL8 promotes pulmonary metastasis of osteosarcoma (Gross et al., 2018), is the prognosis marker of pancreatic cancer (Litman‐Zawadzka, Lukaszewicz‐Zajac, Gryko, Kulczynska‐Przybik, & Mroczko, 2018), and malignant melanoma (Ugurel, Rappl, Tilgen, & Reinhold, 2001) is the recurrence marker of breast cancer (Khazali, Clark, & Wells, 2018). Moreover, CXCL8 is highly expressed in colorectal cancer, and negatively correlated with overall and disease‐free survival (Xiao et al., 2015). CXCL8 has also been reported to play an important role in chemotherapy resistance, antivascular‐targeted therapy, and immunotherapy of tumors. CXCL8 can induce the accumulation of myeloid‐derived suppressor cells (MDSCs), which downregulate the immune activity to tumor cells by inhibiting T cells (Mabuchi, Yokoi, Komura, & Kimura, 2018; Shi et al., 2018). Sanmamed MF et al (Sanmamed et al., 2017) showed that the decrease in baseline level of CXCL8 could predict the better efficacy of anti–pd‐1 treatment in lung cancer and melanoma, and better overall survival, and the pseudo progress of immunotherapy. On the other hand, CXCL8 works in combination with its cognate receptors cxcr1/2. SINGH S et al (Fields & Justilien, 2010; Varney et al., 2011) found that CXCR1 and CXCR2 inhibitors could inhibit the liver metastasis of colon cancer, and the growth of melanoma in mice tumor model. The combination of CXCR1 inhibitors Reparixin and paclitaxel inhibits breast cancer stem cells (CSC) to suppress the brain metastases from breast cancer model (Brandolini et al., 2015), and the first relevant clinical trial is currently underway (NCT02370238) in breast cancer. Lin et al. (2019) indicate that Reparixin can suppress PD‐L1 expression, and improve tumor immune microenvironment. Although the above studies prove the importance of CXCL8 in a variety of tumors, the research and evidence of CXCL8 in cervical cancer still needed to be explored. Our bioinformatics study showed that CXCL8 is a key gene for the carcinogenesis of cervical cancer, and its expression increased in cervical cancer, and might be a marker of poor prognosis. This conclusion shown in Figure 7 has been verified in different databases such as oncomine and UCLCAN. TCGA is the largest cancer gene information database with reliable data sources so that GEPIA using data from TCGA may make the results more reliable. In addition, more basic and clinical studies needed to confirm this conclusion, we could further explore the application of CXCL8/ CXCR1/2 axis inhibitors in the treatment of cervical cancer.
TYMS encodes thymidine synthase (TS), an important rate‐limiting enzyme that catalyzes the synthesis of pyrimidines, is important for DNA synthesis and repair, and also a target of fluorouracil medicines. Multiple studies have shown that the expression level of TYMS may be related to the sensitivity of radiotherapy and chemotherapy for cervical cancer (Saga et al., 2002, 2003). Suzuki, Tsukagoshi, Saga, Ohwada, and Sato (1999) compared the immunohistochemical level of TS and the survival of 66 patients with stage IIIb cervical cancer, showing that the 5‐year survival rate with high and low level of TS was 36.8% and 87.2%, respectively. Therefore, high TS may be associated with poor prognosis of cervical cancer. As for the other two factors, both TOP2A and MCM2 regulate cells in S phase. Studies have indicated that TOP2A and MCM2 involved in the occurrence and development of cervical cancer, and the increased expression of them could be viewed as diagnostic markers of cervical cancer (Peres et al., 2016; Zheng, 2015). ProExC, a cocktail of antibodies developed by becton‐dickinson for TOP2A and MCM2, can be used to identify advanced cervical lesions (Dixon et al., 2017; Tosuner et al., 2017). Our analysis results showed that the expression levels of TYMS, MCM2, HELLS, and TOP2A in cervical cancer increased and positively correlated with the prognosis of cervical cancer. Previous studies have shown that MCM2 and TOP2A may be markers for screening of precancerous lesions and cervical cancer, whereas others believe that the expression level of MCM2 cannot predict tumor stage and posttreatment response (Amaro Filho et al., 2014; Kuku et al., 2015). And few studies have followed patients, and analyzed their survival and prognosis. Thus, our findings may help answer this question. At present, the relevant mechanisms and pathways have not been clearly defined. We consider that in addition to the influence on other genes, they may also play a role in posttranslational modification or activation of other pathways, and ultimately lead to prolonged survival of patients, which needs further research and exploration.
The E6/E7 gene is thought to be involved in tumorigenesis in cells infected with high‐risk HPV. Studies suggest that CDK1 and WDHD1 involved in the mechanisms of the cancer promotion of E6 and E7 genes by evading mitotic checkpoints and inducing polyploid formation, thus resulted in gene instability and cervical cancer (Zhang et al., 2015; Zhou et al., 2016). AURKA is associated with centrosomal expansion, spindle and chromosomal instability, and its high expression promotes tumor cell proliferation and invasion (Zhang, Wang, Liu, Hua, & Xin, 2009). The researchers (Sun et al., 2015) (Ma et al., 2017) found that AURKA is related to the resistance of taxol and radiosensitivity. Currently, AUARK inhibitors are being explored as molecular‐targeted medicines for the treatment of cervical cancer (Gabrielli et al., 2015; Umene et al., 2013). All the above molecules have been proved to play an important role in the occurrence and development of cervical cancer.
Although the other genes, we got by the bioinformatic analysis, have not been found to work in the occurrence and development of cervical cancer, they are still proved to be involved in the carcinogenesis and progression in tumor. ECT2 encodes guanine nucleotide exchange factor, and drives the proliferation and invasion of breast cancer, lung cancer, and gastric cancer. FANCI and FANCD form heterodimers, and participate in DNA repairing (van Twest et al., 2017). However, HPV virus might damage this repair mechanism, leading to the occurrence of tumors and resistance to chemotherapy (Khanal & Galloway, 2019). DTL is involved in cell cycle regulation, and when DNA is damaged, it can stop replication and maintain gene stability (Abbas & Dutta, 2011). HELLS encodes a lymphoid‐specific helicase that involved in DNA replication, transcription, and repair during cell proliferation (Geiman, Durum, & Muegge, 1998). Increased expression of HELLS is detected in various malignant tumors, such as head and neck squamous cell carcinoma, melanoma, and kidney cancer (Chen et al., 2017; Kim, Symanowski, Samlowski, Gonzales, & Ryu, 2010; Waseem, Ali, Odell, Fortune, & Teh, 2010). Through our screening methods, we obtained these genes, and the mechanisms of them in cervical cancer deserve further exploration.
To sum up, we utilized the public online database, and bioinformatics analysis tools to successfully identified 57 DEGs in cervical cancer. Among these, 12 genes may be biomarker of cervical cancer, of which five genes (TYMS, TOP2A, MCM2, HELLS, and CXCL8.) are related to occurrence and prognosis. These results indicated that our methods are meaningful to screen the key genes by using the public database and tools. On the other hand, some genes related to cervical cancer have been estimated by our analysis, and more experiments are needed to explore these genes.
CONFLICT OF INTERESTS
There are no conflicts of interest of the authors.
ETHICAL COMPLIANCE
This article does not contain any studies with human participants or animals performed by any of the authors. The authors are accountable for all aspects of the work.
ACKNOWLEDGMENTS
This study was supported by the Medical Scientific Research Project in Chongqing, China (grant no. 20141003), Chongqing Clinical Oncological Research Center, China (grant no. cstc2015yfpt_gcjsyjzx120010), and the Natural Science Foundation Project of Chongqing Science and Technology Commission (CSTC), China (grant no. cstc2018jcyjAX0012).
Yang H‐J, Xue J‐M, Li J, Wan L‐H, Zhu Y‐X. Identification of key genes and pathways of diagnosis and prognosis in cervical cancer by bioinformatics analysis. Mol Genet Genomic Med. 2020;8:e1200 10.1002/mgg3.1200
REFERENCES
- Abbas, T. , & Dutta, A. (2011). CRL4Cdt2: Master coordinator of cell cycle progression and genome stability. Cell Cycle, 10(2), 241–249. 10.4161/cc.10.2.14530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro Filho, S. M. , Nuovo, G. J. , Cunha, C. B. , De Oliveira Ramos Pereira, L. , Oliveira‐Silva, M. , Russomano, F. , … Nicol, A. F. (2014). Correlation of MCM2 detection with stage and virology of cervical cancer. The International Journal of Biological Markers, 29(4), e363–e371. 10.5301/jbm.5000081 [DOI] [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C. A. , Blake, J. A. , Botstein, D. , Butler, H. , Cherry, J. M. , … Sherlock, G. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics, 25(1), 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, T. , Wilhite, S. E. , Ledoux, P. , Evangelista, C. , Kim, I. F. , Tomashevsky, M. , … Soboleva, A. (2013). NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Research, 41(D1), D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandolini, L. , Cristiano, L. , Fidoamore, A. , De Pizzol, M. , Di Giacomo, E. , Florio, T. M. , … Cimini, A. (2015). Targeting CXCR1 on breast cancer stem cells: Signaling pathways and clinical application modelling. Oncotarget, 6(41), 43375–43394. 10.18632/oncotarget.6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Cerami, E. , Gao, J. , Dogrusoz, U. , Gross, B. E. , Sumer, S. O. , Aksoy, B. A. , … Schultz, N. (2012). The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery, 2(5), 401–404. 10.1158/2159-8290.Cd-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezar‐dos‐Santos, F. , Ferreira, R. S. , Okuyama, N. C. M. , Trugilo, K. P. , Sena, M. M. , Pereira, É. R. , … de Oliveira, K. B. (2019). FOXP3 immunoregulatory gene variants are independent predictors of human papillomavirus infection and cervical cancer precursor lesions. Journal of Cancer Research and Clinical Oncology, 145(8), 2013–2025. 10.1007/s00432-019-02951-x [DOI] [PubMed] [Google Scholar]
- Chandrashekar, D. S. , Bashel, B. , Balasubramanya, S. A. H. , Creighton, C. J. , Ponce‐Rodriguez, I. , Chakravarthi, B. , & Varambally, S. (2017). UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia, 19(8), 649–658. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. , Maruschke, M. , Hakenberg, O. , Zimmermann, W. , Stief, C. G. , & Buchner, A. (2017). TOP2A, HELLS, ATAD2, and TET3 are novel prognostic markers in renal cell carcinoma. Urology, 102, 265.e1–265.e7. 10.1016/j.urology.2016.12.050 [DOI] [PubMed] [Google Scholar]
- den Boon, J. A. , Pyeon, D. , Wang, S. S. , Horswill, M. , Schiffman, M. , Sherman, M. , … Ahlquist, P. (2015). Molecular transitions from papillomavirus infection to cervical precancer and cancer: Role of stromal estrogen receptor signaling. Proceedings of the National Academy of Sciences of the United States of America, 112(25), E3255–E3264. 10.1073/pnas.1509322112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, E. P. , King, L. M. , Nelson, R. , Simkins, S. G. , Knapp, S. L. , Brough, G. H. , … Malinowski, D. P. (2017). Characterization and clinical validation of MCM2 and TOP2A monoclonal antibodies in the BD ProEx C assay: An immunoassay which detects aberrant S‐phase induction in cervical tissue. Journal of Immunological Methods, 442, 35–41. 10.1016/j.jim.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. , & Lash, A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30(1), 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, A. P. , & Justilien, V. (2010). The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Advances in Enzyme Regulation, 50(1), 190–200. 10.1016/j.advenzreg.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli, B. , Bokhari, F. , Ranall, M. V. , Oo, Z. Y. , Stevenson, A. J. , Wang, W. , … McMillan, N. A. J. (2015). Aurora A is critical for survival in HPV‐transformed cervical cancer. Molecular Cancer Therapeutics, 14(12), 2753–2761. 10.1158/1535-7163.Mct-15-0506 [DOI] [PubMed] [Google Scholar]
- Gao, J. , Aksoy, B. A. , Dogrusoz, U. , Dresdner, G. , Gross, B. , Sumer, S. O. , … Schultz, N. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signalling, 6(269), pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman, T. M. , Durum, S. K. , & Muegge, K. (1998). Characterization of gene expression, genomic structure, and chromosomal localization of Hells (Lsh). Genomics, 54(3), 477–483. 10.1006/geno.1998.5557 [DOI] [PubMed] [Google Scholar]
- Geng, F. , Wang, Q. , Li, C. , Liu, J. , Zhang, D. , Zhang, S. , & Pan, Y. (2019). Identification of potential candidate genes of oral cancer in response to chronic infection with porphyromonas gingivalis using bioinformatical analyses. Front Oncol, 9, 91 10.3389/fonc.2019.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, A. C. , Cam, H. , Phelps, D. A. , Saraf, A. J. , Bid, H. K. , Cam, M. , … Roberts, R. D. (2018). IL‐6 and CXCL8 mediate osteosarcoma‐lung interactions critical to metastasis. JCI Insight, 3(16), 10.1172/jci.insight.99791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, M. K. , Kushwah, A. S. , Singh, R. , & Banerjee, M. (2019). Genotypic analysis of XRCC4 and susceptibility to cervical cancer. British Journal of Biomedical Science, 1–6, 10.1080/09674845.2019.1637573 [DOI] [PubMed] [Google Scholar]
- Huang da, W. , Sherman, B. T. , & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , & Sato, Y. (2019). KEGG Mapper for inferring cellular functions from protein sequences. Protein Science, 10.1002/pro.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal, S. , & Galloway, D. A. (2019). High‐risk human papillomavirus oncogenes disrupt the Fanconi anemia DNA repair pathway by impairing localization and de‐ubiquitination of FancD2. PLoS Path, 15(2), e1007442 10.1371/journal.ppat.1007442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazali, A. S. , Clark, A. M. , & Wells, A. (2018). Inflammatory cytokine IL‐8/CXCL8 promotes tumour escape from hepatocyte‐induced dormancy. British Journal of Cancer, 118(4), 566–576. 10.1038/bjc.2017.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. E. , Symanowski, J. T. , Samlowski, E. E. , Gonzales, J. , & Ryu, B. (2010). Quantitative measurement of circulating lymphoid‐specific helicase (HELLS) gene transcript: A potential serum biomarker for melanoma metastasis. Pigment Cell and Melanoma Research, 23(6), 845–848. 10.1111/j.1755-148X.2010.00753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuku, S. , Proctor, I. , Loddo, M. , Kadalayil, L. , KhoshZaban, M. , Ledermann, J. , & McCormack, M. (2015). Do cell‐cycle phase‐specific markers predict disease grade, stage, and outcome in cervical carcinoma? International Journal of Gynecological Cancer: Official Journal of the International Gynecological Cancer Society, 25(6), 1066–1072. 10.1097/IGC.0000000000000356 [DOI] [PubMed] [Google Scholar]
- Kumar, A. , Cherukumilli, M. , Mahmoudpour, S. H. , Brand, K. , & Bandapalli, O. R. (2018). ShRNA‐mediated knock‐down of CXCL8 inhibits tumor growth in colorectal liver metastasis. Biochemical and Biophysical Research Communications, 500(3), 731–737. 10.1016/j.bbrc.2018.04.144 [DOI] [PubMed] [Google Scholar]
- Li, X. , Zheng, R. , Li, X. , Shan, H. , Wu, Q. , Wang, Y. , & Chen, W. (2017). Trends of incidence rate and age at diagnosis for cervical cancer in China, from 2000 to 2014. Chinese Journal of Cancer Research, 29(6), 477–486. 10.21147/j.issn.1000-9604.2017.06.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. , He, H. , Liu, H. , Li, R. , Chen, Y. , Qi, Y. , … Xu, J. (2019). Tumour‐associated macrophages‐derived CXCL8 determines immune evasion through autonomous PD‐L1 expression in gastric cancer. Gut, 68(10), 1764–1773. 10.1136/gutjnl-2018-316324 [DOI] [PubMed] [Google Scholar]
- Litman‐Zawadzka, A. , Lukaszewicz‐Zajac, M. , Gryko, M. , Kulczynska‐Przybik, A. , & Mroczko, B. (2018). Serum chemokine CXCL8 as a better biomarker for diagnosis and prediction of pancreatic cancer than its specific receptor CXCR2, C‐reactive protein, and classic tumor markers CA 19–9 and CEA. Polish Archives of Internal Medicine, 128(9), 524–531. 10.20452/pamw.4307 [DOI] [PubMed] [Google Scholar]
- Liu, F. , Zou, Y. , Wang, F. , Yang, B. , Zhang, Z. , Luo, Y. , … Huang, O. (2019). FBXW7 mutations promote cell proliferation, migration, and invasion in cervical cancer. Genet Test Mol Biomarkers, 23(6), 409–417. 10.1089/gtmb.2018.0278 [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Yang, J. , Wang, R. , Zhang, Z. , Qi, X. , Liu, C. , & Ma, M. (2017). Aurora‐A affects radiosenstivity in cervical squamous cell carcinoma and predicts poor prognosis. Oncotarget, 8(19), 31509–31520. 10.18632/oncotarget.15663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi, S. , Yokoi, E. , Komura, N. , & Kimura, T. (2018). Myeloid‐derived suppressor cells and their role in gynecological malignancies. Tumour Biology, 40(7), 1010428318776485. 10.1177/1010428318776485 [DOI] [PubMed] [Google Scholar]
- Marrero‐Rodriguez, D. , Taniguchi‐Ponciano, K. , Subramaniam, M. , Hawse, J. R. , Pitel, K. S. , Arreola‐De la Cruz, H. , … Salcedo, M. (2018). Kruppel‐Like Factor 10 participates in cervical cancer immunoediting through transcriptional regulation of Pregnancy‐Specific Beta‐1 Glycoproteins. Scientific Reports, 8(1), 9445 10.1038/s41598-018-27711-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres, A. L. , Paz, E. S. K. M. , de Araujo, R. F. , de Lima Filho, J. L. , de Melo Junior, M. R. , Martins, D. B. , & de Pontes Filho, N. T. (2016). Immunocytochemical study of TOP2A and Ki‐67 in cervical smears from women under routine gynecological care. Journal of Biomedical Science, 23(1), 42 10.1186/s12929-016-0258-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyeon, D. , Newton, M. A. , Lambert, P. F. , den Boon, J. A. , Sengupta, S. , Marsit, C. J. , … Ahlquist, P. (2007). Fundamental differences in cell cycle deregulation in human papillomavirus‐positive and human papillomavirus‐negative head/neck and cervical cancers. Cancer Research, 67(10), 4605–4619. 10.1158/0008-5472.Can-06-3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razia, S. , Nakayama, K. , Nakamura, K. , Ishibashi, T. , Ishikawa, M. , Minamoto, T. , … Kyo, S. (2019). Clinicopathological and biological analysis of PIK3CA mutation and amplification in cervical carcinomas. Experimental and Therapeutic Medicine, 18(3), 2278–2284. 10.3892/etm.2019.7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga, Y. , Suzuki, M. , Mizukami, H. , Kohno, T. , Takei, Y. , Fukushima, M. , & Ozawa, K. (2003). Overexpression of thymidylate synthase mediates desensitization for 5‐fluorouracil of tumor cells. International Journal of Cancer, 106(3), 324–326. 10.1002/ijc.11221 [DOI] [PubMed] [Google Scholar]
- Saga, Y. , Suzuki, M. , Mizukami, H. , Urabe, M. , Fukushima, M. , Ozawa, K. , & Sato, I. (2002). Enhanced expression of thymidylate synthase mediates resistance of uterine cervical cancer cells to radiation. Oncology, 63(2), 185–191. 10.1159/000063804 [DOI] [PubMed] [Google Scholar]
- Sanmamed, M. F. , Perez‐Gracia, J. L. , Schalper, K. A. , Fusco, J. P. , Gonzalez, A. , Rodriguez‐Ruiz, M. E. , … Melero, I. (2017). Changes in serum interleukin‐8 (IL‐8) levels reflect and predict response to anti‐PD‐1 treatment in melanoma and non‐small‐cell lung cancer patients. Annals of Oncology, 28(8), 1988–1995. 10.1093/annonc/mdx190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto, L. , Narayan, G. , Nandula, S. V. , Arias‐Pulido, H. , Subramaniyam, S. , Schneider, A. , … Murty, V. V. (2008). Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: Potential role in progression. Genes, Chromosomes and Cancer, 47(9), 755–765. 10.1002/gcc.20577 [DOI] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N. S. , Wang, J. T. , Ramage, D. , … Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Han, X. , Sun, Y. , Shang, C. , Wei, M. , Ba, X. , & Zeng, X. (2018). Chemokine (C‐X‐C motif) ligand 1 and CXCL2 produced by tumor promote the generation of monocytic myeloid‐derived suppressor cells. Cancer Science, 109(12), 3826–3839. 10.1111/cas.13809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. M. , Yang, L. N. , Xu, H. , Chang, B. , Wang, H. Y. , & Yang, G. (2015). Inhibition of Aurora A promotes chemosensitivity via inducing cell cycle arrest and apoptosis in cervical cancer cells. American Journal of Cancer Research, 5(3), 1133–1145. [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M. , Tsukagoshi, S. , Saga, Y. , Ohwada, M. , & Sato, I. (1999). Enhanced expression of thymidylate synthase may be of prognostic importance in advanced cervical cancer. Oncology, 57(1), 50–54. 10.1159/000012000 [DOI] [PubMed] [Google Scholar]
- Szklarczyk, D. , Gable, A. L. , Lyon, D. , Junge, A. , Wyder, S. , Huerta‐Cepas, J. , … Mering, C. (2019). STRING v11: Protein‐protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Research, 47(D1), D607–d613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z. , Li, C. , Kang, B. , Gao, G. , Li, C. , & Zhang, Z. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research, 45(W1), W98–W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium . (2019). The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Research, 47(D1), D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosuner, Z. , Turkmen, I. , Arici, S. , Sonmez, C. , Turna, S. , & Onaran, O. (2017). Immunocytoexpression profile of ProExC in smears interpreted as ASC‐US, ASC‐H, and cervical intraepithelial lesion. Journal of Cytology, 34(1), 34–38. 10.4103/0970-9371.197605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurel, S. , Rappl, G. , Tilgen, W. , & Reinhold, U. (2001). Increased serum concentration of angiogenic factors in malignant melanoma patients correlates with tumor progression and survival. Journal of Clinical Oncology, 19(2), 577–583. 10.1200/jco.2001.19.2.577 [DOI] [PubMed] [Google Scholar]
- Uhlen, M. , Zhang, C. , Lee, S. , Sjöstedt, E. , Fagerberg, L. , Bidkhori, G. , … Ponten, F. (2017). A pathology atlas of the human cancer transcriptome. Science, 357(6352), 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- Umene, K. , Banno, K. , Kisu, I. , Yanokura, M. , Nogami, Y. , Tsuji, K. , … Aoki, D. (2013). Aurora kinase inhibitors: Potential molecular‐targeted drugs for gynecologic malignant tumors. Biomedical Reports, 1(3), 335–340. 10.3892/br.2013.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Twest, S. , Murphy, V. J. , Hodson, C. , Tan, W. , Swuec, P. , O'Rourke, J. J. , … Deans, A. J. (2017). Mechanism of ubiquitination and deubiquitination in the fanconi anemia pathway. Molecular Cell, 65(2), 247–259. 10.1016/j.molcel.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Varney, M. L. , Singh, S. , Li, A. , Mayer‐Ezell, R. , Bond, R. , & Singh, R. K. (2011). Small molecule antagonists for CXCR2 and CXCR1 inhibit human colon cancer liver metastases. Cancer Letters, 300(2), 180–188. 10.1016/j.canlet.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visciano, C. , Liotti, F. , Prevete, N. , Cali', G. , Franco, R. , Collina, F. , … Melillo, R. M. (2015). Mast cells induce epithelial‐to‐mesenchymal transition and stem cell features in human thyroid cancer cells through an IL‐8‐Akt‐Slug pathway. Oncogene, 34(40), 5175–5186. 10.1038/onc.2014.441 [DOI] [PubMed] [Google Scholar]
- Waseem, A. , Ali, M. , Odell, E. W. , Fortune, F. , & Teh, M. T. (2010). Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncology, 46(7), 536–542. 10.1016/j.oraloncology.2010.03.022 [DOI] [PubMed] [Google Scholar]
- Xiao, Y.‐C. , Yang, Z.‐B. , Cheng, X.‐S. , Fang, X.‐B. , Shen, T. , Xia, C.‐F. , … Li, Y.‐F. (2015). CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis. Cancer Letters, 361(1), 22–32. 10.1016/j.canlet.2015.02.021 [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Liu, Y. , Zhao, N. , Chen, H. , Qiao, L. , Zhao, W. , & Chen, J. J. (2015). Role of Cdk1 in the p53‐independent abrogation of the postmitotic checkpoint by human papillomavirus E6. Journal of Virology, 89(5), 2553–2562. 10.1128/jvi.02269-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Wang, J. , Liu, S. J. , Hua, W. , & Xin, X. Y. (2009). Correlation between Aurora‐A expression and the prognosis of cervical carcinoma patients. Acta Obstetricia et Gynecologica Scandinavica, 88(5), 521–527. 10.1080/00016340902835927 [DOI] [PubMed] [Google Scholar]
- Zheng, J. (2015). Diagnostic value of MCM2 immunocytochemical staining in cervical lesions and its relationship with HPV infection. International Journal of Clinical and Experimental Pathology, 8(1), 875–880. [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Zhang, Q. , Gao, G. E. , Zhang, X. , Liu, Y. , Yuan, S. , … Chen, J. J. (2016). Role of WDHD1 in human papillomavirus‐mediated oncogenesis identified by transcriptional profiling of E7‐expressing cells. Journal of Virology, 90(13), 6071–6084. 10.1128/jvi.00513-16 [DOI] [PMC free article] [PubMed] [Google Scholar]


