Abstract
Background
Cobalamin (cbl) C is a treatable rare hereditary disorder of cbl metabolism with autosomal recessive inheritance. It is the most common organic acidemia, manifested as methylmalonic academia combined with homocysteinemia. Early screening and diagnosis are important. The mutation spectrum of the MMACHC gene causing cblC varies among populations. The mutation spectrum in Chinese population is notably different from that in other populations.
Methods
A PCR followed by high‐resolution melting curve analysis (PCR‐HRM) method covering all coding exons of MMACHC gene was designed to verify 14 pathogenic MMACHC gene variants found in patients with cblC, including all common mutations in Chinese patients with cblC.
Result
By PCR‐HRM analysis, 14 pathogenic variants of MMACHC showed distinctly different melting curves, which were consistent with Sanger sequencing. The homozygous type of the most common mutation c.609G > A (p.Trp203Ter) can also be analyzed by specially designed PCR‐HRM.
Conclusion
The established PCR‐HRM method for screening common pathogenic MMACHC variants in Chinese patients with cblC has the advantages of high accuracy, high throughput, low cost, and high speed. It is suitable for the large‐sample screening of suspected children with methylmalonic acidemia and carriers in population.
Keywords: cblC, Chinese population, MMACHC, PCR‐HRM
cblC is a treatable genetic metabolic disease with relatively high morbidity. Early screening and diagnosis are important. The 13 MMACHC gene mutations found in children with clbC in the study covered common mutations in the Chinese population. The established PCR‐HRM analysis method can detect the above mutations, and has the advantages of high accuracy, rapidity, and economy, it can be applied to large sample screening for target population.

1. INTRODUCTION
A kind of disorder in intracellular cobalamin (cbl) metabolism and belonging to organic academia, cblC is a rare genetic metabolic disease with autosomal recessive inheritance (Deodato, Boenzi, Santorelli, & Dionisi‐Vici, 2006). It is the most common type of methylmalonic acidemia (MMA) combined with homocysteinemia (HC); its incidence is between 1:46,000 and 1:200,000 in European and American countries (Cusmano‐Ozog et al., 2007; Weisfeld‐Adams et al., 2010), and varies greatly from 1:3,220 to 1:21,488 in China (Guo et al., 2018; Han et al., 2016; Zhou, Li, Wang, Wang, & Gu, 2019). The clinical phenotypes of cblC are diverse and atypical, have varying degrees of severity and usually involve multiple systems. The common symptoms are feeding difficulties, growth retardation, anemia, thrombocytopenia, microcephaly, epilepsy, dementia, metabolic abnormalities, and ophthalmic abnormalities. More than 90% of children with cblC are severe early onset cases in infancy (Fischer et al., 2014; Rosenblatt et al., 1997), and the most severe cases have severe clinical phenotypes just after birth, which may even cause death; meanwhile, patients with delayed onset may have no morbidity for years (Han et al., 2015).
CblC is directly caused by the mutations in the MMACHC gene (MIM# 609831). In addition, it is reported in a literature that PRDX1 (MIM# 176763), which is adjacent, reverse oriented to and can cause “secondary epigenetic” mutation of MMACHC, can also lead to cblC (Gueant et al., 2018).MMACHC is located on chromosome 1p34.1 and contains five exons. The coding sequence is located in exon 1–4 with a total length of 849 bp, encoding a protein containing 282 amino acids (Lerner‐Ellis et al., 2006). At present, 100 pathogenic MMACHC variants are known (Fischer et al., 2014; Hu, Mei, Liu, & Kong, 2018; Lerner‐Ellis et al., 2009; Liu et al., 2010). However, the mutants of MMACHC greatly vary in incidence. The common mutations of MMACHC in Chinese patients with cblC are c.609G > A (p.Trp203Ter), c.658_660delAAG (p.Lys220del), c.482G > A (p.Arg161Gln), c.80A > G (p.Gln27Arg), and c.609G > A (p.Trp203Ter) mutation alone accounts for almost half of all the mutations (Hu et al., 2018; Liu et al., 2010). The sum of the four most common mutations, c.609G > A (p.Trp203Ter), c.658_660delAAG (p.Lys220del), c.482G > A (p.Arg161Gln), and c.80A > G (p.Gln27Arg), accounts for 72.52% of all pathogenic variants of MMACHC (Our unpublished data).
PCR followed by high‐resolution melting curve analysis (PCR‐HRM/HRMA) is a relatively new method for detecting gene variants. On the basis of the principle of conventional PCR, new DNA saturated fluorescent dyes is added into a reaction system for the detection of fluorescent signal changes in amplified products in real time. The method can distinguish DNA fragments with different GC contents and lengths or with double‐strand complementarity (Montgomery, Sanford, & Wittwer, 2010). At present, HRM can be applied to point mutation, CNV detection, SNP genotyping, identification of bacterial species, screening of antibiotic resistant bacterial strains, and detection of methylation level (Borun et al., 2014; Nagai et al., 2013; Sun et al., 2018; Yin et al., 2013). HRM has a strong competitive advantage and alternative potential in the clinical application of large‐scale population gene variants screening owing to its high throughput, low cost, easy operation, and high sensitivity and specificity. HRM detection methods have been established for the respective causing genes MMAA (MIM# 607481) and MMAB (MIM# 607568) of cblA and cblB that are complementary to cblC diseases (Dempsey‐Nunez et al., 2012; Illson et al., 2013). The aim of this study is to validate and establish a screening method for common pathogenic MMACHC variants in Chinese patients with cblC by PCR‐HRM.
2. MATERIALS AND METHODS
2.1. Sample
Our research has been approved by the ethics committee, under the “Ethical Compliance.” During the screening of suspected children by urine GC/MS and dried blood tablets MS/MS, those identified with abnormal increase in MMA, homocysteine (HCY), and me‐citrate (MCA) levels underwent DNA extraction for genetic testing. Informed consent was provided. PCR primers for the coding region and splicing site of the MMACHC [GenBank, NG_013378.1, NM_015506.2] gene were designed according to the methods described in previous literature (Lerner‐Ellis et al., 2006). PCR amplification and Sanger sequencing were performed. The sequencing results were compared with the normal MMACHC sequence using Chromas software (Version 2.6.4) and NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.2. PCR‐HRM
2.2.1. PCR‐HRM primer design
MMACHC gene sequence [NG_013378.1] was obtained from NCBI genebank database, and the primers covering four coding exons were designed by DNA MAN software (Version 6.0.3.99). The length of all amplification products was between 166 and 260 bp (Table 1). The primers were synthesized by Suzhou Jinweizhi Biotechnology Co., Ltd.
TABLE 1.
PCR‐HRM primers of MMACHC b gene
| Name | Sequence (5′—3′) | Direction | Length of amplicon | Scope of screening a | Mutation screened |
|---|---|---|---|---|---|
| EX1a‐F | TGTCCTTGAGACTTCATTCCC | Forward | 229 bp | c.1‐27 ~ c.81 + 82 (Exon 1) | c.1 A > G (p.Met1Val) c.80A > G (p.Gln27Arg) c.81 + 1G>A (p. ?) |
| EX1a‐R | GCAGGAACCCAGGAGGAT | Reverse | |||
| EX2a‐F | CTGGGGCAAAAGTGTGAG | Forward | 255 bp | c. 82‐89 ~ c.211 (Exon 2) | None |
| EX2a‐R | AGTCAGCATTCGGAGGTG | Reverse | |||
| EX2b‐F | ACTCAGCACGCCTGCCAT | Forward | 180 bp | c. 174 ~ c.276 + 41 (Exon 2) | c.271dupA (p.Arg91Lysfs*14) |
| EX2b‐R | TGGAGGAACTGGAGGCAG | Reverse | |||
| EX3a‐F | TCGGACAAGGTCATAACTCC | Forward | 260 bp | c. 277–41 ~ c.429 + 24 (Exon 3) | c.315C > G (p.Tyr105Ter) c.364dupC (p.His122Profs*17) c.394C > T (p.Arg132Ter) |
| EX3a‐R | GCCTTTACCAGTCTATCTCAGC | Reverse | |||
| EX4a‐F | TGGCAGTTGACTTGGTGC | Forward | 241 bp | c. 430–44 ~ c.590 (Exon 4) | c.445_446insA (p.Cys149Ter) c.482G > A (p.Arg161Gln) c.565C > T (p.Arg189Cys) |
| EX4a‐R | CAATCACGCCAGTGGAAA | Reverse | |||
| EX4b‐F | ACCGTATCGCCCTACTCG | Forward | 166 bp | c. 581 ~ c.710 (Exon 4) | c.609G > A (p.Trp203Ter) c.626dupT (p.Thr210Aspfs*35) c.626−627delTG (p.Val209Aspfs*35) c.658_660del (p.Lys220del) |
| EX4b‐R | GGCTTCTCTGAGGGCTGA | Reverse | |||
| EX4c‐F | GCCTACTTCTCCACTCCACC | Forward | 223 bp | c. 681 ~ c.849 + 13 (Exon 4) | None |
| EX4c‐F | TACCACCATAAATCAGGGTCC | Reverse |
Location of exons of MMACHC in the coding sequence: Exon 1—c.1 ~ 81; Exon 2—c.82 ~ 276; Exon 3—c.277 ~ 429; Exon 4—c.430 ~ 849.
MMACHC [GenBank, NG_013378.1, NM_015506.2]
2.2.2. PCR‐HRM analysis
PCR‐HRM analysis involves three steps: PCR reaction, melting of amplicons, and gene scanning analysis. All the reactions were completed in a closed tube within 2 hr.
The instrument was Roche LightCycler®480 high‐throughput real‐time fluorescence quantitative PCR system (Roche Diagnostics, Penzberg, Germany, 96 wells). The running software was LightCycler® 480 Gene Scanning Software (Version 1.5). Biotium Forget‐Me‐Not™ EvaGreen® qPCR Master Mix Kit (# 31042‐1) was used for the analysis. The total volume of the PCR reaction was 20 μl, including 10‐μl premix (2×), 1‐μl forward and 1‐μl reverse primers (10 μM), and 25‐ng genomic DNA template. The EvaGreen was a saturated dye, which can completely bind to double‐stranded DNA but not to single‐stranded DNA. The reaction conditions were as follows: initial denaturation at 95°C for 5 min; 48 cycles of denaturation at 95°C for 5 s, annealing at 58°C for 10 s, and extension at 72°C for 20 s with fluorescence reading and single‐point acquisition mode. The subsequent melting analysis process of PCR amplification products includes three steps: denaturation at 95°C for 1 min, renaturation at 40°C for 1 min followed by continuous fluorescence reading mode at 65–95°C: the rise rate is 0.02°C/s, and data acquisition is 25 times/°C (once every 0.04°C rise in temperature). The fluorescence data changed with time to form the original melting curve. In the whole PCR‐HRM analysis, the repeatability of the experiment was ensured by setting the three repetitive wells in all the reactions.
Gene scanning analysis performed on the final fluorescence data consisted of three steps: first, the melting curve was standardized. The fluorescence value of dsDNA was set at 100%, and the fluorescence value of ssDNA after complete melting was set at 0%. Second, the complete denaturation point of dsDNA of all the standardized curves was moved to the same location according to the temperature axis. Finally, the differences among the melting curves of the genotypes were enlarged through a differentiation operation performed with the scanning software, and final difference plots were obtained. The experimental groups of different genotypes were distinguished obviously. DNA Sanger sequencing was necessary for the identification of specific mutations in the samples with melting curves different from those of wild‐type and control mutations (Figure S1).
2.2.3. PCR‐HRM analysis of the Homozygous type of the most common mutation c.609G > A (p.Trp203Ter)
The c.609G > A (p.Trp203Ter) mutation accounted for almost half of all MMACHC mutations, but about 40% of them were homozygous. Thus, screening the homozygous c.609G > A (p.Trp203Ter) mutation is necessary. PCR‐HRM analysis cannot distinguish homozygous mutations from wild‐type mutations; this problem is addressed by adding a certain proportion of wild type to the specimen to be examined (Er et al., 2012). Therefore, we added 25% (quality) wild‐type template (homozygous to wild‐type ratio is 3:1) to the homozygous c.609G > A (p.Trp203Ter) mutation template to form a mixed‐type experimental group. The wild‐type, heterozygous, and homozygous c.609G > A (p.Trp203Ter) mutations were used as control groups, and then PCR‐HRM analysis was carried out according to part (2).
3. RESULTS
3.1. Rapid identification of pathogenic MMACHC variants by PCR‐HRM analysis
A total of 62 (14 variants) pathogenic variants were detected in 32 children with cblC, as shown in our previously published data (Wang, Li, et al., 2019) (Table 2). Among them, mutations c.609G > A (p.Trp203Ter), c.658_660delAAG (p.Lys220del), c.482G > A (p.Arg161Gln), and c.80A > G (p.Gln27Arg) were the most common in this study. Except c.364dupC (p.His122Profs*17), which is a novel mutation, the mutations were reported previously, and the mutation spectrum was consistent with previous reports (Hu et al., 2018; Liu et al., 2010).
TABLE 2.
Pathogenic variants of MMACHC b gene verified in this study
| No. | cDNA change | Amino acid change | Exon (EX) | Variant type | Allele frequency a | Percentage a |
|---|---|---|---|---|---|---|
| 1 | c.1 A > G | p.Met1Val | EX1 | Missense | 2 | 3.23% |
| 2 | c.80A > G | p.Gln27Arg | EX1 | Missense | 7 | 11.29% |
| 3 | c.81 + 1G > A | ? | Intron1 | Splicing site | 1 | 1.61% |
| 4 | c.271dupA | p.Arg91Lysfs*14 | EX2 | duplication/frameshift | 1 | 1.61% |
| 5 | c.315C > G | p.Tyr105Ter | EX3 | Nonsense | 2 | 3.23% |
| 6 | c.364dupC | p.His122Profs*17 | EX3 | Duplication | 1 | 1.61% |
| 7 | c.394C > T | p.Arg132Ter | EX3 | Nonsense | 3 | 4.84% |
| 8 | c.445_446insA | p.Cys149Ter | EX4 | Nonsense | 2 | 3.23% |
| 9 | c.482G > A | p.Arg161Gln | EX4 | Missense | 5 | 8.06% |
| 10 | c.565C > T | p.Arg189Cys | EX4 | Missense | 1 | 1.61% |
| 11 | c.609G > A | p.Trp203Ter | EX4 | Nonsense | 26 | 41.94% |
| 12 | c.626dupT | p.Thr210Aspfs*35 | EX4 | Deletion/frameshift | 2 | 3.23% |
| 13 | c.626‐627delTG | p.Val209Aspfs*35 | EX4 | Deletion | 1 | 1.61% |
| 14 | c.658_660del | p.Lys220del | EX4 | Deletion | 8 | 12.90% |
From our previously published data (Wang, Li, et al., 2019), this is the result of 64 alleles from 32 patients with cblC.
MMACHC [GenBank, NG_013378.1, NM_015506.2]
PCR‐HRM analysis of MMACHC gene in this study showed that 14 heterozygous pathogenic variants were significantly different from those of wild‐type control (Figure 1). No mutation was verified in the coverages of the EX2a and EX4c primers, but their difference plots were clearly distinguished from other genotypes in the same group (Figure 1c, d, i and j). PCR‐HRM exhibited 100% accuracy in the screening common pathogenic variants of MMACHC in the Chinese population.
FIGURE 1.
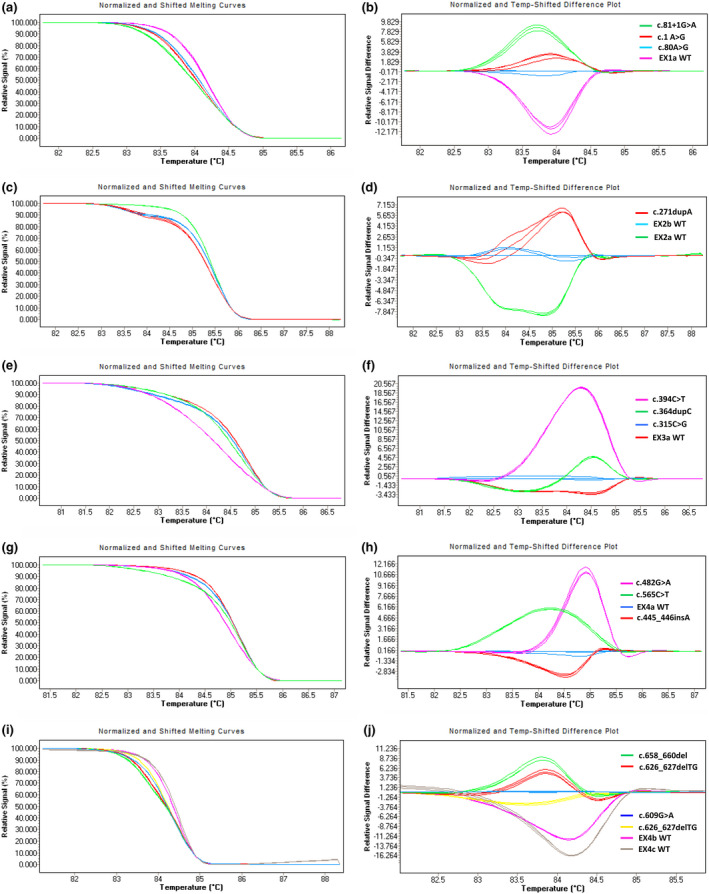
HRM analysis of pathogenic variants of MMACHC gene from different patients with cblC. (a–b) Melting curves and difference plots of c.1A > G (p.Met1Val), c.80A > G (p.Gln27Arg), c.81 + 1G>A (p. ?), and EX1a WT. (c–d) Melting curves and difference plots of c.271dupA (p.Arg91Lysfs*14), EX2a and EX2b WTs. (e–f) Melting curves and difference plots of c.315C > G (p.Tyr105Ter), c.364dupC (p.His122Profs*17), c.394C > T (p.Arg132Ter), and EX3a WT. (g–h) Melting curves and difference plots of c.445_446insA (p.Cys149Ter), c.482G > A (p.Arg161Gln), c.565C > T (p.Arg189Cys), and EX4a WT. (i–j) Melting curves and difference plots of c.609G > A (p.Trp203Ter), c.626dupT (p.Thr210Aspfs*35), c.626‐627delTG (p.Val209Aspfs*35), c.658_660del (p.Lys220del), EX4b WT, and EX4c WT. The mutations represented by different colors in melting curves and difference plots are shown in the upper right corner of the difference graphs in each group of graphs
3.2. Identification of homozygous c.609G > A (p.Trp203Ter) by PCR‐HRM
Heterozygous c.609G > A (p.Trp203Ter) mutation can be distinguished from the wild type, but the homozygous c.609G > A (p.Trp203Ter) mutation showed almost the same difference plot as the wild type. Mixed‐type difference plot is different from heterozygous type, wild type, and homozygous type, but between the two groups (Figure 2). This result showed that 25% (quality) wild‐type samples can be added to the samples to be tested in advance in the screening of homozygous c.609G > A (p.Trp203Ter) mutation in unknown samples. If the samples to be tested are wild type, it will still be wild type after adding the wild‐type samples. If the samples to be tested are homozygous, it will become the above‐mentioned mixed‐type after the addition of the wild‐type samples and become detectable.
FIGURE 2.
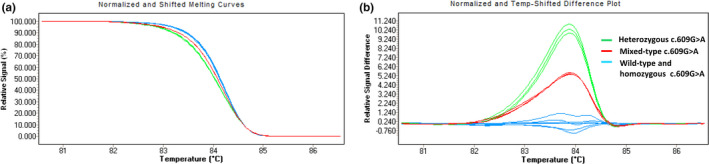
HRM analysis of homozygous c.609G > A (p.Trp203Ter) mutation of MMACHC gene. (a) Normalized and shifted melting curves of EX4b WT, heterozygous c.609G > A (p.Trp203Ter), homozygous c.609G > A (p.Trp203Ter), and mixed‐type c.609G > A (p.Trp203Ter). (b) Normalized and temp‐shifted differentiation plots of the same genotypes. The mutations represented by different colors in the graph are shown in the upper right corner of the difference plots
4. DISCUSSION
The disorders of intracellular cbl metabolism are caused by a variety of protein defects in cbl transport and processing pathways, including MMACHC, MMADHC, LMBRD1, and ABCD4. cblC caused by MMACHC defect is the most common, accounting for 80% of all MMA combined with HC (Sloan, Carrillo, Adams, & Venditti, 2018). According to the age of onset, cblC can be divided into early onset and late onset types. Those less than 1 year are early onset type, and those older than 4 years are late onset type (Rosenblatt et al., 1997). As one of the few treatable genetic diseases, cblC can achieve obvious therapeutic effect by early supplementation of vitamin B12 and L‐carnitine, and some clinical phenotypes can be reversed (Wang, Li, et al., 2019; Wang, Zhao, et al., 2019). Therefore, the establishment of a rapid screening method for the pathogenic variants of MMACHC gene has important clinical significance, especially for patients with late onset cblC, which are difficult to detect because of the absence of typical clinical symptoms.
The mutation spectrum of MMACHC gene varies greatly among different populations. The common mutations of the MMACHC gene in Caucasian patients with cblC are completely different from those in the Chinese population. The most common pathogenic variants are c.271dupA (p.Arg91Lysfs*14), c.394C > T (p.Arg132Ter), and c.331C > T. The c.271dupA (p.Arg91Lysfs*14) mutation accounts for almost half of all mutations (Lerner‐Ellis et al., 2009). The most common mutations in Chinese patients with cblC are c.609G > A (p.Trp203Ter), c.658_660delAAG (p.Lys220del), c.482G > A (p.Arg161Gln), c.80A > G (p.Gln27Arg), and the c.609G > A (p.Trp203Ter) mutation accounts for almost half of all these mutations (Hu et al., 2018; Liu et al., 2010). In this study, a PCR‐HRM method was established for the analysis of the common pathogenic variants of the MMACHC gene in Chinese patients with cblC. It can distinguish the mutants from wild types. The results of HRM were consistent with those of Sanger sequencing, and the accuracy was 100%. When the mutations were added as controls in PCR‐HRM, the specific mutations were determined directly. The MMACHC gene has a relatively short coding sequence, and 849 bp is distributed in four exons. Given that the gene can be fully covered by seven pairs of PCR‐HRM primers, the screening method is efficient, fast and economical. The c.609G > A (p.Trp203Ter) and c.271dupA (p.Arg91Lysfs*14) mutations mainly cause severe early onset cblC (Liu et al., 2010) and are the most common and high‐proportion mutations in Chinese and Caucasian patients with cblC, respectively. Thus, they can be screened separately in clinical practice if necessary. Spanish researchers previously performed HRM analysis on c.271dupA (p.Arg91Lysfs*14) mutation in diagnostics (Richard et al., 2009). In this study, a PCR‐HRM method for detecting heterozygous c.609G > A (p.Trp203Ter) mutation was established for the first time. Given that homozygous c.609G > A (p.Trp203Ter) mutation is difficult to detect and has high proportion, this study verified the PCR‐HRM detection method through a special experimental design (Figure S1).
Instruments for HRM were first developed in 2000 (Wittwer, 2010). Since then, HRM modules have been added to many real‐time quantitative PCR instruments. HRM‐only instruments, such as HR‐1 and LightScanner, have high accuracy, sensitivity, and specificity for mutation scanning. At the same time, new alternative saturated dyes have been introduced, such as LC Green ® Plus (Idaho), Syto9 (Invitrogen) and EvaGreens (Biotum). The development of analysis software has resulted in the improvement of HRM resolution (Montgomery et al., 2010). HRM has high resolution and can accurately distinguish fluorescence signal changes caused by the substitution of a single base in an amplicon (Reed, Kent, & Wittwer, 2007). Therefore, HRM is not limited by mutation base sites and types, it can scan unknown mutations and perform genotyping for known mutations. HRM analysis allows single closed‐tube operation and complete the experiment within 2 hr, showing its significant advantages of high‐throughput and fast operation. Ideal screening methods for genetic diseases in large populations should be highly sensitive, specific, high throughput, fast, inexpensive, easy to implement, and automated. HRM is one of the best screening methods for screening point mutation‐based diseases, especially single‐gene diseases. (Fu et al., 2018). This screening method greatly reduces the need to validate sequencing for patients and help exclude a large number of risk‐free patients (Chambliss, Resnick, Petrides, Clarke, & Marzinke, 2016). HRM is as fast, economical and sensitive as conformation sensitive capillary electrophoresis (CSCE), and has better sensitivity and specificity for mutation scanning than denatured high‐performance liquid chromatography (dHPLC), particularly in the identification of homozygous sequence variants (Simko, 2016). Although next‐generation sequencing (NGS) has become the primary tool for genetic and genomic analysis; this approach is expensive, and the huge data it generates poses great challenges to bioinformatic analysis. The cost of equipment, labor, reagents, and supply for HRM analysis is much lower than that of NGS (Cousins et al., 2012).
HRM has many advantages and is still developing, but it also has shortcomings. For example, studies have shown that PCR‐HRM cannot distinguish some homozygous variants from wild‐type variants (Chambliss et al., 2016) and different heterozygotes may produce similar melting curves (Wittwer, 2009). In the process of establishing the screening method for c.609G > A (p.Trp203Ter) mutation, we found that homozygous c.609G > A (p.Trp203Ter) and its wild type produce the same melting curve. This problem is caused by the defect in the technology itself and can only be improved by changing the experimental design. The solution is to add a certain amount of wild‐type sample into the sample to be tested. If the sample tested is homozygous, the homozygous and heterozygote samples will form heteroduplexes, which will make the melting curve different from the melting curves of the wild‐type and heterozygous samples, but this approach may increase the amount of work (Er et al., 2012). For detecting c.609G > A homozygotes, other detection methods such as restriction fragment length polymorphism (RFLP) analysis and single‐strand conformation polymorphism (SSCP) analysis can also be used, but the labor and time cost of these two methods is greater than PCR‐HRM. Allele‐specific oligonucleotide (ASO) can also be used, but compared with PCR‐HRM, however, its economic cost is much higher due to the probes. HRM analysis can be improved by other approaches, for example, adding a certain proportion of DMSO before or after the PCR. This approach can increase detection sensitivity, and DMSO can be applied to any platform. Organic solvents, such as betaine and formamide used as PCR modifiers, may also improve HRM analysis (Song, Castellanos‐Rizaldos, Bejar, Ebert, & Makrigiorgos, 2015). Because Sanger sequencing is the gold standard of gene detection, and HRM is a screening technology, the positive screening results of HRM for the pathogenic variant need to be confirmed by Sanger sequencing.
In conclusion, this study established a screening method for common MMACHC mutations in Chinese patients with cblC by detecting the DNA samples of children with cblC. This method has the advantages of economy, rapidity, high throughput, and high accuracy. It is suitable for the large‐sample rapid screening of suspected children with methylmalonic acidemia and the rapid screening of population carriers.
CONFLICTS OF INTEREST
Authors declare no conflicts.
ETHICAL APPROVAL
This study was approved by the ethics committee of Tianjin Children's Hospital.
Supporting information
Figure S1
ACKNOWLEDGMENTS
This work was supported by Natural Science Foundation of Tianjin City [grant number: 16JCQNJC11900]; Key Project of Tianjin Health Care Professionals [grant number: 16KG166]; National Natural Science Foundation of China [grant number: 81771589]. The Program of Tianjin Science and Technology Plan [grant number: 18ZXDBSY00170].
Wang C, Liu Y, Cai F, et al. Rapid screening of MMACHC gene mutations by high‐resolution melting curve analysis. Mol Genet Genomic Med. 2020;8:e1221 10.1002/mgg3.1221
Chao Wang and Yang Liu contributed equally to this work.
Contributor Information
Chunquan Cai, Email: tjpns@126.com.
Jianbo Shu, Email: shjb1981@sina.com.
REFERENCES
- Borun, P. , Kubaszewski, L. , Banasiewicz, T. , Walkowiak, J. , Skrzypczak‐Zielinska, M. , Kaczmarek‐Rys, M. , & Plawski, A. (2014). Comparative‐high resolution melting: A novel method of simultaneous screening for small mutations and copy number variations. Human Genetics, 133(5), 535–545. 10.1007/s00439-013-1393-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambliss, A. B. , Resnick, M. , Petrides, A. K. , Clarke, W. A. , & Marzinke, M. A. (2016). Rapid screening for targeted genetic variants via high‐resolution melting curve analysis. Clinical Chemistry & Laboratory Medicine, 55(4), 507–516. 10.1515/cclm-2016-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins, M. M. , Ou, S.‐S. , Wawer, M. J. , Munshaw, S. , Swan, D. , Magaret, C. A. , … Redd, A. D. (2012). Comparison of a high‐resolution melting assay to next‐generation sequencing for analysis of HIV diversity. Journal of Clinical Microbiology, 50(9), 3054–3059. 10.1128/JCM.01460-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusmano‐Ozog, K. , Lorey, F. , Levine, S. , Martin, M. , Nicholas, E. , Packman, S. , … Enns, G. M. (2007). 82 Cobalamin C Disease And Expanded Newborn Screening: The California experience. Journal of Investigative Medicine, 55(1), 227–265. 10.1097/00042871-200701010-00090 [DOI] [Google Scholar]
- Dempsey‐Nunez, L. , Illson, M. L. , Kent, J. , Huang, Q. , Brebner, A. , Watkins, D. , … Rosenblatt, D. S. (2012). High resolution melting analysis of the MMAA gene in patients with cblA and in those with undiagnosed methylmalonic aciduria. Molecular Genetics and Metabolism, 107(3), 363–367. 10.1016/j.ymgme.2012.09.012 [DOI] [PubMed] [Google Scholar]
- Deodato, F. , Boenzi, S. , Santorelli, F. M. , & Dionisi‐Vici, C. (2006). Methylmalonic and propionic aciduria. American Journal of Medical Genetics Part C Seminars in Medical Genetics, 142C(2), 104–112. 10.1002/ajmg.c.30090 [DOI] [PubMed] [Google Scholar]
- Er, T. K. , Kan, T. M. , Su, Y. F. , Liu, T. C. , Chang, J. G. , Hung, S. Y. , & Jong, Y. J. (2012). High‐resolution melting (HRM) analysis as a feasible method for detecting spinal muscular atrophy via dried blood spots. Clinica Chimica Acta, 413(21–22), 1781–1785. 10.1016/j.cca.2012.06.033 [DOI] [PubMed] [Google Scholar]
- Fischer, S. , Huemer, M. , Baumgartner, M. , Deodato, F. , Ballhausen, D. , Boneh, A. , … Dionisi‐Vici, C. (2014). Clinical presentation and outcome in a series of 88 patients with the cblC defect. Journal of Inherited Metabolic Disease, 37(5), 831–840. 10.1007/s10545-014-9687-6 [DOI] [PubMed] [Google Scholar]
- Fu, D.‐M. , Zhou, Y.‐L. , Zhao, J. , Hu, P. , Xu, Z.‐F. , Lv, S.‐M. , … Guo, Q.‐W. (2018). Rapid screening for Klinefelter syndrome with a simple high‐resolution melting assay: A multicenter study. Asian Journal of Andrology, 20(4), 349–354. 10.4103/aja.aja_15_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueant, J.‐L. , Chery, C. , Oussalah, A. , Nadaf, J. , Coelho, D. , Josse, T. , … Rosenblatt, D. S. (2018). A PRDX1 mutant allele causes a MMACHC secondary epimutation in cblC patients. Nature Communications, 9(1), 10.1038/s41467-017-02306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, K. , Zhou, X. , Chen, X. , Wu, Y. , Liu, C. , & Kong, Q. (2018). expanded newborn screening for inborn errors of metabolism and genetic characteristics in a Chinese population. Frontiers in Genetics, 9, 122 10.3389/fgene.2018.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, B. , Cao, Z. , Tian, L. , Zou, H. , Yang, L. , Zhu, W. , & Liu, Y. (2016). Clinical presentation, gene analysis and outcomes in young patients with early‐treated combined methylmalonic acidemia and homocysteinemia (cblC type) Shandong province, China. Brain & Development, 38(5), 491–497. 10.1016/j.braindev.2015.10.016 [DOI] [PubMed] [Google Scholar]
- Han, L. , Wu, S. , Ye, J. , Qiu, W. , Zhang, H. , Gao, X. , … Gu, X. (2015). Biochemical, molecular and outcome analysis of eight Chinese asymptomatic individuals with methyl malonic acidemia detected through newborn screening. American Journal of Medical Genetics Part A, 167(10), 2300–2305. 10.1002/ajmg.a.37147 [DOI] [PubMed] [Google Scholar]
- Hu, S. , Mei, S. , Liu, N. , & Kong, X. (2018). Molecular genetic characterization of cblC defects in 126 pedigrees and prenatal genetic diagnosis of pedigrees with combined methylmalonic aciduria and homocystinuria. BMC Medical Genetics, 19, 154 10.1186/s12881-018-0666-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illson, M. L. , Dempsey‐Nunez, L. , Kent, J. , Huang, Q. , Brebner, A. , Raff, M. L. , … Rosenblatt, D. S. (2013). High resolution melting analysis of the MMAB gene in cblB patients and in those with undiagnosed methylmalonic aciduria. Molecular Genetics and Metabolism, 110(1–2), 86–89. 10.1016/j.ymgme.2013.04.020 [DOI] [PubMed] [Google Scholar]
- Lerner‐Ellis, J. P. , Anastasio, N. , Liu, J. , Coelho, D. , Suormala, T. , Stucki, M. , Loewy, A. D. , … Fowler, B. (2009). Spectrum of mutations in MMACHC, allelic expression, and evidence for genotype‐phenotype correlations. Human Mutation, 30(7), 1072–1081. 10.1002/humu.21001 [DOI] [PubMed] [Google Scholar]
- Lerner‐Ellis, J. P. , Tirone, J. C. , Pawelek, P. D. , Doré, C. , Atkinson, J. L. , Watkins, D. , … Rosenblatt, D. S. (2006). Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nature Genetics, 38(1), 93–100. 10.1038/ng1683 [DOI] [PubMed] [Google Scholar]
- Liu, M.‐Y. , Yang, Y.‐L. , Chang, Y.‐C. , Chiang, S.‐H. , Lin, S.‐P. , Han, L.‐S. , … Liu, T.‐T. (2010). Mutation spectrum of MMACHC in Chinese patients with combined methylmalonic aciduria and homocystinuria. Journal of Human Genetics, 55(9), 621–626. 10.1038/jhg.2010.81 [DOI] [PubMed] [Google Scholar]
- Montgomery, J. L. , Sanford, L. N. , & Wittwer, C. T. (2010). High‐resolution DNA melting analysis in clinical research and diagnostics. Expert Review of Molecular Diagnostics, 10(2), 219–240. 10.1586/erm.09.84 [DOI] [PubMed] [Google Scholar]
- Nagai, Y. , Iwade, Y. , Hayakawa, E. , Nakano, M. , Sakai, T. , Mitarai, S. , Katayama, M. , … Yamaguchi, T. (2013). High resolution melting curve assay for rapid detection of drug‐resistant Mycobacterium tuberculosis. Journal of Infection and Chemotherapy, 19(6), 1116–1125. 10.1007/s10156-013-0636-3 [DOI] [PubMed] [Google Scholar]
- Reed, G. H. , Kent, J. O. , & Wittwer, C. T. (2007). High‐resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics, 8(6), 597–608. 10.2217/14622416.8.6.597 [DOI] [PubMed] [Google Scholar]
- Richard, E. , Jorge‐Finnigan, A. , Garcia‐Villoria, J. , Merinero, B. , Desviat, L. R. , Gort, L. , … Pérez, B. (2009). Genetic and cellular studies of oxidative stress in methylmalonic aciduria (MMA) cobalamin deficiency type C (cblC) with homocystinuria (MMACHC). Human Mutation, 30(11), 1558–1566. 10.1002/humu.21107 [DOI] [PubMed] [Google Scholar]
- Rosenblatt, D. S. , Aspler, A. L. , Shevell, M. I. , Pletcher, B. A. , Fenton, W. A. , & Seashore, M. R. (1997). Clinical heterogeneity and prognosis in combined methylmalonic aciduria and homocystinuria (cblC). Journal of Inherited Metabolic Disease, 20(4), 528–538. 10.1023/a:1005353530303 [DOI] [PubMed] [Google Scholar]
- Simko, I. (2016). High‐resolution DNA melting analysis in plant research. Trends in Plant Science, 21(6), 528–537. 10.1016/j.tplants.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Sloan, J. L. , Carrillo, N. , Adams, D. , & Venditti, P. (2018). Disorders of intracellular cobalamin metabolism. Retrieved from: https://www.ncbi.nlm.nih.gov/books/NBK1328/ [PubMed] [Google Scholar]
- Song, C. , Castellanos‐Rizaldos, E. , Bejar, R. , Ebert, B. L. , & Makrigiorgos, G. M. (2015). DMSO increases mutation scanning detection sensitivity of high‐resolution melting in clinical samples. Clinical Chemistry, 61(11), 1354–1362. 10.1373/clinchem.2015.245357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. , Zhou, H. , Zhang, Y. , Chen, J. , Han, X. , Huang, D. , … Zhao, Y. (2018). Aberrant methylation of and in peripheral blood leukocytes and their association with gastric cancer risk. Journal of Cancer, 9(13), 2275–2283. 10.7150/jca.24797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Li, D. , Cai, F. , Zhang, X. , Xu, X. , Liu, X. , … Shu, J. (2019). Mutation spectrum of MMACHC in Chinese pediatric patients with cobalamin C disease: A case series and literature review. European Journal of Medical Genetics, 62(10), 103713 10.1016/j.ejmg.2019.103713 [DOI] [PubMed] [Google Scholar]
- Wang, S. J. , Zhao, Y. Y. , & Yan, C. Z. (2019). Reversible encephalopathy caused by an inborn error of cobalamin metabolism. Lancet, 393(10172), e29 10.1016/S0140-6736(19)30043-1 [DOI] [PubMed] [Google Scholar]
- Weisfeld‐Adams, J. D. , Morrissey, M. A. , Kirmse, B. M. , Salveson, B. R. , Wasserstein, M. P. , McGuire, P. J. , … Diaz, G. A. (2010). Newborn screening and early biochemical follow‐up in combined methylmalonic aciduria and homocystinuria, cblC type, and utility of methionine as a secondary screening analyte. Molecular Genetics & Metabolism, 99(2), 116–123. 10.1016/j.ymgme.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer, C. T. (2009). High‐resolution DNA melting analysis: Advancements and limitations. Human Mutation, 30(6), 857–859. 10.1002/humu.20951 [DOI] [PubMed] [Google Scholar]
- Wittwer, C. T. (2010). Making DNA melting useful. Clinical Chemistry, 56(9), 1500–1501. 10.1373/clinchem.2010.146175 [DOI] [PubMed] [Google Scholar]
- Yin, X. , Zheng, L. , Liu, Q. , Lin, L. , Hu, X. , Hu, Y. , & Wang, Q. (2013). High‐resolution melting curve analysis for rapid detection of rifampin resistance in Mycobacterium tuberculosis: A meta‐analysis. Journal of Clinical Microbiology, 51(10), 3294–3299. 10.1128/JCM.01264-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. , Li, H. , Wang, C. , Wang, X. , & Gu, M. (2019). Newborn screening for methylmalonic acidemia in a Chinese population: Molecular genetic confirmation and genotype phenotype correlations. Frontiers in Genetics, 9, 726 10.3389/fgene.2018.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1


