Key Points
Subjects with CHIP have higher level of hs-CRP.
Abstract
Clonal hematopoiesis of indeterminate potential (CHIP) is predictive of hematological cancers and cardiovascular diseases, but the etiology of CHIP initiation and clonal expansion is unknown. Several lines of evidence suggest that proinflammatory cytokines may favor mutated hematopoietic stem cell expansion. To investigate the potential link between inflammation and CHIP, we performed targeted deep sequencing of 11 genes previously implicated in CHIP in 1887 subjects aged >70 years from the Montreal Heart Institute Biobank, of which 1359 had prior coronary artery disease (CAD), and 528 controls did not. We assessed association of CHIP with log transformed high-sensitivity C-reactive protein (hs-CRP), a validated biomarker of inflammation. CHIP was identified in 427 of the 1887 subjects (22.6%). CHIP mutations were more frequently identified in DNMT3A (11.6%) and TET2 (6.1%), with a higher proportion of TET2 mutations occurring in controls than in patients with CAD (9.0% vs 4.9%, P < .001). CHIP carriers had 21% higher hs-CRP levels compared with their noncarrier counterparts (eβ = 1.21, 95% confidence interval [CI]: 1.08 to 1.36; P = .001). A similar effect was observed in the subgroup of patients with known CAD (eβ = 1.22, 95% CI: 1.06 to 1.41; P = .005). These findings confirm the association between inflammation and CHIP. This association may open investigational avenues aimed at documenting mechanisms linking inflammation to clonal progression and ultimately supports prevention interventions to attenuate CHIP’s impact on cardiovascular disease and cancer.
Visual Abstract
Introduction
Clonal hematopoiesis (CH) occurring in normally aging subjects, initially suggested by X-chromosome inactivation studies,1-3 is caused by acquired mutations in genes recurrently mutated in hematological cancers,4-7 and in nondriver candidates.6,8 CH prevalence increases significantly in patients aged >60 years old and confers an increased risk of progression to hematological cancers and cardiovascular diseases.5,6,8,9 The precise risk associated with the presence of CH in healthy individuals is uncertain; hence, the creation of a clinical entity named clonal hematopoiesis of indeterminate potential (CHIP).10
Little is known about the etiology of clone initiation and clonal expansion. Genetic predisposition is controversial. Zink et al showed an association between a small germline deletion in intron 3 of the telomerase reverse transcription gene (TERT)8 and CH. Studying a cohort of sib-ships, we identified a significant 2.7-fold increase in the familial risk for mutation in TET2 but not in DNMT3A.11 However, a strong genetic contribution to CH was recently refuted when the concordance of CH was studied in monozygotic and dizygotic twin pairs.12,13
The association between CHIP and both cardiovascular5,6,9 and chronic pulmonary disease8,11 raised the possibility that age-associated chronic inflammation14 may be a key common denominator between these medical conditions.15 Studies in mice have supported the role of inflammation in clonal expansion of mutated hematopoietic stem cells (HSCs). Abegunde et al demonstrated that a proinflammatory environment supported by tumor necrosis factor-α (TNF-α) promotes the expansion of Tet2 mutant clones in mice,16 and we found that inflammation was a key driver of preleukemic myeloproliferation in Tet2-deficient mice (Tet2−/−).17 In humans, Jaiswal et al demonstrated higher interleukin-6 (IL-8) levels in patients with CHIP and cardiovascular disease in a subset of 12 individuals with TET2 mutations.9 A trend toward increased levels of IL-6 was also observed in patients with CHIP.18
We report here a statistically significant correlation between high-sensitivity C-reactive protein (hs-CRP), a validated and routinely available biomarker of inflammation,19 and CHIP.
Methods
Study population
We selected all subjects aged 70 years old or older (1940) from participants of the Montreal Heart Institute biobank, an ongoing prospective cohort including 23 000 individuals for the purpose of clinical and genetic research,20 that had hs-CRP level tested. Participants were recruited on a voluntary basis during any hospital visit, regardless of the presence or stage of heart disease. All subjects underwent a medical questionnaire by a research nurse, and their electronic chart was reviewed. DNA, plasma, and serum were collected at baseline.
For the purpose of the current study, patients with coronary artery disease (CAD) were defined as those with a prior history of myocardial infarction (MI), percutaneous coronary intervention (PCI), or coronary artery bypass graft (CABG) surgery. Subjects also needed to be aged 70 years old or older. The protocol was approved by Montreal Heart Institute’s ethics committee and performed in accordance with the Declaration of Helsinki.
hs-CRP
hs-CRP concentration was measured by quantitative immunonephelometric analysis on a Dimension Vista 500 Intelligent Laboratory System (Siemens Healthineers).
CHIP determination by next-generation sequencing
The subject’s DNA (n = 1940) was sequenced at high coverage (95% >500×) on an Ion Proton sequencer using a custom Ampliseq “CHIP” panel (Thermo Fisher Scientific) designed to target the top 11 genes reported in CHIP (ASXL1, CBL, DNMT3A, GNAS, GNB1, JAK2 [chr9:5073674- 5073808], PPM1D, SF3B1 [exons 14 to 16], SRSF2, TET2, and TP53)5-7,11 with 202 amplicons covering 38.49 kb. The panel coverage, specificity, and sensitivity were validated (supplemental Figure 1). Mutations were considered present if the variant allele frequency (VAF) was ≥2% as defined by Steensma et al.10 Base calling, alignment (hg19), and variant calling were performed in instrument by TorrentServer v5.8.0 (Thermo Fisher Scientific). Subsequently, mutations were annotated and filtered using IonReporter v5.8 (Thermo Fisher Scientific), and only exonic and splice site mutations with a minor allele frequency ≤0.001 were kept for further annotation. Frameshift, nonsense, in-frame deletions or insertions, splice sites, and predicted consequential missense mutations (based on ClinVar, FATHMM, or PolyPhen) were considered significant. In the absence of nonhematological tissue for germline status confirmation, mutations with a VAF of 50% or 100% (±4%) were considered potentially germline and excluded (n = 9) except if they cooccurred with other somatic mutations (n = 9) (supplemental Table 1).
Statistical analyses
Bivariate associations were evaluated using the Fisher’s exact test and the Kruskal-Wallis test for categorical and continuously coded variables, respectively. hs-CRP was modeled as log (ln)(hs-CRP) due to its nonnormal distribution. The geometric mean was calculated by taking the antilog of the mean of the log-transformed hs-CRP data, as previously described.21 Normality of residuals from a generalized linear regression model adjusted for age, sex, body mass index (BMI), and previous history of CAD with ln(CRP) was confirmed. Generalized linear regression models tested the associations of CHIP-associated mutations and levels of ln(hs-CRP) after adjusting for age, sex, BMI, and CAD status at baseline. Stratified analyses were performed according to CAD status. Back transformation of the regression β term for ln(CRP) was derived as eβ, and the 95% confidence interval (CI) was derived as eβ±1.96×standard error. The percent difference in hs-CRP between CHIP+ and CHIP− was derived as eβ − 1 × 100. Additional analyses were performed according to the CHIP VAF and categorized as VAF ≥ 0.10 vs no mutation. Analyses were performed using SAS v.9.4 and R version 3.5.1. In sensitivity analyses, we adjusted for additional confounding factors, such as diabetes mellitus at baseline, as well as statin, aspirin, and beta-blocker use at baseline. In subgroup analyses, we focused on those without a history of cancer at study entry.
Results
Study population
The population comprised 1940 subjects, of which 9 were excluded because of potential germline mutations and another 44 were excluded because they had cardiovascular disease without CAD at baseline. The remaining 1887 participants included 1359 patients with at least 1 previous account of MI, PCI, or CABG, and 528 patients without a previous history of CAD.
CHIP-associated mutations
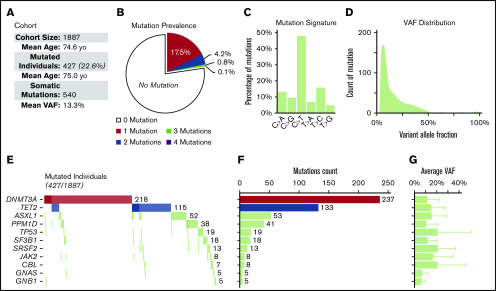
We identified 427 CHIP carriers among the 1887 participants (22.6%; Figure 1). Of these, 331 (17.5%) had a single mutation, and 96 (5.1%) had >1 mutation. The prevalence and the relative proportion between the 11 candidate genes were similar to previous reports.5-8,11 Mutations in DNMT3A, TET2, and ASXL1 accounted for the majority of mutations (82.9%). The mean VAF was 13.3%. The prevalence of CHIP carriers was slightly higher in the non-CAD than in the CAD cohort (25.3% vs 21.5%, P = .075). The relative prevalence between the different genes was similar between the 2 cohorts, except for TET2, whose prevalence was higher in the non-CAD cohort (9.0% vs 4.9%; P < .001). All mutations are described in supplemental Table 1.
Figure 1.
Prevalence and distribution of somatic mutations. (A) Description of the cohort (n = 1887). (B) Prevalence of the 540 somatic mutations identified in 427 individuals of the cohort. (C) Mutation signature of all single nucleotide substitutions (supplemental Figure 2). (D) VAF distribution of all somatic mutations. (E) Cooccurrence of the all somatic mutations in the 427 mutated individuals. Darker shades represent double mutation in the same gene. (F) Contribution of individual genes to the total number of observed somatic mutations. (G) Average VAF of somatic mutation for each gene.
Population characteristics according to CHIP carrier status
Subjects were segregated according to their CHIP carrier status (CHIP− or CHIP+) in the 3 following groups: (1) all subjects, (2) subjects with CAD, and (3) subjects without CAD. Table 1 describes the baseline characteristics of the 3 groups. Univariable analyses showed that CHIP carriers were significantly older (mean: 75.0 vs 74.4 years old; P = .002), were less affected by dyslipidemia (85% vs 88%; P = .041), and had fewer previous PCI (30% vs 38%; P = .006) than CHIP noncarriers. hs-CRP was significantly higher in CHIP carriers compared with noncarriers (median: 1.60 vs 1.41 mg/L; P = .009) (Figure 2). Smoking, past or current, was not associated with CHIP.
Table 1.
Baseline characteristics of the study population, according to CHIP status (univariate analysis)
| All patients | Without baseline CAD | With baseline CAD | ||||
|---|---|---|---|---|---|---|
| CHIP+ | CHIP− | CHIP+ | CHIP− | CHIP+ | CHIP− | |
| n (%) | 427 (22.6) | 1460 (77.4) | 134 (25.3) | 394 (74.7) | 293 (21.5) | 1066 (78.5) |
| Age, mean (SD), y | 75.0 (3.4)* | 74.4 (3.3) | 74.9 (3.2)* | 74.1 (3.3) | 75.0 (3.5)* | 74.583 (3.3) |
| Weight, mean (SD), kg | 77.5 (15.5) | 78.2 (14.4) | 77.0 (16.5) | 75.0 (14.9) | 77.8 (15.0) | 79.4 (14.1) |
| Height, mean (SD), m | 1.65 (0.09) | 1.66 (0.08) | 1.64 (0.10) | 1.63 (0.09) | 1.66 (0.08) | 1.67 (0.08) |
| BMI, mean (SD) | 28.197 (5.041) | 28.245 (4.423) | 28.390 (5.265) | 27.915 (4.446) | 28.108 (4.942) | 28.367 (4.410) |
| Male, n (%) | 292 (68.4) | 1044 (71.5) | 67 (50.0) | 183 (46.4) | 225 (76.8) | 861 (80.8) |
| Cardiovascular risk factors, n (%) | ||||||
| Dyslipidemia | 361 (84.5)* | 1287 (88.3) | 84 (62.7) | 267 (67.9) | 277 (94.5) | 1020 (95.8) |
| Hypertension | 327 (76.8) | 1095 (75.3) | 82 (61.7) | 239 (60.7) | 245 (83.6) | 856 (80.7) |
| Diabetes | 120 (28.1) | 408 (28.0) | 26 (19.4) | 73 (18.5) | 94 (32.1) | 335 (31.5) |
| Ever smoker | 301 (70.7) | 988 (67.7) | 79 (59.4) | 218 (55.3) | 222 (75.8) | 770 (72.3) |
| Current smoker | 21 (4.9) | 71 (4.9) | 1 (0.8)* | 18 (4.6) | 20 (6.8) | 53 (5.0) |
| CVD history, n (%) | ||||||
| Coronary heart disease | 293 (68.6) | 1066 (73.0) | — | — | — | — |
| Previous MI | 193 (45.3) | 649 (44.5) | 0 (0.0) | 0 (0.0) | 193 (66.1) | 649 (60.9) |
| Previous PCI | 130 (30.4)* | 550 (37.7) | 0 (0.0) | 0 (0.0) | 130 (44.4)* | 550 (51.6) |
| Previous CABG | 163 (38.2) | 601 (41.2) | 0 (0.0) | 0 (0.0) | 163 (55.6) | 601 (56.4) |
| Stroke | 42 (9.9) | 154 (10.6) | 0 (0.0) | 0 (0.0) | 42 (14.4) | 154 (14.5) |
| Angina | 250 (58.7) | 924 (63.5) | 0 (0.0) | 0 (0.0) | 250 (85.6) | 924 (87.1) |
| PVD | 85 (19.9) | 306 (21.0) | 0 (0.0) | 0 (0.0) | 85 (29.0) | 306 (28.8) |
| Angiography | 279 (65.3) | 1018 (69.7) | 15 (11.2) | 52 (13.2) | 264 (90.1) | 966 (90.6) |
| CHF | 72 (16.9) | 195 (13.4) | 1 (0.7) | 2 (0.5) | 71 (24.3)* | 193 (18.2) |
| Medication, n (%) | ||||||
| Aspirin | 310 (72.6) | 1123 (76.9) | 66 (49.3) | 193 (49.0) | 244 (83.3) | 930 (87.2) |
| Antiplatelet | 329 (77.0) | 1164 (79.7) | 67 (50.0) | 199 (50.5) | 262 (89.4) | 965 (90.5) |
| Statin | 360 (84.3) | 1276 (87.4) | 83 (61.9) | 259 (65.7) | 277 (94.5) | 1017 (95.4) |
| ACE | 144 (33.7) | 538 (36.8) | 24 (17.9) | 77 (19.5) | 120 (41.0) | 461 (43.2) |
| ARB | 157 (36.8) | 496 (34.0) | 43 (32.1) | 130 (33.0) | 114 (38.9) | 366 (34.3) |
| Beta-blockers | 287 (67.2) | 970 (66.4) | 55 (41.0) | 143 (36.3) | 232 (79.2) | 827 (77.6) |
| Lipids, mean (SD), mmol/L | ||||||
| Total cholesterol | 3.89 (1.00) | 3.86 (0.98) | 4.44 (1.02) | 4.59 (1.11) | 3.63 (0.88) | 3.59 (0.76) |
| Triglycerides | 1.92 (1.03) | 1.99 (1.02) | 1.92 (1.08) | 2.04 (1.03) | 1.93 (1.00) | 1.97 (1.01) |
| HDL | 1.23 (0.37) | 1.22 (0.36) | 1.32 (0.36) | 1.34 (0.42) | 1.19 (0.36) | 1.18 (0.33) |
| LDL | 1.80 (0.84) | 1.75 (0.83) | 2.28 (0.89) | 2.36 (0.99) | 1.58 (0.71) | 1.52 (0.62) |
| hs-CRP mg/L, median (IQR), min-max | ||||||
| hs-CRP | 1.60 (2.96),* 0.09-115.0 | 1.41 (2.29), 0.08-80.70 | 1.55 (2.99), 0.09-92.80 | 1.45 (2.23), 0.08-61.30 | 1.60 (2.88),* 0.09-115.0 | 1.39 (2.30), 0.08-80.70 |
| Cancer history, n (%) | ||||||
| Any cancer | 89 (20.8) | 257 (17.6) | 26 (19.4) | 67 (17.0) | 63 (21.5) | 190 (17.8) |
| Hematological cancer | 6 (1.4) | 17 (1.1) | 3 (2.2)* | 0 (0.0) | 3 (1.0) | 17 (1.5) |
| Incident/recurrent cancer, n (%) | ||||||
| Any cancer | 34 (7.9) | 111 (7.6) | 11 (8.2) | 25 (6.3) | 23 (7.8) | 86 (8.0) |
| Hematological cancer | 7 (1.6) | 21 (1.4) | 0 (0.0) | 2 (0.5) | 2 (0.6) | 4 (0.3) |
ACE, angiotensin-converting enzyme; ARB, angiotensin receptor blockers; CVD, cardiovascular disease; HDL, high-density lipoprotein; IQR, interquartile range; LDL, low-density lipoprotein; SD: standard deviation.
P < .05 per Fisher’s exact test or Kruskal-Wallis 2-sided significance test.
Figure 2.

hs-CRP concentration (mg/L) in CHIP− and CHIP+ individuals. Box-and-whisker plot of the hs-CRP concentration (mg/L) in all CHIP− (n = 1460) and CHIP+ (n = 427) individuals of the cohort. The center line denotes the statistical median, while the box contains the first (Q1) to third (Q3) quartiles. Outliers are not represented. *P < .05 per Kruskal-Wallis 2-sided significance test.
Primary analysis (entire cohort)
Our primary analysis focused on all patients, regardless of CAD status, and consisted of performing a multivariable generalized linear regression analysis for prediction of hs-CRP (n = 1887). CHIP carriers of any gene mutations had significantly higher hs-CRP than non-CHIP carriers in adjusted analyses (eβ = 1.21, 95% CI: 1.08 to 1.36; P = .001; Table 2). Focusing on mutations in individual genes, CHIP carriers of mutations in DNMT3A had higher hs-CRP than those without mutations in that gene (eβ = 1.17, 95% CI: 1.01 to 1.36; P = .04). No other significant effect was observed for any of the remaining individual genes.
Table 2.
Association between CHIP and hs-CRP at study entry (multivariate analysis)
| All patients (N = 1887) | Without baseline CAD (n = 528) | With baseline CAD (n = 1359) | ||||
|---|---|---|---|---|---|---|
| CHIP+ | CHIP− | CHIP+ | CHIP− | CHIP+ | CHIP− | |
| Any mutations | ||||||
| n | 427 | 1460 | 134 | 394 | 293 | 1066 |
| hs-CRP GM (IQR), mg/L | 1.85 (0.81-3.77) | 1.5 (0.71-3.00) | 1.86 (0.83-3.82) | 1.51 (0.77-3.02) | 1.84 (0.81-3.72) | 1.49 (0.69-3.01) |
| Back-transformed β (95% CI)* | 1.21 (1.08-1.36) | 1.15 (0.94-1.40) | 1.22 (1.06-1.41) | |||
| % difference | 21 | 15 | 22 | |||
| P* | .001 | .178 | .005 | |||
| DNMT3A | ||||||
| n | 218 | 1669 | 66 | 462 | 152 | 1207 |
| hs-CRP GM (IQR), mg/L | 1.82 (0.80-3.79) | 1.54 (0.73-1.43) | 1.91 (0.81-4.72) | 1.55 (0.77-3.10) | 1.79 (0.78-3.66) | 1.54 (0.71-3.13) |
| Back-transformed β (95% CI)* | 1.17 (1.01-1.36) | 1.23 (0.95-1.59) | 1.15 (0.95-1.38) | |||
| % difference | 17 | 23 | 15 | |||
| P* | .04 | .125 | .148 | |||
| TET2 | ||||||
| n | 115 | 1772 | 48 | 480 | 67 | 1292 |
| hs-CRP GM (IQR), mg/L | 1.78 (0.81-3.46) | 1.56 (0.73-3.19) | 1.77 (0.82-2.77) | 1.58 (0.77-3.27) | 1.79 (0.76-4.01) | 1.55 (0.71-3.17) |
| Back-transformed β (95% CI)* | 1.13 (0.92-1.38) | 0.92 (0.68-1.24) | 1.24 (0.94-1.62) | |||
| % difference | 13 | −8 | 24 | |||
| P* | .253 | .585 | .128 | |||
| ASXL1 | ||||||
| n | 52 | 1835 | 14 | 514 | 38 | 1321 |
| hs-CRP GM (IQR), mg/L | 1.93 (0.88-3.64) | 1.56 (0.73-3.19) | 1.77 (0.96-2.45) | 1.59 (0.77-3.27) | 1.99 (0.83-4.25) | 1.55 (0.71-3.17) |
| Back-transformed β (95% CI)* | 1.26 (0.94-1.69) | 1.09 (0.64-1.86) | 1.31 (0.92-1.87) | |||
| % difference | 26 | 9 | 31 | |||
| P | .128 | .751 | .138 | |||
| PPM1D | ||||||
| n | 38 | 1849 | 9 | 519 | 29 | 1330 |
| hs-CRP GM (IQR), mg/L | 1.64 (0.67-3.43) | 1.57 (0.74-3.19) | 1.84 (0.60-5.09) | 1.59 (0.78-3.23) | 1.59 (0.74-3.28) | 1.56 (0.71-3.18) |
| Back-transformed β (95% CI)* | 0.98 (0.69-1.39) | 1.14 (0.59-2.20) | 0.95 (0.63-1.43) | |||
| % difference | −2 | 14 | −5 | |||
| P | .909 | .705 | .805 | |||
The sample size related to carriers of mutations in TP53, SF3B1, SRSF2, CBL, and JAK2 was insufficient to perform analyses in these subgroups. The presented P values are for mutations yes vs no status differences in log-transformed changes in hs-CRP.
CHIP+, carrier of mutation(s); CHIP−, noncarrier of mutation; GM, geometric mean.
Generalized linear regression model-derived ln(hs-CRP) with adjustment for age, sex, and CAD status at baseline-adjusted where CHIP− was the referent category.
Secondary analyses (CAD and non-CAD subgroups)
Among the patients with CAD at baseline (n = 1359), CHIP carriers were older (mean: 75.1 vs 74.5 years), had fewer previous PCI (44% vs 52%), had more congestive heart failure (24% vs 18%), and had higher hs-CRP (median: 1.60 vs 1.39 mg/L) than non-CHIP carriers (all P < .05; Table 1). In the non-CAD cohort (n = 528), CHIP carriers were also significantly older than noncarriers (74.9 vs 74.1 years) but had lower rates of current smoking (0.8% vs 4.6%), and hs-CRP levels were not significantly different between those 2 groups. In multivariable generalized linear regression analyses focusing on patients with a history of CAD at baseline, CHIP carriers of any mutations had higher hs-CRP than non-CHIP carriers after adjusting for covariates (eβ = 1.22, 95% CI: 1.06 to 1.41; P = .005; Table 2). This association was not significant in the smaller subgroup of patients without a history of CAD at baseline (eβ = 1.15, 95% CI: 0.94 to 1.40; P = .178).
Sensitivity, subgroup, and additional analyses
When limiting analyses to individuals with VAF ≥ 10%, as compared with those without CHIP, carriers of any gene mutation also had higher ln(CRP) (eβ = 1.23, 95% CI: 1.05 to 1.45; P=.038) in adjusted analyses (data not shown). This was not different from the entire CHIP cohort (eβ = 1.21). However, we could not document a quantitative correlation between ln(CRP) and CHIP due to limited sample size in patients with VAF ≥ 10% (n = 191). In sensitivity analyses, we adjusted for additional confounders (statin, aspirin, beta-blocker use at baseline, as well as diabetes mellitus) that could have influenced the results of our analyses. The additional adjustment of these confounding factors in our multivariable analyses did not affect the magnitude of our findings. Finally, because patients with a cancer diagnosis at study entry are likely to have received chemotherapy or radiation therapy, which could result in clonal expansion, we also conducted a subgroup analysis in patients without cancer at baseline (n = 1541). The relationship between CHIP and ln(CRP) in this cohort remained unchanged.
Discussion
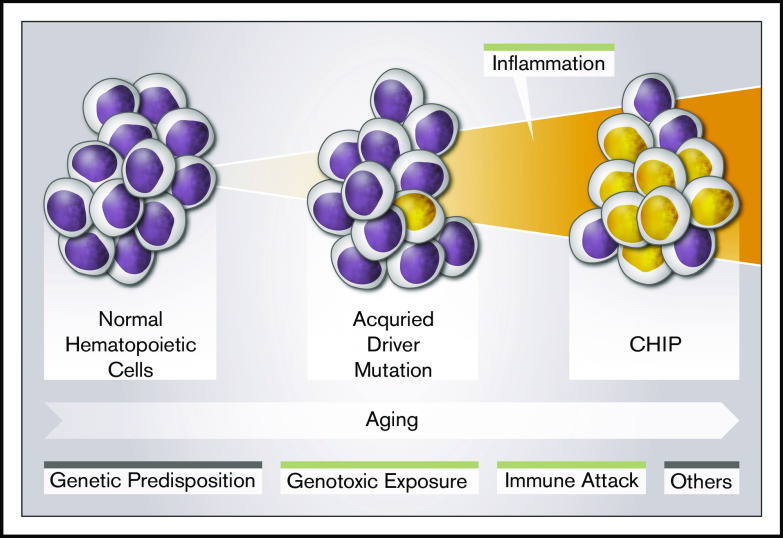
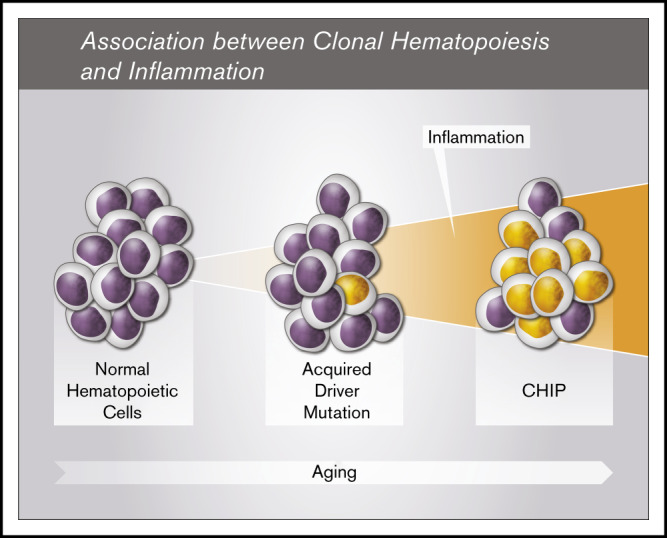
Young et al documented mutations in TET2 or DNMT3A at very low frequencies in 95% of individuals aged 50 to 60 years, suggesting that these genes are almost ubiquitously mutated after the age of 50.22 However, only a fraction of mutated individuals will have clonal expansion of a magnitude corresponding to CHIP,10 and an associated increased risk of hematological cancers and cardiovascular disease. It is therefore of prime importance to identify the factors associated with clonal progression. Thus, we evaluated the hypothesis that inflammation is associated with clonal development of CHIP-related mutations in a single-center cohort of 1887 individuals aged >70, comprising individuals with (1359) and without (528) CAD at baseline. We showed that hs-CRP is significantly higher in subjects with CHIP than in those without CHIP, supporting the hypothesis that there is a link between inflammation and CHIP’s pathogenesis in aging individuals (Figure 3).
Figure 3.
Risk factors for CHIP. CHIP is age dependent. Several factors may influence its development. These include genetic predisposition, genotoxic exposure, and immune attack (reviewed in Busque et al15). This study further confirms the potential role of an inflammatory state in CHIP development.
We selected patients aged >70 to maximize the prevalence of CHIP, which was indeed 22.6% in this cohort. Surprisingly, patients without CAD at baseline had slightly higher prevalence of CHIP than those with CAD at baseline (25.3% vs 21.5%; P < .075). Because CHIP is associated with an increased risk of CAD, we expected a higher prevalence of CHIP in this subgroup. We speculate that selecting patients aged >70 years old may have systematically introduced a survival bias, where CHIP carriers with CAD may have been left out due to earlier death. It is also intriguing that the prevalence of TET2 mutations was significantly lower in the CAD cohort than in the non-CAD subgroup. It is possible that specific CHIP-associated genes have a different survival impact. Expanding the cohort to include younger patients may ultimately allow us to answer this question.
The primary objective of this study was to document an association between inflammation using hs-CRP and CHIP carrier status. CRP was discovered in 1930 in the serum of patients with pneumococcal pneumonia.23 It is a pentraxin produced by the liver and an acute phase reactant,24 and IL-6, IL-1, and TNF-α are the main inducers of CRP.25,26 CRP measurement is commonly used to evaluate tissue injury, infection, and inflammatory diseases.27 The development of high-sensitivity assays28 has been invaluable for the investigation of low-grade chronic inflammation in different human diseases.27 The value of hs-CRP as a predictor of cardiovascular events was demonstrated 20 years ago.29 For example, in a cohort of 28 263 normal women, hs-CRP measured only once at baseline was the strongest predictor of coronary heart disease death, nonfatal MI, stroke, or the need for coronary revascularization procedures among other biomarkers, including IL-6. We adopted a similar approach using baseline hs-CRP measurement in a cohort of patients with and without CAD.
The association between hs-CRP and CHIP was demonstrated in both univariable and multivariable analyses for the entire study population and for the cohort with CAD. This demonstration was made using an adequately large, single-center, cohort of older individuals, and a commonly available biomarker of inflammation. The differences in hs-CRP levels between CHIP and non-CHIP carriers within non-CAD cohorts were slightly lower, and the difference did not reach statistical significance. This might be related to the fact that our non-CAD cohort was smaller than the CAD counterpart (528 vs 1359) or that the influence of inflammation as measured by hs-CRP has a greater influence on CHIP development in CAD patients. Interestingly, 2 recent studies did not identify a clear relationship between hs-CRP and CHIP. Bick et al analyzed exome sequences from 35 416 individuals from the UK biobank without prevalent cardiovascular disease (similar to our non-CAD cohort).30 They reported a modest and significant increase in hs-CRP (2.49 vs 2.85; P = .03) in univariate analysis, but this association was lost in the covariate-adjusted model (P = .08). Furthermore, in a recent TOPMed communication,31 Bick et al identified in data from diverse genetic cohorts (n = 32) that CHIP was associated with IL-6 (P = .0035; n = 11762), IL-1b (P = 2.4 × 10−4; n = 598), but not with hs-CRP (P = .10; n = 22092). These discordant results are surprising as both IL-6 and IL-1 are inducers of CRP production by the liver.25 It is possible that CRP is not a sensitive or specific inflammation biomarker of CHIP compared with IL-6 or IL-1, and/or that our single-center and laboratory approach increased the capacity to identify a true relationship in contrast to large heterogeneous cohorts. Nevertheless, these studies confirm the association between biomarkers of inflammation and CHIP.
The demonstration of an association between inflammation and CHIP in human subjects does not address the causal relationship between the 2 entities. Does inflammation promote clonal expansion of CHIP or do CHIP clones drive inflammatory response? Zhang et al demonstrated that myeloid-derived cells with loss of TET2 maintain higher expression of IL-6, and that TET2 is required to resolve inflammation and specifically repressed IL-6.32 Fuster et al studied clonal expansion of tet2-mutant cells in atherosclerosis-prone mice and demonstrated that TET2-deficient macrophages exhibited an increase in NLRP inflammasome-mediated IL-1 β secretion.33 Interestingly, Bick et al demonstrated that a relatively common coding mutation in the IL6-Receptor (pAsp358Ala), which significantly reduces signalization of IL-6, attenuated cardiovascular risk of CHIP carriers vs non-CHIP carriers,30 supporting a causal link between CHIP, inflammation, and CAD. Taken together, this indicates that CHIP’s mutation increases inflammation cytokines profile, at least at the cellular level. However, there are also several lines of evidence that suggest that increased inflammation promotes clonal expansion of mutated HSCs (reviewed in King et al34). In a key experiment, Cai et al showed a rapid increase in the frequency and absolute number of Tet2-KO mature myeloid cells and HSCs in response to inflammatory stress and demonstrated an enhanced production of inflammatory cytokines, including IL-6 by these cells.35 Taken together, these studies suggest that extrinsic inflammation mediators may selectively support clonal expansion of mutated HSCs, which will eventually contribute to excessive cytokine release perpetuating a feedback loop.34
The difference of hs-CRP levels between CHIP carriers and non-CHIP carriers was relatively modest (±20%). However, a small quantitative difference in inflammatory cytokines may have a lever effect if it is linked with increase receptor expression. Such a model has been demonstrated for chronic myelogenous leukemia in which stem cells have increased IL-1 receptor activity,36 and increased proliferation and survival in a proinflammatory environment.37 Furthermore, in a recent meta-analysis of inflammatory cytokine profile of 697 individuals with myelodysplastic syndromes vs controls, Shi et al demonstrated a small but significant increase in levels of TNF, IL-8, and IL-6.38 In a cohort of patients with ulcerative colitis, Zhang et al documented a small but significantly increased level of serum IFN-γ but not TNF-α in subjects with DNMT3A mutation.39 Therefore, it is possible that the small but long-standing difference in inflammation level may have a biological impact. Interestingly, treatment with the anti-IL1b antibody canakinumab led to significant reductions of not only cardiovascular disease40 but also incident lung cancer in patients with CAD.41 Furthermore, the recent demonstration that utilization of a low dose of the anti-inflammatory drug colchicine reduces cardiovascular events after MI may further justify the prospective evaluation of such approaches in CHIP carriers.42
This study also allowed us to make additional observations. CHIP regroups alterations occurring in several different genes, but clinical outcomes have been usually estimated according to the CHIP-carrier status5,6 rather than gene-specific estimation. Recently, some studies have demonstrated gene specificities in regards to risk of progression to acute myeloid leukemia,43 genetic predisposition,11 or lineage restriction.44 We wanted to study the relationship between inflammation and specific CHIP-associated genes. We were able to document a positive association between DNMT3A and hs-CRP in the entire cohort. No other association was statistically significant, although quantitative differences were similar for DNMT3A, TET2, and ASXL1. A significantly larger cohort would be necessary to address this question. We documented a significant association between heart failure and CHIP carrier status. This is in line with several previous observations in mice45 and humans.46,47 In contrast, we did not find an association with smoking in this cohort, where prevalence was very low.
In conclusion, this study highlights the role of inflammation in CHIP. The etiology of CHIP is probably multifactorial, and several other factors need to be identified. Clinical trials should test whether anti-inflammatory therapy can reduce CHIP progression and related diseases.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) and by the Leukemia Lymphoma Society of Canada (LLSC).
Footnotes
For inquiries concerning data related to this paper, e-mails may be sent to the corresponding authors, Lambert Busque (lbusque.hmr@ssss.gouv.qc.ca), Marie-Pierre Dubé (marie-pierre.dube@mhi-rc.org), and Jean-Claude Tardif (jean-claude.tardif@icm-mhi.org).
Authorship
Contribution: L.B., J.-C.T., M.-P.D. conceived the study; M.B. performed and analyzed the sequencing data with the support of S.A. and V.B.; M.B. generated the figures; M.S. performed statistical analyses with the support of Y.F.Z. and S.P. under the supervision of M.-P.D.; and all authors revised the manuscript and provided insightful suggestions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lambert Busque, Hôpital Maisonneuve-Rosemont, 5415 Boulevard de l’Assomption, Montreal, QC H1T 2M4, Canada; e-mail: lbusque.hmr@ssss.gouv.qc.ca; Jean-Claude Tardif, Montreal Heart Institute, 5000 Belanger St, Montreal, QC H1T 1C8, Canada; e-mail: jean-claude.tardif@icm-mhi.org; and Marie-Pierre Dubé, Montreal Heart Institute, 5000 Belanger St, QC H1T 1C8, Canada; e-mail: marie-pierre.dube@mhi-rc.org.
References
- 1.Fey MF, Liechti-Gallati S, von Rohr A, et al. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 beta DNA probe. Blood. 1994;83(4):931-938. [PubMed] [Google Scholar]
- 2.Busque L, Mio R, Mattioli J, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88(1):59-65. [PubMed] [Google Scholar]
- 3.Ayachi S, Buscarlet M, Busque L. 60 Years of clonal hematopoiesis research: from X-chromosome inactivation studies to the identification of driver mutations. Exp Hematol. 2020;83:2-11. [DOI] [PubMed] [Google Scholar]
- 4.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44(11):1179-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zink F, Stacey SN, Norddahl GL, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130(6):742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buscarlet M, Provost S, Zada YF, et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130(6):753-762. [DOI] [PubMed] [Google Scholar]
- 12.Hansen JW, Pedersen DA, Larsen LA, et al. Clonal hematopoiesis in elderly twins: concordance, discordance and mortality. Blood. 2020;135(4):261-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabre MA, McKerrell T, Zwiebel M, et al. Concordance for clonal hematopoiesis is limited in elderly twins. Blood. 2020;135(4):269-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franceschi C, Garagnani P, Vitale G, Capri M, Salvioli S. Inflammaging and “Garb-aging”. Trends Endocrinol Metab. 2017;28(3):199-212. [DOI] [PubMed] [Google Scholar]
- 15.Busque L, Buscarlet M, Mollica L, Levine RL. Concise review: age-related clonal hematopoiesis: stem cells tempting the devil. Stem Cells. 2018;36(9):1287-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abegunde SO, Buckstein R, Wells RA, Rauh MJ. An inflammatory environment containing TNFα favors Tet2-mutant clonal hematopoiesis. Exp Hematol. 2018;59:60-65. [DOI] [PubMed] [Google Scholar]
- 17.Meisel M, Hinterleitner R, Pacis A, et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 2018;557(7706):580-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook EK, Izukawa T, Young S, et al. Comorbid and inflammatory characteristics of genetic subtypes of clonal hematopoiesis. Blood Adv. 2019;3(16):2482-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridker PM. From C-reactive protein to interleukin-6 to interleukin-1: moving upstream to identify novel targets for atheroprotection. Circ Res. 2016;118(1):145-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montreal Heart Institute Hospital Biobank https://www.icm-mhi.org/en/research/infrastructures-services/mhis-hospital-biobank. Accessed 11 December 2018. [Google Scholar]
- 21.Tardif JC, Rhainds D, Brodeur M, et al. Genotype-dependent effects of dalcetrapib on cholesterol efflux and inflammation: concordance with clinical outcomes. Circ Cardiovasc Genet. 2016;9(4):340-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7(1):12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tillett WS, Francis T. Serological Reactions in pneumonia with a non-protein somatic fraction of Pneumococcus. J Exp Med. 1930;52(4):561-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira EB, Gotschlich C, Liu TY. Primary structure of human C-reactive protein. J Biol Chem. 1979;254(2):489-502. [PubMed] [Google Scholar]
- 25.Moshage HJ, Roelofs HM, van Pelt JF, et al. The effect of interleukin-1, interleukin-6 and its interrelationship on the synthesis of serum amyloid A and C-reactive protein in primary cultures of adult human hepatocytes. Biochem Biophys Res Commun. 1988;155(1):112-117. [DOI] [PubMed] [Google Scholar]
- 26.Yoshizaki K. Pathogenic role of IL-6 combined with TNF-α or IL-1 in the induction of acute phase proteins SAA and CRP in chronic inflammatory diseases. Adv Exp Med Biol. 2011;691:141-150. [DOI] [PubMed] [Google Scholar]
- 27.Pepys MB, Berger A. The renaissance of C reactive protein. BMJ. 2001;322(7277):4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rifai N, Ridker PM. High-sensitivity C-reactive protein: a novel and promising marker of coronary heart disease. Clin Chem. 2001;47(3):403-411. [PubMed] [Google Scholar]
- 29.Ridker PM, Hennekens CH, Buring JE, Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N Engl J Med. 2000;342(12):836-843. [DOI] [PubMed] [Google Scholar]
- 30.Bick AG, Pirruccello JP, Griffin GK, et al. Genetic interleukin-6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141(2):124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bick AG, Weinstock JS, Nandakumar SK, et al. Inherited causes of clonal hematopoiesis of indeterminate potential in TOPMed whole genomes. bioRxiv. doi: 10.1101/782748 [DOI] [Google Scholar]
- 32.Zhang Q, Zhao K, Shen Q, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525(7569):389-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355(6327):842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King KY, Huang Y, Nakada D, Goodell MA. Environmental influences on clonal hematopoiesis. Exp Hematol. 2020;83:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Z, Kotzin JJ, Ramdas B, et al. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 2018;23(6):833-849.e835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Li M, McDonald T, et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-β-catenin signaling. Blood. 2013;121(10):1824-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Chu S, Agarwal P, et al. Inhibition of interleukin-1 signaling enhances elimination of tyrosine kinase inhibitor-treated CML stem cells. Blood. 2016;128(23):2671-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi X, Zheng Y, Xu L, Cao C, Dong B, Chen X. The inflammatory cytokine profile of myelodysplastic syndromes: a meta-analysis. Medicine (Baltimore). 2019;98(22):e15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang CRC, Nix D, Gregory M, et al. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp Hematol. 2019;80:36-41.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridker PM, Everett BM, Thuren T, et al. ; CANTOS Trial Group . Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119-1131. [DOI] [PubMed] [Google Scholar]
- 41.Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ; CANTOS Trial Group . Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390(10105):1833-1842. [DOI] [PubMed] [Google Scholar]
- 42.Tardif JC, Kouz S, Waters DD, et al. Efficacy and safety of low-dose colchicine after myocardial infarction. N Engl J Med. 2019;381(26):2497-2505. [DOI] [PubMed] [Google Scholar]
- 43.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buscarlet M, Provost S, Zada YF, et al. Lineage restriction analyses in CHIP indicate myeloid bias for TET2 and multipotent stem cell origin for DNMT3A. Blood. 2018;132(3):277-280. [DOI] [PubMed] [Google Scholar]
- 45.Sano S, Oshima K, Wang Y, et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J Am Coll Cardiol. 2018;71(8):875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorsheimer L, Assmus B, Rasper T, et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 2019;4(1):25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorsheimer L, Assmus B, Rasper T, et al. Hematopoietic alterations in chronic heart failure patients by somatic mutations leading to clonal hematopoiesis [published online ahead of print 7 November 2019]. Haematologica. doi: 10.3324/haematol.2019.224402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.