Abstract
The global health emergency of novel COVID-19 is due to severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Currently there are no approved drugs for the treatment of coronaviral disease (COVID-19), although some of the drugs have been tried. Chloroquine is being widely used in treatment of SARS-CoV-2 infection. Hydroxychloroquine, the derivative of Chloroquine shows better inhibition than Chloroquine and has in vitro activity against SARS-CoV-2 also used to treat COVID-19. To study the interactions of Chloroquine and derivatives of Chloroquine with SARS-CoV-2, series of computational approaches like pharmacophore model, molecular docking, MM_GBSA study and ADME property analysis are explored. The pharmacophore model and molecular docking study are used to explore the structural properties of the compounds and the ligand-receptor (PDB_ID: 6LU7) interactions respectively. MM_GBSA study gives the binding free energy of the protein-ligand complex and ADME property analysis explains the pharmacological property of the compounds. The resultant best molecule (CQD15) further subjected to molecular dynamics (MD) simulation study which explains the protein stability (RMSD), ligand properties as well as protein-ligand contacts. Outcomes of the present study conclude with the molecule CQD15 which shows better interactions for the inhibition of SARS-CoV-2 in comparison to Chloroquine and Hydroxychloroquine.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, SARS-CoV-2, pharmacophore model, molecular docking, molecular dynamics simulations
Introduction
Coronavirinae family is made up of four genera based on their genetic material, including Alpha, Beta, Gamma and Delta coronavirus genus. The coronavirus family is a positive-stranded RNA virus, which mainly causes respiratory and central nervous system disease (Perlman & Netland, 2009). Human coronaviruses namely HCoV-OC43, HCoV-229E, HCoV-NL63 and HCoV-HKU1 are responsible for mild respiratory diseases (Owusu et al., 2014). The SARS-CoV outbreak in 2002 demonstrated that coronaviruses would cross the barrier to species and emerge as highly pathogenic viruses (Perlman, 2020). The disease caused by SARS-CoV-2 is called COVID-19. The coronavirus has been named SARS-CoV-2 (Gorbalenya et al., 2020), because the RNA genome is about 82% identical to the SARS coronavirus (SARS-CoV); both viruses belong to the genus beta-coronavirus (Wu et al., 2020; Zhou et al., 2020). The novel COVID-2019 was first reported in Wuhan, China in December 2019. The outbreak has been rapidly spread all over the world. More than 200 countries were affected by this COVID-19. The world health Organization (WHO) announced on 11th march 2020 that the COVID-19 as pandemic disease. Currently, more than 4,310,786 number (as on 12th May 2020) of people were infected with COVID-19 which is increasing by approximately 100000+ cases per day and more than 290,455 people have died. Unfortunately, to date, there is no medication or vaccine approved by the World Health Organization (WHO), the Centers for Disease Control and Prevention (CDC), and the FDA for the effective treatment of COVID-19 (CDC, 2020 WHO, 2020).
Many drugs have been tried recently in the treatment of COVID-19 due to the situation of “public-health emergency”, that includes antimalarial drugs (Chloroquine and its derivative Hydroxychloroquine) (Colson et al., 2020; Gao et al., 2020; Wang et al., 2020; Yao et al., 2020) antiviral drugs (HIV Protease Inhibitors i.e. Lopinavir and Ritonavir) (Chu et al., 2004; Yao et al., 2020), investigational nucleoside analogue (Remdesivir (GS-5734)) (Agostini et al., 2018; Wang et al., 2020) and investigational RNA-dependent RNA polymerase inhibitor (Favipiravir) (Beijing Chao Yang Hospital, 2020; Peking University First Hospital, 2020; Wang et al., 2020). Some Adjunctive therapeutic drugs are also used for the treatment of COVID-19, which includes macrolide antibacterial drug (Azithromycin) (Amsden, 2005; Beigelman et al., 2010; Gautret et al., 2020), Interleukin-6 (IL-6) Receptor-Inhibiting Monoclonal Antibody (Tocilizumab, Sarilumab) (Assistance Publique – Hopitaux de Paris, 2020; Xu et al., 2020; Zhou et al., 2020) and Investigational Humanized Monoclonal Antibody to the Chemokine Receptor CCR5 (Leronlimab) (CytoDyn, 2020; Zhou et al., 2020). The detailed outcomes of the clinical trials are still awaited. However, among all these reported drugs Chloroquine and Hydroxychloroquine were used/being used widely for the treatment of COVID-19.
Many research institutes as well as pharmaceutical companies are working to identify potential candidate against SARS-CoV-2 worldwide. Some researcher reported that Chloroquine has the potential to treat COVID-19 with clinical study as well as in silico study (Adeoye et al., 2020) Boopathi et al., 2020; Gao et al., 2020). Department of Pathology, Vanderbilt University Medical Center, USA reported that antiviral Remdesivir (GS-5734) can treat SARS-CoV-2 (Agostini et al., 2018). The investigation of COVID-19 with Plasma therapy and its clinical trials are being conducted to evaluate the use of COVID-19 to treat patients with severe or immediately life-threatening COVID-19 infections (Duan et al., 2020). Hasan et al. reviewed the ongoing challenges and future prospects for the development of potential drugs to promote targeting specific activities of the CoV (Hasan et al., 2020). Some research groups are also working in the field of computational approach to find a solution for COVID-19. Khan et al. worked on a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-Oribose methyltransferase and proposed that Raltegravir, Paritaprevir, Bictegravir and Dolutegravir are excellent lead candidates for these crucial proteins and they could become potential therapeutic drugs against SARS-CoV-2 (Khan et al., 2020). 3,4 Dihydropyrimidone class molecule identified as a drug candidate against 2019-nCoV N-protein (N terminal domain) (Sarma et al., 2020). Enayatkhani et al. worked on reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19 and conclude with NOM-TLR4 and NOM-HLA-A11:01 docked model (Enayatkhani et al., 2020). Elfiky et al. suggested guanosine derivatives (Guanosine triphosphate (GTP), Uridine triphosphate (UTP), IDX-184 (GTP derivative), sofosbuvir (UTP derivative), ribavirin (wide acting antiviral drug) can be the potential candidate against COVID-19 (Elfiky & Azzam, 2020). Zhou et al. and Joshi et al. reported the network-based methodologies for rapid identification of candidate repurposable drugs and potential drug combinations targeting 2019-nCoV/SARS-CoV-2 (Joshi et al., 2020; Zhou et al., 2020). Zhang et al. from Institute of Biochemistry, Center for Structural and Cell Biology in Medicine, University of Lübeck, Germany designed the improved α-ketoamide inhibitors for the inhibition of SARS-CoV-2 (Zhang et al., 2020). Yogi Vemana University, India, Government Medical College, Anantapur, India and ICAR-National Rice Research Institute, India collaboratively worked on In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel (Gupta et al., 2020). A few in silico research has also been done by considering FDA approved drugs and concludes with Lopinavir (Pant et al., 2020), Setrobuvir (Elfiky, 2020a), Saquinavir (Al-Khafaji et al., 2020) Remdesivir, Saquinavir, Darunavir, (Khan et al., 2020) and Disulfiram (Lobo-Galo et al., 2020) which have better interactions with SARS CoV-2. Some compounds like Crocin, Digitoxigenin, b-Eudesmol, (Aanouz et al., 2020) Andrographolide, (Enmozhi et al., 2020) phytoestrogens (Diadiazin, Genistein, Formontein and Biochanin A), (Elfiky, 2020b) phytochemicals (hypericin, cyanidin 3-glucoside, baicalin, glabridin, and α-ketoamide-11r), (Islam et al., 2020) Resveratrol, (Wahedi et al., 2020) Rutin, (Das et al., 2020) Ammoides verticillata, (Abdelli et al., 2020) alkaloids (10–Hydroxyusambarensine, and Cryptoquindoline) and terpenoids (6-Oxoisoiguesterin and 22-Hydroxyhopan-3-one) (Gyebi et al., 2020) also reported as a better candidate against COVID-19. Umesh et al. Worked on Indian spices to study structure based interaction with COVID-19 and conclude with three compounds Carnosol, Arjunglucoside-I and Rosmanol (Umesh et al., 2020). Sinha et al. evaluated different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets (Sinha et al., 2020).
As Hydroxychloroquine is the derivative of Chloroquine and shows better inhibition in comparison to Chloroquine against SARS CoV-2 (Pastick et al., 2020; Yao et al., 2020), the current study is emphasized on the derivatives of Chloroquine against COVID-19. A set of 18 molecules based on Chloroquine scaffold (Scheme 1) are designed and optimized computationally by using Schrodinger software. These molecules were designed by substituting different groups (like, methyl (CH3), hydroxyl group (OH), NO2, Amino group (NH2), halogens (F, Cl and Br) and ring aromatic (benzene, phenol)) at the position of R1 and R2 (Scheme 1). These groups were considered due to its potential to interact with the amino acids (like ‘hydroxyl group, methyl and amine can interact with the amino acid through H-bond’, ‘halogens; Cl, Br, F can interact through halogen bonding’, ‘NO2 can show H-bond as well as salt bridges’ and ‘ring aromatic compounds can show pi-pi and pi-cation interactions. In addition to pi-pi and pi-cation interactions the phenol can also show H-bonding due to OH group’) (Rawel et al., 2005; Zavodszky & Kuhn, 2005). The generated pharmacophore model can efficiently explain the essential structural features present in the molecules having the potency to interact with the target protein. Molecular docking, MM/GBSA study, ADME property and MD simulation analysis have been employed to study and identify the candidate having better interactions and binding affinity with the SARS-CoV-2.
Scheme 1.
Chemical structure of Chloroquine scaffolds. []
Methodology
Molecular simulation studies were performed by using Maestro 12.3 module (Schrodinger Release 2020-1, 2020). All the 18 designed molecules (including chloroquine and hydroxychloroquine) and protein (6LU7) (Jin et al., 2020; Muralidharan et al., 2020; Elmezayen et al., 2020; Kumar et al., 2020) were optimized and followed the methodology of pharmacophore hypothesis, molecular docking, MM-GBSA study, ADME property analysis and molecular dynamics (MD) simulations as per the previous reports (Beura & Chetti, 2020; Ganjoo et al., 2019) and detailed methodologies are given in supporting information. The detailed working methodology flowchart is shown in Scheme 2.
Scheme 2.
Working methodology.
For the validation of theoretical results, a set of different PDB_IDs i.e. 5REB (Fearon et al., 2020), 6W02 (Michalska et al., 2020) were downloaded from protein data bank and re-docking of the self ligand with the protein was performed by using ‘Ligand Docking’ module with extra precision (XP) mode. RMSE values were calculated between crystal and re-dock pose by using the ‘superimpose’ module. RMSD value generated by re-docking the native ligands in the active site of the respective protein was 1.33 Å, and 1.29 Å. As the RMSD values are less than X-ray resolution of the corresponding protein which is 1.68 Å, and 1.5 Å. It indicates that the generated model has a good correlation with the experimental X-ray crystal structures (Figure 1).
Figure 1.
Superimpose of the self-docked ligand with native ligand in the protein grid (a) 5REB and (b) 6W02.
Result and discussion
Pharmacophore model
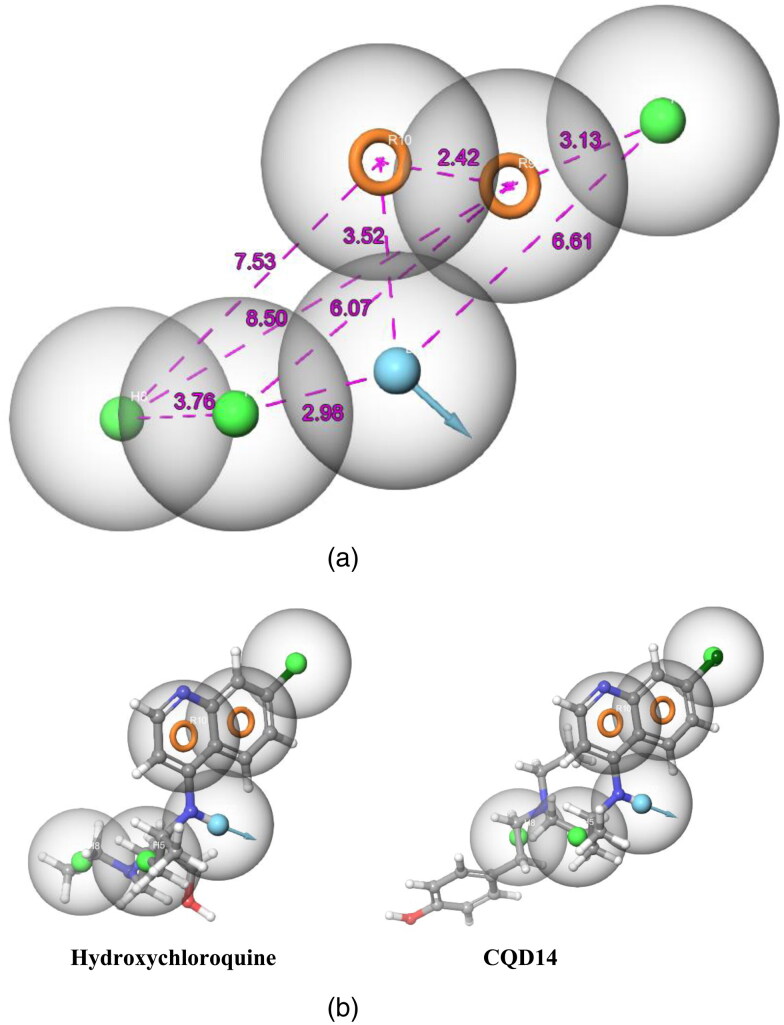
A total of 20 pharmacophore hypothesis models (Table 1) were generated by using the ‘phase module.’ The hypotheses have consisted of all active match molecules. There were 2 types of different models generated like DHHHRR (model 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10) and DHHRR (model 11, 12, 13, 14, 15, 16, 17, 18, 19 and 20). Out of 20 models, DHHHRR_1 was considered as the best model as this six featured models shows the best PhaseHypoScore (1.383), Survival Score (6.392) and good BEDROC score (0.999). Pharmacophore hypothesis model DHHHRR_1 consists of one H-bond donor (blue color), three hydrophobic sites (green color) and two aromatic rings (orange color), shown in Figure 2(a) (also shown the position of features concerning each other in Figure 2(a)). DHHHRR_1 model shows proper fit with all designed ligands. For example; Hydroxychloroquine and CQD14 shows a proper fit with the best pharmacophore model DHHHRR_1 (Figure 2(b)) which indicates all the optimized Chloroquine derivatives are having nearly similar orientations.
Table 1.
Phase generated pharmacophore hypothesis models for chloroquine derivatives (CQD).
| S. No. | Title | BEDROC Score | Survival Score | PhaseHypoScore |
|---|---|---|---|---|
| 1 | DHHHRR_1 | 0.999 | 6.392 | 1.383 |
| 2 | DHHHRR_2 | 1 | 6.374 | 1.382 |
| 3 | DHHHRR_3 | 1 | 6.372 | 1.382 |
| 4 | DHHHRR_4 | 0.998 | 6.364 | 1.380 |
| 5 | DHHHRR_5 | 1 | 6.362 | 1.382 |
| 6 | DHHHRR_6 | 0.995 | 6.341 | 1.376 |
| 7 | DHHHRR_7 | 1 | 6.334 | 1.380 |
| 8 | DHHHRR_8 | 1 | 6.331 | 1.380 |
| 9 | DHHHRR_9 | 1 | 6.299 | 1.378 |
| 10 | DHHHRR_10 | 0.999 | 6.288 | 1.376 |
| 11 | DHHRR_1 | 1 | 5.869 | 1.352 |
| 12 | DHHRR_2 | 0.995 | 5.853 | 1.346 |
| 13 | DHHRR_3 | 1 | 5.814 | 1.349 |
| 14 | DHHRR_4 | 1 | 5.804 | 1.348 |
| 15 | DHHRR_5 | 1 | 5.792 | 1.348 |
| 16 | DHHRR_6 | 0.994 | 5.768 | 1.341 |
| 17 | DHHRR_7 | 0.996 | 5.765 | 1.342 |
| 18 | DHHRR_8 | 0.987 | 5.765 | 1.333 |
| 19 | DHHRR_9 | 1 | 5.759 | 1.346 |
| 20 | DHHRR_10 | 1 | 5.744 | 1.345 |
Figure 2.
(a) Arrangement of individual features in a fixed distance (in Å) of pharmacophore hypothesis model DHHHRR_1 for Chloroquine derivatives. (b) Mapping of molecules on pharmacophore hypothesis model DHHHRR_1.
The Pharmacophore models were generated to study the maximum number of structural features present in the designed molecules. The ligand-receptor interactions depends upon these structural features present in the ligand like, H-bond acceptor (A, red color ball), H-bond donor (D, blue color ball), Aromatic rings (R, orange color ring) and Hydrophobic region (H, green color ball) (Figure 3). The selected molecules CQD-15, CQD-14 and CQD-16 shows 13, 12 and 13 number of structural features whereas Chloroquine and Hydroxychloroquine shows 9 and 10 number of structural features. It indicates that the selected molecules CQD-15, CQD-14 and CQD-16, have the potential to interact with the enzyme (Figure 3 and Figure S1).
Figure 3.
Pharmacophre model generated structural features present in the ligands (Hydroxychloroquine and CQD15).
Molecular docking
The generated active site of SARS-CoV-2 main protease (PDB ID: 6LU7) is occupied by charged amino acids (ARG-298, LYS-102, ASP-153 and ASP-295), polar amino acids (THR-111, GLN-110, GLN-127, THR-292, ASN-151 and SER-158) and hydrophobic amino acid (ILE-152, TYR-154, PRO-9, PHE-8, PHE-112, PHE-294, VAL-104, VAL-297 and ILE-106) residue. To ensure the excavation of the best candidates, molecular interaction study was performed through ligand docking into the active site of the SARS-CoV-2. The designed 18 molecules were visually inspected based on the docking score, ligand-receptor interactions, H-bonding, hydrophobic potential, electrostatic rewards and rewards for the ligand of low molecular weight. This deep study concludes with 3 best molecules i.e. CQD15, CQD14 and CQD16 and are showing docking scores of -6.17, −5.14 and -4.19 respectively (Table 2). CQD15 shows three H-bonds with two polar amino acids (THR-111 and GLN-110) and one charged amino acid (ASP-153), two H-bonds due to the H-bond donor group and one due to H-bond acceptor site present in the ligand. Additionally, CQD15 shows one pi-pi interaction with hydrophobic amino acid (PHE-8) (Figure 4). CQD14 shows 2 H-bond interactions due to two H-bond donor groups present in the ligand along with one pi-pi interaction. The H-bond interactions take place with one polar amino acid (THR-111) and one charged amino acid (GLN-110) present in the active site (Figure 4). CQD16 shows similar interactions like CQD15 and additionally, it shows one pi-pi interaction with hydrophobic PHE-294 amino acid (Figure 4). All 3 new ligands showing a good range of solvent exposures (Figure 4) and the docked pose shows the best fit of the ligand in the active site cavity of the enzyme (Figure 5).
Table 2.
Docking score and functional parameters of all 18 molecules including Chloroquine and Hydroxychloroquine with SARS-CoV-2 main protease (PDB ID: 6LU7).
| S. No. | Ligand | DockScore | LipophilicEvdW | PhobEnHB | HBond | Electro | LowMW |
|---|---|---|---|---|---|---|---|
| 1 | CQD15 | −6.17 | −2.24 | −1.5 | −1.87 | −1.1 | −0.12 |
| 2 | CQD14 | −5.14 | −1.93 | −1.5 | −1.35 | −0.77 | −0.13 |
| 3 | CQD16 | −4.19 | −2.78 | 0 | −1.11 | −0.63 | −0.12 |
| 4 | CQD6 | −4.15 | −2.57 | 0 | −1.15 | −0.63 | −0.38 |
| 5 | CQD5 | −3.92 | −1.45 | −1.5 | −0.7 | −0.42 | −0.28 |
| 6 | Hydroxychloroquine | −3.48 | −2.38 | 0 | −1.28 | −0.48 | −0.38 |
| 7 | CQD13 | −3.44 | −2.68 | 0 | −1.15 | −0.6 | −0.18 |
| 8 | CQD12 | −3.41 | −2.49 | 0 | −1.25 | −0.47 | −0.17 |
| 9 | CQD9 | −3.39 | −3.26 | 0 | −0.04 | −0.14 | −0.37 |
| 10 | CQD10 | −3.3 | −2.64 | 0 | −0.52 | −0.21 | −0.38 |
| 11 | Chloroquine | −3.27 | −2.86 | 0 | −0.34 | −0.15 | −0.43 |
| 12 | CQD7 | −3.08 | −3.36 | 0 | 0 | −0.14 | −0.17 |
| 13 | CQD4 | −3.04 | −2.46 | 0 | −1.68 | −0.62 | −0.38 |
| 14 | CQD1 | −3.01 | −2.78 | 0 | −0.45 | −0.14 | −0.17 |
| 15 | CQD8 | −2.93 | −3.24 | 0 | −0.02 | −0.12 | −0.32 |
| 16 | CQD2 | −2.9 | −2.84 | 0 | 0 | −0.14 | −0.32 |
| 17 | CQD11 | −2.78 | −2.83 | 0 | 0 | −0.26 | −0.18 |
| 18 | CQD3 | −2.73 | −2.75 | 0 | 0 | −0.24 | −0.37 |
LipophilicEvdW: Lipophilic term derived from hydrophobic grid potential and fraction of the total protein-ligand vdW energy
PhobEnHB: Reward for hydrophobicallypacked H-bond
HBond: ChemScore H-bond pair term
Electro: Electrostatic rewards
LowMW: Reward for ligands with low
Figure 4.
Ligand-receptor interaction diagram of chloroquine derivative molecules (CQD14, CQD15 and CQD16) with the SARS-CoV-2 (PDB ID: 6LU7).
Figure 5.
Molecular docking pose of Chloroquine, Hydroxychloroquine, CQD14, CQD15 and CQD16 with the SARS-CoV-2 (PDB ID: 6LU7). (a) Complete image of ligands binding with the protein (b) Magnified image of ligands binding at the active site.
The comparative analysis of selected molecules (CQD15, CQD14 and CQD16) with Chloroquine and Hydroxychloroquine were studied. Chloroquine and Hydroxychloroquine are showing a docking score of -3.27 and -3.48 respectively (Table 2). Chloroquine shows one pi-pi stacking with hydrophobic region (PHE-294) along with a good range of solvent exposure where as Hydroxychloroquine shows two H-bond interactions, one is due to H-bond acceptor and another is due to H-bond donor group present in the active site of SARS-CoV-2. The selected molecules (CQD15, CQD14 and CQD16) were showing better docking scores (-6.17, −5.14 and -4.19 respectively) as well as better ligand-receptor interactions in comparison to chloroquine and hydroxychloroquine (Figure 6).
Figure 6.
Ligand-receptor interaction diagram of chloroquine and hydroxychloroquine with the SARS-CoV-2(PDB ID: 6LU7).
Further, the designed ligands were also docked with coronavirus SARS Spike Glycoprotein - human ACE2 complex (PDB ID: 6CS2) (Kirchdoerfer et al., 2018) to study the impact of ligands on SARS Spike Glycoprotein - human ACE2 complex. The docking study of “ligand and SARS Spike Glycoprotein - human ACE2 complex” shows similar result like “ligand and COVID-19 main protease”. The ligands CQD-15, CQD-14 and CQD-16 showing docking score of -6.47, −5.48, and 5.35 respectably where as Choloroquine and Hydroxychloroquine shows -3.56 and -3.6 respectively (Table 3). The detailed interaction diagrams are shown in Figure S2a–S2e. Docking study is also performed with SARS-CoV-2 main protease (PDB ID: 7BUY) (Zhao et al., 2020) to validate the ligand-main protease interactions. The study also concludes with same ligands CQD-16, CQD-15 and CQD-14 which shows better docking score and ligand-receptor interactions in comparison to Chloroquine and Hydroxychloroquine which follows the trend of ligand-6LU7 docking study. The detailed interactions and docking score are shown in Figure S3a–S3e and Table S1 respectively.
Table 3.
Docking score and functional parameters of all 18 molecules including Chloroquine and Hydroxychloroquine with coronavirus SARS Spike Glycoprotein - human ACE2 complex (PDB ID: 6CS2).
| Ligand | DockScore | LipophilicEvdW | PhobEnHB | HBond | Electro | LowMW |
|---|---|---|---|---|---|---|
| CQD-15 | −6.47 | −3.77 | −1.37 | −1.25 | −0.55 | −0.12 |
| CQD-14 | −5.48 | −3.74 | −1.5 | −1.28 | −0.38 | −0.13 |
| CQD-16 | −5.35 | −3.32 | −1.5 | −2.2 | −0.81 | −0.12 |
| CQD-11 | −5.33 | −4.56 | −0.75 | −0.7 | −0.38 | −0.18 |
| CQD-12 | −4.91 | −3.21 | −1.5 | −0.94 | −0.44 | −0.17 |
| CQD-13 | −4.88 | −3.93 | 0 | −1.2 | −0.39 | −0.18 |
| CQD-6 | −4.78 | −2.79 | −1.5 | −0.63 | −0.34 | −0.38 |
| CQD-7 | −4.68 | −2.69 | −1.5 | −0.7 | −0.22 | −0.17 |
| CQD-5 | −4.44 | −3.51 | −0.75 | −0.7 | −0.49 | −0.28 |
| CQD-4 | −4.06 | −3.38 | 0 | −0.7 | −0.41 | −0.38 |
| CQD-9 | −3.9 | −3.38 | 0 | −0.65 | −0.32 | −0.37 |
| CQD-10 | −3.7 | −3.44 | 0 | −0.11 | −0.25 | −0.38 |
| CQD-2 | −3.6 | −3.53 | 0 | 0 | −0.14 | −0.32 |
| Hydroxychloroquine | −3.6 | −3.28 | 0 | 0 | −0.19 | −0.38 |
| Chloroquine | −3.56 | −3.48 | 0 | 0 | −0.16 | −0.43 |
| CQD-1 | −3.54 | −3.62 | 0 | 0 | −0.07 | −0.17 |
| CQD-3 | −3.52 | −3.62 | 0 | 0 | −0.14 | −0.37 |
| CQD-8 | −3.51 | −3.81 | 0 | 0 | −0.05 | −0.32 |
MM-GBSA study
All 18 molecules were subjected to ligand-receptor binding energy, MM-GBSA analysis. The MM-GBSA analysis computed the binding free energy of docked ligand-receptor complex which confirms the stability of the ligand after binding to the active site of the enzyme. The generated ‘MM-GBSA_Dg_Bind’ energy by using the MM-GBSA module shows the energy difference between prime energy (optimized ligand-receptor complex) and the combined energy of optimized free ligand and optimized free receptor. The MM-GBSA_dG_Bind energies for all 18 compounds are shown in Table 4. MM-GBSA study concludes that all the molecules except CQD1 and CQD5 shows better binding energy in comparison to Choloroquine and Hydroxychloroquine.
Table 4.
MM-GBSA_dG_Bindvalues of 18 molecules in (kcal/mol) obtained through MM-GBSA analysis.
| Ligand | r_psp_MMGBSA_dG_Bind |
|---|---|
| CQD15 | −47.74711519 |
| CQD14 | −58.40628355 |
| CQD16 | −41.70415476 |
| CQD6 | −51.56851776 |
| CQD5 | −39.24253856 |
| Hydroxychloroquine | −40.34126469 |
| CQD13 | −52.76079182 |
| CQD12 | −49.54062105 |
| CQD9 | −53.07817314 |
| CQD10 | −43.20765873 |
| Chloroquine | −40.46204749 |
| CQD7 | −55.4049401 |
| CQD4 | −58.3743558 |
| CQD1 | −32.47031282 |
| CQD8 | −52.27239411 |
| CQD2 | −51.74337235 |
| CQD11 | −51.76783519 |
| CQD3 | −48.48654391 |
ADME analysis
The drug-likeness properties of the molecules studied by using well-known ADME analysis i.e. Absorption (A), Distribution (D), Metabolism (M) and Excretion (E) which explains the disposition of pharmaceutical compound inside an organism and therefore, influences the pharmacological activity of it. The screening was performed based on a violation of ‘Lipinski’s rule of five’, QPPCaco and QPlogS. In the ADME property analysis out of 18 molecules, 3 were showing violation of ‘Lipinski’s rule of five’ and (QPlogS) (shown in the bold in Table 5) whereas remaining 15 molecules were following the standard parameters. All the molecules show percentage of oral absorption more than 70%. ADME property analysis concludes that all molecules except CQD1, CQD7 and CQD11 show drug-likeness properties.
Table 5.
ADME properties of all 18 ligands to determine their ‘drug-likeness’.
| Ligand | mol_MW | donorHB | accptHB | QPlogPo/w | RuleOfFive | QPlogS | %Human Oral Absorption |
|---|---|---|---|---|---|---|---|
| CQD15 | 413.946 | 3 | 6.45 | 4.094 | 0 | −4.705 | 91.032 |
| CQD14 | 411.973 | 2 | 4.75 | 4.998 | 0 | −4.7 | 100 |
| CQD13 | 396.962 | 3 | 5 | 4.313 | 0 | −3.387 | 92.144 |
| CQD12 | 397.947 | 2 | 5.7 | 4.235 | 0 | −3.367 | 100 |
| CQD11 | 395.974 | 1 | 4 | 5.569 | 1 | −4.44 | 100 |
| CQD10 | 334.891 | 3 | 5 | 2.496 | 0 | −0.975 | 78.096 |
| CQD9 | 337.867 | 1 | 4 | 4.661 | 0 | −4.601 | 100 |
| CQD8 | 354.322 | 1 | 4 | 4.991 | 0 | −4.997 | 100 |
| CQD7 | 398.773 | 1 | 4 | 5.066 | 1 | −5.083 | 100 |
| CQD6 | 335.876 | 2 | 5.7 | 3.352 | 0 | −3.13 | 95.059 |
| CQD5 | 364.874 | 1 | 6 | 3.505 | 0 | −3.524 | 92.172 |
| CQD4 | 334.891 | 3 | 5 | 2.968 | 0 | −2.604 | 79.338 |
| CQD3 | 337.867 | 1 | 4 | 4.777 | 0 | −4.824 | 100 |
| CQD2 | 354.322 | 1 | 4 | 4.983 | 0 | −4.944 | 100 |
| CQD1 | 398.773 | 1 | 4 | 5.047 | 1 | −4.798 | 100 |
| Hydroxy-chloroquine | 335.876 | 2 | 5.7 | 3.329 | 0 | −3.335 | 93.491 |
| Chloroquine | 319.876 | 1 | 4 | 4.559 | 0 | −4.582 | 100 |
| CQD16 | 412.961 | 4 | 5.75 | 3.619 | 0 | −3.619 | 76.602 |
Molecular dynamics simulations
Protein RMSD
MD simulations were performed to study the physical movements of atoms & molecules and the dynamic evolution for the best complex (6LU7-CQD15). The RMSD is a quantitative parameter to estimate the stability of the protein system (Figure 7). The protein RMSD trajectory of SARS-CoV-2 (6LU7) shows a fluctuation up to 5 ns simulation time then gradually tends to equilibrium (Figure 7). The RMSD average value of SARS-CoV-2 (6LU7) after reaching equilibrium was 1.6 Å (Figure 7) as for the small globular protein the deviation within 1-3 Å is acceptable. Based on the stability, 6LU7-CQD15 was considered for further study like Protein-Ligand contact study (Histograms and Graph).
Figure 7.
The Protein RMSD trajectory of the (CQD15-6LU7) complex during the 100 ns simulation.
Ligand properties
Ligand property study includes Ligand RMSD, Radius of Gyration (rGyr), Molecular Surface Area (MolSA), Solvent Accessible Surface Area (SASA) and Polar Surface Area (PSA). The root mean squre deviation (RMSD) of the ligand fluctuates up to 40 ns simulation and then gradually comes to equilibrium. The ligand is showing range of RMSD around 1 to 3 Å and the equilibrium around 2.5 Å (Figure 8). The rGyr is the radial distance to a point which would have a moment of inertia the same as the body’s (ligand) actual distribution of mass, if the total mass of the body (ligand) were concentrated. rGyr of the ligand also showing a heavy fluctuate up to 65 ns simulation and then gradually comes to equilibrium. The ligand is showing range of rGyr around 4 to 6.4 Å and the equilibrium around 5.5 Å (Figure 8). MolSA is surface calculation with 1.4 Å probe radius. This value is equivalent to a van der Waals surface area. The MolSA of the ligand is constant throughout the simulation but there is some fluctuation in 42 ns to 65 ns simulation period. The ligand is showing range of MolSA around 360 to 400 Å2 and the equilibrium around 400 Å2 (Figure 8). SASA is surface area of a molecule accessible by a water molecule. SASA of the ligand also showing a heavy fluctuate up to 23 ns simulation and then gradually comes to equilibrium. The ligand is showing range of SASA around 300 to 600 Å2 and the equilibrium around 300 Å2 (Figure 8). PSA is solvent accessible surface area in a molecule contributed only by oxygen and nitrogen atoms. The PSA of the ligand is constant throughout the simulation but there is some fluctuation in 41 ns to 68 ns simulation period. The ligand is showing range of PSA around 80 to 120 Å2 and the equilibrium around 120 Å2 (Figure 8). All the ligand properties are showing some fluctuation in the initial simulation period or intermediate simulation period but gradually come to equilibrium at the end of the simulation which conforms that the stability of the ligand in the active site of the protein.
Figure 8.
The ligand property trajectory of the (CQD15-6LU7) complex during the 100 ns simulation.
Protein-ligand contacts
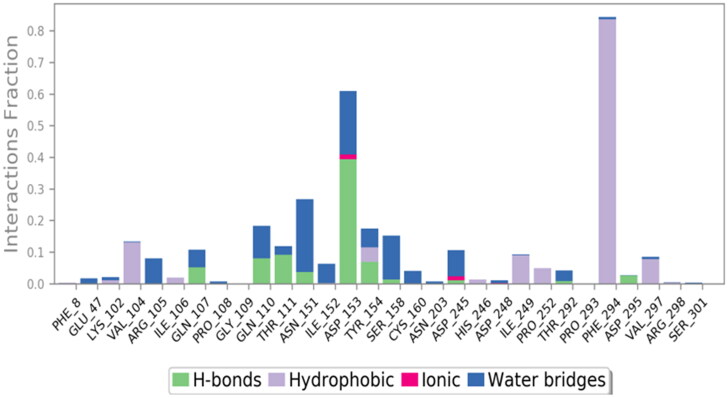
The protein-ligand contacts of the stable ligand-receptor complex (CQD15-6LU7) was studied by using the protein-ligand contact histograms (Figure 9) (Dariya & Nagaraju, 2020). Protein-ligand interactions (or ‘contacts’) are categorized into four types: hydrogen honds, hydrophobic, ionic and water bridge. Hydrogen bonds play a significant role in ligand binding. Consideration of hydrogen-bonding properties for drug design is important because of their strong influence on drug specificity, metabolization and adsorption. Hydrogen bonds between a protein and a ligand can be further broken down into four sub-types: backbone acceptor, backbone donor, side-chain acceptor and side-chain donor. The geometric criteria for protein-ligand hydrogen bond is of 2.5 Å between the donor and acceptor atoms (D—H···A), a donor angle of ≥120° between the donor-hydrogen-acceptor atoms (D—H···A) and an acceptor angle of ≥90° between the hydrogen-acceptor-bonded_atom (H···A—X). The histogram shows the active site amino acids (mainly GLN-107, GLN-110, THR-111, ASN-151, ASP-153, TYR-154, ASP-295) interacting through hydrogen bonding (Figure 9). Hydrophobic contacts fall into three subtypes like π-cation, π-π and other non-specific interactions. These types of interactions involve a hydrophobic amino acid and an aromatic or aliphatic group on the ligand. Some hydrophobic amino acids like VAL-104, ILE-106, TYR-154, ILE-249, PRO-252, PHE-294 and VAL-297 showing hydrophobic interactions with the ligand (Figure 9). Ionicinteractions or polar interactions, are between two oppositely charged atoms that are within 3.7 Å of each other and do not involve a hydrogen bond. All ionic interactions are broken down into two subtypes: those mediated by a protein backbone or side chains. ASP-153 and ASP-245 are showing minimal ionic interaction with the ligand (Figure 9). Water bridges are hydrogen-bonded protein-ligand interactions mediated by a water molecule. The hydrogen-bond geometry is slightly relaxed from the standard hydrogen bond definition. The current geometric criteria for a protein-water or water-ligand hydrogen bonds are: a distance of 2.8 Å between the donor and acceptor atoms (D—H···A); a donor angle of ≥110° between the donor-hydrogen-acceptor atoms (D—H···A); and an acceptor angle of ≥90° between the hydrogen-acceptor-bonded_atom (H···A—X). Almost all the major interacting amino acids are showing water bridges (Figure 9). In this protein-ligand contact histograms some amino acids were showing highly effective interactions like ‘ASP-153 and PHE-294’ having 62% and 83% time interactions in 6LU7-CQD15 complex of 100 ns simulation.
Figure 9.
The histogram of protein-ligand contact over the course of the trajectory.
A timeline representation of the interactions and contacts (Hydrogen bonds, Hydrophobic, Ionic and Water bridges) summarized in the ligand-receptor interaction (histogram) study analysed in the following two panels in Figure 10(a, b). The top panel shows the total number of specific contacts the protein makes with the ligand in each and every trajectory frame. The number of contact varies zero to nine over the course of the trajectory (Figure 10(a)). The contribution of amino acids in each trajectory frame of 100 ns MD simulation was studied from ligand-protein interaction (bottom panel) (Figure 10(b)). The bottom panel shows, which amino acid residues interact with the ligand in each trajectory frame. Some residues make more than one specific contact with the ligand in a particular trajectory frame, which is represented by a darker shade of orange, according to the scale to the bellow the plot. The 6LU7-CQD15 receptor-ligand complex shows two deep bands (PHE-294 and ASP-153 row), which explains that the above amino acid have more interactions with the ligands in almost all possible orientations (geometry) which is exactly similar as histogram results.
Figure 10.
(a) Total number of contacts/interaction in each trajectory frame of 6LU7-CQD15 complex. (b) Interaction shown by the active site amino acids in each trajectory frame of 6LU7-CQD15 complex.
Conclusion
A series of computational approaches used to identify more effective drug candidate against SARS-CoV-2. The pharmacophore modelling, molecular docking, MM_GBSA study and ADME property analysis combinedly concluded with 3 ligands (CQD15, CQD14 and CQD16) which have good docking score, ligand-receptor interactions, pharmacophore-based structural features and drug likeness property in comparison to chloroquine and hydroxychloroquine. The ligand-receptor MD simulation study validates the molecular docking study by exploring the protein stability (RMSD), various ligand property and protein-ligand contacts. Further, in vitro analysis followed by its in vivo testing may help in proving CQD15 ligand as a better inhibitor of SARS-CoV-2. The whole study concludes that derivatives of chloroquine may play a prominent role for the treatment of COVID-19.
Supplementary Material
Acknowledgments
Mr. Satyajit Beura is grateful to MHRD for fellowship. Authors also thank to Mr. Vinod Deveraji (Application Scientist from Schrodinger, Bangalore) for their technical assistance.
Disclosure statement
The authors declare no competing financial interest.
References
- Aanouz I., Belhassan A., El-Khatabi K., & Lakhlifi T. (2020). Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelli I., Hassani F., Brikci S. B., & Ghalem S. (2020). Insilico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoidesverticillata components harvested from western Algeria. Journal of Biomolecular Structure andDynamics, [Epub ahead of print]. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeoye A. O., Oso B. J., Olaoye I. F., & Tijjani H. (2020). Repurposing of chloroquine and some clinically approvedantiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1765876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M. L., Andres E. L., Sims A. C., Graham R. L., Sheahan T. P., Lu X., Smith E. C., Case J. B., Feng J. Y., Jordan R., Ray A. S., Cihlar T., Siegel D., Mackman R. L., Clarke M. O., Baric R. S., & Denison M. R. (2018). Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio, 9(2), e00221–18. 10.1128/mBio.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khafaji K., Al-DuhaidahawiL D., & Tok T. T. (2020). Using Integrated Computational Approaches to Identify Safe and Rapid Treatment for SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1764392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsden G. W. (2005). Anti-inflammatory effects of macrolides-an underappreciated benefit in the treatment of community-acquired respiratory tract infections and chronic inflammatory pulmonary conditions? The Journal of Antimicrobial Chemotherapy, 55(1), 10–21. 10.1093/jac/dkh519 [DOI] [PubMed] [Google Scholar]
- Assistance Publique – Hopitaux de Paris (2020). Cohort multiple randomized controlled trials open label of immune modulatory drugs and other treatments in COVID-19 patients – sarilumab trial – CORIMUNO-19-SARI (CORIMUNO-SARI) Retrieved April 2, 2020, from https://clinicaltrials.gov/ct2/show/NCT04324073?term=sarilumab&draw=3&rank=2
- Beigelman A., Mikols C. L., Gunsten S. P., Cannon C. L., Brody S. L., & Walter M. J. (2010). Azithromycin attenuates airway inflammation in a mouse model of viral bronchiolitis. Respiratory Research, 11(1), 90 10.1186/1465-9921-11-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijing Chao Yang Hospital (2020). Clinical trial of favipiravir tablets combine with chloroquine phosphate in treatment of novel coronavirus pneumonia Retrieved April 2, 2020, from https://clinicaltrials.gov/ct2/show/NCT04319900?term=favipiravir&draw=2&rank=9
- Beura S., & Chetti P. (2020). Identification of potential human COX-2 inhibitors using computational modeling and molecular dynamics simulations. Journal of Molecular Structure, 1216, 128271 10.1016/j.molstruc.2020.128271 [DOI] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against itstreatment. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC ). (2020). https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidancemanagement-patients.html.
- Chu C. M., Cheng V. C. C., & Hung I. F. N. (2004). Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax, 59(3), 252–256. 10.1136/thorax.2003.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Rolain J.-M., Lagier J.-C., Brouqui P., & Raoult D. (2020). Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. International Journal of Antimicrobial Agents, 55(4), 105932 [Epub ahead of print]. 10.1016/j.ijantimicag.2020.105932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CytoDyn (2020). Press release Retrieved April 2, 2020, from https://www.cytodyn.com/newsroom/press-releases/detail/405/treatment-with-cytodynsleronlimab-indicates-significant
- Dariya B., & Nagaraju G. P. (2020). Understanding novel COVID-19: Its impact on organ failure and risk assessment for diabetic and cancer patients. Cytokine and Growth Factor Reviews, [Epub ahead of print]. 10.1016/j.cytogfr.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Sarmah S., Lyndem S., & Roy A. S. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., … Yang X. (2020). Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proceedings of the National Academy of Sciences of the United States of America, 117(17), 9490–9496. 10.1073/pnas.2004168117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). Natural products may interfere with SARSCoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). SARS-CoV-2 RNA dependent RNA polymerase(RdRp) targeting: An insilico perspective. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An insilico perspective. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): insilico screening of known drugs againstcoronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure andDynamics, [Epub ahead of print]. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., & Guklani H. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidateagainst COVID-19: An insilico study. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon D., Owen C. D., Douangamath A., & Lukacik P. (2020). PanDDA analysis group deposition - Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434899.[To be published]. 10.2210/pdb5REB/pdb [DOI]
- Ganjoo A., Tripathi A., & Chetti P. (2019). Structural assessment and identification of 11β-hydroxysteroid dehydrogenase type 1 inhibitors. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2019.1688193 [DOI] [PubMed] [Google Scholar]
- Gao J., Tian Z., & Yang X. (2020). Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience Trends, 14(1), 72–73. 10.5582/bst.2020.01047 [DOI] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Hoang V. T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V. E., Dupont H. T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., & Raoult D. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. International Journal of Antimicrobial Agents, [Epub ahead of print]. 10.1016/j.ijantimicag.2020 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gorbalenya A. E., Baker S. C., Baric R. S., & de Groot R. J. (2020). Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. bioRxiv, [To be published] 10.1101/2020.02.07.937862 [DOI] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi G. A., Ogunro O. B., Adegunloye A. P., & Ogunyemi O. M. (2020). Potential Inhibitors of Coronavirus 3-Chymotrypsin-LikeProtease (3CLpro): An insilico screening of Alkaloids and Terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., & Qadir F. A. (2020). A review on thecleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., & Uddin N. (2020). A molecular modeling approach to identify effective antiviral phytochemicals againstthe main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., & Deng Y. (2020). Structure of M pro From COVID-19 virus and discovery of its inhibitors. Nature, [Epub ahead of print]. 10.1038/s41586-020-2223-y [DOI] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., & Shankar S. S. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G. M., & Jain M. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identifypromising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal ofBiomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., & Uddin R. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 viaintegrated computational approach. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R. N., Wang N., Pallesen J., Wrapp D., Turner H. L., Cottrell C. A., Corbett K. S., Graham B. S., McLellan J. S., & Ward A. B. (2018). Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Scientific Reports, 8(1), 15701–15701. 10.1038/s41598-018-34171-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Kumari K., Jayaraj A., Kumar V., et al. (2020). Understandingthe binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MDsimulations at different temperatures. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Raja K., Sebastine I., Joseph J., et al. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silicoapproach. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Galo N., Terrazas-López M., Martínez-Martínez A., & Díaz-Sánchez Á. G. (2020). FDA-approved thiol-reacting drugs that potentially bind into theSARS-CoV-2 main protease, essential for viral replication. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1764393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K., Kim Y., Jedrzejczak R., Maltseva N., Endres M., Mececar A., & Joachimiak A. (2020). Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose. bioRxiv [To be published]. 10.2210/pdb6W02/pdb [DOI] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & GromihaM M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir andritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structureand Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Owusu M., Annan A., Corman V. M., Larbi R., Anti P., Drexler J. F., Agbenyega O., Adu-Sarkodie Y., & Drosten C. (2014). Human coronaviruses associated with upper respiratory tract infections in three rural areas of Ghana. PLoS One, 9(7), e99782 10.1371/journal.pone.0099782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., MurtyU S. N., et al. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal ofBiomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastick K. A., Okafor E. C., Wang F., Lofgren S. M., Skipper C. P., Nicol M. R., Pullen M. F., Rajasingham R., McDonald E. G., Lee T. C., Schwartz I. S., Kelly L. E., Lother S. A., Mitjà O., Letang E., Abassi M., & Boulware D. R. (2020). Review: Hydroxychloroquine and Chloroquine for treatment of SARS-CoV-2 (COVID-19). Open Forum Infectious Diseases, 7(4), 1–22. 10.1093/ofid/ofaa130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peking University First Hospital (2020). Favipiravir combined with tocilizumab in the treatment of Corona Virus Disease 2019 Retrieved March 24, 2020, from https://clinicaltrials.gov/ct2/show/NCT04310228?cond=Coronavirus&intr=Tocilizumab&draw=2&rank=1
- Perlman S. (2020). Another decade, another Coronavirus. The New England Journal of Medicine, 382(8), 760–762. 10.1056/NEJMe2001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., & Netland J. (2009). Coronaviruses post-SARS: Update on replication and pathogenesis. Nature reviews. Nature Reviews. Microbiology, 7(6), 439–450. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawel H. M., Meidtner K., & Kroll J. (2005). Binding of selected phenolic compounds to proteins. Journal of Agricultural and Food Chemistry, 53(10), 4228–4235. 10.1021/jf0480290 [DOI] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., et al. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger Release 2020-1 (2020). Glide, phase, ligprep. Schrodinger, LLC. [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., et al. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh D. K., Selvaraj C., Singh S. K., et al. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal ofBiomolecular Structure and Dynamics, [Epub ahead of print]. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., & Xiao G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO ). (2020). Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected.
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., & Zhang Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Han M., Li T., et al. (2020). Effect treatment of severe COVID-19 patients with tocilizumab. ChinaXiv.20200300026.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T. ‐T., Qian J. ‐D., Zhu W. ‐Y., Wang Y., & Wang G. ‐Q. (2020). A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. Journal of Medical Virology, 92(6), 556–563. [Epub ahead of print]. 10.1002/jmv.25729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., & Liu D. (2020). In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clinical Infectious Diseases, [Epub ahead of print]. 10.1093/cid/ciaa237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky M. I., & Kuhn L. A. (2005). Side-chain flexibility in protein-ligand binding: The minimal rotation hypothesis. Protein Science: A Publication of the Protein Society, 14(4), 1104–1114. 10.1110/ps.041153605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368, 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet, [Epub ahead of print] 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., … Shi Z.-L., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., & Cheng F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6, 14 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang B., Jin Z., Liu X., Yang H., & Rao Z. (2020). The crystal structure of COVID-19 main protease in complex with carmofur. Protein DataBank; [To be published]. 10.2210/pdb7buy/pdb [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.