Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a novel corona virus that causes corona virus disease 2019 (COVID-19). The COVID-19 rapidly spread across the nations with high mortality rate even as very little is known to contain the virus at present. In the current study, we report novel natural metabolites namely, ursolic acid, carvacrol and oleanolic acid as the potential inhibitors against main protease (Mpro) of COVID-19 by using integrated molecular modeling approaches. From a combination of molecular docking and molecular dynamic (MD) simulations, we found three ligands bound to protease during 50 ns of MD simulations. Furthermore, the molecular mechanic/generalized/Born/Poisson-Boltzmann surface area (MM/G/P/BSA) free energy calculations showed that these chemical molecules have stable and favourable energies causing strong binding with binding site of Mpro protein. All these three molecules, namely, ursolic acid, carvacrol and oleanolic acid, have passed the ADME (Absorption, Distribution, Metabolism, and Excretion) property as well as Lipinski’s rule of five. The study provides a basic foundation and suggests that the three phytochemicals, viz. ursolic acid, carvacrol and oleanolic acid could serve as potential inhibitors in regulating the Mpro protein’s function and controlling viral replication.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, protease, ursolic acid, carvacrol, oleanolic acid
1. Introduction
Coronavirus disease (COVID-19) is a respiratory infectious disease caused by a novel virus strain, SARS-CoV-2 (Boopathi et al., 2020; Hemida & Ba Abduallah, 2020; Salata et al., 2019; Sarma et al., 2020; Seah & Agarwal, 2020; Su et al., 2016). In the past two decades, two other coronavriruses have caused global outbreaks, namely SARS-CoV (2002–2003) and Middle East respiratory syndrome coronavirus (2012–present) (de Wit et al., 2016; Gupta et al., 2020; Wang et al., 2020; Wu et al., 2020; Yuan et al., 2020). The Coronavirus Study Group (CSG) taxonomists working under the aegis of International Committee on Taxonomy of Viruses (ICTV) coined the nomenclature of SARS-COV-2 based on its 82% identity to the SARS coronavirus (SARS-CoV) genome (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020; Hasan et al., 2020). Whole genome functional analysis revealed that both viruses phylogenetically belong to clade b of the genus Betacoronavirus (Chan et al., 2020; Muralidharan et al., 2020; WHO, 2020). The very first case of the novel COVID-19 was originally reported in Wuhan, Hubei Province, China, and has quickly spread over the 212 countries of the world (Elmezayen et al., 2020; Enayatkhani et al., 2020; Mackenzie & Smith, 2020; Xu et al., 2020). This contagious disease has led to over 4, 098, 018 confirmed cases and 283,271fatalities as on May 12, 2020; https://covid19.who.int/). The number of cases across the globe is increasing abruptly, and so far, there is no standard drug has proved to be effective for COVID-19 disease that have high mortality rate in immunocompromised patients (Aanouz et al., 2020; Bhatraju et al., 2020; Weiss & Murdoch, 2020). On 30 January, this respiratory infectious disease has been declared as a Public Health Emergency of International Concern by the World Health Organization (WHO) (Mackenzie & Smith, 2020; https://www.who.int/).
The SARS-CoV-2 genome is about 30 kb in size (29, 903 nucleotides) and encodes as many as 14 open reading frames (ORFs) (Chen et al., 2020; Elfiky & Azzam, 2020; Gorden et al., 2020; Woo et al., 2005). At the 5′ end of the viral genome, Orf1a/Orf1ab encodes several proteins, which are auto-proteolytically processed into 16 non-structural proteins (Nsp1-16) and form the replicase/transcriptase complex (RTC). Whereas, 3′ endencode structural viral proteins; spike (S), membrane (M), envelope (E) and nucleocapsid (N), and nine putative accessory factors. On the other hand, protease enzyme called the Mpro or also 3CLpro, play a crucial role in the life cycle of COVID-19 replication and maturation (Joshi et al., 2020; Khan et al., 2020; Pant et al., 2020; Wu et al., 2020). It has been demonstrated that no part of COVID-19 is more exposed than its main protease, as belongs to non-structural class proteins of the viral genome (Enmozhi et al., 2020; Zhang et al., 2020). Zhang et al., 2020 determined the three-dimensional crystal structure, at 1.75 Å resolution, of the Mpro of SARS-CoV-2 using x-ray crystallography.
Like other coronaviruses, the 3D structure of 3CLpro of SARS-COV-2 possesses three functional domains (Jin et al., 2020; Khan et al., 2020). The length of domain I, II, and III ranges from 8-101, 102-184, 201-306 amino acid residues, respectively. Among them, domain I and II are essentially beta-barrels and similar to the chymotrypsin. While, structural composition of domain III mainly consists of alpha-helices (Anand et al., 2005; Jin et al., 2020; Lu et al., 2006; Yang et al., 2006), protease has been characterized as one of the potential drug targets among coronaviruses (Anand et al., 2003). In this sense, many recent studies suggested that the selection of various FDA-approved antiviral compounds may yield promising results against COVID-19 infection (Chang et al., 2016; Chang et al., 2020; Contini, 2020; Gonzalez-Paz et al., 2020; Khan et al., 2020; Elfiky, 2020a, 2020b; Islam et al., 2020; Sinha et al., 2020; Wahedi et al., 2020; Umesh et al., 2020; Das et al., 2020; Abdelli et al., 2020). Chang et al., 2020 reported that chloroquine, an older antimalarial drug, has ability to inhibit the viral 3CL-protease activity. As crystal structure of protease provides a foundation for design of improved α-ketoamide inhibitors (Zhang et al., 2020), the capability of chloroquine to inhibit the protease activity its uses has been recommended in different countries including China, USA and India for the treatment of COVID-19 (Devaux et al., 2020; Gonzalez-Paz et al., 2020; Wang et al., 2020). However, many studies questioned the safety and reported the severe adverse effects of chloroquine (Kaisari & Borruat, 2020; Wang et al., 2020).
Ancient Indian scriptures including Rig-Veda, Atherveda, and Charka Sanhita demonstrated abundant benefits of plants for the treatment of various human aliments (Kumar et al., 2018). Plants are a remarkable natural source of high value alkaloids, flavonoids, phenols, chalcones, coumarines, lignans, polyketides, alkanes, alkenes, alkynes, simple aromatics, peptides, terpenes, and steroids. In the current era of drug discovery, enormous medicinal properties of plants allows the researchers to exclusively use them for the discovery of drug-like natural molecules (Jee et al., 2018; Kumar et al., 2018; Panchangam et al., 2016).
Ursolic acid (3-β-3-hydroxy-urs-12-ene-28-oic-acid) and oleanolic acid (3β-hydroxyolean-12-en-28-oic acid) are pentacyclic triterpenoid compounds with a widespread occurrence throughout the plant kingdom (Pollier & Goossens, 2012; Woźniak et al., 2015). Both molecules enrich various therapeutic properties such as antibacterial, antiviral, anticancer, antioxidant and tantimycotic activity (Jesus et al., 2015). Previous in vitro studies reported that these molecules exhibit antiviral activity against rotavirus, HIV, the influenza virus, hepatitis B and C viruses (Jesus et al., 2015; Khwaza et al., 2018; Tohmé et al., 2019).
Carvacrol (2-Methyl-5-(propan-2-yl)phenol) is a monoterpenoid phenol, possesses a wide range of strong antimicrobial and antiviral activity (Gilling et al., 2014; Kamalabadi et al., 2018; Marinelli et al., 2019). It is a major constituent of essential oil of plants of Labiatate family including oregano and thyme, and has been emerged as active molecule for therapeutic purpose (Hyldgaard et al., 2012). Using in-vitro methods, antiviral effects of carvacrol has been tested and validated on herpes simplex virus type 1, retrovirus and human respiratory syncytial virus (Kamalabadi et al., 2018).
Besides the uses of various FDA-approved antiviral compounds as mentioned above, there are many in-silico studies have been performed to screen the novel phytochemical molecules as a potential inhibitors of main protease of SARS-CoV-2 or develop new drugs against COVID-19 (Adem et al., 2020; Chandel et al., 2020; Gentile et al., 2020; Gonzalez-Paz et al., 2020; Khaerunnisa et al., 2020; Khan et al., 2020; Qamar et al., 2020; Sharma & Kaur, 2020; Sun et al., 2020). But there is not a single report available for the function of phytochemicals namely, ursolic acid, carvacrol and oleanolic acid derived from the antiviral herbs for the treatment of COVID-19. With the advent of molecular modeling approaches, targeted drug design may be possible and functional proteins of SARS-CoV-2 could be targeted with natural compounds to develop an effective treatment for COVID-19.
In the present study, we have targeted the protease of SARS-CoV-2 virus using available molecular modelling based methods and studied the interactions with selected natural compounds (ursolic acid, carvacrol and oleanolic acid) by molecular docking and molecular dynamics simulations followed by molecular mechanic/generalized Born/Poisson-Boltzmann surface area (MM/G/P/BSA) validation.
2. Material and methods
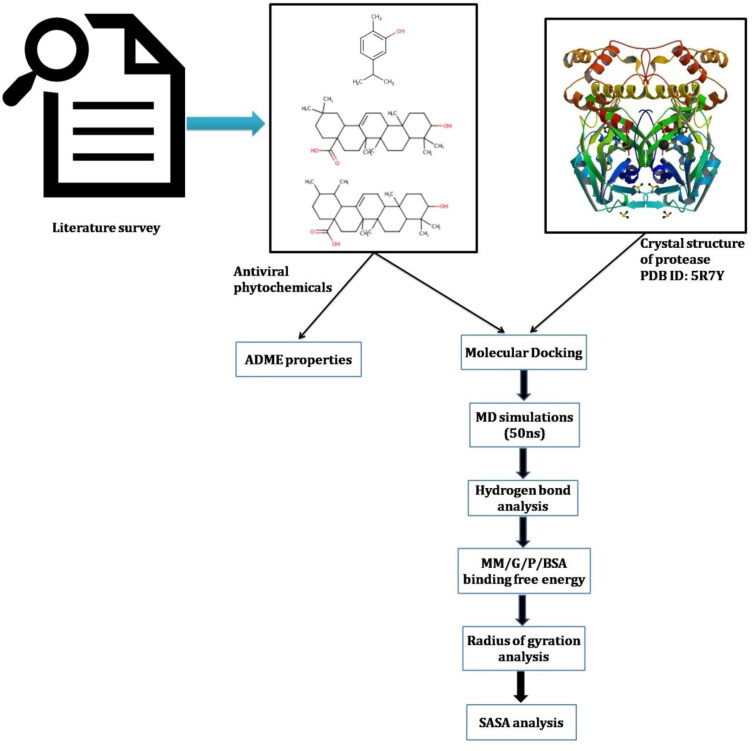
A flow chart of pipeline used in present study is summarized in Figure 1.
Figure 1.
Flowchart of pipeline used in present study to identify the phytochemical based inhibitors of Mpro.
2.1. Preparation of protease
The crystal structure of COVID-19 virus main protease (Mpro) in complex with Z45617795 (PDB ID: 5R7Y) was solved by PanDDA analysis group (https://www.rcsb.org/structure/5R7Y). This solved protein structure (PDB ID: 5R7Y) was extracted from the RCSB-Protein Data Bank (Berman et al., 2000; Burley et al., 2019). Crystal structure of Mpro have dimer in form with fragment of N-(2-phenylethyl) methanesulfonamide (Z45617795) compound provides a model for identifying potent inhibitors to target COVID-19 virus Mpro through in-silico study. The protein structure was prepared by removing water atoms, hetero atoms and adding polar hydrogen atom and kollman charges on it.
2.2. Ligand selection
Chemical structure of carvacrol (CID_10364), oleanolic acid (CID_10494) and ursolic acid (CID_64945) were extracted from PubChem database (Kim et al., 2019).
2.3. Molecular docking
In order to find out the potential drug targets, ligand based molecular docking between phytochemical compounds i.e. (carvacrol, oleanolic acid and ursolic acid) and Mpro protein were performed using AutoDock v4.2 (Morris et al., 2009). For docking experiments, the amino acid residues including Thr24, Thr26, Asn119, Phe140, Gly143, Cys145, His163, His164, Glu166, Gln189, and Thr190 were used as the active sites. Default mode parameters were selected in AutoDock during docking analysis.
2.4. Drug likeness
Absorption, distribution, metabolism, and excertion (ADME) calculations are important aspects of drug designing. All three ligand molecules were analysed based on the Lipinski’s rule of five using (Lipinski et al., 2001), Veber’s rule (Veber et al., 2002), Egan’s rule (Veber et al., 2002) and polar surface area (TPSA), and number of rotatable bonds (Daina et al., 2017), were calculated using SWISSADME tool (Daina et al., 2017).
2.5. Molecular dynamics (MD) simulations
The structural and dynamics transition at atomistic level in the Mpro of COVID-19 upon binding of small molecules were investigated by using MD simulations. MD simulations were performed using GROMOS96 43a1 force field embedded in GROMACS 5.1.1 suite on LINUX based platform (Berendsen et al., 1995). For the MD simulation, we followed the protocol described by Gajula, 2008; Gajula et al., 2016; Kumar et al., 2018. PRODRG server (Schüttelkopf & van Aalten, 2004) was used to generate the topology files of small molecules. The protein complexes were solvated in the dodecahedron box with simple point charge (SPC) waters, and 4 Na+ were added for the overall electrostatic neutrality of the system. Energy minimization of the system was performed by using the steepest descent algorithm for 50,000 iteration steps and cut-off up to 1000 kJmol−1 to reduce the steric clashes. The minimization of the system was performed at two phases for 50,000 steps, where the first phase equilibrated in two different phases for 50,000 steps. The first phase of equilibration was performed with a constant number of particles, volume, and temperature (NVT), each step 2 fs, the fallowed second phase of equilibration was performed with a constant number of particles, pressure, and temperature (NPT), the ensemble at 300 K. LINCS algorithm was utilized for covalent bond constraints in the equilibration steps. For the calculation of Lennard-Jones and Coulomb interactions, a 1.4 nm radius cut-off was used. Long-range electrostatics was calculated by using the Particle Mesh Ewald (PME) method with a Fourier grid spacing of 1.6 Å. The temperature inside the box was regulated by using V-rescale, a modified Berendsen temperature coupling method. Parrinello-Rahman pressure coupling method was utilized in NPT equilibration. The final production step of molecular dynamics simulation was carried out for 50 ns, each step of 2 fs. Trajectories were saved, and results were analyzed using XMGRACE. Root mean square deviation (RMSD) variation in protein backbone was calculated by using g RMS tool, which utilizes the least-square fitting method. Overall root mean square fluctuation (RMSF) in the atomic positions of protein C backbone was calculated by using the g rmsf tool. A rough measure of the compactness factor of protein during the course of the simulation was estimated by using the g gyrate tool of GROMACS. Gmxsasa was used for computation of the total solvent accessible surface area (SASA). Hydrogen bonds were calculated with 3.5 Å distance cut-off by using g h bond, and the distribution of intermolecular hydrogen bond lengths throughout the simulation was also analyzed.
2.6. MM/G/P/BSA binding free energy calculations
Molecular Mechanic/Poisson-Boltzmann Surface Area (MMPBSA) method was used to obtain the binding free energy of the interaction between ligand-protein complexes (Aldeghi et al., 2017). This employs ensembles derived from MD simulation. The g mmpbsa application of GROMACS module was used for the calculation of different components of the binding free energy of the Mpro and ligand complexes. Here, the binding energy is an average of three energetic terms, i.e. potential energy in the vacuum, polar-solvation energy, and non-polar solvation energy, respectively. The snapshots at every 100ps between 40 and 50 ns were collected, and MMPBSA was performed to predict the binding energy.
In the MMPBSA calculation, the binding free energy between a receptor and a ligand was calculated using following equations:
| (1) |
| (2) |
| (3) |
| (4) |
2.7. Computational facility details
The MD simulations and corresponding energy calculations were carried out on HP Gen7 server with 48 Core AMD processors and 32GB of RAM.
3. Results and discussion
3.1. Molecular docking
The molecular docking approach to identify the drug targets has become one of the most popular methods for ligand-based computer-aided drug discovery (LB-CADD). In current era, with this approach, big data of drug libraries can be analyzed and annotated quickly and immense amount of energy, time, and costs related to CADD can be saved (Wadood et al., 2013; Yu & MacKerell, 2017). Currently, there are no effective treatments available to cure COVID-19 virus, and hence, identification of potential drug targets is urgently needed.
We used molecular modeling approach with molecular docking and MD simulation to identify potential phytochemcials active against the Mpro protein of COVID-19. These screened natural compounds may pave way for the development of drugs against COVID-19. On the basis of AutoDock binding affinity, phytochemicals namely, carvacrol, oleanolic acid and ursolic acid have shown satisfactory interactions with active site residues. These compounds have been found to have binding energy of −4.0 kcal/mol, −6.0 kcal/mol, −5.9 kcal/mol, respectively. The docked ligand molecules with protease are shown in Figure 2. Table 1 represented the 2D and 3D structures of docked ligand molecules along with AutoDock score. Hydrogen bonds play an important role in determining its specificity and affinity within protein-ligand complexes (Sakkiah et al., 2012).
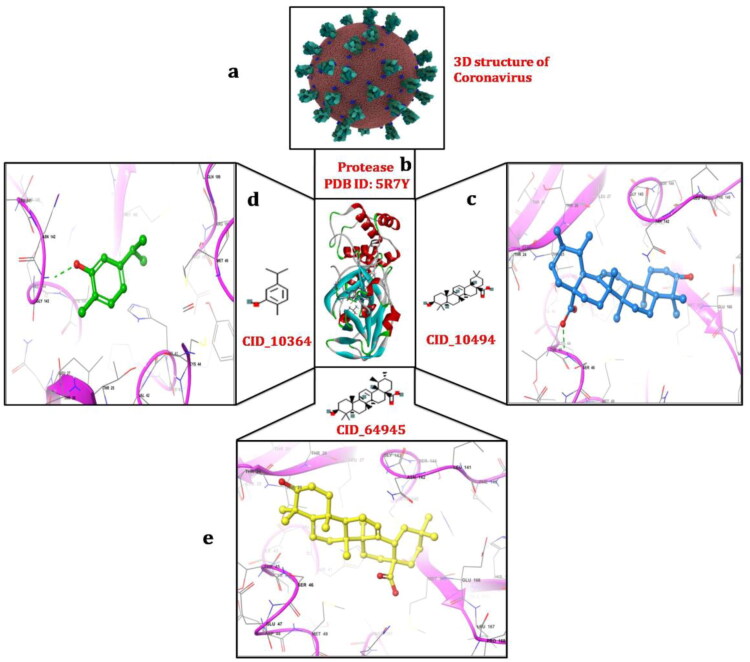
Figure 2.
Schematic representation of molecular docking between Mpro and phytochemicals; (a) 3D structure of coronavirus derived from the RCSB-Protein Data Bank (PDB); (b) crystal structure of main protease of COVID-19 obtained from the PDB with PDB ID:5R7Y (c)interaction between Mpro and oleanolic acid with −6.0 kcal/mol docking energy; (d) interaction between Mpro and carvacrol with docking energy −4.0 kcal/mol; (e) interaction between Mpro and ursolic acid with −5.9 kcal/mol docking energy. Interactions were visualized using maestro and discovery studio programs.
Table 1.
Molecular docking results of carvacrol, oleanolic acid and ursolic acid.
| Chemical Structure |
||||
|---|---|---|---|---|
| Compound Name | PubChem ID | 2D | 3D | Docking Energy (AutoDock) |
| Carvacrol | CID_10364 | 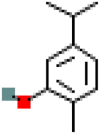 |
 |
−4.0 kcal/mol |
| Oleanolic acid | CID_10494 |  |
 |
−6.0 kcal/mol |
| Ursolic acid | CID_64945 |  |
 |
−5.9 kcal/mol |
In order to evaluate the affinity of the ligand molecules with the Mpro protein, we also studied the residues of protease involved in forming hydrogen bonds with the phytochemicals and the strength of these bonds. Amino acids residues participated in hydrophobic interactions between protease and ligand molecules were also investigated. Molecular interactions (hydrogen bonds and hydrophobic interactions) play a key role in giving shape and stabilizing the docking complexes (Wade & Goodford, 1989).
With AutoDock binding energy −4.0 kcal/mol, Gly143 amino acid was found to be involved in forming hydrogen bond with carvacrol (Figure 2). In addition to this, six other residues (Leu27, His41, Met49, Asn142, Cys145, Met165) are involved in forming hydrophobic interactions. Oleanolic acid compound make complex using AutoDock with binding energy −6.0 kcal/mol. The Gln189 amino acid formed the hydrogen bond, and two amino acids (Cys145, His163) were involved in hydrophobic interactions. Autodock binding energy of ursolic acid compound was −5.9 kcal/mol. The Ser46 amino acid at the active site of protease formed hydrogen bond. Nine amino acids (Thr24, Thr25, Thr26, Cys44, Thr45, Asn142, Gly143, Cys145, Glu166) participated in the formation of van der Waals interactions (Figure 2). Carvacrol having less binding energy as compared to oleanolic acid and ursolic acid molecules, the binding mode of interaction was found to be reasonably good. Previous studies reported a similar trend of the presence of binding pockets in main Mpro of SARS-CoV, which confirms our study (Aanouz et al., 2020; Khan et al., 2020; Li et al., 2020; Muralidharan et al., 2020). Aanouz et al. (2020) reported the β–Eudesmol molecule as a potential inhibitor of Mpro of COVID-19 based on its significant antibacterial and antiviral power well documented in literatures. However, this compound showed the low affinity interaction with Mpro and formed the hydrogen only with Thr111.
In our earlier study we have reported the ursolic acid carvacrol molecules as potential inhibitors of Small heat shock protein16.3 (sHSP16.3) of Mycobacterium tuberculosis (MTB) (Jee et al., 2018). Docking study of ursolic acid with sHSP16.3 formed two strong hydrogen bonds with Glu56 and Pro58, while carvacrol was found to be bound with only one hydrogen bond Glu92. Binding affinity of both compounds with sHSP16.3 confirms the present study results. From a biological point of view, reported phytochemicals, carvacrol, oleanolic acid and ursolic acid which are proposed as potential inhibitors of the Mpro having a significant antiviral activity with evident to bibliographical research and performed in-vitro studies (Gilling et al., 2014; Jesus et al., 2015; Kamalabadi et al., 2018; Khwaza et al., 2018; Marinelli et al., 2019; Tohmé et al., 2019).
3.2. Evaluation of drug likeness
Carvacrol, oleanolic acid and ursolic acid have respectively the following molecular weights (150.22, 456.70, 456.70) g/mol, all three molecules have molecular weight ≤ 500 g,/mol which follows the Lipinski’s rule. The carvacrol molecule has been found to have topological polar surface (TPSA) as 20.23 Å2, while oleanolic acid and ursolic acid molecules were found to have 57.53 Å2. According to the rule lowest TPSA values always produce good results; therefore, we noted that selected molecules are better behaved than the co-crystallized ligand (Daina et al., 2017; Lipinski et al., 2001). The (LogP) values of carvacrol, oleanolic acid and ursolic acid were found with the range of 2.82, 6.06, 5.94, respectively. Predicted LogP values depict that these molecules can be absorbed in the body. All three molecules present number of hydrogen bond donors: ≤5, a number of hydrogen bond acceptors ≤10 and also molar refractivity values between 48.01, 136.65, 136.91, respectively. The hydrogen bonding and molar refractivity calculations showed that these molecules validate the five Lipinski’s rule. These molecules also follow the Veber’s rule which denotes the oral bioavailability of drug molecules. Since in some CAD molecules cannot be synthesized, synthetic accessibility (SA) is a key aspect of drug designing. The carvacrol, oleanolic acid and ursolic acid molecules were found to have SA score 1, 6.08, 6.21, respectively. According to SA method molecules having score 1 is easy to synthesize it, on the other hand, score 10 represent difficulties to synthesize the molecules. All three molecules having less than 10 score, so they can be easily synthesized. The results of the ADME calculations are listed in the Table 2.
Table 2.
Results of the phytochemical molecules druglikeness properties.
| Drug Likeness Properties | Carvacrol | Oleanolic acid | Ursolic acid |
|---|---|---|---|
| Molecular weight g/mol | 150.22 | 456.70 | 456.70 |
| Concensus Log Po/w | 2.82 | 6.06 | 5.94 |
| Num. H-bond acceptors | 1 | 3 | 3 |
| Num. H-bond donors | 1 | 2 | 2 |
| Molar Refractivity | 48.01 | 136.65 | 136.91 |
| Lipinski | Yes | Yes | Yes |
| Veber | Yes | Yes | Yes |
| Bioavailability score | 0.55 | 0.56 | 0.56 |
| Synthetic accessibility (SA) | 1.00 | 6.08 | 6.21 |
| TPSA (Ų) | 20.23 | 57.53 | 57.53 |
| No of rotatable bonds | 1 | 1 | 1 |
| Solubility | 1.46e-01 | 3.45e-04 | 9.72e-04 |
3.3. MD Simulations
MD simulations are one of proven in-silico methods for obtaining dynamic data at atomic spatial resolution and picoseconds or finer temporal resolution (Benson & Daggett, 2012; Gajula et al., 2016). The main protease docked complexes with phytochemical compounds; carvacrol, ursolic acid and oleanolic acid simulation study done for 50 ns simulation period to analyze the stability of these docked phytochemical compounds in binding region of Mpro.
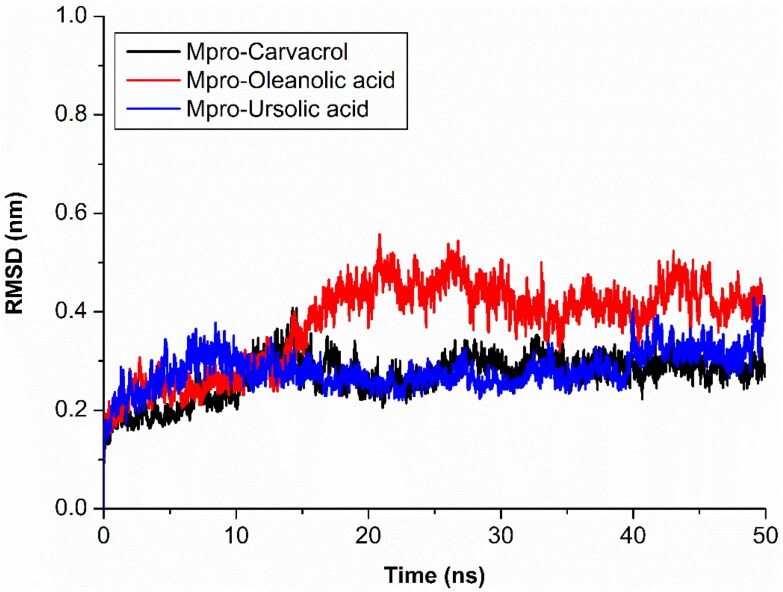
The Mpro with compound carvacrol (black) showed stable and constant range of RMSD between 0.23 nm to 0.30 nm and similar results showed by ursolic acid (blue), only slight changes showed in starting of the simulation. The oleanolic acid (red) showed stable RMSD between 4 ns to 20 ns at RMSD range between 0.21 nm to 0.38 nm, after 20 ns RMSD increases and constant at 0.41 nm (Figure 3). The differences of backbone RMSD with carvacrol, ursolic acid and oleanolic acid complex in protein backbone RMSD suggest that Mpro have conformational changes in oleanolic acid while no significant change shown in carvacrol and ursolic acid during 50 ns simulation.
Figure 3.
RMSD analysis of protein backbone during MD simulation.
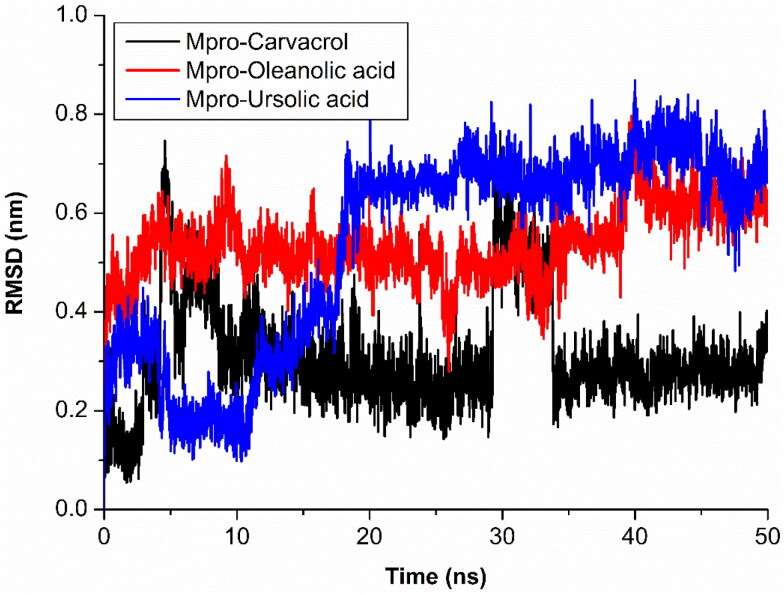
The compound carvacrol (red) has shown fluctuations at two different intervals on 50 ns time scale as it may has changed a conformation in binding region of Mpro. The first stable conformation between 6 ns to 29 ns and second stable conformation is between 34 ns to 50 ns. The RMSD remain constant at 0.3 nm and large fluctuation observed between 28 ns to 33 ns RMSD > 0.4 nm. As large fluctuation in compound, no effect on protein structure was found. Similar changes also seen in ursolic acid (magenta) RMSD fluctuations, it shown stable RMSD after 18 ns between 0.58 nm to 0.66 nm while in starting period between 4 ns to 17 ns, RMSD consistently fluctuated (Figure 4). It also not affects the fluctuation of protein backbone which depicts that binding region of protein has some fluctuation that causes the compound fluctuations during simulation. It may cause by large binding region and presence of loop at binding region.
Figure 4.
Analysis of RMSD of ligands and Mpro complexes; Mpro-carvacrol complex (Black), Mpro-oleanolic acid complex (Red), Mpro-ursolic acid complex (Blue).
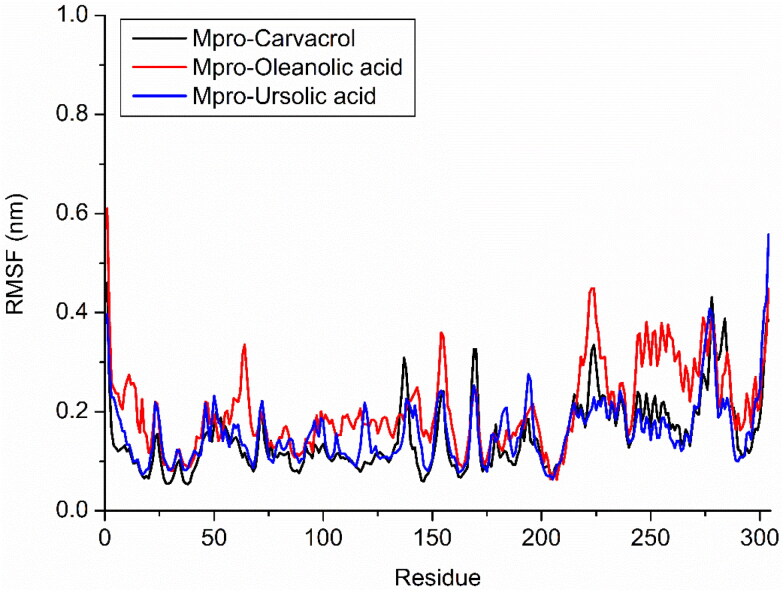
The oleanolic acid (blue) showed higher but stable RMSD between 0.44 nm to 0.61 nm throughout simulation. Oleanolic acid binding affects the RMSD of protein backbone but it has stable till end of the simulation. Two similar conformations of ursolic and oleanolic acid showed different behaviour with protein binding region during simulation analysis. This observation we also confirm by their local changes in residues level by RMSF plot. Protein backbone with carvacrol (black) and ursolic acid (blue ) not affect the behaviour of protein or structure of protein during simulation while oleanolic acid (red) affects the structure of protein during simulation. RMSF confirms the changes in structure with binding of these phytochemical compounds (Figure 5).
Figure 5.
Analysis of RMSF of Cα during MD simulation; Mpro-carvacrol complex (Black), Mpro-oleanolic acid complex (Red), Mpro-ursolic acid complex (Blue).
3.4. Hydrogen bond analysis
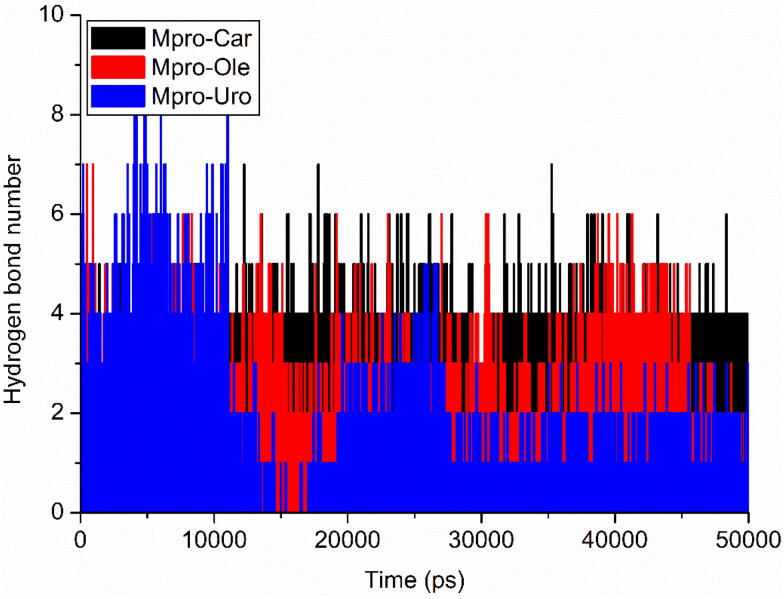
Hydrogen bonding plays a crucial role in determining the binding strength in between ligands and protein. The carvacrol (red) and ursolic acid (black) have constant range of hydrogen bond between 2 to 4 in whole simulation while oleanolic acid showed changes in bonding. More hydrogen (>5) bonds between 0 to 12 ns, after 12 ns the hydrogen bond are <4 and last 10 ns, the hydrogen bond between 2 to 3. This may suggests that there is a conformational change around oleanolic acid in the binding site during simulation (Figure 6). Over all observation suggested that all three protein-ligand complexes are stable during simulation.
Figure 6.
Intermolecular hydrogen bonds between the ligands and Mpro protein; Mpro-carvacrol complex (Black), Mpro-oleanolic acid complex (Red), Mpro-ursolic acid complex (Blue).
3.5. Free binding energy analysis/Poisson − Boltzmann surface area (MM-PBSA)
The pre docking suggest the binding energy of complex while binding free energy (ΔG bind) analysis after simulation suggest the consistency of non-bonding interactions energy of binding region with compound also called post docking. In previous reports, it has been shown that 100–200 snapshots are enough to calculate the binding free energies (Khan et al., 2020; Sarma et al., 2020). The van der waal energy component compared with each complex, the carvacrol (−93.913 +/−9.776) has less effect on binding affinity while ursolic (−189.889 +/−12.027) and oleanolic acid (−197.509 +/−11.086) has strong binding affinity. Similar results are shown in binding energy, ursolic (−168.918 +/−13.703) and oleanolic acid (−158.999 +/−15.306) showed similar effect of binding energy while moderate in carvacrol (−82.781 +/−9.401). The electrostatic energy doesn’t show significant in ursolic and oleanolic acid while moderate in carvacrol. The polar solvation and SASA energy are shown moderate effects on binding energy component in each compound (Table 3).
Table 3.
Free binding energy calculations of each Mpro and ligand complexes.
| Complexes | Binding energy (kJ/mol) | van der Waal energy (ΔEvdW)(kJ/mol) | Electrostattic energy (ΔEelec), (kJ/mol) | Polar solvation energy (ΔG polar) (kJ/mol) | SASA energy (kJ/mol) |
|---|---|---|---|---|---|
| Carvacrol | −82.781 +/− 9.401 | −93.913 +/− 9.776 | −37.778 +/− 6.738 | 58.079 +/− 6.712 | −9.169 +/− 0.708 |
| Oleanolic acid | −158.999 +/− 15.306 | −197.509 +/− 11.086 | −5.129 +/− 5.617 | 59.634 +/− 8.719 | −15.995 +/− 0.944 |
| Ursolic acid | −168.918 +/− 13.703 | −189.889 +/− 12.027 | −1.985 +/− 2.485 | 37.781 +/− 9.198 | −14.825 +/− 1.721 |
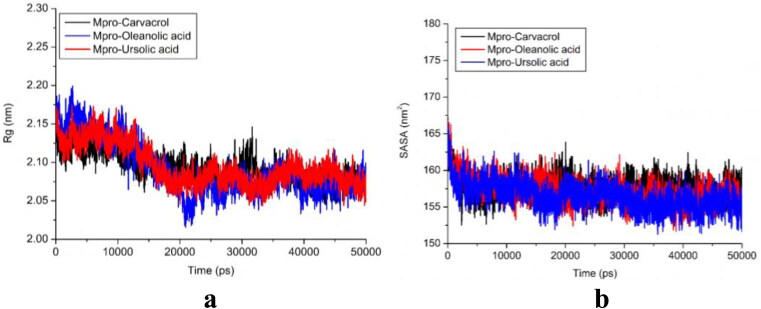
3.6. Radius of gyration analysis
In order to determine the compactness of the system with the time, Rg was calculated, in which higher Rg values depict less compactness (more unfolded) with conformational entropy, while low Rg values explain high compactness with more stability in the structure (more folded). As evident from Figure 7(a), the simulation Rg values of all three cases reported as 2.05–2.15 nm. The fewer changes in the Rg value exhibited the stability of the protein in the complex, which showed less difference. Rg results revealed that the binding of these three molecules does not induce structural changes. The Rg values of all three protein-ligand complexes (Figure 7(a)) support their condensed architecture as well as size. In a recent study, Khan et al. (2020) calculated the Rg values for Remdesivir Saquinavir and Darunavir drugs (reported as inhibiting drugs against chymotrypsin-like protease of SARS-CoV-2). The average gyration score of Remdesivir was found to be 22.25 ± 0.1 Å, while Saquinavir and Darunavir durgs prompted to have average Rg scores of 22.32 ± 0.4 Å and 22.29 ± 0.6 Å, respectively.
Figure 7.
(a) Radius of gyration (Rg); Mpro-carvacrol complex (Black), Mpro-oleanolic acid complex (Blue), Mpro-ursolic acid complex (Red); (b) Solvent surface accessible area (SASA); Mpro-carvacrol complex (Black), Mpro-oleanolic acid complex (Red), Mpro-ursolic acid complex (Blue).
3.7. SASA analysis
Additionally, we also conducted the solvent accessible surface area (SASA) analysis of all of the three protein-ligand complexes. SASA is important to measure of the receptor exposed to the solvents during the simulation. The exposures of the hydrophobic residues by the binding of the ligand molecules contribute to the values of the SASA. The SASA value exhibited between 155–165 nm2 (Figure 7(b)) showed that the binding of ligands does not affect the folding of the protein. Khan et al. (2020) have performed the SASA analysis for 3CLpro and its protein-drug complexes. They reported that SASA values for the 3CLpro, 3CLpro-Paritaprevir complex and 3CLpro-Raltegravir complex were 133.89 nm2, 131.79 nm2 and 131.26 nm2, respectively. The results reported in the present study suggest that all three of the complexes were impressively stable after the binding of ligands to active sites of Mpro protein.
4. Conclusion
The Mpro protein has shown to be crucial and highly potent target for the inhibition of novel COVID-19. This study concludes three natural compounds (ursolic acid, carvacrol and oleanolic acid) as potential inhibitors against Mpro. Molecular docking analysis revealed that Carvacrol molecule having less binding energy as compared to other two molecules, oleanolic acid and ursolic acid. The binding mode of interaction was found to be reasonably good. MD simulations revealed that all three docking complexes showed stability at 50 ns. These inhibitors also fulfil the ADME parameters as well as Lipinski’s rule of five. All these reported compounds are natural and also commercially available for further in vivo/in vitro validations. The information generated from this present study may be utilized in future for the development of more phytochemical based therapeutics against COVID-19.
Glossary
Abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
- COVID-19
Corona virus disease 2019
- MD
Molecular dynamic
- Mpro
protease
- MM/G/P/BSA
Molecular mechanic/generalized/Born/Poisson-Boltzmann surface area
- CSG
Coronavirus study group
- ICTV
International committee on taxonomy of viruses
- WHO
World health organization
- ORFs
Open reading frames
- RTC
Replicase/transcriptase complex
- PDB
Protein data bank
- ADME
Absorption, Distribution, Metabolism, and Excretion
- RMSD
Root mean square deviation
- RMSF
Root mean square fluctuation
- SASA
Solvent accessible surface area
- LB-CADD
Ligand-based computer-aided drug discovery
- Rg
Radius of gyration
Acknowledgements
Authors are thankful to Dr. Prashanth N Suravajhala, Birla Institute of Scientific Research (BISR). Jaipur, Rajasthan and Dr. MNV Prasad Gajula, Institute of Biotechnology, Hyderabad for critical proofreading of final manuscript. AK thanks the Director, UCB for encouragement, suggestions and timely help.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical standards
Ethical standards are compulsory for studies relating to human and animal subjects.
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelli I., Hassani F., Brikci S. B. & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoidesverticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adem S., Eyupoglu V., Sarfraz I., Rasul A., & Ali M. (2020). Identification of potent COVID–19 main protease (Mpro) inhibitors from natural polyphenols: An in silico strategy unveils a hope against CORONA. Preprints. 10.20944/preprints202003.0333.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldeghi M., Bodkin M. J., Knapp S., & Biggin P. C. (2017). Statistical analysis onthe performance of molecular mechanics poissonboltzmann surface area versus absolute binding free energy calculations: Bromodomains as a case study. Journal of Chemical Information and Modeling, 57(9), 2203–2221. 10.1021/acs.jcim.7b00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J. R., & Hilgenfeld R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Anand K., Yang H., Bartlam M., Rao Z., & Hilgenfeld R. (2005). Coronavirus main proteinase: Target for antiviral drug therapy In Schmidt A., Weber O., & Wolff M. H. (Eds.), Coronaviruses with special emphasis on first insights concerning SARS (pp. 173–199). Birkhäuser Basel; 10.1007/3-7643-7339-3_9. [DOI] [Google Scholar]
- Benson N. C., & Daggett V. (2012). A comparison of multiscale methods for the analysis of molecular dynamics simulations. The Journal of Physical Chemistry. B, 116(29), 8722–8731. 10.1021/jp302103t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H. J. C., van der Spoel D., & van Drunen R. (1995). GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications, 91(1–3), 43–56. 10.1016/0010-4655(95)00042-E [DOI] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., & Bourne P. E. (2000). The protein data bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatraju P. K., Ghassemieh B. J., Nichols M., Kim R., Jerome K. R., Nalla A. K., Greninger A. L., Pipavath S., Wurfel M. M., Evans L., Kritek P. A., West T. E., Luks A., Gerbino A., Dale C. R., Goldman J. D., O’Mahony S., & Mikacenic C. (2020). Covid-19 in critically ill patients in the Seattle Region—Case Series. The New England Journal of Medicine, 382(21), 2012–2022. 10.1056/NEJMoa2004500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K., Berman H. M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J. M., Dutta S., Feng Z., Ghosh S., Goodsell D. S., Green R. K., Guranović V., Guzenko D., Hudson B. P., Kalro T., Liang Y., … Zardecki C. (2019). RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Research, 47(D1), D464–D474. 10.1093/nar/gky1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W., Yuan S., Kok K.-H., To K. K.-W., Chu H., Yang J., Xing F., Liu J., Yip C. C.-Y., Poon R. W.-S., Tsoi H.-W., Lo S. K.-F., Chan K.-H., Poon V. K.-M., Chan W.-M., Ip J. D., Cai J.-P., Cheng V. C.-C., Chen H., Hui C. K.-M., & Yuen K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel V., Raj S., Rathi B., & Kumar D. (2020). In silico identification of potent COVID–19 main protease inhibitors from FDA approved antiviral compounds and active phytochemicals through molecular docking: A drug repurposing approach. Preprints. [Google Scholar]
- Chang C., Lo S.-C., Wang Y.-S., & Hou M.-H. (2016). Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discovery Today, 21(4), 562–572. 10.1016/j.drudis.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Tung Y., Lee K., Chen T., Hsiao Y., Chang H., & Shih S. (2020). Potential therapeutic agents for COVID–19 based on the analysis of protease and RNA polymerase docking. Priprints. 10.20944/preprints202002.0242.v1 [DOI] [Google Scholar]
- Chen Y., Liu Q., & Guo D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contini A. (2020). Virtual screening of an FDA approved drugs database on two COVID-19 coronavirus proteins. ChemRxiv. Preprint. 10.26434/chemrxiv.11847381.v1 [DOI] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020). The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5(4), 536 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., & Zoete V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Sarmah S., Lyndem S., & Roy A. S. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., & Munster V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews Microbiology, 14(8), 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C., Rolain J., Colson P., & Raoult D. (2020). New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID–19? International Journal of Antimicrobial Agents, 55(5), 105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in-silico perspective. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Natural products may interfere with SARS-CoV-2attachment to the host cell. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.17618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in-silico perspective. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekci K. (2020). Drug repurposing for coronavirus (COVID-19): In silicoscreening of knowndrugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in-silico approach. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajula M. N. P. (2008). Computer simulation meets experiment: molecular dynamics simulations of spin labeled proteins MNV Prasad Gajula [PhD dissertation]. [Google Scholar]
- Gajula M. N. P., Kumar A., & Ijaq J. (2016). Protocol for molecular dynamics simulations of proteins. BIO-PROTOCOL, 6(23), 1–11. 10.21769/BioProtoc.205127642615 [DOI] [Google Scholar]
- Gentile D., Patamia V., Scala A., Sciortino M. T., Piperno A., & Rescifina A. (2020). Inhibitors of SARS-CoV–2 main protease from a library of marine natural products: A virtual screening and molecular modeling study. Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilling D. H., Kitajima M., Torrey J. R., & Bright K. R. (2014). Antiviral efficacy and mechanisms of action of oregano essential oil and its primary component carvacrol against murine norovirus. Journal of Applied Microbiology, 116(5), 1149–1163. 10.1111/jam.12453 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Paz L. A., Lossada C. A., Moncayo L. S., Romero F., Paz J. L., Vera-Villalobos J., Pérez A. E., San-Blas E., & Alvarado Y. J. (2020). Theoretical molecular docking study of the structural disruption of the viral 3CL-protease of COVID19 induced by binding of capsaicin, piperine and curcumin part 1: A comparative study with chloroquine and hydrochloroquine two antimalaric drugs. Research Square, (Preprint). 10.21203/rs.3.rs-21206/v1 [DOI] [Google Scholar]
- Gorden D. E., Gwendolyn M. J., & Bouhaddou M. (2020). A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv. 10.1101/2020.03.22.002386 [DOI] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A. A., & Falahati M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M. G., & Ba Abduallah M. M. (2020). The SARS-CoV-2 outbreak from a one health perspective. One Health. 10.1016/j.onehlt.2020.100127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyldgaard M., Mygind T., & Meyer R. L. (2012). Essential oils in food preservation: Mode of action, synergies, and interactions with food matrix components. Frontiers in Microbiology, 3, 12 10.3389/fmicb.2012.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee B., Kumar S., Yadav R., Singh Y., Kumar A., & Sharma M. (2018). Ursolic acid and carvacrol may be potential inhibitors of dormancy protein small heat shock protein16.3 of Mycobacterium tuberculosis. Journal of Biomolecular Structure and Dynamics, 36(13), 3434–3443. 10.1080/07391102.2017.1389305 [DOI] [PubMed] [Google Scholar]
- Jesus J. A., Lago J. H., Laurenti M. D., Yamamoto E. S., & Passero L. F. (2015). Antimicrobial activity of oleanolic and ursolic acids: An update. Evidence-Based Complementary and Alternative Medicine, 2015, 620472 10.1155/2015/620472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., & Zhang B. (2020). Structure of Mpro from COVID-19 virus and discovery of itsinhibitors. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.02.26.964882v2 [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisari E., & Borruat F. (2020). Keeping an eye on hydroxychloroquine retinopathy. KlinischeMonatsblätterfürAugenheilkunde, Efirst. [DOI] [PubMed] [Google Scholar]
- Kamalabadi M., Astani A., & Nemati F. (2018). Anti-viral effect and mechanism of carvacrol on herpes simplex virus type 1. International Journal of Molecular Laboratory, 5(2), 113–122. [Google Scholar]
- Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., & Soetjipto S. (2020). Potential inhibitor of COVID–19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Preprints. [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ui-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Khwaza V., Oyedeji O. O., & Aderibigbe B. A. (2018). Antiviral activities of oleanolic acid and its analogues. Molecules, 23(9), 2300 10.3390/molecules23092300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B. A., Thiessen P. A., Yu B., Zaslavsky L., Zhang J., & Bolton E. E. (2019). PubChem 2019 update: improved accesses to chemical data. Nucleic Acids Research, 47(D1), D1102–D1109. 10.1093/nar/gky1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Kumar R., Sharma M., Kumar U., Gajula M. N. P., & Singh K. P. (2018). Uttarakhand medicinal plants database (UMPDB): A platform for exploring genomic, chemical, and traditional knowledge. Data (Data), 3(1), 7 10.3390/data3010007 [DOI] [Google Scholar]
- Kumar A., Kumar S., Kumar A., Sharma N., Sharma M., Singh K. P., Rathore M., & Gajula M. N. V. P. (2018). Homology modeling, molecular docking and molecular dynamics based functional insights into rice urease bound to urea. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences, 88(4), 1539–1548. 10.1007/s40011-017-0898-0 [DOI] [Google Scholar]
- Li Y., Zhang J., Wang N., Li H., Shi Y., Guo G., & Zou Q. (2020). Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. bioRxiv. 10.1101/2020.01.28.922922 [DOI] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W., & Feeney P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 46(1–3), 3–26. 10.1016/j.addr.2012.09.019 [DOI] [PubMed] [Google Scholar]
- Lu I.-L., Mahindroo N., Liang P.-H., Peng Y.-H., Kuo C.-J., Tsai K.-C., Hsieh H.-P., Chao Y.-S., & Wu S.-Y. (2006). Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. Journal of Medicinal Chemistry, 49(17), 5154–5161. 10.1021/jm060207o [DOI] [PubMed] [Google Scholar]
- Mackenzie J. S., & Smith D. W. (2020). COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and what we don’t. Microbiology Australia, 41(1), 45 10.1071/MA20013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli L., Fornasari E., Eusepi P., Ciulla M., Genovese S., Epifano F., Fiorito S., Turkez H., Örtücü S., Mingoia M., Simoni S., Pugnaloni A., Di Stefano A., & Cacciatore I. (2019). Carvacrol prodrugs as novel antimicrobial agents. European Journal of Medicinal Chemistry, 178, 515–529. 10.1016/j.ejmech.2019.05.093 [DOI] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., & Olson A. J. (2009). Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. Journal of Computational Chemistry, 16, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Panchangam S. S., Vahedi M., Megha M. J., Kumar A., Raithatha K., Suravajhala P., & Reddy P. (2016). Saffron‘omics’: The challenges of integrating omic technologies. Avicenna Journal of Phytomedicine, 6(6), 604–620. [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollier J., & Goossens A. (2012). Oleanolic acid. Phytochemistry, 77, 10–15. 10.1016/j.phytochem.2011.12.022 [DOI] [PubMed] [Google Scholar]
- Qamar M., Alqahtani S., Alamri M., & Chen L. (2020). Structural basis of SARS-CoV–2 3CLpro and anti-COVID–19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis. 10.1016/j.jpha.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkiah S., Chandrasekaran M., Lee Y., Kim S., & Lee K. W. (2012). Molecular modeling study for conformational changes of Sirtuin 2 due to substrate and inhibitor binding. Journal of Biomolecular Structure & Dynamics, 30(3), 235–254. 10.1080/07391102.2012.680026 [DOI] [PubMed] [Google Scholar]
- Salata C., Calistri A., Parolin C., & Palù G. (2019). Coronaviruses: A paradigm of new emerging zoonotic diseases. Pathogens and Disease, 77(9), ftaa006 10.1093/femspd/ftaa006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüttelkopf A. W., & van Aalten D. M. F. (2004). PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Seah I., & Agarwal R. (2020). Can the coronavirus disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals. Ocular Immunology and Inflammation, 28(3), 391–395. 10.1080/09273948.2020.1738501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. D., & Kaur I. (2020). Eucalyptol (1,8 cineole) from eucalyptus essential Oil a potential inhibitor of COVID 19 corona virus infection by molecular docking studies. Preprints. [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A. C. K., Zhou J., Liu W., Bi Y., & Gao G. F. (2016). Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends in Microbiology, 24(6), 490–502. 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Wong W., & Guo J. (2020). Prediction of potential 3CLpro-targeting anti-SARS-CoV–2 compounds from Chinese medicine. Preprints. [Google Scholar]
- Tohmé M. J., Giménez M. C., Peralta A., Colombo M. I., & Delgui L. R. (2019). Ursolic acid: A novel antiviral compound inhibiting rotavirus infection in vitro. International Journal of Antimicrobial Agents, 54(5), 601–609. 10.1016/j.ijantimicag.2019.07.015 [DOI] [PubMed] [Google Scholar]
- Umesh D. K., Selvaraj C., Singh S. K., & Dubey V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber D. F., Johnson S. R., Cheng H. Y., Smith B. R., Ward K. W., & Kopple K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. Journal of Medicinal Chemistry, 45(12), 2615–2623. 10.1021/jm020017n [DOI] [PubMed] [Google Scholar]
- Wade R. C., & Goodford P. J. (1989). The role of hydrogen-bonds in drug binding. Progress in Clinical and Biological Research, 289, 433–444. [PubMed] [Google Scholar]
- Wadood A., Ahmed N., Shah L., Ahmad A., Hassan H., & Shams S. (2013). In-silico drug design: An approach which revolutionarised the drug discovery process. OA Drug Design & Delivery, 1(1), 3. [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang B., Guo H., Ling L., Ji J., Niu J., & Gu Y. (2020). The chronic adverse effect of chloroquine on kidney in rats through an autophagy dependent and independent pathways. Nephron, 144(2), 96–64. 10.1159/000503882 [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., & Xiao G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P. W., Hayden F. G., & Gao G. F. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P., & Murdoch D. R. (2020). Clinical course and mortality risk of severe COVID-19. The Lancet, 395(10229), 1014–1015. 10.1016/S0140-6736(20)30633-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. Y., Lau S. K. P., Chu C-m., Chan K-h., Tsoi H-w., Huang Y., Wong B. H. L., Poon R. W. S., Cai J. J., Luk W-k., Poon L. L. M., Wong S. S. Y., Guan Y., Peiris J. S. M., & Yuen K.-Y. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, 79(2), 884–895. 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO ). (2020). Naming the coronavirus disease (Covid-19) and the virus that causes it. World Health Organization. [Google Scholar]
- Woźniak Ł., Skąpska S., & Marszałek K. (2015). Ursolic Acid-A Pentacyclic Triterpenoid with a wide spectrum of pharmacological activities. Molecules (Basel, Switzerland), 20(11), 20614–20641. 10.3390/molecules201119721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., Hu Y., Tao Z. W., Tian J. H., Pei Y. Y., Yuan M. L., Zhang Y. L., Dai F. H., Liu Y., Wang Q. M., Zheng J. J., Xu L., Holmes E. C., & Zhang Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., & Hao P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China. Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bartlam M., & Rao Z. (2006). Drug design targeting the main protease, the Achilles' heel of coronaviruses. Current Pharmaceutical Design, 12(35), 4573–4590. 10.2174/138161206779010369 [DOI] [PubMed] [Google Scholar]
- Yu W., & MacKerell, A. D. Jr. (2017). Computer-aided drug design methods. Methods in Molecular Biology (Clifton, N.J.), 1520, 85–106. 10.1007/978-1-4939-6634-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Yin W., Tao Z., Tan W., & Hu Y. (2020). Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan, China. PLoS One, 15(3), e0230548 10.1371/journal.pone.0230548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten U., Sauerhering L., Becker S., Rox K., & Hilgenfeld R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]