Abstract
The recent novel coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2/2019-nCoV) has caused a large number of deaths around the globe. There is an urgent need to understand this new virus and develop prophylactic and therapeutic drugs. Since drug development is an expensive, intense and time-consuming path, timely repurposing of the existing drugs is often explored wherein the research avenues including genomics, bioinformatics, molecular modeling approaches offer valuable strengths. Here, we have examined the binding potential of Withaferin-A (Wi-A), Withanone (Wi-N) (active withanolides of Ashwagandha) and Caffeic Acid Phenethyl Ester (CAPE, bioactive ingredient of propolis) to a highly conserved protein, Mpro of SARS-CoV-2. We found that Wi-N and CAPE, but not Wi-A, bind to the substrate-binding pocket of SARS-CoV-2 Mpro with efficacy and binding energies equivalent to an already claimed N3 protease inhibitor. Similar to N3 inhibitor, Wi-N and CAPE were interacting with the highly conserved residues of the proteases of coronaviruses. The binding stability of these molecules was further analyzed using molecular dynamics simulations. The binding free energies calculated using MM/GBSA for N3 inhibitor, CAPE and Wi-N were also comparable. Data presented here predicted that these natural compounds may possess the potential to inhibit the functional activity of SARS-CoV-2 protease (an essential protein for virus survival), and hence (i) may connect to save time and cost required for designing/development, and initial screening for anti-COVID drugs, (ii) may offer some therapeutic value for the management of novel fatal coronavirus disease, (iii) warrants prioritized further validation in the laboratory and clinical tests.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2 coronavirus, Ashwagandha, Withanone, Withaferin-A, honeybee propolis, caffeic acid phenethyl ester, molecular docking, binding, main protease (Mpro)
1. Introduction
Coronaviruses, discovered in 1960, are infectious strains of viruses originally named on the basis of their crown like appearance, due to the glycoprotein projections on its envelope, under the electron microscope and grouped into the family Coronaviridae; order Nidovirales. They invade the respiratory tract via the nose. After an incubation period of about 3–7 days, they cause the symptoms of a mild common cold/bronchitis (nasal obstruction, sneezing, runny nose, cough, headache, fever, pneumonia, asthenia and inflammation in airway) in avian and mammalian species. In contrast to animals, wherein they have been shown to infect several tissues causing a large variety of diseases, mainly respiratory infections with mild common cold like symptoms, occasional gastrointestinal and diarrhea have been reported for humans. The infected individuals shed virus in nasal secretions and mucosa resulting in disease transmission that can often be controlled, at least partially, by following hygienic measures. Vaccines for coronaviruses are not available and treatment remains symptomatic.
Designing and development of antiviral medicine requires understanding of the molecular mechanisms of viral replication and packaging into the infectious particles in host cells, their release, selection of antiviral target proteins and development of their inhibitors. Coronaviruses have been shown to invade and replicate in differentiated respiratory epithelial cells resulting in their vacuolation, damaged cilia, local inflammation, swelling, sneezing and fever. Among the several strains of coronaviruses known so far, including HCoV-229E, HCoV-OC43, HCoV-NL63, SARS-CoV, MERS-CoV and 2019-nCoV/SARS-CoV-2 (Graham et al., 2013; van der Hoek et al., 2004; Woo et al., 2010), the latter was designated as a novel strain of coronavirus that caused pneumonia outbreak in Wuhan city of China in December 2019 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020; Wu et al., 2020; Zhou et al., 2020). As of May 14, 2020, it has infected over 4,258,666 individuals globally with 294,190 deaths, as reported to WHO (WHO, 2020). It has been declared as international public health emergency and advocated rapid research efforts. Genomic characterization of the SARS-CoV-2, its variance, evolution, transmission dynamics (human to human transmission by droplets or direct contact) have been reported recently (Benvenuto et al., 2020; Ceraolo & Giorgi, 2020; Li, Wang, et al., 2020; Li, Zai, et al., 2020; Lu et al., 2020; Paraskevis et al., 2020). It is an enveloped single-stranded RNA β-coronavirus similar to the SARS and MERS viruses and closely related to two bat-derived SARS-like coronaviruses (CoVZC45 and bat-SL-CoVZXC21) (FDA News Release, 2020). Severity of the infections and fatality caused by SARS-CoV-2 has evoked urgency for devising novel drug discovery strategies to find potential prophylactic and therapeutic drugs. The entry of SARS-CoV-2 into the host cell depends on the interaction with ACE-2 receptor of the host cell surface. Upon attachment with ACE-2, the S protein of the virus gets cleaved and activated for its fusion into the host cell membrane, which is facilitated by the TMPRSS-2 and furin (Hasan et al., 2020; Hoffmann et al., 2020). The viral genome codes for non-structural proteins including 3-chymotrypsin-like protease, RNA-dependent RNA polymerase and its helicase, papain-like protease, the structural glycoprotein and accessory proteins (Boopathi et al., 2020). In the absence of complete understanding of the detailed molecular structure, function, survival and replication of novel strain of virus, repurposing of the existing drugs has been adopted as a quick and hopeful track. Kaletra (an inhibitor of 3-chymotrypsin-like protease of the SARS and MERS), and Ascletis (HIV protease inhibitor) were shown to improve clinical outcomes of COVID-19 (Wang et al., 2020). In view of this, use of computational approaches for quick screening of available compounds is gaining attention of the scientific community (Aanouz et al., 2020; Abdelli et al., 2020; Al-Khafaji et al., 2020; Chang et al., 2020; Das et al., 2020; Elfiky, 2020; Elmezayen et al., 2020; Enmozhi et al., 2020; Gupta et al., 2020; Gupta & Vadde, 2020; Gyebi et al., 2020; Islam et al., 2020; Joshi et al., 2020; Khan, Jha, et al., 2020; Khan, Zia, et al., 2020; Kumar et al., 2020; Lobo-Galo et al., 2020; Muralidharan et al., 2020; Pant et al., 2020; Sarma et al., 2020; Sinha et al., 2020; Thuy et al., 2020; Umesh et al., 2020; Wahedi et al., 2020). Using molecular docking tools, Indinavir and Remdesivir were investigated for their docking potential to SARS-CoV-2 protease. Neither of these docked on any active sites of the protease. However, the active form (CHEMBL2016761) of Remdesivir showed perfect docking in the overlapping region of the NTP binding motif urging its validation through clinical trials for COVID-19 infection (Chang et al., 2020). The U.S. Food and Drug Administration has now issued an emergency use authorization for Remdesivir for the treatment of suspected or laboratory-confirmed adults and children hospitalized with severe symptoms of SARS-CoV-2 disease (FDA News Release, 2020). In another recent study employing an in silico approach, it has been reported that Belachinal, Macaflavanone E and Vibsanol B phytochemicals may inhibit the functional activity of SARS-CoV-2 E protein (Gupta et al., 2020). In line with this, several studies have recently explored the repurposing of drugs to find an immediate therapeutic strategy for the deadly COVID-19, by mainly targeting SARS-CoV-2 main protease (Aanouz et al., 2020; Al-Khafaji et al., 2020; Das et al., 2020; Enmozhi et al., 2020; Gyebi et al., 2020; Islam et al., 2020; Joshi et al., 2020; Khan, Jha, et al., 2020; Khan, Zia, et al. 2020; Kumar et al., 2020; Lobo-Galo et al., 2020; Muralidharan et al., 2020; Pant et al., 2020; Umesh et al., 2020), NSP15 and prefusion 2019-nCoV spike glycoprotein (Sinha et al., 2020), RNA-dependent RNA polymerase (Elfiky et al., 2020), N and E protein (Gupta et al., 2020; Sarma et al., 2020), and cell surface receptors (ACE-2 and TMPRSS2) of host cells (Abdelli et al., 2020; Elmezayen et al., 2020; Thuy et al., 2020; Wahedi et al., 2020).
Indian Ayurvedic herb, Ashwagandha (Withania sominifera) and honeybee propolis have been heavily used in the traditional home medicine systems. Both are known to boost the immune function, possess a variety of prophylactic and therapeutic activities (Akyol et al., 2013; Anjaly & Tiku, 2018; Erdemli et al., 2015; Kaul & Wadhwa, 2017; Murtaza et al., 2015). Besides, Withaferin-A , one of the withanolides from Ashwagandha, was shown to possess inhibitory activity for HPV and influenza viruses (Cai et al., 2015; Latheef et al., 2017; Munagala et al., 2011). Several studies have also provided evidence that the honeybee propolis inhibits variety of viruses including herpes simplex virus, sindbis virus, parainfuenze-3 virus, human cytomegalovirus, dengue virus type-2, influenza virus A1 and rhinovirus (Amoros et al., 1994; Kwon et al., 2019; Lyu et al., 2005; Serkedjieva et al., 1992; Zandi et al., 2011). In light of these literature, we examined the potential of three natural compounds, i.e. Withaferin A (Wi-A) and Withanone (Wi-N) derived from Ashwagandha and Caffeic acid phenethyl ester (CAPE) from honeybee propolis, against SARS-CoV-2 by molecular docking tools. Based on the fact that the main viral proteases isolated from different coronaviruses are mainly conserved in terms of sequence and structure, we used a key viral enzyme Main Protease (Mpro - 306 amino acids, essential for the functional polypeptide generation and virus survival in host cells) as a target. The 3D crystal structure of the SARS-CoV-2 Mpro in complex with an inhibitor N3 has recently been released in PDB (6LU7) (Jin et al., 2020). We demonstrate that Wi-N and CAPE, but not Wi-A, are capable of binding to the substrate-binding pocket of SARS-CoV-2 Mpro. Wi-N and CAPE showed interactions with the highly conserved residues of the coronavirus proteases similar to N3 inhibitor, demonstrating their inhibitory potential for SARS-CoV-2 protease, and therapeutic significance for novel SARS-CoV-2 coronavirus.
2. Methodology
2.1. Preparation of structure of SARS-CoV-2 Mpro and natural compounds for molecular docking
The structure of SARS-CoV-2 Mpro in complex with N3 inhibitor, resolved using X-ray diffraction (resolution of 2.16 Å), was downloaded from RCSB Protein Data Bank (PDB ID: 6LU7). The crystal structure was prepared for docking using protein preparation wizard of the maestro (version 2019-3, Schrodinger suite). This mainly involved addition of missing disulfide bonds, removal of water molecules, addition of missing hydrogen atoms, filling of missing amino acids side chains and optimization of hydrogen bonds (Schrodinger, 2020). OPLS3e forcefield was then used for restrained minimization until the average root mean square deviation (RMSD) of the non-hydrogen atoms converged to 0.30 Å. This allowed sufficient movement of heavy atom to relax strained bonds, angles and clashes.
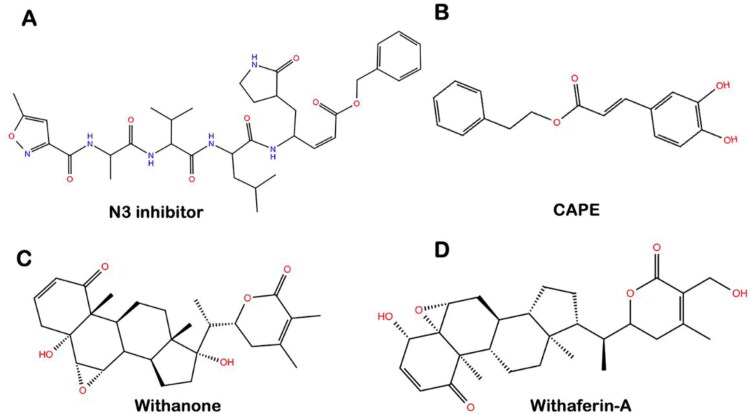
The structure of Wi-A (CID 265237), Wi-N (CID 21679027) and CAPE (CID 5281787) were downloaded from the PubChem database, while the structure of the N3 inhibitor was extracted from SARS-CoV-2 Mpro-inhibitor complex downloaded from PDB (PDB ID: 6LU7). The structure of all the ligands used in this study is shown in Figure 1.
Figure 1.
Molecular structure of (A) N3 protease inhibitor, (B) CAPE, (C) Withanone, and (D) Withaferin-A.
LigPrep module of the Schrodinger suite (Sastry et al., 2013; Schrodinger, 2020) was used to prepare these structures for docking studies. It desalts the ligands, generates tautomer and stereoisomers (while retaining specified chiralities) in all possible ionization states (Greenwood et al., 2010). The energy of the generated ligands was then minimized using OPLS3e force field (Harder et al., 2016).
2.2. Molecular docking of natural compounds with SARS-CoV-2 Mpro
The grid for docking studies was generated to enclose all the residues of SARS-CoV-2 Mpro making polar interactions with the atoms of N3 inhibitor (deduced from PDB structure 6LU7). These residues mainly involved Phe140, Asn142, Gly143, His164 and Glu166. Extra precision flexible docking protocol of Glide was used for all dockings (Friesner et al., 2006; Schrodinger, 2020). All the protein-ligand complexes were obtained. Glide has been shown to provide better prediction in comparison to that of the other docking software; because it uses both empirical as well as force field terms to calculate the best binding pose and binding energy (Friesner et al., 2004).
2.3. Explicit-solvent molecular dynamics (MD) simulations
Desmond with OPLS3e force field from Schrodinger was used to study the dynamic behavior of the all protein-ligand complexes in the presence of explicit water molecules (Harder et al., 2016; Schrodinger, 2020). Each protein-ligand complex was solvated with TIP4P water model in an orthorhombic periodic boundary box. To prevent interaction of the protein complex with its own periodic image, the distance between the complex and the box wall was kept 10 Å. The system was then neutralized by addition of ions. Energy of the prepared systems was minimized for 5000 steps using steepest descent method or until a gradient threshold of 25 kcal/mol/Å was achieved. It was followed by L-BFGS (Low-memory Broyden- Fletcher- Goldfarb Shanno quasi-Newtonian minimizer) until a convergence threshold of 1 kcal/mol/Å was met. The equilibrated system was then subjected to 50 ns simulation in NPT ensemble with 300 K temperature, constant pressure of 1 atm and time step of 2fs.
2.4. Analysis of simulation trajectories
The simulation trajectories were analyzed using the simulation event analysis tool available in Desmond module (Bowers et al., 2006). Overall changes in the configuration of the protein-ligand complexes were first studied in term of root mean square deviation (RMSD) in the coordinates of protein backbone and ligand from the coordinates in the initial docked pose. The interaction pattern of all three ligands- Wi-A, Wi-N and CAPE was compared with the interactions of N3 inhibitor within the catalytic domain of SARS-CoV-2 Mpro. Various residues residing in the catalytic domain of the protein were investigated for their time of contact with the different ligands throughout the simulation to calculate their occupancy percentage. The radius of gyration, solvent accessible surface area and RMSD of ligands along the simulation trajectory was calculated and analyzed. All the systems under study, including apo enzyme or the docked complexes, were getting stabilized within initial 5–10 ns of the production simulation runs. No significant deviation was observed in the RMSD trajectory throughout the course of simulation thereafter. An extensive analysis of Mpro-N3 inhibitor complex, retrieved from Protein Data Bank did not show much conformational changes in the protein after the binding of the inhibitor. Based on these parameters, we decided the simulation time to be 50 ns to observe convergence in the RMSD trajectory and account for the stability of the docked complexes.
2.5. MM/GBSA free energy calculations
A total of 100 frames representing the protein-ligand conformations spanned in between 10 and 50 ns of simulation run were extracted to calculate the MM/GBSA free binding energy using the Prime module of Schrodinger suite (Jacobson et al., 2004; Schrodinger, 2020).
The equation used for the calculation is:
ΔGcomplex, Greceptor, and Gligand represent the free energies of the complex, receptor and ligand respectively.
3. Results and discussion
3.1. SARS-CoV-2 Mpro enzyme as a candidate therapeutic target
Coronaviruses (CoV) are enveloped positive-strand RNA viruses that mainly cause respiratory and enteric diseases. Six human coronaviruses - SARS-CoV, MERS-CoV, HCoV-229E, HCoV-OC43, HCoV-NL63 and HCoV-HKU1 have been reported previously (Graham et al., 2013; van der Hoek et al., 2004; Woo et al., 2010). Along with various structural proteins, all the CoV genomes encode for a critical viral component called Mpro. The latter is a 306 amino acids long enzyme that mainly helps in the replication of the virus through proteolytic processing of its RNA replicase machinery. Mpro from different human and animal CoVs have been shown to possess high similarity in terms of primary amino acid sequence as well as the functional tertiary confirmation of the enzyme. All these proteins comprise of a highly conserved substrate binding pocket that serves as an attractive therapeutic target for the prevention and treatment of these coronaviruses (Woo et al., 2005, 2010). The recently discovered SARS-CoV-2 also shares this homology in its Mpro enzyme. Several key residues at the various sub-sites of substrate binding pocket in all these seven coronaviruses are identical. Though not an exhaustive list, these conserved residues broadly include His41, Tyr54, His163, Glu166, His172, Asp187 and Gln192 (Figure 2). N3, a synthetic peptidomimetic compound has been reported to target the substrate binding pocket of this protease across different variants of coronavirus including SARS-CoV-2 (Wang et al., 2016). As Ashwagandha-derived withanolides (Wi-A and Wi-N) and propolis-derived CAPE has been previously shown to possess anti-microbial and antiviral activity, we have tried to explore the potential of these natural compounds to interact within the catalytic pocket of SARS-CoV-2 Mpro in comparison to the already claimed inhibitor (N3).
Figure 2.
Sequence alignment showing high homology among proteases of various human CoVs strains. The residues conserved among the substrate-binding pocket of various CoV proteases and interacting with all the ligands in this study, namely N3 inhibitor, Wi-N and CAPE are highlighted for reference.
3.2. Wi-N and CAPE predicted to bind to Mpro and mimic the binding pattern of N3 inhibitor
The structure of N3 inhibitor co-crystalized to the Mpro of SARS-CoV-2, recently deposited to Protein Data Bank, gave us insights into the molecular mechanism of the action of N3 inhibitor against the new coronavirus (Jin et al., 2020). N3 inhibitor showed interaction within the substrate binding pocket of the main SARS-CoV-2 protease (Figure 3(A)). The EC50 of N3 inhibitor has been reported to be 50 µM and its mechanism of action has also been investigated. The residues of Mpro mainly interacting with N3 inhibitor were found to be Cys145, Phe140, Asn142, Glu166, His163 and His172. Among these, Cys and His were reported to be crucial for the catalytic activity of the enzyme. N3 was found to be forming multiple hydrogen bonds with the main chain of the residues in the substrate-binding pocket of Mpro, thereby tightly locking the inhibitor inside the pocket (Jin et al., 2020). We firstly re-docked N3 inhibitor in the same configuration as to get a docking score for the natural binding. This docking score was found to be −5.68 Kcal/mol and was used to compare the binding of Withanolides and CAPE with the Mpro of SARS-CoV-2. The two main residues forming polar interaction with the inhibitor were Thr190 and Glu166. The other residues involved in non-bonded interactions making the binding of N3 inhibitor stable within the pocket included His41, Cys44, Met49, Pro52, Tyr54, Phe140, Leu141, Asn142, His164, Met165, Leu167, Pro168, Thr169, Gly170, His172, Asp187, Arg188, Gln189, Ala191 and Gln192 (Figure 3(B)). A grid was generated around the conserved residues of the substrate-binding pocket with main emphasis on the residues making polar contacts with N3 inhibitor. The three test compounds- Wi-A, Wi-N and CAPE were then docked using the same grid. All the molecules were able to dock into the same site and scores were found to be −4.42 Kcal/mol for Wi-N, −4.79 Kcal/mol for CAPE and −1.11 Kcal/mol for Wi-A. The docking score of Wi-N and CAPE was found comparable to that of N3 inhibitor and were hence studied further to get more insights into the interaction pattern of these molecules. Of note, although Wi-A and Wi-N are highly similar in their molecular structure, Wi-A did not show good affinity for Mpro. It can be attributed to the torsional constraint at atom C20, because of which the Wi-A was not in its relaxed state in the best docked pose. These data suggested that the predicted SARS-CoV-2 Mpro-Wi-N as well as SARS-CoV-2 Mpro-CAPE interactions may not be the false positives. As shown in Figure 3(C), similar to N3 inhibitor, CAPE also formed polar contact with Glu166. The other residue involved in hydrogen bond formation between SARS-CoV-2-Mpro and CAPE was Asn142. In case of Wi-N, the binding was due to the polar interaction with Cys145 (Figure 3(D)), a residue important for the catalytic activity of Mpro. However, the other non-boned interactions involved many other conserved residues of the substrate-binding pocket. The complete list of interacting residues in each case is summarized in Table 1.
Figure 3.
(A) Molecular representation of N3 inhibitor within the substrate-binding pocket of SARS-CoV-2 main protease (Mpro) (PDB: 6LU7). (B) Molecular interaction pattern of N3 inhibitor with the conserved residues of Mpro. N3 inhibitor was found to be interacting with Glu166 and Thr190 of Mpro by the formation of hydrogen bonds. Conserved residues participating in other non-bonded interactions are shown in line representation. (C) Molecular interactions of CAPE with SARS-CoV-2 Mpro after docking. Hydrogen bond interactions between CAPE and protease were found to be through Glu166 and Asn142. The other residues participating in other non-bonded interactions are highlighted in orange. The residues conserved in the substrate-binding pocket of various coronaviruses and involved in interaction with all three ligands in this study- N3 inhibitor, CAPE and Wi-N are highlighted in B, C and D using line representation. (D) Molecular interactions of Withanone with SARS-CoV-2 Mpro in the best docking pose. Wi-N was forming one hydrogen bond with Cys145, however many other residues (highlighted in orange) were participating in other non-bonded interactions with the small molecule.
Table 1.
Residues of SARS-CoV-2 Mpro interacting with the three ligands before and during the simulation run.
| Types of interactions and residues of main protease involved |
|||||
|---|---|---|---|---|---|
| Pre-MD interactions |
Interactions during MD simulations |
||||
| Ligand bound | Docking score (Kcal/mol) | H-bonds | Hydrophobic interactions and pi-pi stacking | H-bonds | Hydrophobic interactions and pi-pi stacking |
| N3 inhibitor | −5.68 | Glu166, Thr190 | His41, Cys44, Met49, Pro52, Tyr54, Phe140, Leu141, Asn142, His164, Met165, Leu167, Pro168, Thr169, Gly170, His172, Asp187, Arg188, Gln189, Ala191, Gln192 | Asn142, Glu166, Arg188, Gln189, Thr190, Gln192 | His41, Ser46, Met49, Lys138, Ser139, Phe140, Asn142, Gly143, Ser144, Cys145, His163, His163, His164, Met165, Glu166, Leu167, Pro168, Thr169, Gly170, His172, Val186, Arg188, Gln189, Thr190, Ala191, Gln192 |
| Wi-N | −4.42 | Cys145 | Thr24, Thr25, Thr26, Leu27, His41, Met49, Tyr54, Asn142, Gly143, His164, Met165, Glu166, Arg188, Asp188, Gln189 | Thr26, His41, Cys44, Asn119, Asn142, Gly143, Gln189, Thr190, Gln192 | Gln19, Thr24, Thr25, Thr26, His41, Cys44, Thr45, Ser46, Glu47, Met49, Pro52, Tyr54, Asn119, Asn142, Gly143, Ser144, Cys145, His164, Met165, Glu166, Leu167, Pro168, Gly170, Asp187, Arg188, Gln189, Thr190, Gln192 |
| CAPE | −4.79 | Asn142, Glu166 | Leu27, Cys44, Met49, Pro52, Tyr54, Phe140, Leu141, Ser144, Cys145, His163, His164, Met165, Glu166, His172, Asp187, Arg188, Gln189 | Thr25, Cys44, Ser46, Phe140, Asn142, Cys145, Glu166, His172, Gln189, Thr190, Ala191, Gln192 | Thr24, Thr25, Thr26, Pro39, His41, Cys44, Thr45, Ser46, Glu47, Met49, Leu50, Asn51, Cys85, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His164, Met165, Glu166, Leu167, Pro168, His172, Phe181, Val186, Asp187, Arg188, Gln189, Thr190, Ala191, Gln192, Ala193, Ala194 |
3.3. MD Simulations predicted stable binding of Wi-N and CAPE to Mpro
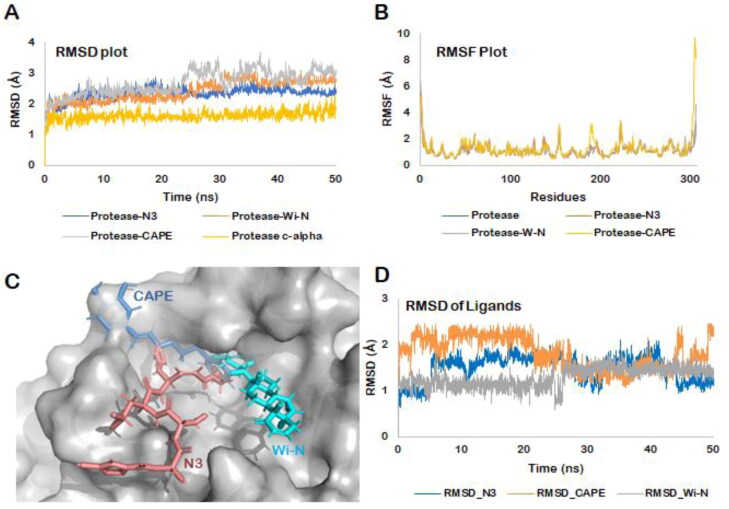
All biological systems can be viewed as dynamic networks of molecular interactions, while molecular docking represents only a single snapshot of the interaction between the protein and ligands. Therefore, to sample the myriad conformations that these complexes might acquire in solvated state, we simulated the dynamics behavior of all the three docked complexes using explicit water models for 50 ns each. The main aim was to confirm that the major contacts found in the best docked pose were maintained during the MD simulation as well keeping the ligand bound in the substrate-binding pocket of SARS-CoV-2 protease. We restricted our simulation runs to 50 ns because all the systems included in this study, apo enzyme or the docked complexes, got stabilized within 5–10 ns of the production runs (Figure 4(A)). No significant deviation was observed in the RMSD trajectory throughout the course of simulation thereafter. Though a little deviation was observed in the simulation trajectory of Mpro-CAPE around the 25–30 ns interval, it finally converged in the last 20 ns time window of the simulation. When we inspected the trajectory, it was found that the catechol ring of CAPE, which was moving freely, was the main reason behind this fluctuation.
Figure 4.
(A) RMSD of the protein backbone along the simulation trajectory for the protein alone and all the docked complexes. The overall structure of Mpro did not change much after the binding of Wi-N or CAPE when compared to N3 inhibitor. (B) RMSF of the amino acids comprising the Mpro. No abrupt fluctuations were observed in any region of the protein with or without the three ligands. (C) Superimposition of the three docked complexes. All the three small molecules- N3 protease inhibitor, Wi-N and CAPE were bound in the same site suggesting their similar mechanism of action. (D) RMSD plot for all the three ligands over the entire simulation trajectory. Similar to N3 inhibitor, Wi-N and CAPE stayed bound in almost the same docked pose throughout the simulation run.
The overall confirmation of all the docked complexes- SARS-CoV-2 Mpro-N3, SARS-CoV-2 Mpro-Wi-N and SARS-CoV-2 Mpro-CAPE was found to be stable with not much deviation from the docked pose. As shown in Figure 4(A), the average RMSD of SARS-CoV-2 Mpro-N3 complex was 2.32 ± 0.09 Å, SARS-CoV-2 Mpro-Wi-N was 2.41 ± 0.33 Å and for SARS-CoV-2 Mpro-CAPE complex was 2.67 ± 0.39 Å. The RMSF plots for all the three complexes also showed that the residues lining the substrate binding pocket in each case were interacting quite stably with the bound ligands as indicated by the low fluctuating values (Figure 4(B)).
To evaluate the global changes in the structure of the Mpro upon binding with Wi-N and CAPE, we superimposed the average structure of the complexes computed from the stable simulation trajectory with the PDB structure of SARS-CoV-2 Mpro-N3. The RMSD was calculated to be 0.84 Å (SARS-CoV-2 Mpro-N3: SARS-CoV-2 Mpro-CAPE) and 0.52 Å (SARS-CoV-2 Mpro-N3: SARS-CoV-2 Mpro-Wi-N). The superimposition further confirmed that all the ligands were binding within the same pocket of the Mpro suggesting similar mechanism of action (Figure 4(C)). The stable RMSD of the coordinates of the ligands with respect to the coordinated in the docked pose further ensured that Wi-N and CAPE were bound at the same site through-out the simulation trajectory (Figure 4(D)).
3.4. Interaction profile of Wi-N and CAPE in comparison to N3 inhibitor
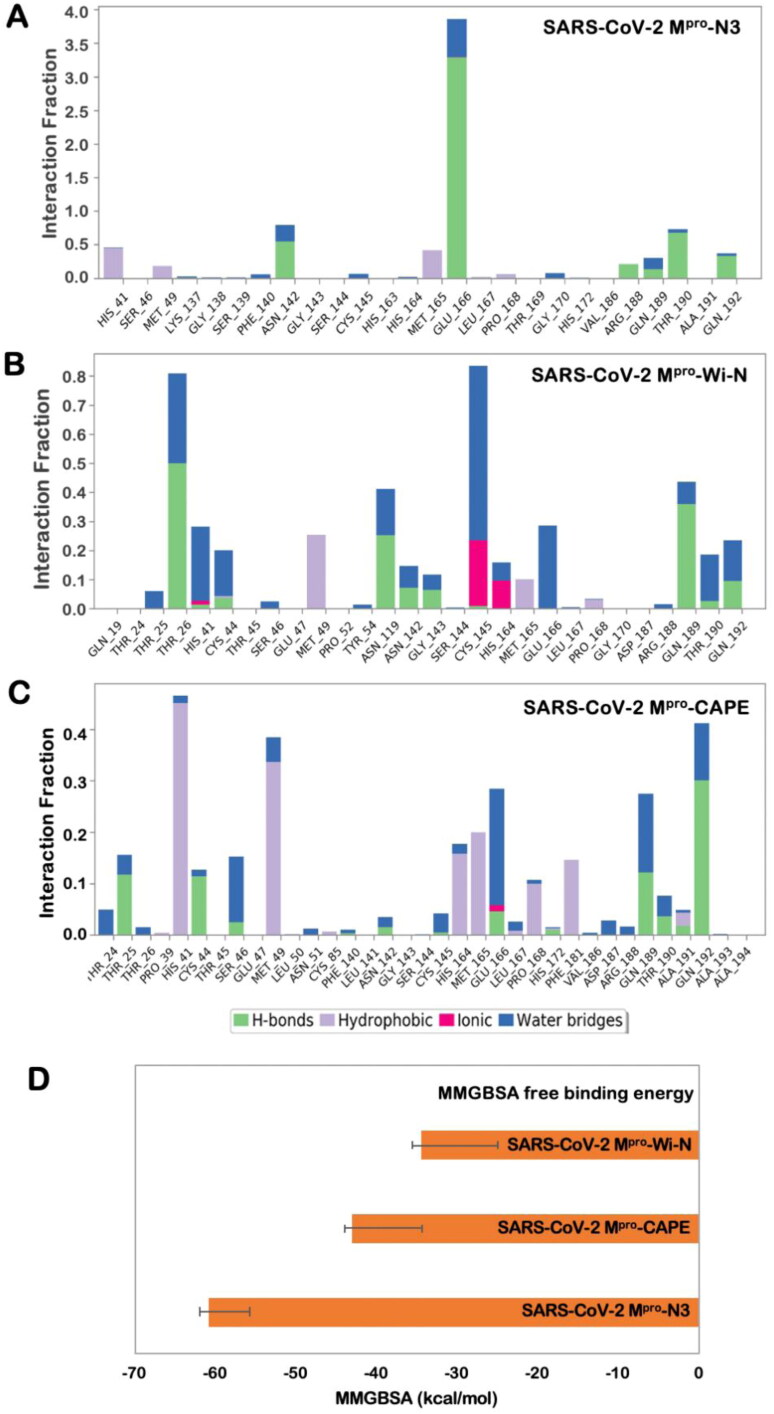
Although weaker than ionic and covalent interactions, hydrogen bonds are exploited the most for the design of effective drug molecules. Hydrogen bonds are the predominant contributors to the specificity of molecular recognition. The free energy for hydrogen bonding usually ranges from of −12 to −20 kJ/mol, and the binding affinity of a ligand increases by almost one order of magnitude per hydrogen bond. Hence, we looked into the hydrogen bonding pattern of N3 inhibitor, Wi-N and CAPE over the entire 50 ns simulation trajectory. Hydrogen bonds were calculated taking the criteria that the distance between hydrogen bond donor and acceptor should be less than 2.5 Å, the donor angle should be less than or equal to 120˚ between donor-hydrogen acceptor atoms, and the acceptor angle should be less than or equal to 90˚ between the hydrogen acceptor-bonded atoms. As shown in Figure 5(A–C), Wi-N was making more hydrogen bonds in comparison to CAPE (Figure 5(C)) and N3 inhibitor (Figure 5(A)) over the entire simulation trajectory. However, the main residues participating in hydrogen bond formation were slightly different. Wi-N was interacting with Glu166 mainly through water mediated interactions in contrast to the interaction profile of N3 inhibitor, where Glu166 had the highest hydrogen bond occupancy. Overall, the interactions analysis showed that at any fraction of time, Wi-N and CAPE were making better contacts while in terms of consistency N3 inhibitor was better. Also altogether, the significant polar interactions as well as specific hydrogen bond interactions (> 30% of occupancy) were more in case of Wi-N than in CAPE (Figure 5(B, C)).A detailed comparison of the residues interacting with the ligands is shown in Table 1. This analysis taking N3 inhibitor as a reference molecule thus suggests that Wi-N and CAPE has good affinity towards the substrate-binding pocket of SARS-CoV-2 Mpro and could probably be natural and readily available drugs for the inhibition of SARS-CoV-2 functional activity.
Figure 5.
Hydrogen bond occupancy of various important residues of the main protease during the simulation run in case of binding with N3 inhibitor (A), Wi-N (B) and CAPE (C). (D) Binding free energy calculated using MM/GBSA for the three protein-ligand complexes- SARS-CoV-2 Mpro-N3, SARS-CoV-2 Mpro-Wi-N and SARS-CoV-2 Mpro-CAPE.
3.5. MM/GBSA free binding energy
The MM/GBSA (molecular mechanics energies combined with the generalized Born and surface area continuum solvation) is a method commonly used or estimating ligand-binding affinities in protein systems. Here, the net free energy change is treated as the sum of a comprehensive set of individual energy components.
In order to overcome the potential limitations of molecular docking, we had further subjected our protein-ligand complexes to MD simulations and MM/GBSA calculations. For MM/GBSA calculations, instead of using a single representative structure, we used hundred frames equally spanned over the stable trajectory for each protein-ligand complex in between 10 to 50 ns for calculating the MMGBSA free binding energy. The values of free binding energy were following the same trend as the docking score. The average MMGBSA free binding energy of N3 inhibitor, CAPE and Wi-N with SARS-CoV-2 Mpro was −60.80 ± 5.04 Kcal/mol, −43.00 ± 8.72 Kcal/mol and −34.51 ± 9.63 Kcal/mol, respectively (Figure 5(D)). Though the free binding energy of N3 inhibitor was more in comparison to Wi-N and CAPE, the values were still good enough to suggest good binding affinity of these natural compounds towards SARS-CoV-2 protease.
Natural resources of Wi-N and CAPE (Ashwagandha and honeybee propolis, respectively) have long history of use in traditional home medicine systems, are generally recognized as safe, easily available and affordable. There are several reports that provide evidence that both CAPE and Ashwagandha derived withanolides (Akyol et al., 2013; Amoros et al., 1994; Anjaly & Tiku, 2018; Cai et al., 2015; Erdemli et al., 2015; Kaul & Wadhwa, 2017; Kwon et al., 2019; Latheef et al., 2017; Lyu et al., 2005; Munagala et al., 2011; Murtaza et al., 2015; Serkedjieva et al., 1992; Zandi et al., 2011) possess antiviral activity. Mechanism of action of antiviral activities has not been completely understood. The propolis extract has been reported to be effective against herpes simplex virus type 1; it significantly reduced the viral titer and also inhibited its replication (Amoros et al., 1994). In light of the published literature on antiviral properties of CAPE, the anti- SARS-CoV-2 activity may be mediated by inhibition of virus adsorption, virus-cell fusion, viral DNA/RNA synthesis or inhibition of viral essential enzymes such as protease and neuraminidase. Shen et al., 2013 attributed anti-HCV activity of CAPE to its unique structure and length of the n-alkyl side chain and catechol moieties. It was shown that CAPE fails to bind to DNA (De Clercq, 2002) and strongly inhibits viral integration and fusion by inhibition of viral integrase (an established target for antiviral drugs) (Costi et al., 2004; De Clercq, 2002; Fesen et al., 1993; Sud'ina et al., 1993). It was also shown to inhibit some endogenous and viral proteins as well as a transcription of NF-κB (Norris et al., 2010). Norris et al., 2010 demonstrated that CAPE caused dose dependent downregulation of a viral protease (HCV-NS3) at protein level that translated to inhibition of viral replication suggesting that CAPE is a promising therapeutic drug for HCV treatment. Therapeutic efficacy of CAPE and its four analogues (methyl caffeate, ethyl 3-(3,4-dihydroxyphenyl) acrylate, phenethyl dimethyl caffeate and phenethyl 3-(4-bromophenyl) acrylic) were also investigated for HIV in in vitro and in vivo wherein significant inhibition of HIV replication was detected (Ho et al., 2005). It was shown to modify protein synthesis profile in type 5 adenovirus-transformed cloned rat embryo fibroblast cells (Lefkovits et al., 1997) and inhibited the growth of Type A and B influenza virus by 95% and 92%, respectively (Kishimoto et al., 2005). These data have firmly suggested that the antiviral activity of CAPE is mediated by multiple pathways. Interestingly, among the chemical drugs that have recently been tested against coronaviral infection in cell culture models, Chloroquine (malaria drug) and Remdesivir (RNA-dependent RNA polymerase inhibitor) have shown some therapeutic response. Both malaria and viral infection require host cell PAK1 (Pathogenic kinase, an oncogenic Rac/CDC42-dependent Ser/Thr kinase) (Maruta, 2020) and hence the latter has been considered as a valid target for anti-malaria drugs. Intriguingly, CAPE has been shown to be a strong inhibitor of PAK1 (Coleman et al., 2016; Demestre et al., 2009). On the other hand, Ashwagandha-derived bioactive withanolide, Wi-A was also found to be effective against nuraminidase (a membrane antigen) that helps in the release of H1N1 influenza virus from host cell after replication (Cai et al., 2015). It has been reported to inhibit the expression of human papilloma virus oncogenes E6/E7 (Munagala et al., 2011). Interestingly, an independent study used bioinformatics screening of thousands of phytochemicals against ACE2 protein, a cellular target involved in SARS-CoV-2 infection, and selected Wi-N as a potential inhibitor (Balkrishna et al., 2020). Of note, our analyses predicted that Wi-N, but not its closely related withanolide Wi-A, is capable of interacting with and inhibiting viral Mpro endorsing its prioritized biological validation.
4. Conclusion
The present study predicted a strong possibility that Wi-N and CAPE possess inhibitory potential for SARS-CoV-2 protease Mpro. Based on the data, although the natural resources of these compounds (such as Ashwagnadha and honeybee propolis) are deemed helpful, preparation of quality-controlled extracts to possess high level of Wi-N and CAPE and their experimental validation in the laboratory and clinical studies are warranted. On the other hand, the data may also provide leads for drug-designing/development for the treatment of COVID-19. This would serve an important purpose of saving time and cost in initiating and implementing the drug screenings in the current scenario of international health emergency caused by COVID-19 epidemic and the lack of treatment modalities.
Author contributions
Conceptualization, V.K., D.S.; Formal analysis V.K., J.K.D., D.S.; Funding acquisition S.C.K., R.W., D.S.; Writing - review & editing V.K., J. K.D., S.C.K., R.W., D.S. All authors contributed to the development of this manuscript and read and approved the final version.
Funding Statement
This study was supported by the funds granted by AIST (Japan) and Department of Biotechnology (Govt. of India). The computations were performed at the Bioinformatics Centre at IIT Delhi.
Data availability statement
All data generated or analyzed during this study are included in this published article.
Disclosure statement
No potential conflict of interest is reported by the authors.
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelli I., Hassani F., Bekkel Brikci S., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyol S., Ozturk G., Ginis Z., Armutcu F., Yigitoglu M. R., & Akyol O. (2013). In vivo and in vitro antineoplastic actions of caffeic acid phenethyl ester (CAPE): therapeutic perspectives. Nutrition and Cancer, 65(4), 515–526. 10.1080/01635581.2013.776693 [DOI] [PubMed] [Google Scholar]
- Al-Khafaji K., Al-DuhaidahawiL D., & Taskin Tok T. (2020). Using integrated computational approaches to identify safe and rapid treatment for SARS -CoV- 2. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1764392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoros M., Lurton E., Boustie J., Girre L., Sauvager F., & Cormier M. (1994). Comparison of the anti-herpes simplex virus activities of propolis and 3-methyl-but-2-enyl caffeate. Journal of Natural Products, 57(5), 644–647. 10.1021/np50107a013 [DOI] [PubMed] [Google Scholar]
- Anjaly K., & Tiku A. B. (2018). Radio-modulatory potential of caffeic acid phenethyl ester: A therapeutic perspective. Anti-Cancer Agents in Medicinal Chemistry, 18(4), 468–475. 10.2174/1871520617666171113143945 [DOI] [PubMed] [Google Scholar]
- Balkrishna A., Pokhrel S., Singh J., & Varshney A. (2020). Withanone from Withania somnifera may inhibit novel Coronavirus (COVID-19) entry by disrupting interactions between viral S-protein receptor binding domain and host ACE2 receptor. Virology Journal, 1–26. 10.21203/rs.3.rs-17806/v131906972 [DOI] [Google Scholar]
- Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., & Ciccozzi M. (2020). The 2019-new coronavirus epidemic: Evidence for virus evolution. Journal of Medical Virology, 92(4), 455–459. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K. J., Chow D. E., Xu H., Dror R. O., Eastwood M. P., Gregersen B. A., Klepeis J. L., Kolossvary I., Moraes M. A., Sacerdoti F. D., & Salmon J. K. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters [Paper presentation]. SC ‘06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, Florida: (pp. 43–44). Association for Computing Machinery; 10.1109/SC.2006.54 [DOI] [Google Scholar]
- Cai Z., Zhang G., Tang B., Liu Y., Fu X., & Zhang X. (2015). Promising anti-influenza properties of active constituent of withania somnifera ayurvedic herb in targeting neuraminidase of H1N1 influenza: Computational study. Cell Biochemistry and Biophysics, 72(3), 727–739. 10.1007/s12013-015-0524-9 [DOI] [PubMed] [Google Scholar]
- Ceraolo C., & Giorgi F. M. (2020). Genomic variance of the 2019-nCoV coronavirus. Journal of Medical Virology, 92(5), 522–528. 10.1002/jmv.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Tung Y. A., Lee K. H., Chen T. F., Hsiao Y. C., Chang H. C., Hsieh T. T., Su C. H., Wang S. S., Yu J. Y., & Shih S. S. (2020). Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. Preprints, 1–11. 10.20944/preprints202002.0242.v1 [DOI] [Google Scholar]
- Coleman J. J., Komura T., Munro J., Wu M. P., Busanelli R. R., Koehler A. N., Thomas M., Wagner F. F., Holson E. B., & Mylonakis E. (2016). Activity of caffeic acid phenethyl ester in Caenorhabditis elegans. Future Medicinal Chemistry, 8(17), 2033–2046. 10.4155/fmc-2016-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020). The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5(4), 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costi R., Di Santo R., Artico M., Massa S., Ragno R., Loddo R., La Colla M., Tramontano E., La Colla P., & Pani A. (2004). 2,6-Bis(3,4,5-trihydroxybenzylydene) derivatives of cyclohexanone: Novel potent HIV-1 integrase inhibitors that prevent HIV-1 multiplication in cell-based assays. Bioorganic & Medicinal Chemistry, 12(1), 199–215. 10.1016/j.bmc.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Das S., Sarmah S., Lyndem S., & Singha Roy A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–18. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. (2002). Strategies in the design of antiviral drugs. Nature Reviews. Drug Discovery, 1(1), 13–25. 10.1038/nrd703 [DOI] [PubMed] [Google Scholar]
- Demestre M., Messerli S. M., Celli N., Shahhossini M., Kluwe L., Mautner V., & Maruta H. (2009). CAPE (caffeic acid phenethyl ester)-based propolis extract (Bio 30) suppresses the growth of human neurofibromatosis (NF) tumor xenografts in mice. Phytotherapy Research: PTR, 23(2), 226–230. 10.1002/ptr.2594 [DOI] [PubMed] [Google Scholar]
- Elfiky A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdemli H. K., Akyol S., Armutcu F., & Akyol O. (2015). Antiviral properties of caffeic acid phenethyl ester and its potential application. Journal of Intercultural Ethnopharmacology, 4(4), 344–347. 10.5455/jice.20151012013034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA News Release (2020). Coronavirus (COVID-19) update: FDA Issues emergency use authorization for potential COVID-19 treatment Retrieved May 01, 2020, from https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment.
- Fesen M. R., Kohn K. W., Leteurtre F., & Pommier Y. (1993). Inhibitors of human immunodeficiency virus integrase. Proceedings of the National Academy of Sciences, 90(6), 2399–2403. 10.1073/pnas.90.6.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., Sanschagrin P. C., & Mainz D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., & Shenkin P. S. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749. 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- Graham R. L., Donaldson E. F., & Baric R. S. (2013). A decade after SARS: Strategies for controlling emerging coronaviruses. Nature Reviews. Microbiology, 11(12), 836–848. 10.1038/nrmicro3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J. R., Calkins D., Sullivan A. P., & Shelley J. C. (2010). Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. Journal of Computer-Aided Molecular Design, 24(6–7), 591–604. 10.1007/s10822-010-9349-1 [DOI] [PubMed] [Google Scholar]
- Gupta M. K., & Vadde R. (2020). Insights into the structure-function relationship of both wild and mutant zinc transporter ZnT8 in human: A computational structural biology approach. Journal of Biomolecular Structure & Dynamics, 38(1), 137–151. 10.1080/07391102.2019.1567391 [DOI] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi G. A., Ogunro O. B., Adegunloye A. P., Ogunyemi O. M., & Afolabi S. O. (2020). Potential inhibitors of Coronavirus 3-Chymotrypsin-like protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J. Y., Wang L., Lupyan D., Dahlgren M. K., Knight J. L., Kaus J. W., Cerutti D. S., Krilov G., Jorgensen W. L., Abel R., & Friesner R. A. (2016). OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. Journal of Chemical Theory and Computation, 12(1), 281–296. 10.1021/acs.jctc.5b00864 [DOI] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., & Shahpasand K. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. C., Lin S. S., Chou M. Y., Chen F. L., Hu C. C., Chen C. S., Lu G. Y., & Yang C. C. (2005). Effects of CAPE-like compounds on HIV replication in vitro and modulation of cytokines in vivo. The Journal of Antimicrobial Chemotherapy, 56(2), 372–379. 10.1093/jac/dki244 [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., & Pöhlmann S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. P., Pincus D. L., Rapp C. S., Day T. J., Honig B., Shaw D. E., & Friesner R. A. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins, 55(2), 351–367. 10.1002/prot.10613 [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., … Yang H. (2020). Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature, 1–33. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S. C., & Wadhwa R. (Eds.). (2017). Science of ashwagandha: Preventive and therapeutic potentials. Springer International Publishing. [Google Scholar]
- Khan R. J., Jha R. K., Amera G., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2’-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kishimoto N., Kakino Y., Iwai K., Mochida K., & Fujita T. (2005). In vitro antibacterial, antimutagenic and anti-influenza virus activity of caffeic acid phenethyl esters. Biocontrol Science, 10(4), 155–161. 10.4265/bio.10.155 [DOI] [Google Scholar]
- Kumar D., Kumari K., Jayaraj A., Kumar V., Kumar R. V., Dass S. K., Chandra R., & Singh P. (2020). Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M. J., Shin H. M., Perumalsamy H., Wang X., & Ahn Y. J. (2019). Antiviral effects and possible mechanisms of action of constituents from Brazilian propolis and related compounds. Journal of Apicultural Research, 1–13. 10.1080/00218839.2019.1695715 [DOI] [Google Scholar]
- Latheef S. K., Dhama K., Samad H. A., Wani M. Y., Kumar M. A., Palanivelu M., Malik Y. S., Singh S. D., & Singh R. (2017). Immunomodulatory and prophylactic efficacy of herbal extracts against experimentally induced chicken infectious anaemia in chicks: Assessing the viral load and cell mediated immunity. VirusDisease, 28(1), 115–120. 10.1007/s13337-016-0355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkovits I. V. A. N., Su Z., Fisher P., & Grunberger D. (1997). Caffeic acid phenethyl ester profoundly modifies protein synthesis profile in type 5 adenovirus-transformed cloned rat embryo fibroblast cells. International Journal of Oncology, 11(1), 59–67. 10.3892/ijo.11.1.59 [DOI] [PubMed] [Google Scholar]
- Li X., Wang W., Zhao X., Zai J., Zhao Q., Li Y., & Chaillon A. (2020). Transmission dynamics and evolutionary history of 2019-nCoV. Journal of Medical Virology, 92(5), 501–511. 10.1002/jmv.25701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zai J., Wang X., & Li Y. (2020). Potential of large “first generation” human-to-human transmission of 2019-nCoV. Journal of Medical Virology, 92(4), 448–454. 10.1002/jmv.25693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo-Galo N., Terrazas-López M., Martínez-Martínez A., & Díaz-Sánchez Á. G. (2020). FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1764393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., … Tan W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu S. Y., Rhim J. Y., & Park W. B. (2005). Antiherpetic activities of flavonoids against herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) in vitro. Archives of Pharmacal Research, 28(11), 1293–1301. 10.1007/BF02978215 [DOI] [PubMed] [Google Scholar]
- Maruta H. (2020). Tackling the Coronaviral infection: Blocking either the “pathogenic” kinase PAK1 or polymerase RdRP. Journal of Infectious Diseases and Therapy, 8(2), 418. [Google Scholar]
- Munagala R., Kausar H., Munjal C., & Gupta R. C. (2011). Withaferin A induces p53-dependent apoptosis by repression of HPV oncogenes and upregulation of tumor suppressor proteins in human cervical cancer cells. Carcinogenesis, 32(11), 1697–1705. 10.1093/carcin/bgr192 [DOI] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Murtaza G., Sajjad A., Mehmood Z., Shah S. H., & Siddiqi A. R. (2015). Possible molecular targets for therapeutic applications of caffeic acid phenethyl ester in inflammation and cancer. Journal of Food and Drug Analysis, 23(1), 11–18. 10.1016/j.jfda.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris P. J., Hirschkorn D. F., DeVita D. A., Lee T. H., & Murphy E. L. (2010). Human T cell leukemia virus type 1 infection drives spontaneous proliferation of natural killer cells. Virulence, 1(1), 19–28. 10.4161/viru.1.1.9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E. G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., & Tsiodras S. (2020). Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 79, 104212 10.1016/j.meegid.2020.104212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., & Sherman W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Schrodinger Release 2020-1. (2020). Maestro 019-3 SR, glide, ligprep, protein preparation wizard, prime, Desmond molecular dynamics system, Maestro-Desmond interoperability tools. Schrödinger. [Google Scholar]
- Serkedjieva J., Manolova N., & Bankova V. (1992). Anti-influenza virus effect of some propolis constituents and their analogues (esters of substituted cinnamic acids). Journal of Natural Products, 55(3), 294–297. 10.1021/np50081a003 [DOI] [PubMed] [Google Scholar]
- Shen H., Yamashita A., Nakakoshi M., Yokoe H., Sudo M., Kasai H., Tanaka T., Fujimoto Y., Ikeda M., Kato N., Sakamoto N., Shindo H., Maekawa S., Enomoto N., Tsubuki M., & Moriishi K. (2013). Inhibitory effects of caffeic acid phenethyl ester derivatives on replication of hepatitis c virus. PloS One, 8(12), e82299 10.1371/journal.pone.0082299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud'ina G. F., Mirzoeva O. K., Pushkareva M. A., Korshunova G. A., Sumbatyan N. V., & Varfolomeev S. D. (1993). Caffeic acid phenethyl ester as a lipoxygenase inhibitor with antioxidant properties. FEBS Letters, 329(1–2), 21–24. 10.1016/0014-5793(93)80184-V [DOI] [PubMed] [Google Scholar]
- Thuy B. T. P., My T. T. A., Hai N. T. T., Hieu L. T., Hoa T. T., Thi Phuong Loan H., Triet N. T., Anh T. T. V., Quy P. T., Tat P. V., Hue N. V., Quang D. T., Trung N. T., Tung V. T., Huynh L. K., & Nhung N. T. A. (2020). Investigation into SARS-CoV-2 resistance of compounds in garlic essential oil. ACS Omega, 5(14), 8312–8320. 10.1021/acsomega.0c00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh K. D., Selvaraj C., Singh S. K., & Dubey V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M. F., Vermeulen-Oost W., Berkhout R. J., Wolthers K. C., Wertheim-van Dillen P. M., Kaandorp J., Spaargaren J., & Berkhout B. (2004). Identification of a new human coronavirus. Nature Medicine, 10(4), 368–373. 10.1038/nm1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang F., Chen C., Tan W., Yang K., & Yang H. (2016). Structure of main protease from human Coronavirus NL63: Insights for wide spectrum anti-coronavirus drug design. Scientific Reports, 6, 22677 10.1038/srep22677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chen X., Lu Y., Chen F., & Zhang W. (2020). Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Bioscience Trends, 14(1), 64–68. 10.5582/bst.2020.01030 [DOI] [PubMed] [Google Scholar]
- WHO (2020). Coronavirus disease (COVID-19) dashboard Retrieved May 12, 2020, from https://covid19.who.int.
- Woo P. C., Huang Y., Lau S. K., & Yuen K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses, 2(8), 1804–1820. 10.3390/v2081803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. Y., Lau S. K. P., Chu C-m., Chan K-h., Tsoi H-w., Huang Y., Wong B. H. L., Poon R. W. S., Cai J. J., Luk W-k., Poon L. L. M., Wong S. S. Y., Guan Y., Peiris J. S. M., & Yuen K-y. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, 79(2), 884–895. 10.1128/JVI.79.2.884-895.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., & Zhang Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi K., Teoh B. T., Sam S. S., Wong P. F., Mustafa M. R., & AbuBakar S. (2011). Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virology Journal, 8(1), 560 10.1186/1743-422X-8-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., … Shi Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.