Abstract
Sever acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a single-stranded RNA (ssRNA) virus, responsible for severe acute respiratory disease (COVID-19). A large number of natural compounds are under trial for screening compounds, possessing potential inhibitory effect against the viral infection. Keeping in view the importance of marine compounds in antiviral activity, we investigated the potency of some marine natural products to target SARS-CoV-2 main protease (Mpro) (PDB ID 6MO3). The crystallographic structure of Mpro in an apo form was retrieved from Protein Data Bank and marine compounds from PubChem. These structures were prepared for docking and the complex with good docking score was subjected to molecular dynamic (MD) simulations for a period of 100 ns. To measure the stability, flexibility, and average distance between the target and compounds, root mean square deviations (RMSD), root mean square fluctuation (RMSF), and the distance matrix were calculated. Among five marine compounds, C-1 (PubChem CID 11170714) exhibited good activity, interacting with the active site and surrounding residues, forming many hydrogen and hydrophobic interactions. The C-1 also attained a stable dynamic behavior, and the average distance between compound and target remains constant. In conclusion, marine natural compounds may be used as a potential inhibitor against SARS-CoV-2 for better management of COVID-19.
Keywords: Marine drugs, COVID-19, dock, interactions, molecular dynamics
1. Introduction
Coronavirus pandemic-19 (COVID‑19) is an ongoing disease caused by severe acute respiratory syndrome (SARS-CoV-2). According to the WHO latest report, 3,442,234 are confirmed SARS-CoV-2 infected people including 239,740 deaths. Among the six WHO regions, the largest number of cases has been reported from Europe (1,544,145) followed by Americas (1,433,756), Eastern Mediterranean (211,555), Western Pacific (152,774), South-East Asia (68,756), and Africa (30,536) (WHO COVID-19 Dashboard, n.d.). Since it was first identified in December 2019, COVID-19 has infected a large population of people around the world (Coronavirus Disease 2019 (COVID-19) Situation Report-35, n.d.).
The SARS-CoV-2, previously known as 2019-nCoV, is a single-stranded RNA (ssRNA) betacoronavirus, responsible for a severe pathological condition (Guarner, 2020). The COVID-19 is expanding rapidly as compared with previous coronaviruses (SARS-CoV and MERS-CoV) with the absence of therapeutic agents (Heymann et al., 2020; Zhang & Liu, 2020). On January 2020, the International Health Regulations Emergency Committee of the World Health Organization declared the outbreak as ‘public health emergency’ in responding to SARS-COVID-19.
Unfortunately, the timeline for characterizing a typical drug discovery process badly couples with the urgency of finding a therapy. It is important to accelerate the early stages of the drug discovery for all possible future emergencies (Mani et al., 2019). The early extraction of the COVID-19 genome to highlight sequence identity (∼80% of conserved nucleotides) with respect to the original SARS-CoV (Gralinski & Menachery, 2020) has paved the way for rapid research.
Although commercially synthetic sources prepared many drugs but the major hurdles, drug side effects, resistance, cell toxicity, and long-term treatment, were some factors behind the failure. The potential marine products are playing a pivotal role in the identification of novel prototypes and also developing drugs using natural products of the marine environment (Vo & Kim, 2010; Wittine et al., 2019). Over two-thirds of the planet has been covered by marine species, making them a major source for novel drug-like compounds (Aneiros & Garateix, 2004; Mayer et al., 2019). Further, a possible vaccine target is viral structural proteins, the development of which is desirable and it is foreseen that the first candidates will be advanced to clinical phase I around mid-2020 (Boopathi et al., 2020; Keener, 2020; Khan, Jha et al., 2020; Letko & Munster, 2020; Sarma et al., 2020; Wrapp et al., 2020). In the meantime, however, a great effort involves the targeting of nonstructural viral proteins which are instead essential for the viral replication and the maturation processes, representing a specific target for anti-COVID-19 drug development (Ahmed et al., 2020; Anand et al., 2005; Gan et al., 2006; Gupta et al., 2020; Hasan et al., 2020; Khan, Zia et al., 2020; Sirois et al., 2007; Wei et al., 2006; Zhang & Liu, 2020). In the current scenario, the crystallographic structure of the SARS-CoV-2 main protease (Mpro) also called 3CL hydrolase or C30 endopeptidase, was made available to the scientific community, just a few weeks after the first COVID-19 outbreak (PDB ID: 6LU7 in complex, 6MO3 apo). The structural characterization of the main protease (Mpro) shares 96.1% of its sequence with those of previous SARS-CoV, contained a highly conserved architecture of the catalytic binding site. We took advantage of the recently solved crystallographic structure of SARS-COVID-19 to perform a cutting edge in-silico investigation.
Once the cell is infected with COVID-19, the existing molecular machinery of the host cell is taken over by the virus to translate its RNA into long chains of proteins, producing more copies. These long viral proteins are activated when cut into smaller pieces by proteases. Hence, viral proteases have a critical role in the propagation of the virus. Identification of specific inhibitors from natural products against the COVID-19 Mpro might be of great importance in terms of proposing the treatment regimen. Here in the current study, we searched some marine compounds and docked into the Mpro, shows a good binding interactions that might be useful against COVID-19.
2. Material and methods
2.1. Protein preparation
The recently submitted crystal structure of COVID-19 Mpro in an apo form (PDB ID: 6M03; Berman et al., 2000) was extracted from Protein Data Bank. The structure was subjected to preparation by Protein Preparation Wizard in molecular operating environment (MOE; Vilar et al., 2008). The missing hydrogens were added, and partial charges were assigned.
2.2. Ligand preparation
The 2 D structures of marine compounds (Table 1) from PubChem converted to 3 D structure via the Ligprep module in MOE. The protonation and ionization states of the compounds were corrected, and proper bond orders were assigned. Afterward, the tautomeric and ionization states were created for each ligand.
Table 1.
Marine compounds docked against SARS-COV-2 main protease.
| Compound no. | Formula | Molecular mass (kDa) | PubChem CID | Source |
|---|---|---|---|---|
| 1 | C31H30Br6N4O11 | 1114.02a | 11170714 | Family Aplysinidae |
| 2 | C19H40O3 | 316.53 | 21646261 | Family Aplysinidae |
| 3 | C16H30O2 | 254.41 | 445638 | Soft coral Pterogorgia citrina |
| 4 | C22H32O4 | 360.49 | 21591485 | Petrosia strongylophora sp. |
| 5 | C21H26O3 | 326.44 | 460087 | Petrosia strongylophora sp. |
awww.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation/; DeGoey et al. (2018).
2.3. Molecular docking
Five marine compounds reported earlier in the study (Felix et al., 2017) were docked using rigid receptor docking protocol in MOE. During the process of docking, the protein was fixed, while ligands were kept flexible. Residue selenomethionines were converted into methionine and side-chain polar hydrogen were refined. Molecular docking grid was specified and centered using 20 × 20 × 20 with 0.375 grid spacing. A total of 50 runs were performed to observe a wide range of conformational orientations.
2.4. Molecular dynamics (MD) simulation
MD simulation was carried out via Gromacs 5.1 [54] for a period of 100 ns. The system was stabilized by adding Na+/Cl− ions. Energy minimization (NVT and NPT) was performed in two-step for a duration of 50,000, continued till the maximum force reached below 1000 kJ/mol/nm. An overall pressure and temperature equal to 1 bar and 300 K were kept with a time gap of 2 fs to achieve a stable state. To maintains a constant temperature inside the box, the v-rescale, an optimized Berendsen thermostat temperature coupling technique, was used. Once the MD was completed, all the obtained trajectories were examined for conformational drifts. The root mean square deviation (RMSD) and root mean square fluctuation (RMSF) were calculated to measure the stability and flexibility of protein and compound. Cpptraj was used to calculate the average distance between marine natural compound and proteins during the simulation period (Bernardi et al., 2019; Gajula et al., 2016; Roe & Cheatham, 2013). Radius of gyration (Rg) was calculated to infer the stable protein folding.
2.5. ADME prediction
To analyze the pharmacodynamics of the marine compounds, Adsorption, Distribution, Metabolism, and Excretion (ADME) is important which could be used as a drug. SWISS-ADME (https://www.swissadme.ch) allows the user to include SMILES data from PubChem and provides lipophilicity, water solubility, and drug likeness rules. SMILES files of all the marine compounds retrieved from the PubChem was entered into the search bar and the results were analyzed.
3. Result and discussion
3.1. Marine drug and mpro interactions
In the current study, five marine compounds, designated as C-1, C-2, C-3, C-4, and C-5 (Table 1) have been docked in the crystal structure of viral Mpro. The compounds (Figure 1) exhibited a good interaction with viral Mpro, forming many hydrogen bonds (Figure 2). The ADME properties (supplementary data S1) shows that these compounds may be applied in the therapy of SARS-CoV-2. Although the molecular weight of C1 is very high but the new FDA approval seems beyond the Lipinski’s rule of five (www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation/). This may due be the increasing focus that offer potential for promising new therapeutic compounds for the treatment of diseases, particularly in the areas of virology and oncology. However, conducting drug discovery ‘beyond rule of 5’ chemical space offerings important drug design and challenges to medicinal scientist to achieve oral pharmacokinetics. In some cases, including HCV NS3/4A protease, and hepatitis C virus (HCV) NS5A inhibitors the Lipinski’s rule of five has not been considered (DeGoey et al., 2018).
Figure 1.
Structure of marine compounds. C-1 CID 11170714 containing halogen group (Br). These marine drugs have been identified in the previous study (Felix et al., 2017), active against latent Mycobacterium tuberculosis isolates. The drug has been observed as potent against Mpro, forming many hydrogen and hydrophobic interactions.
Figure 2.
Interaction between C-1 CID 11170714 and Mpro apo. Drug has been shown, enclosed in black color blanket, signifying the binding pocket (Vilar et al., 2008).
In the last 20 years, SARS and MERS have been found as new infectious agents, emerged to cause epidemics (de Wit et al., 2016; Guarner, 2020). Conventional drug development methods take years and costly, offering more time for transmission of pathogens. The appropriate and timely development of potent antiviral agents for clinical use is of central interest, using cost-effective and fast computational approaches. Moreover, the approved pharmaceutical drugs may be repurposed as alternative method to screen for rapid identification of potential leads (Chu et al., 2006; Enayatkhani et al., 2020; Muralidharan et al., 2020; Pillaiyar et al., 2016; Yang et al., 2005). In this regard, recently a large number of in-silico studies have been performed on medicinal plants, drug designing, and vaccine development (Aanouz et al., 2020; Elfiky, 2020a, 2020b; Elfiky & Azzam, 2020; Elmezayen et al., 2020; Enmozhi et al., 2020; Joshi et al., 2020; Pant et al., 2020).
Hundreds and thousands of humans have been died in many epidemics, broken out over the centuries. Some infections have been found, more deadly, especially viral pathogens. These pathogens have resisted in majority of cases to all kinds of medical treatment. Synthesizing drugs against rapidly replicated viruses resulting in acute syndromes is a laborious and time-consuming procedure, requires a lot of financial aid. However, the natural compounds are lying around on the earth on land and water (Abdelli et al., 2020; Das et al., 2020; Islam et al., 2020; Kumar et al., 2020; Sinha et al., 2020; Umesh et al., 2020; Wahedi et al., 2020) that could be screened for potential compounds against SARS-CoV-2 main targets.
Over a 1000 of novel marine compounds isolated from marine organisms are being pharmacologically tested, and over 40 are being existed in the medicine market. In modern pharmacological industry, marine products are paving the way for a new trend (Ahmadi et al., 2015; Che, 1991; Gogineni et al., 2015; Khan et al., 2019; Moghadamtousi et al., 2015; Raveh et al., 2013; Sagar et al., 2010; Uzair et al., 2011; Vijayakumar & Menakha, 2015).
Interactions of five marine natural products have been shown (Figures 2 and 3). Residues Thr24, Leu27, His41, Phe140, Cys145, His163, Met165, Pro168, and His172 are present in the active site and its surrounding (Wu et al., 2020). The drug C1 shows good binding affinity, forming many hydrogen and hydrophobic interactions. Compound C2 also exhibited interactions with active site of Mpro, creating a catalytic dyad, consist of Cys145 and His41, where the cysteine is a nucleophile in the proteolytic process (Figure 3). The best interacting pose was selected based on E_refine and E_score2. The more negative score shows a good ligand and protein complex (Table 2).
Figure 3.
COVID-19 Mpro residues forming hydrogen and hydrophobic interactions with five marine compounds. Compound C1 CID 11170714 exhibited more hydrogen and hydrophobic interactions. C2 interaction with Cys145, the active site of SARS-CoV 3CLpro creating a catalytic dyad (Cys145 and His41).
Table 2.
Docking score of marine drugs and COVID-19 Mpro.
| S | E_place | E_score1 | E_refine | E_score2 |
|---|---|---|---|---|
| −7.58 | −84.29 | −7.82 | −46.55 | −7.58 |
| −7.55 | −85.64 | −8.22 | −44.93 | −7.55 |
| −7.54 | −77.91 | −8.73 | −44.70 | −7.54 |
| −7.26 | −62.71 | −8.39 | −43.95 | −7.26 |
| −7.22 | −77.08 | −8.60 | −43.06 | −7.22 |
| −5.86 | −55.39 | −6.97 | −28.48 | −5.86 |
| −5.73 | −62.11 | −7.06 | −28.53 | −5.73 |
| −5.50 | −70.48 | −7.93 | −25.24 | −5.50 |
| −5.22 | −70.33 | −8.13 | −22.25 | −5.22 |
| −5.21 | −71.26 | −6.90 | −26.29 | −5.21 |
| −5.16 | −37.99 | −6.90 | −23.32 | −5.16 |
| −5.09 | −41.22 | −7.20 | −23.20 | −5.09 |
| −5.09 | −35.11 | −6.61 | −21.29 | −5.09 |
| −5.08 | −29.77 | −6.34 | −24.76 | −5.08 |
| −5.06 | −31.38 | −7.00 | −21.46 | −5.06 |
| −5.42 | −32.23 | −7.39 | −28.69 | −5.42 |
| −5.39 | −25.44 | −7.81 | −26.96 | −5.39 |
| −5.29 | −24.91 | −7.40 | −23.56 | −5.29 |
| −5.22 | −33.38 | −7.69 | −24.96 | −5.22 |
| −5.03 | −35.70 | −8.80 | −27.37 | −5.03 |
| −5.27 | −28.55 | −7.35 | −26.48 | −5.27 |
| −5.13 | −42.11 | −7.75 | −25.53 | −5.13 |
| −5.07 | −37.36 | −6.29 | −20.07 | −5.07 |
| −5.04 | −34.24 | −8.03 | −24.74 | −5.04 |
| −4.96 | −20.93 | −7.26 | −24.26 | −4.96 |
Natural products may provide lead compounds, especially as antimicrobial agents (Dias et al., 2012; Hu et al., 2015; Newman & Cragg, 2016). A large range of marine products displays chemical structures with good biological activities to discover drug like for various human diseases caused by virus, including COVID-19. An additional advantage of marine products, as most of them has the property of drug-likeness with high degrees of bioavailability, and effective drugs against viral diseases shortly.
Mpro (3CLpro) monomer has three domains: domain I, domain II, and domain III containing residues 8–101, residues 102–184, and residues 201–303 respectively, and a long loop (residues 185–200) connects domains III and II (Wu et al., 2020). The active site (Cys145 and His41) is located in the gap between domains I and II, while hydrophobic amino acids, T24, L27, H41, F140, C145, H163, M165, P168, and H172 also form a hydrophobic surrounding in the pocket (Yang et al., 2003). The identification of compounds fitting in the pockets is one of the fundamental step in structure-based drug design. The recent progress and developments of the computational analysis of pockets have been found useful to screen potent inhibitors (Zheng et al., 2013). Analysis of Mpro complex with C1 shows many hydrogen and hydrophobic interaction (Figure 4). The compound exhibited affinity with SARS-CoV-2 Mpro from all sides, showing its best fitting in the pocket. Drug interactions and fitting in the pocket is essential for drug designing and lead optimization. it It is also important to identify the locations of binding sites to infer protein–ligand binding or protein–protein interaction.
Figure 4.
Interaction of C1 after MD simulation. Residues, Ser46, Met49, Asp187, Gln192, Ala194, Thr169, and Gln189, are involved in hydrogen bonding.
In addition to hydrogen bonds, hydrophobic and electrostatic interactions are also important. The hydrogen bonds may play as an ‘anchoring’ role, defining the spatial location of the druggable compounds in the binding pocket, facilitating the electrostatic and hydrophobic interactions. In rational drug design, it is equally essential to recognize the hydrophobic groups of the compound and receptor, facing to each other upon binding. These interactions have been detected while analyzing the Connolly surface (Connolly, 1993) of the complex of SARS-CoV Mpro and marine compounds. It is the steric complementarity between the ligand and receptor site that performs the role of the principal driving force for mechanical interlocking (Chou et al., 2009; Sirois et al., 2007; Wei et al., 2006).
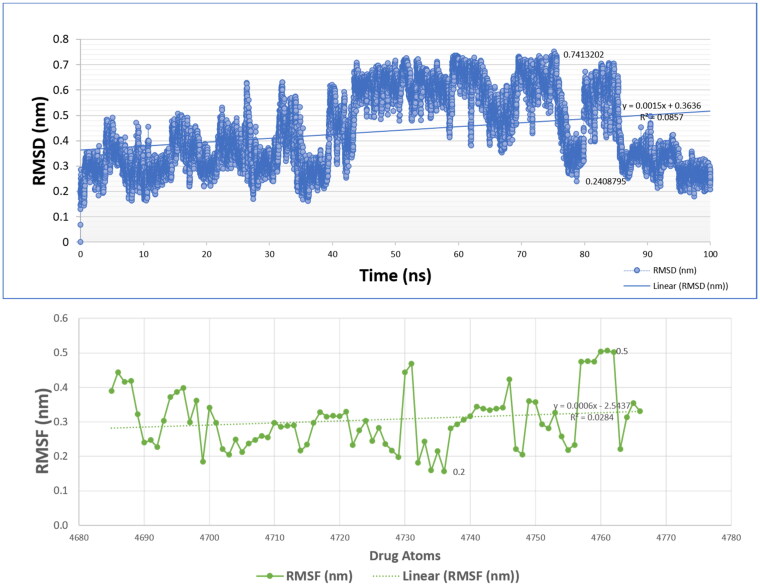
RMSD and RMSF are calculated in MD simulations to infer the stability and flexibility, a fundamental property of biomolecules. High deviation and fluctuation of proteins during a simulation may show weak stability and stability in thermodynamics (Chen & Shen, 2009). SARS-CoV-2 Mpro in complexed with C1 exhibited a stable RMSD between 0.2 and 0.45 nm (Figure 5) and the initial and final RMSDs during the whole simulation period were not found in the significance difference (0.2 and 0.3 nm). This shows a stable binding of C1 with Mpro that might be a useful as a good inhibitor. Moreover, residues fluctuations were also observed, not too flexible in motion (0.05–0.36 nm). Both, RMSD and RMSF stabilities are essential to infer good binding affinities (Doniach & Eastman, 1999; Dubey et al., 2013; Figure 6).
Figure 5.
RSMD and RMSF of Mpro in complex with C1 compound. The complex exhibited a stable RMSD and RMSF during a 100 ns MD simulation period.
Figure 6.
RMSD and RMSF of compound C1.
The initial and final RMSD of C1 atoms is almost similar. The residues atoms fluctuations have been detected in a range of 0.2–0.5 nm. However, the majority of C1 atoms exhibited RMSF below 0.3 nm.
The average distance of C1 and Mpro is approximately in range, with little fluctuation during the simulation period. However, the final and initial distance is almost similar (Figure 7). The distant matrix signifies the C1 and Mpro distance stability during the simulation period. This approach might be useful to infer the strong binding affinity during the simulation period (Ernst et al., 2015; Khan, Ashfaq-Ur-Rehman et al., 2020). The average distance is commonly affected when a variant occurs at the active site of target proteins during the course of therapy, causing drug resistance (Figure 8).
Figure 7.
Distance matrix between C1 and Mpro during 100 ns (100,000 ps).
Figure 8.
Radius of gyration. The degree of folding is constant during the whole simulation period.
The degree of compactness and folding is plotted against time, which is commonly measured through the radius of gyration (Rg). A long range variations in proteins show their weak folding (Lobanov et al., 2008; Smilgies & Folta-Stogniew, 2015). A stable Rg value shows compactness and stable folding maintains a steady value of Rg, required for proper function, whereas in case of misfolding, the Rg will show a long range of variation over time.
In conclusion, marine natural product is the most diverse group, containing potential inhibitors against RNA viruses. Among marine natural products, C1 forming many interactions with Thr24, Leu27, His41, Phe140, Cys145, His163, Met165, Pro168, and His172, present in the active site and its surrounding. These compounds have been observed as best fitting in the binding pocket, that might be good inhibitor of SARS-CoV-2 Mpro for better management of COVID-19.
Glossary
Abbreviations
- ADME
adsorption, distribution, metabolism and excretion
- HCV
Hepatitis C virus
- MD
molecular dynamic simulations
- MPro
Main protease
- MOE
molecular operating environment
- PDB
Protein Data Bank
- RMSD
Root mean square deviation,
- Rg
radius of gyration
- RMSF
root mean square fluctuation
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
Funding Statement
Dong-Qing Wei is supported by the grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Natural Science Foundation of China (Contract no. 61832019, 61503244), the Science and Technology Commission of Shanghai Municipality (Grant: 19430750600), the Natural Science Foundation of Henan Province (162300410060), and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2017ZD14). The computations were partially performed at the Peng Cheng Lab and the Center for High-Performance Computing, Shanghai Jiao Tong University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Acknowledgements
The current study as technically supported by Director PTRL KP, Peshawar, Dr. Sajid Ali, Molecular biologist, and Anwar Sheed Khan, microbiologist. Conceptualization was carried out by DQW, GS, and MTK; data curation by MTK, AA, QW, and SC; experimental work by MTK, QW, AA, and AK; formal analysis by AA, SC, and MTK; funding acquisition by DQW and approval by DQW and GS.
References
- Aanouz I., Belhassan A., Khatabi K. E., Lakhlifi T., Idrissi M. E., & Bouachrine M. (2020). Moroccan medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–12. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelli I., Hassani F., Brikci S. B., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–17. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi A., Zorofchian Moghadamtousi S., Abubakar S., & Zandi K. (2015). Antiviral potential of algae polysaccharides isolated from marine sources: A review. BioMed Research International, 2015, 825203 10.1155/2015/825203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S. F., Quadeer A. A., & McKay M. R. (2020). Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses, 12(3), 254 10.3390/v12030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Yang H., Bartlam M., Rao Z., & Hilgenfeld R. (2005). Coronavirus main proteinase: Target for antiviral drug therapy In Coronaviruses with special emphasis on first insights concerning SARS (pp. 173–199). Birkhäuser Basel; 10.1007/3-7643-7339-3_9 [DOI] [Google Scholar]
- Aneiros A., & Garateix A. (2004). Bioactive peptides from marine sources: Pharmacological properties and isolation procedures . Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 803(1), 41–53. 10.1016/j.jchromb.2003.11.005 [DOI] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., & Bourne P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi A., Faller R., Reith D., & Kirschner K. N. (2019). ACPYPE update for nonuniform 1–4 scale factors: Conversion of the GLYCAM06 force field from AMBER to GROMACS. SoftwareX, 10, 100241 10.1016/j.softx.2019.100241 [DOI] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 0(0), 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che C.-T. (1991). Marine products as a source of antiviral drug leads. Drug Development Research, 23(3), 201–218. 10.1002/ddr.430230302 [DOI] [Google Scholar]
- Chen J., Shen B. (2009). Computational analysis of amino acid mutation: A proteome wide perspective [Text]. https://www.ingentaconnect.com/content/ben/cp/2009/00000006/00000004/art00004
- Chou K.-C., Wei D.-Q., Du Q.-S., Sirois S., Shen H.-B., & Zhong W.-Z. (2009). Study of inhibitors against SARS coronavirus by computational approaches In Lendeckel U. & Hooper N. M. (Eds.), Viral proteases and antiviral protease inhibitor therapy: Proteases in biology and disease (pp. 1–23). Springer; 10.1007/978-90-481-2348-3_1 [DOI] [Google Scholar]
- Chu L.-H M., Choy W.-Y., Tsai S.-N., Rao Z., & Ngai S.-M. (2006). Rapid peptide-based screening on the substrate specificity of severe acute respiratory syndrome (SARS) coronavirus 3C-like protease by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein Science: A Publication of the Protein Society, 15(4), 699–709. 10.1110/ps.052007306 [pubmedMismatch]7Connolly M. L. (1993). The molecular surface package. Journal of Molecular Graphics, 11(2), 139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus disease 2019 (COVID-19) Situation Report-35 (n.d.). Retrieved February 26, 2020, from https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200224-sitrep-35-covid-19.pdf
- Das S., Sarmah S., Lyndem S., & Roy A. S. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–18. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., & Munster V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews. Microbiology, 14(8), 523–534. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGoey D. A., Chen H.-J., Cox P. B., & Wendt M. D. (2018). Beyond the Rule of 5: Lessons learned from AbbVie’s drugs and compound collection. Journal of Medicinal Chemistry, 61(7), 2636–2651. 10.1021/acs.jmedchem.7b00717 [DOI] [PubMed] [Google Scholar]
- Dias D. A., Urban S., & Roessner U. (2012). A historical overview of natural products in drug discovery. Metabolites, 2(2), 303–336. 10.3390/metabo2020303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniach S., & Eastman P. (1999). Protein dynamics simulations from nanoseconds to microseconds. Current Opinion in Structural Biology, 9(2), 157–163. 10.1016/S0959-440X(99)80022-0 [DOI] [PubMed] [Google Scholar]
- Dubey K. D., Tiwari R. K., & Ojha R. P. (2013). Recent advances in protein-ligand interactions: Molecular dynamics simulations and binding free energy. Current Computer-Aided Drug Design, 9(4), 518–531. 10.2174/15734099113096660036 [DOI] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–15. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–16. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 0(0), 1–9. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 0(0), 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 0(0), 1–16. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–10. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M., Sittel F., & Stock G. (2015). Contact- and distance-based principal component analysis of protein dynamics. The Journal of Chemical Physics, 143(24), 244114 10.1063/1.4938249 [DOI] [PubMed] [Google Scholar]
- Felix C. R., Gupta R., Geden S., Roberts J., Winder P., Pomponi S. A., Diaz M. C., Reed J. K., Wright A. E., & Rohde K. H. (2017). Selective killing of dormant mycobacterium tuberculosis by marine natural products. Antimicrobial Agents and Chemotherapy, 61(8), e00743–17. 10.1128/AAC.00743-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajula M., Kumar A., & Ijaq J. (2016). Protocol for molecular dynamics simulations of proteins. Bio-Protocol, 6(23), e2051. 10.21769/BioProtoc.2051 [DOI] [Google Scholar]
- Gan Y.-R., Huang H., Huang Y.-D., Rao C.-M., Zhao Y., Liu J.-S., Wu L., & Wei D.-Q. (2006). Synthesis and activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase. Peptides, 27(4), 622–625. 10.1016/j.peptides.2005.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogineni V., Schinazi R. F., & Hamann M. T. (2015). Role of marine natural products in the genesis of antiviral agents. Chemical Reviews, 115(18), 9655–9706. 10.1021/cr4006318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L. E., & Menachery V. D. (2020). Return of the coronavirus: 2019-nCoV. Viruses, 12(2), 135 10.3390/v12020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner J. (2020). Three emerging coronaviruses in two decades: The story of SARS, MERS, and Now COVID-19. American Journal of Clinical Pathology, 153(4), 420–421. 10.1093/ajcp/aqaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 0(0), 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A. A., & Falahati M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 0(0), 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann D. L., & Shindo N, & WHO Scientific and Technical Advisory Group for Infectious Hazards (2020). COVID-19: What is next for public health? The Lancet, 395(10224), 542–545. 10.1016/S0140-6736(20)30374-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Chen J., Hu G., Yu J., Zhu X., Lin Y., Chen S., & Yuan J. (2015). Statistical research on the bioactivity of new marine natural products discovered during the 28 years from 1985 to 2012. Marine Drugs, 13(1), 202–221. 10.3390/md13010202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener A. B. (2020). Four ways researchers are responding to the COVID-19 outbreak. Nature Medicine, 10.1038/d41591-020-00002-4 [DOI] [PubMed] [Google Scholar]
- Khan A., Ashfaq-Ur-Rehman, Junaid M., Li C.-D., Saleem S., Humayun F., Shamas S., Ali S. S., Babar Z., & Wei D.-Q. (2020). Dynamics insights into the gain of flexibility by Helix-12 in ESR1 as a mechanism of resistance to drugs in breast cancer cell lines. Frontiers in Molecular Biosciences, 6 10.3389/fmolb.2019.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. T., Kaushik A. C., Bhatti A. I., Zhang Y.-J., Zhang S., Wei A. J., Malik S. I., & Wei D. Q. (2019). Marine natural products and drug resistance in latent tuberculosis. Marine Drugs, 17(10), 549 10.3390/md17100549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G. M., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 0(0), 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 0(0), 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kumar D., Kumari K., Jayaraj A., Kumar V., Kumar R. V., Dass S. K., Chandra R., & Singh P. (2020). Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics, 0(0), 1–14. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., & Munster V. (2020). Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV. BioRxiv, 202001.22.915660. 10.1101/2020.01.22.915660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov M. Y., Bogatyreva N. S., & Galzitskaya O. V. (2008). Radius of gyration as an indicator of protein structure compactness. Molecular Biology, 42(4), 623–628. 10.1134/S0026893308040195 [DOI] [PubMed] [Google Scholar]
- Mani D., Wadhwani A., & Krishnamurthy P. T. (2019). Drug repurposing in antiviral research: A current scenario. Journal of Young Pharmacists, 11(2), 117–121. 10.5530/jyp.2019.11.26 [DOI] [Google Scholar]
- Mayer A. M. S., Guerrero A. J., Rodríguez A. D., Taglialatela-Scafati O., Nakamura F., & Fusetani N. (2019). Marine pharmacology in 2014–2015: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, antiviral, and anthelmintic activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Marine Drugs, 18(1), 5 10.3390/md18010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadamtousi S. Z., Nikzad S., Kadir H. A., Abubakar S., & Zandi K. (2015). Potential antiviral agents from marine fungi: An overview. Marine Drugs, 13(7), 4520–4538. 10.3390/md13074520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 0(0), 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Newman D. J., & Cragg G. M. (2016). Natural products as sources of new drugs from 1981 to 2014 . Journal of Natural Products, 79(3), 629–661. 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–15. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., & Jung S.-H. (2016). An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. Journal of Medicinal Chemistry, 59(14), 6595–6628. https://www.ncbi.nlm.nih.gov/pubmed/26878082 [DOI] [PMC free article] [PubMed]
- Raveh A., Delekta P. C., Dobry C. J., Peng W., Schultz P. J., Blakely P. K., Tai A. W., Matainaho T., Irani D. N., Sherman D. H., & Miller D. J. (2013). Discovery of potent broad spectrum antivirals derived from marine actinobacteria. PLoS One, 8(12), e82318 10.1371/journal.pone.0082318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe D. R., & Cheatham T. E. (2013). PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Sagar S., Kaur M., & Minneman K. P. (2010). Antiviral lead compounds from marine sponges. Marine Drugs, 8(10), 2619–2638. 10.3390/md8102619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 0(ja), 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois S., Zhang R., Gao W., Gao H., Li Y., Zheng H., Wei D.-Q. (2007). Discovery of Potent Anti-SARS-CoV MPro Inhibitors. Bentham Science Publishers. https://www.ingentaconnect.com/content/ben/cad/2007/00000003/00000003/art00002
- Smilgies D.-M., & Folta-Stogniew E. (2015). Molecular weight-gyration radius relation of globular proteins: A comparison of light scattering, small-angle X-ray scattering and structure-based data. Journal of Applied Crystallography, 48(Pt 5), 1604–1606. 10.1107/S1600576715015551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh K. D., Selvaraj C., Singh S. K., & Dubey V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzair B., Mahmood Z., & Tabassum S. (2011). Antiviral activity of natural products extracted from marine organisms. BioImpacts, 1(4), 203–211. 10.5681/bi.2011.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar S., & Menakha M. (2015). Pharmaceutical applications of cyanobacteria—A review. Journal of Acute Medicine, 5(1), 15–23. 10.1016/j.jacme.2015.02.004 [DOI] [Google Scholar]
- Vilar S., Cozza G., & Moro S. (2008). Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Current Topics in Medicinal Chemistry, 8(18), 1555–1572. 10.2174/156802608786786624 [DOI] [PubMed] [Google Scholar]
- Vo T.-S., & Kim S.-K. (2010). Potential Anti-HIV Agents from Marine Resources: An Overview. Marine Drugs, 8(12), 2871–2892. 10.3390/md8122871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–16. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wei D.-Q., Zhang R., Du Q.-S., Gao W.-N., Li Y., Gao H., Wang S.-Q., Zhang X., Li A.-X., Sirois S., & Chou K.-C. (2006). Anti-SARS drug screening by molecular docking. Amino Acids, 31(1), 73–80. 10.1007/s00726-006-0361-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO COVID-19 Dashboard. (n.d.). Retrieved May 5, 2020, from https://covid19.who.int/
- Wittine K., Saftić L., Peršurić Ž., & Kraljević Pavelić S. (2019). Novel antiretroviral structures from marine organisms. Molecules, 24(19), 3486 10.3390/molecules24193486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., & McLellan J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., & Li H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K. Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., & Rao Z. (2005). Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biology, 3(10), e324. 10.1371/journal.pbio.0030324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G. F., Anand K., Bartlam M., Hilgenfeld R., & Rao Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of America, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., & Liu Y. (2020). Potential interventions for novel coronavirus in China: A systemic review. Journal of Medical Virology, 92(5), 479–490. 10.1002/jmv.25707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Gan L., Wang E., & Wang J. (2013). Pocket-based drug design: Exploring pocket space. AAPS Journal, 15(1), 228–241. 10.1208/s12248-012-9426-6 [DOI] [PMC free article] [PubMed] [Google Scholar]