Abstract
Current SARS-CoV-2 pandemy mortality created the hypothesis that some populations may be more susceptible to SARS-CoV-2. TMPRSS2 encodes a transmembrane serine protease which plays a crucial role in SARS-CoV-2 cell entry. Single nucleotide polymorphisms (SNPs) in TMPRSS2 might influence SARS-CoV2 entry into the cell. This study aimed to investigate the impact of SNPs on TMPRSS2 function and structure. In silico tools such as Ensembl, Gtex, ExPASY 2, GEPIA, CCLE, KEGG and GO were engaged to characterize TMPRSS2 and its expression profile. The functional effects of SNPs were analyzed by PolyPhen-2, PROVEN, SNAP2, SIFT and HSF. Also, Phyre2, GOR IV and PSIPRED were used to predict the secondary structure of TMPRSS2. Moreover, post-translational modification (PTM) and secretory properties were analyzed through Modpredand Phobius, respectively. Finally, miRNA profiles were investigated by PolymiRTS and miRSNPs. Out of 11,184 retrieved SNPs from dbSNP, 92 showed a different frequency between Asians and other populations. Only 21 SNPs affected the function and structure of TMPRSS2 by influencing the protein folding, PTM, splicing and miRNA function. Particularly, rs12329760 may create a de novo pocket protein. rs875393 can create a donor site, silencer and broken enhancer motifs. rs12627374 affects a wide spectrum of miRNAs profile. This study highlighted the role of TMPRSS2 SNPs and epigenetic mechanisms especially non-coding RNAs in appearance of different susceptibility to SARS-CoV-2 among different populations. Also, this study could pave the way to potential therapeutic implication of TMPRSS2 in designing antiviral drugs.
Communicated by Ramaswamy H. Sarma
Keywords: In silico, TMPRSS2, SARS-CoV-2, single nucleotide polymorphisms, SNPs
Introduction
In December 2019 a novel coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported in some patients with fever, cough, expectoration, headache, fatigue, diarrhea and hemoptysis in Wuhan, China (Aanouz et al., 2020; Khan et al., 2020; Xu et al., 2020). SARS-CoV-2 rapidly disseminated all over the world with 1,909,804 infected cases and 118,507 deaths until April 13, 2020 (Anonymous, 2020a). Increasing body of evidence showed that SARS-CoV-2 as well as SARS-CoV that caused global outbreak of severe acute respiratory syndrome from 2002 to 2003 (Elfiky & Azzam, 2020; Pant et al., 2020), enters cells through a receptor called angiotensin-converting enzyme (ACE) (Boopathi et al., 2020; Muralidharan et al., 2020; Zhang et al., 2020). ACE is expressed in several tissues including lung, heart, kidney and gastrointestinal tract, and has a key role in blood pressure regulation in renin angiotensin axis (Chen et al., 2020; Pieruzzi et al., 1995). It’s noteworthy that the ACE is not sufficient to SARS-CoV-2 infection. The transmembrane serine protease 2 (TMPRSS2) is another vital component to viral infection (Elmezayen et al., 2020; Hasan et al., 2020; Hoffmann et al., 2020). TMPRSS2 facilitates SARS-CoV-2 membrane fusion via cleavage of spike (S) protein in several residues. S protein is a high glycosylated protein that covers SARS-CoV-2, and is assembled into corona shape (Gupta et al., 2020; Iwata-Yoshikawa et al., 2019; Sarma et al., 2020). ACE expression evaluation in cell line and knocked-out (KO) mice demonstrated that ACE expression levels are significantly correlated with susceptibility to SARS-CoV-2 (Hamming et al., 2004; Hofmann et al., 2004; Kuba et al., 2005). Also, current study conducted on VeroE6 cell line have revealed that the engineered VeroE6/TMPRSS2 cell line is 10 times more susceptible to SARS-CoV-2 infection in comparison with parental VeroE6 cells (Matsuyama et al., 2020). Accordingly, results of study on Tmprss2-KO mice infected with SARS-CoV illustrated a lower viral replication in lungs, mild lung pathology and no weight loss in Tmprss2-KO mice (Iwata-Yoshikawa et al., 2019). The higher rate of morbidity and mortality of SARS-CoV2 in Asian population in comparison with other populations mentioned the possibility that susceptibility to SARS-CoV-2may be influenced by ethnicity (Cao et al., 2020). Accumulating evidence showed that the rate of infectivity is associated with age, and elderly persons are more susceptible to SARS-CoV-2 (Chen et al., 2020; Huang et al., 2020; Khan et al., 2020). Intriguingly, increasing evidence suggests that mRNA levels of TMPRSS2 are influenced by androgen hormone. Androgen regulates the expression levels of TMPRSS2 by binding to androgen response element (ARE) which is located in TMPRSS2 promoter (Clinckemalie et al., 2013; Nickols & Dervan, 2007). Achieved results from several studies on prostate cancer disclosed that overexpression of TMPRSS2 induced by transactivation of androgen receptor caused growth, invasion and metastasis of prostate cancer stem cells (Chen et al., 2019; Ko et al., 2015). Recently, a large amount of evidence showed that single nucleotide polymorphisms (SNPs) in TMPRSS2 may be involved in several disorders including prostate and breast cancers via modulation of TMPRSS2 expression (Bhanushali et al., 2018; Luostari et al., 2014; Maekawa et al., 2014). Given that the TMPRSS2 plays an essential role in cell entry of SARS-CoV-2, and regarding its potential therapeutic implication in designing antiviral drugs, in this study we exploited several bioinformatics tools and databases for the first comprehensive computational analysis of TMPRSS2 to investigation of pathways, expression profile, epigenetic mechanisms, and SNPs of TMPRSS2.
Materials and methods
Analysis of gene position and its variation within genome by Ensembl
Ensembl available at https://asia.ensembl.org/index.html is a genome browser which provides useful information about many genes, molecular biology pathways, regulatory features and genetic variation in vertebrates and model organisms. Inputs for Ensembl are including gene symbol, protein, UniProt ID, etc.
Analysis of the effect of genetic variations on tissue-specific gene expression levels and exon expression by GTEx
Genotype-Tissue Expression (GTEx) (https://gtexportal.org/home/) is a powerful bioinformatics database that analyzes tissue-specific gene expression levels upon genetic variation, and thereby predicts inherited susceptibility to diseases. Also, GTEx determines expression quantitative trait loci (eQTL) which categorizes genetic variants (including millions of SNPs) and their effects on several genes expression profile. Therefore, GTEx can predict susceptibility to diseases resulting from genetic variations in different populations.
Identification of amino acid sequence and peptide mass by ExPASY 2
Expert protein analysis system 2 (ExPASY2) (https://www.expasy.org/) is an informative bioinformatics resource that prepares comprehensive data to multiple different domains, such as proteomics (protein characterization, post-translational modifications, etc.), genomics, phylogenetics/evolution, systems biology, population genetics, transcriptomics, etc.
Analysis of gene expression profiling between tumor samples and paired normal tissues, and cell lines
Gene expression profiling interactive analysis (GEPIA) (http://gepia.cancer-pku.cn/) is a user-friendly bioinformatics website which has provided a gene expression profile to a wide spectrum of cancers. GEPIA presents the RNA sequencing expression information of 9,736 tumors and 8,587 normal samples from cancer genome atlas (TCGA) project. GEPIA compares the expression levels of a specific RNA between normal and tumor tissues in boxplot format. Moreover, it can analyze the survival and tumor stages of patients with high confidence. Cancer cell line encyclopedia (CCLE) (https://portals.broadinstitute.org/ccle) is another web server to prediction of RNA expression levels which analyzes expression levels of 84,434 genes in 1457 cancer cell lines.
Investigation of biological pathways related to TMPRSS2 through KEGG, GO
Kyoto Encyclopedia of Genes and Genomes (KEGG) available at https://www.genome.jp/kegg/ is a powerful pathway predictor bioinformatics tool that can study genomes, biological pathways, diseases, drugs and chemical substances. GO (gene ontology) available at webserver http://geneontology.org/ is a comprehensive computational program to analyze and predict the pathways related to many essential genes.
Databases and characterization of SNPs
The sequence and SNPs of TMPRSS2 were obtained-from National Center for Biotechnology Information (NCBI) website browser (https://www.ncbi.nlm.nih.gov/) and dbSNP, respectively. NCBI comprises comprehensive information about SNPs, microsatellites, mutation types, population frequency and clinical variations. We used universal protein resource (UniProtKB) database (https://www.uniprot.org/) to retrieve the sequence of TMRSS2 isoforms. Analysis of allelic frequency in different populations was conducted through Ensembl 1000 genome browser (http://www.ensembl.org/Homo_sapiens/Info/Index?db=core).
Prediction of functional consequences of SNPs by SIFT
Sorting intolerant from tolerant (SIFT) is a bioinformatics tool which predicts the effects of amino acids substitution (non-synonymous polymorphisms) on protein structure based on sequence homology and physical properties of amino acids. SIFT results are divided into deleterious and tolerated phenotypes with scores range from 0 (deleterious) to 1 (tolerated). The score ranges of 0 to 0.05 and 0.05 to 1 are considered as deleterious and benign substitutions, respectively. This database is available at (https://sift.bii.a-star.edu.sg/).
Analysis of functional consequences of SNPs by POLYPHEN-2
Polymorphism phenotyping v2 (PolyPhen-2), is a useful database that predicts the possible consequences of amino acid substitution on functional and structural proteins. The necessary input to Polyphen-2 is protein sequence in FASTA format and single point substitution. The output information is along with a score range from 0.0 (benign) to 1.0 (damaging). Score range of 0.0–0.15, 0.15–0.85 and 0.85–1.0 are considered benign, possibly damaging and damaging, respectively. This tool is retrievable at (http://genetics.bwh.harvard.edu/pph2/).
Analysis of functional effects of SNPs by PROVEAN
Protein variation effect analyzer (PROVEAN) is a bioinformatics tool to predict the effects of amino acid substitutions and indels on biological functions of proteins. The predefined threshold for PROVEAN is −2.5 (cutoff). PROVEAN score < −2.5 is considered as a ‘deleterious’ variant, and the PROVEAN score > −2.5 is considered as a ‘neutral’ variant. This software is accessible at http://provean.jcvi.org/index.php.
Analysis of functional effects of SNPs by SNAP2
Analysis of functional impacts of SNPs by Screening for Non-acceptable Polymorphisms (SNAP2) available at https://www.rostlab.org/services/snap/ is an advantageous bioinformatics program which predicts the functional effects of sequence variations on protein function and phenotypic properties. It has an advantage over other tools because this server gives an informative heat map besides the protein functional effects. In heat map, the score equal to −100 (dark blue) indicates that amino acid substitution is completely neutral, while the score equal to +100 (dark red) is highly predicted to be pathogenic. For instance, the dark red (score > 50) for a specific amino acid substitution in heat map shows the powerful pathogenicity impacts.
Prediction of functional impacts of SNPs on splicing
Human splicing finder (HSF) is an in silico software (http://www.umd.be/HSF/) which combines 12 different algorithms to prediction of splicing motifs affected by mutation including donor and acceptor splicing site, branch point, exonic splicing enhancers (ESE) and exonic splicing silencers (ESS). For prediction of donor and acceptor splicing site HSF applies ‘position weight matrices’ algorithm with consensus values (CV) range from 0 to 100. CVs higher than CV threshold (65) are considered as acceptor or donor splicing site. Moreover, the wild type sequence score higher than threshold along with variation score under −10% indicates that the mutation creates a new splice site. On the other hand, the wild type sequence score under the threshold along with variation score higher than +10%, discloses that the mutation creates a new splice site.
Prediction of molecular effects of TMPRSS2 related-SNPs on protein secondary and tertiary structures
Protein homology/analogy recognition engine 2.0 (Phyre2) (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index) is a useful web server which determines the protein structure, function and mutations. Phyre2 predicts ligand binding sites, protein secondary structure (α-helices, β-strands and coils) and analyzes the effects of amino acid variations (e.g. non-synonymous SNPs (nsSNPs)) on secondary structure. It also analyzes several processes including prediction of disorder, domain structure, transmembrane helix and homology through providing an alignment algorithm. GOR IV (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html) (Enayatkhani et al., 2020) is one of the protein secondary structure prediction tools which represents two outputs, eye-friendly native sequence with a predicted secondary structure in H = helix, E = extended or beta-strand and C = coil; and presents a probability value for secondary structure of each amino acid position. PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) is another bioinformatics database to secondary and tertiary structure prediction besides other functions such as structural contact prediction, topology and helix packing, protein domain fold recognition, and eukaryotic protein function prediction.
Prediction of post-translational modifications (PTM) using Modpred
Predictor of post-translational modification (PTM) sites in proteins (Modpred) available at http://www.modpred.org/ predicts different types of PTMs such as acetylation, phosphorylation, proteolytic cleavage, methylation, O-linked glycosylation, N-linked glycosylation and carboxylation. Modpred predicts these modifications with scores ranging from 0 to 1 with confidence rate divided into low, medium and high-confidence. The modification sites with scores at least 0.5 are labeled as low confidence sites.
Analysis of functional impacts of SNPs on secretory characteristics through phobius
Phobius is a reliable tool to prediction of transmembrane topology and signal peptides alteration upon amino acid substitution. Phobius available at http://phobius.sbc.su.se/ has been designed to predict secretion, transmembrane helix domains, cytoplasmic and non-cytoplasmic domains based on homology supported predictions along with useful information such as plot and topology data.
Analysis of influence of polymorphisms on miRNAs function and development of severe disease by PolymiRTS and miRSNPs
Investigation of the effects of polymorphisms in microRNAs (miRNAs) and their target sites (PolymiRTS) was conducted by PolymiRTS accessible at http://compbio.uthsc.edu/miRSNP/. PolymiRTS is an online database that predicts changes of miRNAs profile upon their targeting alterations created by SNPs. Also, PolymiRTS predicts the effect of polymorphisms in miRNA seed regions on miRNA targeting profiling. One of other databases to analyzing the effect of polymorphisms on alteration of miRNAs profile is miRSNPs (http://bioinfo.bjmu.edu.cn/mirsnp/search/). miRSNPs provides results with a specific miRNA-mRNA binding energy and a precise score with higher scores representing a more stable miRNA-mRNA binding.
Results
TMPRSS2 and TMPRSS2 characterization
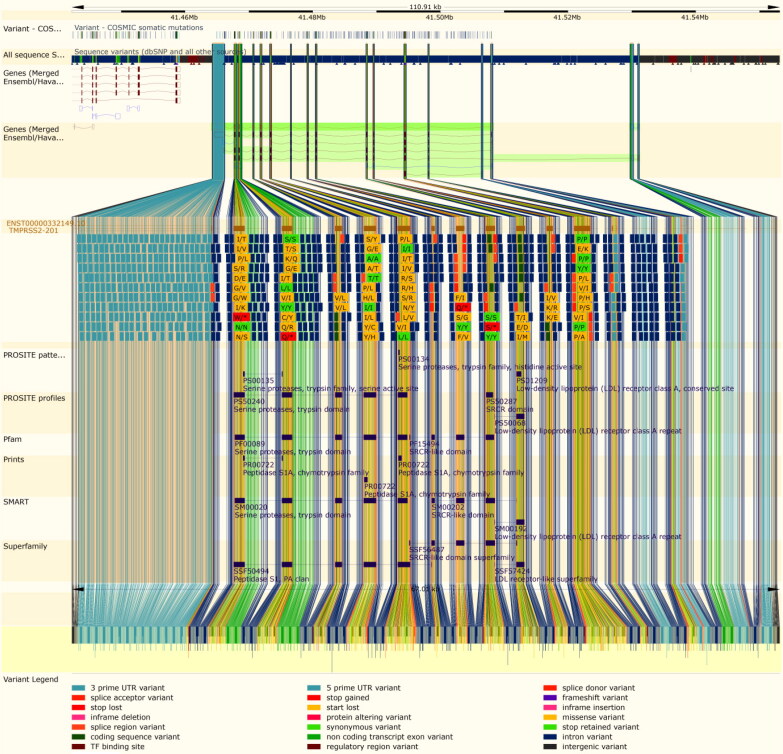
Extracted data from Ensembl revealed that TMPRSS2 is located on 21q22.3 and contains 15 exons. Also,Ensembl showed the distributionof TMPRSS2 variation types and their position including intronic variants, missense variants, frameshift variants, etc. (Figure 1).
Figure 1.
Ensembl gene map of TMPRSS2 and its variants. Distribution of types of TMPRSS2 variants which most of them are intronic variants are shown.
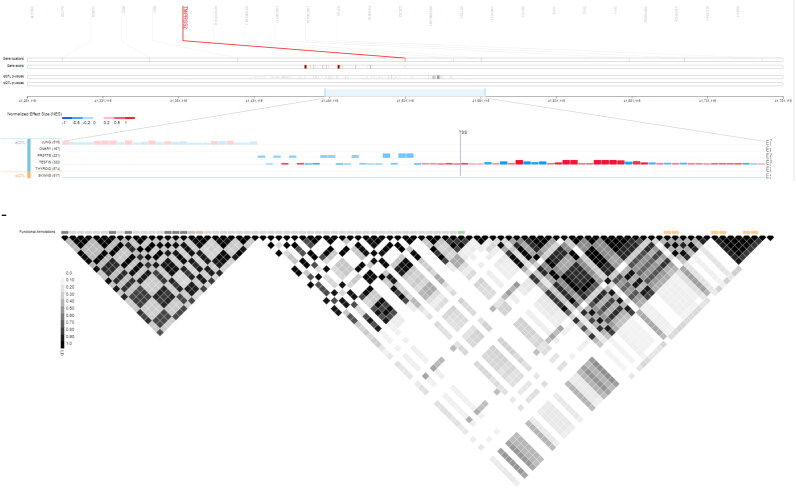
Similar to Ensembl, Gtex showed the location of TMPRSS2 along with eQTL which categorized all SNPs in TMPRSS2 and their effects on expression levels of TMPRSS2 with normalized effect size (NES) for several tissues including lung, testis, thyroid, etc. (Figure 2).
Figure 2.
Position of TMPRSS2 along with eQTL mapping in Gtex. Expression quantitative trait loci (eQTL) categorizes genetic variants of TMPRSS2 and their effects on its expression profile.
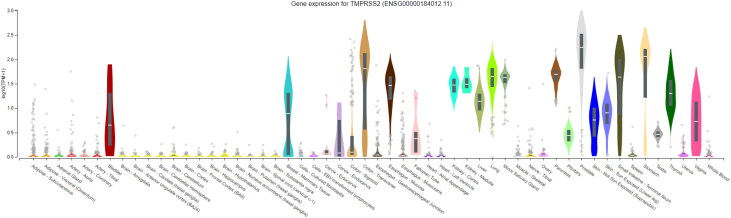
Moreover, Gtex showed that TMPRSS2 is highly expressed in prostate, colon-transverse, stomach and lung (Figure 3).
Figure 3.
Comparison of TMPRSS2 expression levels in several tissues. 54 tissues with different expression levels of TMPRSS2 compared with each other. Only some of them show a significant expression level of TMPRSS2.
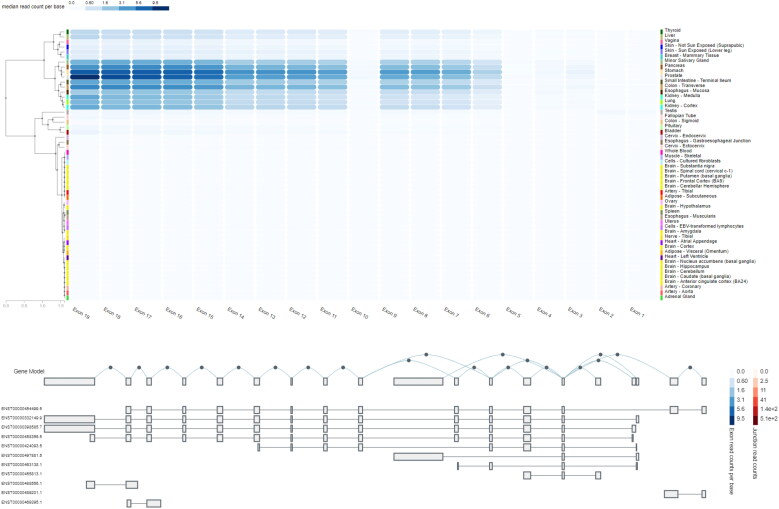
Results from Gtex demonstrated that median read count per base for exons 15 in prostate and stomach is significantly higher than other tissues. It’s not surprising that median read count per base for almost all exons was higher in prostate and stomach in comparison with other tissues (Figure 4).
Figure 4.
Exon expression of TMPRSS2 in several tissues with median read count per base score. Read count was used to quantify gene expression (by RNA-seq) by counting the number of reads that map (i.e. align) to each gene. Raw read counts are affected by factors such as transcript length (longer transcripts have higher read counts, at the same expression level) and total number of reads.
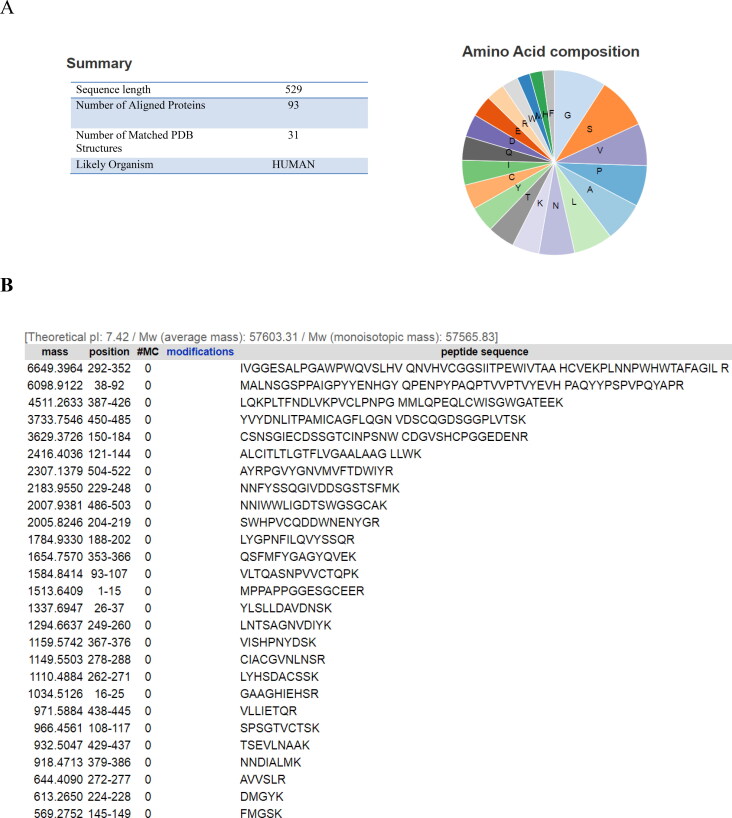
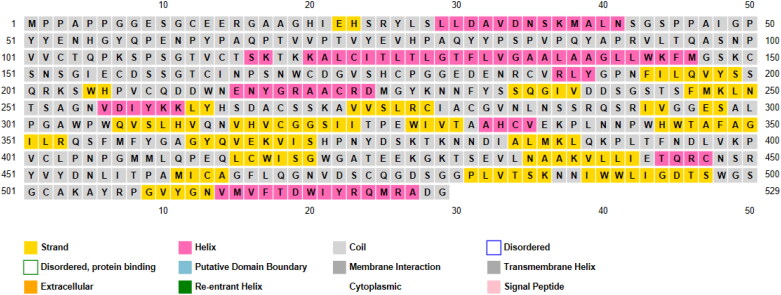
The sequences of two TMPRSS2 isoforms (A and B) were achieved from UniProt. Amino acid composition of TMPRSS2 retrieved from ExPASY2 revealed that TMPRSS2 mostly comprises glycine, serine, valine, proline and leucine (Figure 5(A)). TMPRSS2 can be cleaved by trypsin into 27 fragments. Peptide mass analyzed for each fragment by ExPASY2 is shown in Figure 5(B).
Figure 5.
Characterization of isoform B of TMPRSS2. (A) Properties of isoform B of TMPRSS2 and its amino acid composition; (B) measurement of TMPRSS2 fragments mass.
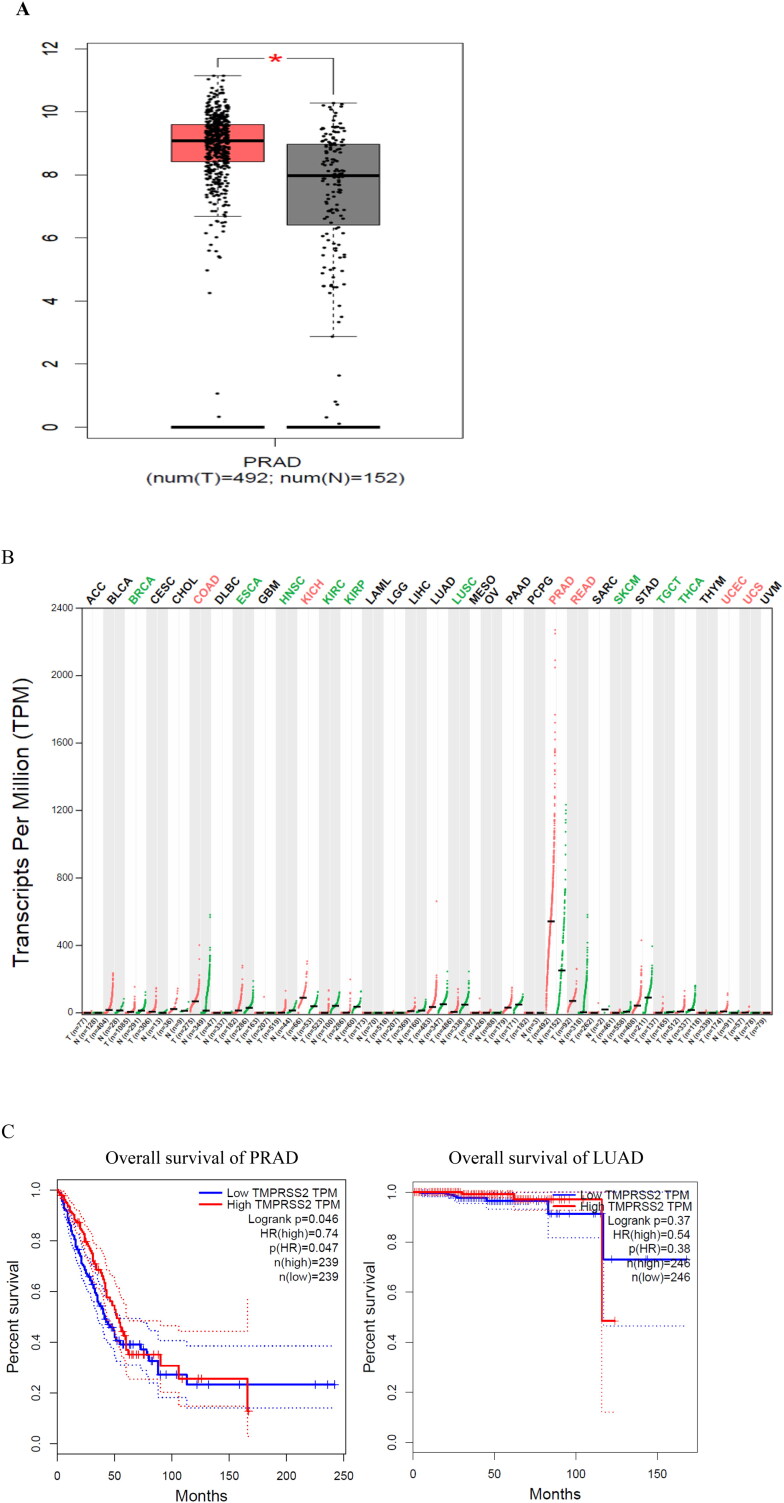
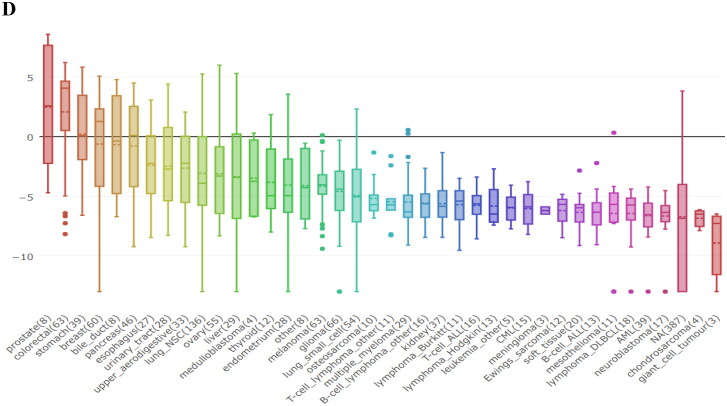
The expression levels of TMPRSS2 in several tumor samples and paired normal tissues were investigated via GEPIA which demonstrated that the expression level of TMPRSS2 in prostate adenocarcinoma (PRAD) and paired normal tissues was higher than other tumor and paired normal tissues. Furthermore, some tumor tissues including PRAD, relative afferent pupillary defect (RAPD) and rectum adenocarcinoma (READ) presented higher expression levels of TMPRSS2 in comparison with normal paired samples (Figure 6(A)). In contrast to PRAD, RAPD and READ some tumor samples including kidney renal clear cell carcinoma (KIRC), sarcoma (SARC) and skin cutaneous melanoma (SKCM) showed significant lower levels of TMPESS2 in comparison with paired normal tissues (Figure 6(B)). Moreover, survival analysis in two groups including patients with PRAD and lung adenocarcinoma (LUAD) conducted by GEPIA revealed that overall survival rate in patients with high TMPRSS2 transcripts per million (TPM) decreased with age in both groups. Also, survival rate in subjects with low TMPRSS2 TPM decreased slowly with age, and became stable after some time (Figure 6(C)). Achieved results from study of TMPRSS2 expression levels in a wide spectrum of cell lines by cancer cell line encyclopedia (CCLE) disclosed that prostate, colorectal, stomach, bile duct, pancreas, urinary tract, and lung cell lines showed significantly greater levels of TMPRSS2 in comparison with other cell lines such as chondrosarcoma and neuroblastoma (Figure 6(D)).
Figure 6.
Analysis of TMPRSS2 expression levels. (A) Comparison of 492 prostate adenocarcinoma (PRAD) tissues with 152 normal tissues (T: tumor; N: normal); (B) TMPRSS2 expression profiles across all tumor samples and paired normal tissues (dot plot); (C) overall survival of PRAD and lung adenocarcinoma (LUAD) patients with different transcripts per million (TPM) of TMPRSS2; (D) mRNA expression (RNAseq) for TMPRSS2 in various cell lines.
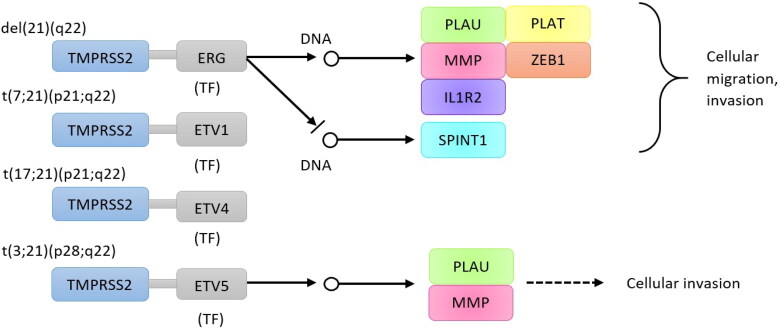
The biological pathways related to TMPRSS2 were retrieved from KEGG (Figure 7), and GO (Table 1).
Figure 7.
The biological pathway related to TMPRSS2 predicted by KEGG. Chromosome rearrangements between chromosome 21 which contains TMPRSS2 and other chromosomes lead to cellular migration, and invasion in prostate cancer through facilitating several transcription factors. ERG: ETS (erythroblast transformation-specific)-related gene; ETV1: ets translocation variant 1; ETV4: ets translocation variant 4; ETV5: ets translocation variant 5; PLAU: urokinase plasminogen activator; MMP: matrix metalloproteinase-3; IL1R2: interleukin 1 receptor type II; SPINT1: kunitz-type protease inhibitor 1; PLAT: tissue plasminogen activator; ZEB1: zinc finger homeobox protein 1.
Table 1.
The biological pathways related to TMPRSS2 predicted by GO.
| GO class |
Reference |
GO class |
Reference |
GO class |
Reference |
GO class |
Reference |
| 1. Serine-type endopeptidase activity | GO_REF:0000002 | 5. Integral component of plasma membrane | PMID:9325052 | 9. Serine-type peptidase activity | PMID:9325052 | 13. Extracellular exosome | PMID:19056867 |
| 2. Scavenger receptor activity | GO_REF:0000002 | 6. Proteolysis | PMID:21068237 | 10. Protein autoprocessing | PMID:21068237 | 14. Extracellular exosome | PMID:19199708 |
| 3. Protein binding | PMID:21068237 | 7. Proteolysis | PMID:24227843 | 11. Positive regulation of viral entry into host cell | PMID:21068237 | 15. Extracellular exosome | PMID:23533145 |
| 4. Plasma membrane | PMID:21068237 | 8. Endocytosis | GO_REF:0000108 | 12. Extracellular exosome | PMID:24227843 | 16. Protein autoprocessing | PMID:21873635 |
Retrieval of the SNPs related to TMPRSS2
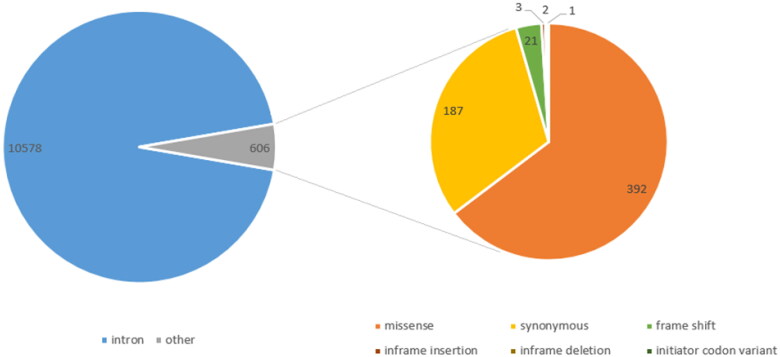
NCBI was applied to retrieve the sequence and all SNPs ofTMPRSS2. Results from dbSNP revealed 11,184 SNPs within TMPRSS2 including intronic (10,578), missense (392), synonymous (187), frameshift (21), inframe insertion (3), inframe deletion (2) and initiator codon (1) variants (Figure 8).
Figure 8.
Pie chart of TMPRSS2 SNPs distribution.
In the next step we limited our study to those SNPs with minor allele frequency (MAF) between 0.01 and 0.95; therefore 493 SNPs remained. Obtained results from investigation of 493 SNPs in 1000 genome browser demonstrated that out of 493 SNPs only the frequency of 92 SNPs (87 intronic, 3 synonymous and 2 missense variants) were significantly different between Asian population and other populations. Taken together, out of 92 SNPs only 21 influenced the function of protein (Table 2).
Table 2.
TMPRSS2 SNPs with different frequency between different populations in 1000 genome project.
| SNP | Function class | Allele | Global | AFR | EAS | EUR | SAS | AMR |
|---|---|---|---|---|---|---|---|---|
| 1. rs386416 | Intron | G > C | G = 0.444 | G = 0.431 | G = 0.698 | G = 0.300 | G = 0.354 | G = 0.439 |
| 2. rs402197 | Intron | T > C | T = 0.126 | T = 0.018 | T = 0.349 | T = 0.021 | T = 0.072 | T = 0.236 |
| 3. rs112467088 | Intron | A > T | A = 0.814 | A = 0.743 | A = 0.981 | A = 0.7187 | A = 0.867 | A = 0.771 |
| 4. rs422761 | Intron | G > A | G = 0.775 | G = 0.707 | G = 0.682 | G = 0.981 | G = 0.759 | G = 0.764 |
| 5. rs423596 | Intron | C > T | C = 0.904 | C = 0.994 | C = 0.750 | C = 0.961 | C = 0.833 | C = 0.977 |
| 6. rs456016 | Intron | T > C | T = 0.125 | T = 0.019 | T = 0.349 | T = 0.018 | T = 0.075 | T = 0.231 |
| 7. rs461194 | Intron | C > G | C = 0.131 | C = 0.004 | C = 0.347 | C = 0.030 | C = 0.113 | C = 0.228 |
| 8. rs8134203 | Intron | C > T | C = 0.464 | C = 0.506 | C = 0.741 | C = 0.256 | C = 0.326 | C = 0.477 |
| 9. rs464431 | Intron | A > G | A = 0.126 | A = 0.019 | A = 0.349 | A = 0.019 | A = 0.075 | A = 0.232 |
| 10. rs2298662 | Intron | G > C | G = 0.123 | G = 0.006 | G = 0.346 | G = 0.020 | G = 0.082 | G = 0.231 |
| 11. rs7364088 | Intron | G > A | G = 0.695 | G = 0.675 | G = 0.604 | G = 0.736 | G = 0.736 | G = 0.751 |
| 12. rs875393 | Intron | G > A | G = 0.944 | G = 0.998 | G = 0.822 | G = 0.942 | G = 0.970 | G = 0.986 |
| 13. rs2094881 | Intron | T > C | T = 0.470 | T = 0.524 | T = 0.744 | T = 0.252 | T = 0.324 | T = 0.491 |
| 14. rs75603675 | Exon G > D | C > A | C = 0.756 | C = 0.705 | C = 0.983 | C = 0.595 | C = 0.777 | C = 0.728 |
| 15. rs12329760 | Exon V > M | C > T | C = 0.738 | C = 0.738 | C = 0.637 | C = 0.764 | C = 0.774 | C = 0.846 |
| 16. rs456142 | 3′UTR | T > A | T = 0.370 | T = 0.3722 | T = 0.6339 | T = 0.1690 | T = 0.317 | T = 0.352 |
| 17. rs462574 | 3′UTR | A > G | A = 0.2570 | A = 0.1778 | A = 0.5903 | A = 0.0338 | A = 0.254 | A = 0.252 |
| 18. rs456298 | 3′UTR | T > A | T = 0.3718 | T = 0.3722 | T = 0.6359 | T = 0.1690 | T = 0.321 | T = 0.353 |
| 19. rs12627374 | 3′UTR | C > T | C = 0.940 | C = 0.995 | C = 0.850 | C = 0.998 | C = 0.863 | C = 0.996 |
| 20. rs12473206 | 3′UTR | C > G | C = 0.861 | C = 0.959 | C = 0.955 | C = 0.740 | C = 0.800 | C = 0.800 |
| 21. rs75036690 | 3′UTR | G > A | G = 0.989 | G = 1.000 | G = 0.955 | G = 1.000 | G = 0.993 | G = 1.000 |
AFR: African; EAS: Asian; EUR: European; SAS: South Asian; AMR: American.
Out of 21 SNPs 9 (rs423596, rs8134203, rs464431, rs2298662, rs2094881, rs75603675, rs456142, rs462574 and rs456298) showed a significant difference in frequency between Asian population and other populations. Also, the frequency of 2 SNPs (rs402197 and rs456016) were similar between Asian and American populations whereas they were different in comparison with other populations. Surprisingly, one SNP (rs461194) revealed a considerable different frequency between African population and others. Additionally, 8 SNPs (rs422761, rs8134203, rs2094881, rs75603675, rs456142, rs462574, rs456298 and rs12473206) revealed a notable different frequency between European and others. Astonishingly, comparison of European and African populations demonstrated that 5 SNPs (rs402197, rs456016, rs461194, rs464431 and rs2298662) showed almost equal frequencies.
Prediction of functionally significant consequences of SNPs on protein function and stability
To analysis the effects of selected SNPs (92) on protein function and stability, we exploited several bioinformatics tools comprising SIFT, PolyPhen-2, PROVEAN, SNAP2 and HSF. Achieved results from PolyPhen-2 showed that both missense SNPs (rs12329760 and rs75603675) affected the TMPRSS2 function. SIFT similar to SNAP2 suggested that only rs12329760 influenced protein function whereas PROVEAN predicted that neither of SNPs do influence the function of TMPRSS2 (Table 3).
Table 3.
Prediction of functional effects of SNPs on protein structure.
| SNP | Substitution | SIFT |
Polyphen |
Provean |
SNAP-2 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | Score | Prediction | Score | Prediction | Score | Prediction | Score | Accuracy | ||
| rs12329760 | V197M | Deleterious | 0.006 | Probably damaging | 0.999 | Neutral | −1.891 | Effect | 49 | 71% |
| rs75603675 | G8V | Tolerated | 0.201 | Benign | 0.386 | Neutral | 0.401 | Neutral | −16 | 57% |
| rs75603675 | G8D | Not found | Possibly damaging | 0.815 | Neutral | 0.222 | Neutral | −8 | 53% | |
HSF which is a powerful predictor of splice site (new site or site broken) upon SNPs, predicted that 7 SNPs caused new donor splice sites, 3 SNPs caused broken donor splice sites, 3 SNPs created new acceptor splice site, and 1 SNP caused new enhancer splice site along with broken enhancer splice site, and 1 SNP broke 3 enhancer sites (Table 4).
Table 4.
Prediction of splice sites modifications by TMPRSS2 SNPs.
| SNP | Donor-site | Score | Acceptor-site | Score | Enhancer motif | Silencer motif |
|---|---|---|---|---|---|---|
| 1. rs386416 | New site | +19.55 | NA | NA | New site Site broken | 2 New sites |
| 2. rs402197 | New site | +58.05 | NA | NA | New site | Site broken |
| 3. rs112467088 | Site broken | −31.22 | New site | +79.29 | Site broken | NA |
| 4. rs422761 | New site | +52.14 | NA | NA | New site | NA |
| 5. rs423596 | NA | NA | NA | NA | Site broken | 2 Sites broken |
| 6. rs456016 | New site | +56.84 | NA | NA | 2 New sites | New site |
| 7. rs461194 | NA | NA | NA | NA | NA | Site broken |
| 8. rs8134203 | NA | NA | New site | +52.57 | New site | NA |
| 9. rs464431 | NA | NA | NA | NA | New site Site broken | New site |
| 10. rs2298662 | NA | NA | NA | NA | 3 sites broken | NA |
| 11. rs7364088 | New site | +54.29 | NA | NA | NA | NA |
| 12. rs875393 | New site | +62.69 | NA | NA | 3 sites broken | 2 new sites |
| 13. rs2094881 | New site | +20.19 | NA | NA | Site broken | NA |
| 14. rs75603675 | Site broken | −32.49 | New site | +3.4 | NA | NA |
| 15. rs12329760 | Site broken | −35.46 | NA | NA | 2 new sites | Site broken |
NA: not available.
Prediction of functional impacts of SNPs on TMPRSS2 secondary structure
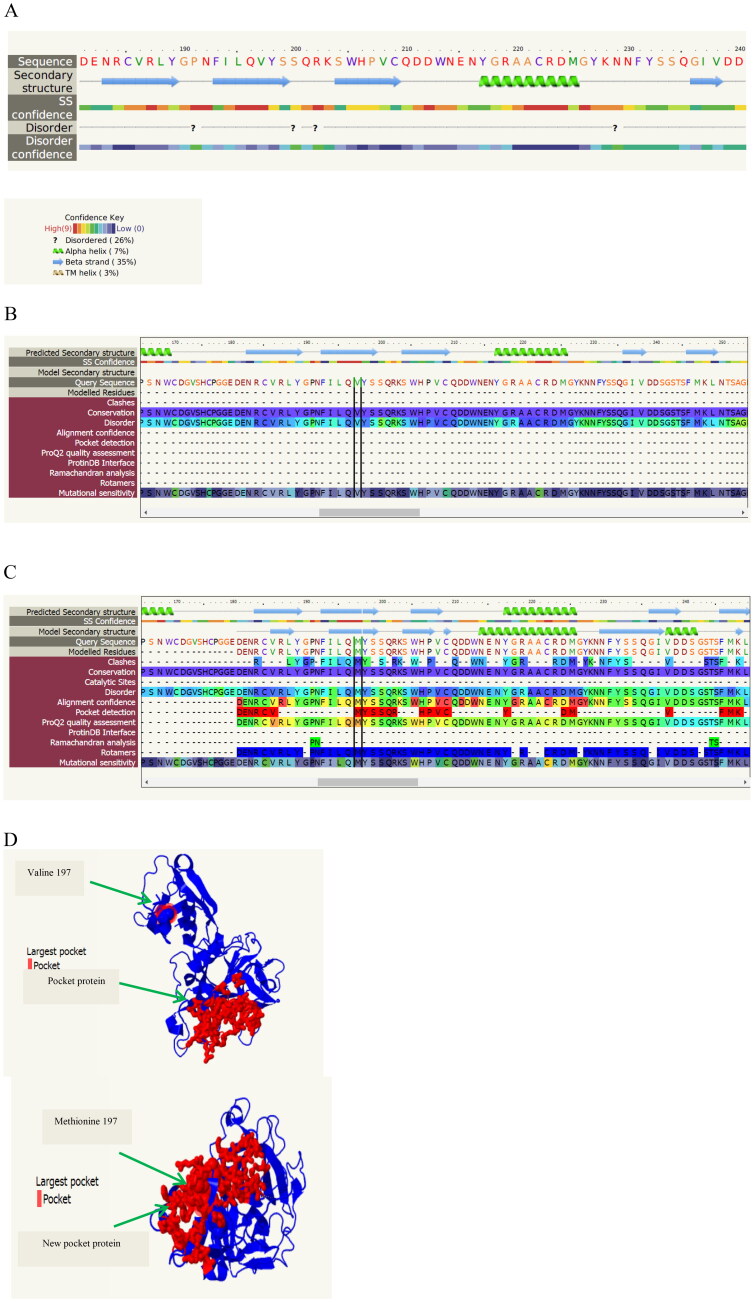
Phyre2, GOR IV and PSIPRED were conducted to investigate the probable effects of SNPs on TMPRSS2 secondary structure. Phyre2 predicted that alteration of valine to methionine in position 197 (V197M) due to C > T conversion (rs12329760) is located in beta strand of TMPRSS2. Also, phyre2 suggested that methionine relative to valine created a pocket protein via influencing several residues (red residues) along with a new rotamer (Figure 9(A–D)). Furthermore, it showed that glycine to valine alteration (G8V) resulting from rs75603675 increased the probability of disorder whereas glycine to aspartate (G8D) did not show any significant change in probability of disorder.
Figure 9.
Prediction of TMPRSS2 secondary structure by Phyre2. A: secondary structure of TMPRSS2 in position 197; B: secondary structure of TMPRSS2 with valine in position 197; C: secondary structure of TMPRSS2 with methionine in position 197; D: position of pocket protein in TMPRSS2 with two different residues (Valine 197, Methionine 197).
GOR IV predicted that most parts of TMPRSS2 are constituted from random coil (56.71%) whereas extended strand (30.06%) and alpha helix (13.23%) made up other parts of TMPRSS2 (Figure 10(A)). Besides, results from GOR IV showed that rs75603675 and rs12329760 were located in random coil and extended strand regions, respectively (Figure 10(B)).
Figure 10.
Prediction of TMPRSS2 secondary structure by GOR IV. A:secondary structure distribution of TMPRSS2; B: secondary structure of TMPRSS2.
Finally, analysis of secondary structure of TMPRSS2 through PSIPRED indicated that rs75603675 and rs12329760 are posited in coil and strand of TMPRSS2, respectively (Figure 11).
Figure 11.
Analysis of the secondary structure of TMPRSS2 by PSIPRED.
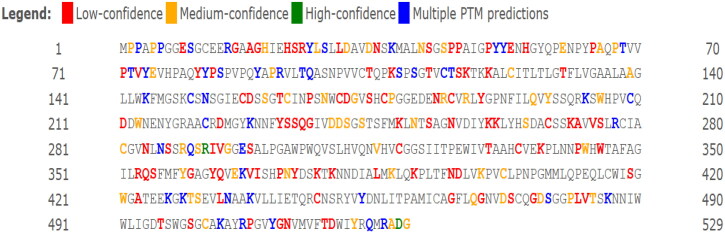
Prediction of post-translational modifications (PTM) and secretory characteristics of TMPRSS2 relative to SNPs
Investigation of TMPRSS2 PTM through Modpred presented probable modifications for each of amino acid residues in TMPRSS2. Furthermore, Modpred illustrated that rs12329760 (V197M) has no effect on TMPRSS2 PTM but rs75603675 (G8D) caused a de novo proteolytic site in this position (Figure 12).
Figure 12.
Prediction of post-translational modifications (PTM) of TMPRSS2.
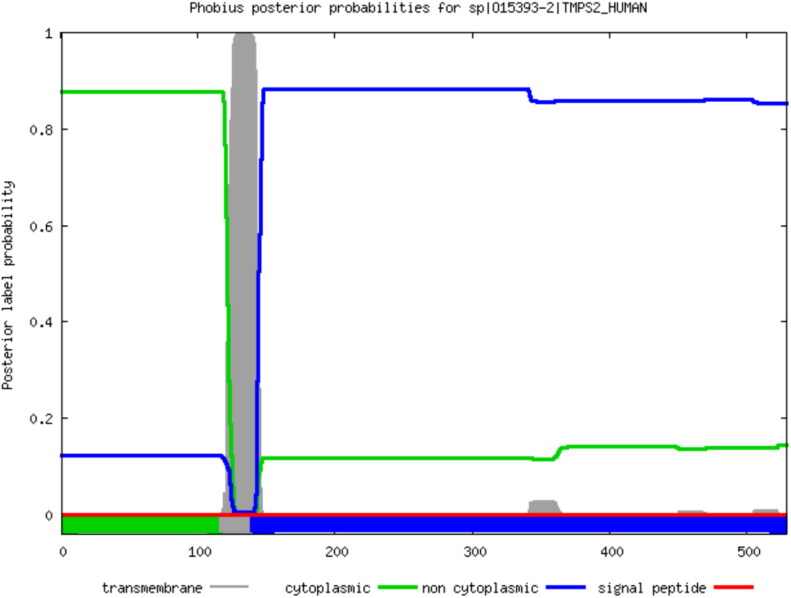
Further, phobius was undertaken in order to analysis of TMPRSS2 secretion alterations resulting from SNPs of TMPRSS2. Reached results from phobius demonstrated that no significant changes in secretion of TMPRSS2 arise out of SNPs in TMPRSS2 (Figure 13).
Figure 13.
Analysis of transmembrane topology and signal peptides of TMPRSS2. The plot is obtained by calculating the total probability that a residue belongs to a helix, cytoplasmic, or noncytoplasmic summed over all possible paths through the model and shows the posterior probabilities of cytoplasmic, noncytoplasmic, TM helix and signal peptide.
Functional effects of TMPRSS2 SNPs on the miRNA profile of different populations
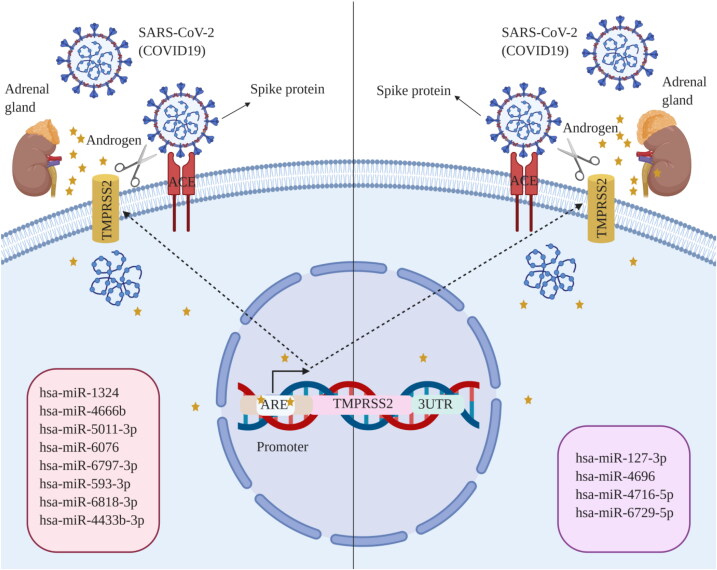
PolymiRTS and miRSNPs were conducted to investigate the effects of SNPs within TMPRSS2 on miRNAs biogenesis and function. PolymiRTS showed that 26 SNPs posited in miRNA target sites, 15 SNPs located in miRNA seed which disrupted miRNA target sites, and 26 SNPs located in miRNA seed which created miRNA target site. Taken together, out of 67 SNPs only the frequency of 6 SNPs including rs456142, rs462574, rs456298 and rs12627374 that are located in miRNA target sites, and rs12473206 and rs75036690 that are located in miRNA seed creating and disrupting miRNA target sites, respectively were different between Asian and other populations. Correspondingly, miRSNPs predicted that the frequency of 4 SNPs including rs456142, rs462574, rs456298 and rs12627374 are different between Asian populations relative to other populations; nevertheless 3 of them (rs456142, rs462574 rs456298) have shown more significant different frequency (Table 5 and Figure 14).
Table 5.
Prediction of miRNA profile through SNPs by PolymiRTS and miRSNPs.
| A. Prediction of SNPs in miRNA target site by PolymiRTS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Location | Variant type | Wobble base pair | Ancestral allele | Allele | miR ID | Conservation | miR site | Function class | Exp support | Context + score change |
| 1. rs456142 | 42836496 | SNP | Y | A | A | hsa-miR-153-3p hsa-miR-448 | 2 2 | tTATGCAAttttt tTATGCAAttttt | D D | N N | −0.179 − 0.17 |
| G | hsa-miR-548c-3p | 2 | ttatgcGATTTTT | C | N | −0.057 | |||||
| 2. rs462574 | 42836729 | SNP | N | C | T | hsa-miR-1324 | 5 | aggatCTGTCTGt | C | N | −0.152 |
| C | hsa-miR-127-3p | 5 | aGGATCCGtctgt | D | N | −0.299 | |||||
| 3. rs456298 | 42836751 | SNP | N | A | A | hsa-miR-4666b hsa-miR-5011-3p hsa-miR-6076 hsa-miR-6797-3p | 2 2 2 2 | aaagtCATGCAAt aaagtCATGCAAt aaaGTCATGCAat aaaGTCATGCAat | D D D D | N N N N | −0.101 − 0.106 − 0.397 − 0.428 |
| T | hsa-miR-4696 | 2 | aaaGTCTTGCAat | C | N | −0.363 | |||||
| 4. rs12627374 | 42837691 | SNP | Y | G | G | hsa-miR-593-3p hsa-miR-6818-3p | 2 2 | aggAGAGACAtgg aggAGAGACAtgg | D D | N N | −0.078 − 0.09 |
| A | hsa-miR-4716-5p | 5 | aggagaAACATGG | C | N | −0.147 | |||||
| B. Prediction of SNPs in miRNA seed causing disruption or creating target site by PolymiRTS | ||||||||
| SNP |
Location |
miR ID |
miR seed |
Allele |
Wobble base pair |
miR site |
Conservation |
Context + score change |
| 1. rs12473206 created miRNA seeds target | 42837404 | hsa-miR-4433b-3p | AGGAGU[G/C] | G/C | 0 | CACUCCU | 2 | 0.055 |
| 2. rs75036690 disrupt miRNA seeds target | 42837415 | hsa-miR-6729-5p | GGGC[G/A]AG | G/A | 1 | CUUGCCC | 2 | −0.149 |
| C. Prediction of miRNAs profile by miRSNPs | ||||||
|---|---|---|---|---|---|---|
| SNP | miRNA | Effect | Allele | Score | Energy | Conservation |
| 1. rs456142 | hsa-miR-548c-3p | Break | G | 158 | −8.18 | 0.557 |
| A | NA | NA | NA | |||
| 2. rs462574 | hsa-miR-127-3p | Create | T | NA | NA | NA |
| C | 146 | −17.49 | 0 | |||
| hsa-miR1324 | Break | T | 154 | −16.86 | 0.002 | |
| C | NA | NA | NA | |||
| 3. rs456298 | hsa-miR-5089 | Enhance | T | 144 | −12.55 | 0.001 |
| A | 150 | −14.2 | 0.001 | |||
| 4. rs12627374 | hsa-miR-204-5p | Decrease | G | 149 | −19.29 | 0 |
| A | 147 | −17.05 | 0 | |||
| hsa-miR-211-5p | Enhance | G | 154 | −21.16 | 0 | |
| A | 158 | −20.63 | 0 | |||
| hsa-miR-4685-3p | Decrease | G | 147 | −18.52 | 0.001 | |
| A | 144 | −18.92 | 0.001 | |||
| hsa-miR-4716-5p | Create | G | NA | NA | NA | |
| A | 153 | −18.1 | 0.006 | |||
NA: not available; D: the derived allele disrupts a conserved miRNA site; C: the derived allele creates a new miRNA site; N: predicted target site with no experimental support.
Figure 14.
Analysis of miRNAs profile upon TMPRSS2 SNPs by PolymiRTS and miRSNPs. (A) miRNAs profile of Asian population; (B) miRNAs profile of global population.
Discussion
The current outbreak of SARS-CoV-2 strongly emphasized on human to human transmission which rapidly spread throughout the world. The high frequency of infected subjects in China despite severe isolation strategies highlighted the probable role of host genome variations in susceptibility to wide spectrum of diseases. Increasing body of evidence clarified the fundamental role of TMPRSS2 in cell entry of SARS-CoV-2. Given the crucial role of TMPRSS2 in cell entry of SARS-CoV-2, TMPRSS2 variation or dysregulation may influence individuals’ susceptibility to SARS-CoV-2 infection. Obtained results from Gtex revealed that TMPRSS2 potentially was expressed in prostate, colon-transverse and stomach. Also, GEPIA suggested that the TMPRSS2 TPM was significantly higher in PRAD in comparison with paired normal tissues. Investigation by CCLE demonstrated higher levels of TMPRSS2 expression in several cell lines such as prostate, colorectal, stomach and bile duct in comparison with other cell lines. These findings were supported by studies conducted on prostate cancer patients that showed higher levels of TMPRSS2 expression in comparison with normal subjects (Emami et al., 2019). Also, results from an investigation of Tmprss2 expression in mice showed that Tmprss2 was considerably expressed in epithelia of the gastrointestinal, urogenital and respiratory tracts (Vaarala et al., 2001). Analysis of overall survival of patients with PRAD and LUAD through GEPIA illustrated that decreased overall survival was positively associated with higher levels of TMPRSS2 expression. This outcome is in accordance with a cohort study including comparison of survival of patients with TMPRSS2-ERG overexpression and patients with lack of TMPRSS2-ERG which showed the lower survival of the first group in comparison with the second group (Hägglöf et al., 2014). Accumulating evidence suggested that the higher levels of TMPRSS2 in prostate tissue and its cell lines might be due to androgen-dependent expression of TMPRSS2 (Graff et al., 2015; Lin et al., 1999). Several studies disclosed that androgen amplified the expression levels of TMPRSS2 via interaction with ARE located in TMPRSS2 promoter (Clinckemalie, 2013; Clinckemalie et al., 2013). Strikingly, a recent study performed on 99 patients with SARS-CoV2 revealed that men (68%) were more susceptible to SARS-CoV2 (Chen et al., 2020). Also, a study performed on mice demonstrated that males were significantly more prone to SARS-CoV than females (Channappanavar et al., 2017). The primary analysis to retrieve all SNPs related to TMPRSS2 demonstrated 11,184 SNPs throughout the TMPRSS2. Out of 11184 SNPs only 92 (with MAF between 0.01 and 0.95) showed different frequencies between Asian and other populations. Analyzing two missense variants including rs12329760 and rs75603675 by SIFT, PolyPhen-2, PROVEAN and SNAP2 revealed that rs12329760 (V197M) was considered deleterious by three tools whereas rs75603675 (G8D) was considered deleterious only by polyphen-2. Correspondingly, a study conducted on 162 patients with prostate cancer revealed that 44 and 35 patients presented rs12329760 and rs75603675, respectively (García-Perdomo et al., 2018). Besides, another study performed on 214 patients affected by prostate cancer showed that the T allele of rs12329760 was related to TMPRSS2-ERG fusion and prostate cancer pathogenesis (FitzGerald et al., 2008). These findings suggest that the high frequency of SARS-CoV2 infection in Chinese population may probably be due partly to their SNPs profile (Anonymous, 2020b; Pant et al., 2020). SNPs were analyzed by HSF in order to prediction of their effects on splice site. Results illustrated that 15 SNPs caused to disruption in splicing processing through several mechanisms such as creation of new splice site, breaking site and broken or new site in enhancer or silencer of splicing. Correspondingly, a case report on two siblings with complete androgen insensitivity syndromes revealed the presence of a point mutation (G > A) at the exon 7/intron 7 splice junction of the AR gene. This splice mutation caused a truncated protein (which is 94 amino acids shorter than wild type) through deletion of exon 7 and thereby splicing ofexon 6 to exon 8 (Lim et al., 1997). Collectively, these results showed the key role of splicing processes in gene expression regulation. Therefore, splice variations due to SNPs might have affected the expression levels of TMPRSS2 and thereby changed the susceptibility of individuals to SARS-CoV2 infection. To investigation of the secondary structure of TMPRSS2 three tools including Phyre2, GOR IV and PSIPRED were engaged. Analysis of rs12329760 (V197M) through Phyre2, GOR IV and PSIPRED revealed that this position is located in beta strand, extended strand region and strand, respectively. Furthermore, analyzing rs75603675 (G8V, G8D) through Phyre2, GOR IV and PSIPRED suggested that this position is located in disordered region, random coil and coil, respectively. Result from phyre2 for rs75603675 showed that G > V might increase the possibility of disorder which could influence the function of TMPRSS2 in facilitating SARS-CoV-2 cell entry. Moreover, all three databases predicted that rs12329760 (V197M) is located in strand structure of TMPRSS2. Strikingly, Phyre2 predicted a new largest pocket protein upon V197M conversion in a wide region which probably affected TMPRSS2 structure and thereby affecting probably its role in SARS-CoV2 cell entry. Investigation of PTM of TMPRSS2 by Modpred showed that a change in position 8 (G8D) upon rs75603675 caused a de novo proteolytic cleavage site in this position. Regarding to prediction of phyre2 for rs75603675 (G8V, G8D) this position located in disordered region which a de novo proteolytic cleavage site probably may influence the efficiency of TMPRSS2 in facilitating SARS-CoV2 infection. Subsequently, secretory properties of TMPRSS2 analyzed by phobius showed no significant change in transmembrane, cytoplasmic, non-cytoplasmic topology and signal peptide of TMPRSS2 due to SNPs. Finally, analyzes of miRNAs profile alteration upon SNPs was carried out by PolymiRTS and miRSNPs. Altogether, 6 SNPs were predicted by these tools which influenced miRNA target site and miRNA seed region. Similarly, comparison of miRNAs profile related to TMPRSS2-ERG between African Americans (AAs) with more aggressive prostate cancer (which are commonly TMPRSS2 fusion negative tumors) and European Americans (EAs) with TMPRSS2 fusion positive tumors have revealed differences in 18 miRNAs, but two miRNAs (miR-106a and miR-17) were significantly different between AAs and EAs. Furthermore, CpG methylation status analysis showed that miRNA encoding genes were modulated epigenetically through their CpG islands. Hypomethylation or hypermethylation of CpG islands of miRNA genes could influence the miRNAs expression levels. Therefore, difference between AAs and EAs in prostate cancer susceptibility might be due to epigenetic mechanisms such as alteration in methylation profile of miRNA genes which are associated to modulating of miRNAs expression (Yates et al., 2017). Accordingly, growing body of evidence indicated the fundamental role of epigenetic mechanisms in regulating miRNA expression, and thereby development of several diseases including Alzheimer, and especially some types of carcinomas (breast, and colorectal) (Frick et al., 2019; Villela et al., 2016; Wu et al., 2019). Obtained results from PolymiRTS and miRSNPs revealed the vital role of miRNAs profile in regulation of TMPRSS2 expression, and thereby highlighted its possible effect on higher susceptibility of Asian populations especially Chinese (57 cases per million) and Iran in the middle East (873 cases per million), and European populations especially Spain (3625cases per million) to SARS-CoV2 (Anonymous, 2020a). Consequently, the present study emphasized on crucial role of SNPs throughout TMPRSS2 in individuals’susceptibility to SARS-CoV-2 infection via influencing several essential processes such as splicing, miRNA expression, epigenetic mechanisms, PTM, protein structure and gene expression. Investigation on the effect of camostatmesylate, a serine protease inhibitor, on several SARS-CoV-2-infected cell lines showed that camostatmesylate significantly reduced viral infection, especially in Calu-3 cell lines. Also, co-treatment of cell lines with camostatmesylate and E64-d, a cathepsin L and cathepsin B inhibitor, led to complete inhibition of SARS-CoV-2s’ cell entry (Hoffmann et al., 2020). Accordingly, a study conducted on HeLa cells expressing both ACE2 and TMPRSS2 which were infected with SARS-CoV illustrated that co-treatment with serine (camostatmesylate) and cysteine protease (EST, as a cathepsin inhibitor) potentially inhibits SARS-CoVs’ cell entry (Kawase et al., 2012). Camostatmesylate blocks the proteolytic cleavage of S protein, and thereby SARS-CoV-2 cell entry by inhibiting TMPRSS2. Camostatmesylate have been long administrated to treatment of pancreatic inflammation (Yamauchi et al., 2001). Correspondingly, treatment of SARS-CoV-infected mice and cell lines with camostatmesylate showed an increased survival rate of mice (about 60%) (Zhou et al., 2015). Moreover, camostatmesylate was shown to inhibit influenza virus cell entry via impeding of proteolytic cleavage of influenza hemagglutinin (HA) which is a key process to virus cell entry. Besides, it was shown that camostatmesylate decreases the levels of cytokines such as interleukin 6 and tumor necrosis factor-α in cell culture supernatants by inhibiting TMPRSS2 and HAT (human trypsin-like protease TMPRSS11D) which cleave HA and activate influenza (Yamaya et al., 2015). Taken together, camostatemesylate might be a hopeful agent to combat several viruses especially SARS-CoV-2. Nonetheless, more clinical trials are needed to determining camostat effectiveness in counteracting the SARS-CoV-2. Also, it is probable that individuals’ response to camostatmesylate treatment may be influenced by their TMPRSS2 SNPs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan Medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–12 (just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous (2020a). Retrieved from https://www.worldometers.info/coronavirus/coronavirus-cases/
- Anonymous (2020b). Retrieved from https://www.who.int/
- Bhanushali A., Rao P., Raman V., Kokate P., Ambekar A., Mandva S., Bhatia S., & Das B. R. (2018). Status of TMPRSS2-ERG fusion in prostate cancer patients from India: Correlation with clinico-pathological details and TMPRSS2 Met160Val polymorphism. Prostate International, 6(4), 145–150. 10.1016/j.prnil.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14 (just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., & Wang W. (2020). Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discovery, 6(1), 1–4. 10.1038/s41421-020-0147-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Mack M., Ten Eyck P. P., Meyerholz D. K., & Perlman S. (2017). Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. Journal of Immunology, 198(10), 4046–4053. 10.4049/jimmunol.1601896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., & Zhang L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shan K., & Qian W. (2020). Asians and other races express similar levels of and share the same genetic polymorphisms of the SARS-CoV-2 cell-entry receptor. [Google Scholar]
- Chen Z., Song X., Li Q., Xie L., Guo T., Su T., Tang C., Chang X., Liang B., & Huang D. (2019). Androgen receptor-activated enhancers simultaneously regulate oncogene TMPRSS2 and lncRNA PRCAT38 in prostate cancer. Cells, 8(8), 864 10.3390/cells8080864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinckemalie L. (2013). The TMPRSS2 gene: study of the androgen regulation and the effects of genetic polymorphisms on androgen receptor binding. [Google Scholar]
- Clinckemalie L., Spans L., Dubois V., Laurent M., Helsen C., Joniau S., & Claessens F. (2013). Androgen regulation of the TMPRSS2 gene and the effect of a SNP in an androgen response element. Molecular Endocrinology, 27(12), 2028–2040. 10.1210/me.2013-1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. (just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami N. C., Kachuri L., Meyers T. J., Das R., Hoffman J. D., Hoffmann T. J., Hu D., Shan J., Feng F. Y., Ziv E., Van Den Eeden S. K., & Witte J. S. (2019). Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms. Nature Communications, 10(1), 1–11. 10.1038/s41467-019-10808-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald L. M., Agalliu I., Johnson K., Miller M. A., Kwon E. M., Hurtado-Coll A., Fazli L., Rajput A. B., Gleave M. E., Cox M. E., Ostrander E. A., Stanford J. L., & Huntsman D. G. (2008). Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: Results from a population-based study of prostate cancer. BMC Cancer, 8(1), 230 10.1186/1471-2407-8-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick E., Gudjonsson T., Eyfjord J., Jonasson J., Tryggvadóttir L., Stefansson O., & Sigurdsson S. (2019). CpG promoter hypo-methylation and up-regulation of microRNA-190b in hormone receptor-positive breast cancer. Oncotarget, 10(45), 4664–4678. 10.18632/oncotarget.27083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Perdomo H. A., Zamora-Segura B. D., & Sanchez A. (2018). Frequency of allelic variants of the TMPRSS2 gene in a prostate cancer-free Southwestern Colombian population. Revista Mexicana de Urología, 78(5), 354–358. [Google Scholar]
- Graff R. E., Pettersson A., Lis R. T., DuPre N., Jordahl K. M., Nuttall E., Rider J. R., Fiorentino M., Sesso H. D., Kenfield S. A., Loda M., Giovannucci E. L., Rosner B., Nguyen P. L., Sweeney C. J., Mucci L. A., & on behalf of the Transdisciplinary Prostate Cancer Partnership ToPCaP (2015). The TMPRSS2:ERG fusion and response to androgen deprivation therapy for prostate cancer. The Prostate, 75(9), 897–906. 10.1002/pros.22973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–17 (just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägglöf C., Hammarsten P., Strömvall K., Egevad L., Josefsson A., Stattin P., Granfors T., & Bergh A. (2014). TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PLoS One, 9(2), e86824 10.1371/journal.pone.0086824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis G., & van Goor H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology, 203(2), 631–637. 10.1002/path.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., & Shahpasand K. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–13 (just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., & Pöhlmann S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G. H., Gramberg T., & Pöhlmann S. (2004). Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochemical and Biophysical Research Communications, 319(4), 1216–1221. 10.1016/j.bbrc.2004.05.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., … Cao B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., & Nagata N. (2019). TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. Journal of Virology, 93(6), 01818 e01815. 10.1128/JVI.01815-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., & Matsuyama S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. Journal of Virology, 86(12), 6537–6545. 10.1128/JVI.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribosemethyltransferase. Journal of Biomolecular Structure and Dynamics, 1–40 (just-accepted), [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–13 (just-accepted), [DOI] [PubMed] [Google Scholar]
- Ko C.-J., Huang C.-C., Lin H.-Y., Juan C.-P., Lan S.-W., Shyu H.-Y., Wu S.-R., Hsiao P.-W., Huang H.-P., Shun C.-T., & Lee M.-S. (2015). Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Research, 75(14), 2949–2960. 10.1158/0008-5472.CAN-14-3297 [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., … Penninger J. M. (2005). A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nature Medicine, 11(8), 875–879. 10.1038/nm1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Ghadessy F. J., & Yong E. (1997). A novel splice site mutation in the androgen receptor gene results in exon skipping and a non-functional truncated protein. Molecular and Cellular Endocrinology, 131(2), 205–210. 10.1016/S0303-7207(97)00109-3 [DOI] [PubMed] [Google Scholar]
- Lin B., Ferguson C., White J. T., Wang S., Vessella R., True L. D., Hood L., & Nelson P. S. (1999). Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Research, 59(17), 4180–4184. [PubMed] [Google Scholar]
- Luostari K., Hartikainen J. M., Tengström M., Palvimo J. J., Kataja V., Mannermaa A., & Kosma V.-M. (2014). Type II transmembrane serine protease gene variants associate with breast cancer. PLoS One, 9(7), e102519 10.1371/journal.pone.0102519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S., Suzuki M., Arai T., Suzuki M., Kato M., Morikawa T., Kasuya Y., Kume H., Kitamura T., & Homma Y. (2014). TMPRSS2 Met160Val polymorphism: significant association with sporadic prostate cancer, but not with latent prostate cancer in Japanese men. International Journal of Urology, 21(12), 1234–1238. 10.1111/iju.12578 [DOI] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., & Takeda M. (2020). Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proceedings of the National Academy of Sciences of the United States of America, 117(13), 7001–7003. 10.1073/pnas.2002589117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–7 (just-accepted). [DOI] [PubMed] [Google Scholar]
- Nickols N. G., & Dervan P. B. (2007). Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proceedings of the National Academy of Sciences of the United States of America, 104(25), 10418–10423. 10.1073/pnas.0704217104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–15 (just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieruzzi F., Abassi Z. A., & Keiser H. R. (1995). Expression of renin-angiotensin system components in the heart, kidneys, and lungs of rats with experimental heart failure. Circulation, 92(10), 3105–3112. 10.1161/01.CIR.92.10.3105 [DOI] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11 (just-accepted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala M. H., Porvari K. S., Kellokumpu S., Kyllönen A. P., & Vihko P. T. (2001). Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. The Journal of Pathology, 193(1), 134–140. [DOI] [PubMed] [Google Scholar]
- Villela D., Ramalho R. F., Silva A. R. T., Brentani H., Suemoto C. K., Pasqualucci C. A., Grinberg L. T., Krepischi A. C. V., & Rosenberg C. (2016). Differential DNA methylation of microRNA genes in temporal cortex from Alzheimer's disease individuals. Neural Plasticity, 2016, 2584940 10.1155/2016/2584940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Ye S., Tan W., Zhou Y., & Quan J. (2019). Analysis of promoter methylation and epigenetic regulation of miR-32 in colorectal cancer cells. Experimental and Therapeutic Medicine, 17(4), 3209–3214. 10.3892/etm.2019.7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.-W., Wu X.-X., Jiang X.-G., Xu K.-J., Ying L.-J., Ma C.-L., Li S. B., Wang H. Y., Zhang S., Gao H. N., and Sheng J. F. (2020). Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. Bmj, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J., Takeda K., Shibuya K., Sunamura M., & Matsuno S. (2001). Continuous regional application of protease inhibitor in the treatment of acute pancreatitis. An experimental study using closed duodenal obstruction model in dogs. Pancreatology, 1(6), 662–667. 10.1159/000055878 [DOI] [PubMed] [Google Scholar]
- Yamaya M., Shimotai Y., Hatachi Y., Lusamba Kalonji N., Tando Y., Kitajima Y., Matsuo K., Kubo H., Nagatomi R., Hongo S., Homma M., & Nishimura H. (2015). The serine protease inhibitor camostat inhibits influenza virus replication and cytokine production in primary cultures of human tracheal epithelial cells. Pulmonary Pharmacology & Therapeutics, 33, 66–74. 10.1016/j.pupt.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates C., Long M. D., Campbell M. J., & Sucheston-Campbell L. (2017). miRNAs as drivers of TMPRSS2-ERG negative prostate tumors in African American men. Frontiers in Bioscience, 22(2), 212–229. 10.2741/4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J. M., Li Y., Zhong N., & Slutsky A. S. (2020). Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Medicine, 46(4), 586–585. 10.1007/s00134-020-05985-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Nunneley J. W., Barnard D., Pöhlmann S., McKerrow J. H., Renslo A. R., & Simmons G. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Research, 116, 76–84. 10.1016/j.antiviral.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]