Abstract
Recently, a pathogen has been identified as a novel coronavirus (SARS-CoV-2) and found to trigger novel pneumonia (COVID-19) in human beings and some other mammals. The uncontrolled release of cytokines is seen from the primary stages of symptoms to last acute respiratory distress syndrome (ARDS). Thus, it is necessary to find out safe and effective drugs against this deadly coronavirus as soon as possible. Here, we downloaded the three-dimensional model of NSP10/NSP16 methyltransferase (PDB-ID: 6w6l) and main protease (PDB-ID: 6lu7) of COVID-19. Using these molecular models, we performed virtual screening with our anti-viral, inti-infectious, and anti-protease compounds, which are attractive therapeutics to prevent infection of the COVID-19. We found that top screened compound binds with protein molecules with good dock score with the help of hydrophobic interactions and hydrogen bonding. We observed that protease complexed with Cyclocytidine hydrochloride (anti-viral and anti-cancer), Trifluridine (anti-viral), Adonitol, and Meropenem (anti-bacterial), and Penciclovir (anti-viral) bound with a good docking score ranging from −6.8 to −5.1 (Kcal/mol). Further, NSP10/NSP16 methyltransferase complexed with Telbivudine, Oxytetracycline dihydrate (anti-viral), Methylgallate (anti-malarial), 2-deoxyglucose and Daphnetin (anti-cancer) from the docking score of −7.0 to −5.7 (Kcal/mol). In conclusion, the selected compounds may be used as a novel therapeutic agent to combat this deadly pandemic disease, SARS-CoV-2 infection, but needs further experimental research.
Highlights
NSP10/NSP16 methyltransferase and main protease complex of SARS CoV-2 bind with selected drugs.
NSP10/NSP16 methyltransferase and protease interacted with drugs by hydrophobic interactions.
Compounds show good DG binging free energy with protein complexes.
Ligands were found to follow the Lipinski rule of five.
Keywords: SARS CoV-2, COVID-19, NSP10/NSP16 methyltransferase, protease complex, anti-viral/anti-infectious/anti-protease drugs
1. Introduction
A new coronavirus, SARS-CoV-2, was found to cause pulmonary disease in the city of Wuhan, China, in December 2019 [1]. The name SARS-CoV-2 is due to its genomic RNA, which is about 82% similar to the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) [2]. These viruses are found to link with clade b of the Betacoronavirus genus [3]. The disease caused by SARS-CoV-2 is known as coronavirus disease-19 (COVID-19). At the beginning of the COVID-19 outbreak/spread, most of the cases were found to connect to the wild animal, seafood, and animal market in Wuhan [4]. These human foods have been found efficient to enhance the number of transmissions among human and later infected humans promotes human to human transmission, which further leads to the exponential growth of these viruses [5]. The World Health Organization (WHO) declared this outbreak as pandemic on 11 March 2020. As of May 16, there are >4.59 million cases globally, with a ∼3.7% case-fatality rate (https://www.worldometers.info/coronavirus).
Coronaviruses (CoVs) have been found to affect the respiratory tract and lead light to harsh respiratory tract infections in humans [6]. In the past two decades, the SARS-CoV (severe acute respiratory syndrome-coronavirus) and the MERS-CoV (middle east respiratory syndrome-coronavirus) have been found to emerge from animal reservoirs and lead to global epidemics with great anguish and fatality [7].
The coronaviruses have been found to encode two proteases (translated nonstructural proteins), i.e. a papain-like protease (Plpro) and main proteases. The main protease is also known as 3-C-like protease (M-pro) [8]. It has been shown that the main Protease (Mpro, also called 3CLpro) is one of the best-characterized drug targets among coronaviruses [9]. Protease is necessary for the processing of polyproteins, which are translated from the SARS CoV-2 RNA [8]. The main Protease of SARS CoV-2 operates at about 11 sites of cleavage on the large polyprotein 1ab (replicase 1ab, size = ∼790 kDa) and the recognition sequence is found to be Leu-Gln↓Ser-Ala-Gly) (where ↓ marks the site of cleavage). It has been seen that inhibiting the activity of the main protease enzyme could be an effective measure to check viral replication [10].
Various forms of the viruses are found to perform replication in the cytoplasm of eukaryotes through evolving 2′-O-methyltransferases (2′-O-MTase) for modification of their mRNAs and carry a Cap-1 structure such as, m7GpppNm at the 5′ end, which facilitate viral duplication and help to escape recognition against innate immune responses done by some immune cells in the hosts [11]. The stimulatory factor NSP10, along with NSP16, stabilizes the pocket of SAM-binding and continuing the RNA-binding groove of the substrate of NSP16, which is observed by crystallographic and biochemical analysis [12]. It has been observed that interference in the interaction between NSP16 and NSP10 of SARS-CoV by short peptides can inhibit the activity of 2′-O-MTase [13]. 2′-O-MTase was also found to catalyze the transfer of methyl group SAM or AdoMet to RNA substrate and produce S-adenosyl-L-homocysteine as a byproduct [14]. It was evidenced that the stimulation of NSP16 methyltransferase activity by NSP10 is a common and primary mechanism for the coronaviruses. It has been further demonstrated that NSP10 is synonymous in the stimulatory function among distinct coronaviruses, and the peptide, which is derived from the conserved domain of MHV NSP10, shows an inhibitory effect on 2′-O-methyltransferase activity during virus duplication. These results might use to find out specific anti-COVID-19 drugs to control infection [11].
It was proved that the availability of FDA-approved anti-RNA-dependent RNA Polymerase (anti-RdRp) drugs could help in the treatment of patients and reduce the chances of the danger of the viral infection caused by SARS CoV2. These drugs can tightly bind to the RdRp and thus may be useful to treat the disease caused by the SARS CoV2. Furthermore, it has also been shown that Ribavirin, vitamin B12, and nicotinamide may be helpful against COVID 19 infections [12]. Moreover, it has also been shown that the anti-virals Velpatasvir and Ledipasvir are particularly attractive as therapeutics to combat the SARS CoV-2 with minimal side effects (fatigue and headache). The drugs Harvoni (Ledipasvir/Sofosbuvir) and Epclusa (Velpatasvir/Sofosbuvir) with their dual inhibitory actions on two viral enzymes could be very effective [15].
The present study provided a comprehensive target of the resolved COVID19 structure of Mpro and NSP10/NSP16 Methyltransferase and found a suitable approach against this disease. We have performed in silico study on anti-viral, anti-infectious, and anti-protease compounds against methyltransferase-stimulatory factor complex and main protease.
2. Material and methods
The in-silico studies were performed on KBS Desktop having 12 GB RAM, Intel i5 generation with 4 cores in Indian Institute of Information Technology, Allahabad, UP, India. Grid-based Ligand Docking with Energetics (GLIDE) module of maestro 12.0 (Schrodinger LLC 2019, USA) was used for our study.
2.1. Ligand preparation
Anti-viral, anti-infectious, and anti-protease compounds were downloaded from the online database Selleckchem (https://www.selleckchem.com/), in SDF format. LigPrep module of Maestro12.0 (Schrodinger) was used to prepare the ligand library for the Docking purpose. Ionization of ligands was retained in actual states with the realistic length of bonds as well as bond angles, ring conformation and tautomers were generated, using the OPLS-2005 force field.
2.2. Protein preparation
The high-resolution structure of SARS-CoV2 main Protease (PDB ID: 6lu7) and NSP10/NSP16 Methyltransferase (PDB ID: 6w61) were downloaded online from the protein data bank (https://www.rcsb.org/). The protein preparation wizard of Maestro 12.0 was used for the preparation of the protein structure. The selected structure was processed for creating disulfide bonds, assigning proper bond orders, and the addition of missing hydrogens in the raw structure. Hydrogen bonds optimization was assigned using non-hydrogen atoms of protein, and the structure was energy minimized until the RMSD (root mean square deviation) reaches the value of 0.3 Å.
2.3. Generation of receptor grid
Protease and methyltransferase were subjected to the Sitemap module of Maestro 12.0 for the prediction of active binding sites. A total of 4 binding sites was generated, and the top one was selected for receptor grid generation. The grid was generated using the default setting of the receptor grid generation module of maestro 12.0, i.e. 0.25 was partial atomic charge and were 1.0 Å was Van der Waals radii of receptor atoms. The center of the sitemap position was selected to generate a grid box by using the receptor grid generation module of Maestro.
2.4. Virtual screening and molecular docking
Virtual screening of prepared library was operated by using virtual screening workflow (VSW) of GLIDE (maestro 12.0, Schrödinger, LLC, NY, USA). VSW has many tools to calculate the properties of ligands, and filter ligands such as QuikProp and prefilters by Lipinski rule were selected. VWS of Maestro has three type docking precision: (1). HTVS (High Throughput Virtual Screening) for fast screening of huge number ligand’s docking, (2) standard-precision (SP) for screening and docking of ligands of unknown quality in large numbers, and (3) extra-precision (XP) is a powerful and discriminating procedure for docking ligands. Extra precision (XP) docking was operated on prepared ligands by keeping default parameters, i.e. scaling factor at 0.80 and partial charge at 0.15. The binding affinity of docked ligands for the receptor protease at the active site was generated from the docking binding energy.
2.5. Binding free energy calculation
Binding free energy of protease enzyme and docked ligands were calculated using the Prime MM-GBSA (Molecular Mechanics-Generalized Born Surface Area) modules of Maestro 12.0, which run on the OPLS_2005 molecular mechanic’s energies, nonpolar solvation and VSGB solvent model [16]. Prime MM-GBSA module was operated using pose viewer file of ligand and receptor (generated after docking) to generate binding free energy. The MM-GBSA analysis employs molecular mechanics and solvent accessibility methods to depict free energies. This analysis is efficient and quite reliable for studying the energetics of biomolecular systems. Prediction of the binding free energies in biomolecular such as, protein, protein-drug complexes is the application of this approach. The following descriptors were used to generate changes in energy upon binding.
MM-GBSA DG bind = Ligand binding free energy, MM-GBSA E complex = complex free energy, MM-GBSA E protein = free energy of the receptor without the ligand, MM-GBSA E Ligand = unbound ligand-free energy.
2.6. Adme/T studies
Absorption, distribution, metabolism, excretion/toxicity (ADME/T) properties are used to eliminate inappropriate compounds before invested inappropriate compounds, time, and money. QuikProp module in VSW (Maestro 12.0) calculated the ADME information of ligands. QikProp module calculates various properties and provides values for descriptors, which predict drug-likeness of ligands and generated functional groups, rings, and a number of elements used for the prediction of descriptors of the ligand/s.
3. Results
3.1. Analysis of docking results of promising compounds
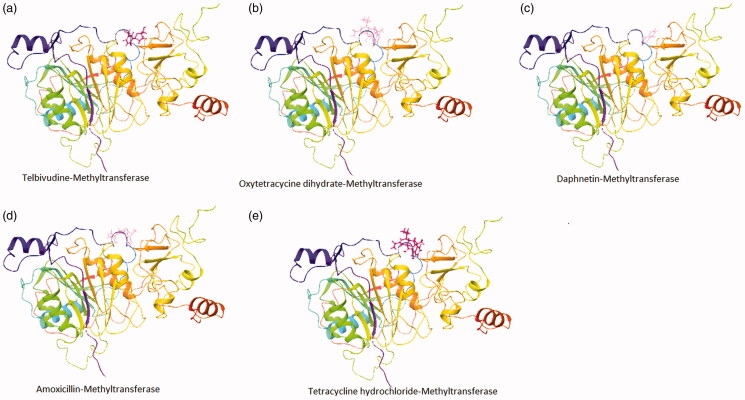
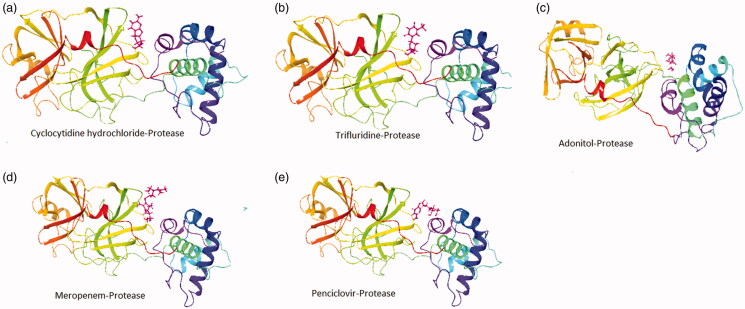
The crystal structure of the NSP10/NSP16 Methyltransferase (PDB ID: 6w6l) and main Protease (PDB ID: 6lu7) of SARS-CoV-2 was used for docking purposes. Molecular docking has been performed using the extra precision (XP) mode of grid-based ligand docking with energetics (GLIDE). We used known anti-viral, anti-infectious, and anti-protease inhibitors for the docking purpose (Figures 1 and 2). We selected the top 5 compounds with good docking scores, which were docked with proteases and NSP10/NSP16 methyltransferase. Our result highlighted that; Cyclocytidine, Hydrochloride, Trifluridine, Adonitol Meropenem, and Penciclovir yielded a preeminent dock score with the main Protease of SARS-COV-2 with docking score of −6.8, −6.0, −5.5, −5.2, and −5.1 Kcal/mol respectively (Table 1) as well as Telbivudine, Oxytetracycline dihydrate, Methylgallate, 2-deoxyglucose, and Daphnetin docked with the NSP10/NSP16 methyltransferase with the docking score of −7.0, −6.0, −6.0, −5.8, and −5.7 Kcal/mol. Cyclocytidine hydrochloride (anti-viral and anti-cancer), Trifluridine (anti-viral), Adonitol (anti-bacterial), Meropenem (antibiotic and anti-bacterial), and Penciclovir (anti-viral) yielded a good docking score with the main Protease and Telbivudine, Oxytetracycline dihydrate (anti-viral), Methylgallate (anti-malarial), 2-deoxyglucose (anti-cancer), and Daphnetin (anti-inflammatory, and antitumor) showed good docking score with methyltransferase-stimulatory factor complex of NSP16 and NSP10 of SARS-CoV-2.
Figure 1.
Ribbon presentation of NSP10/NSP16 methyltransferase complex (PDB-ID: 6w61) with selected compounds. (a) Telbivudine- NSP10/NSP16 methyltransferase; (b) Oxytetracycycine dihydrate- NSP10/NSP16 methyltransferase; (c) Daphnetin- NSP10/NSP16 methyltransferase; (d) Amoxicillin- NSP10/NSP16 methyltransferase; (e) Tetracycline hydrochloride- NSP10/NSP16 methyltransferase.
Figure 2.
Ribbon presentation of Protease complex (PDB-ID: 6lu7) with selected compounds. (a) Cyclocytidine hydrochloride-protease; (b) Trifluridine-protease; (c) Adonitol-protease; (d) Meropenem-protease; (e) Penciclovir-protease.
Table 1.
Docking score (Kcal/mol) of the NSP10/NSP16 methyltransferase (PDB: 6w61) and main protease (PDB: 6lu7) with selected compounds detected by molecular docking.
| NSP10/NSP16 methyltransferase |
Main protease |
|||
|---|---|---|---|---|
| SN | Compounds | Dock score (Kcal/mol) | Compounds | Dock score (Kcal/mol) |
| 1 | TYZEKA (telbivudine) | −7.08 | Cyclocytidine hydrochloride | −6.86 |
| 2 | Oxytetracycline dihydrate | −6.07 | Trifluridine | −6.09 |
| 3 | Methylgallate | −6.02 | Adonitol | −5.58 |
| 4 | 2-deoxy-D-glucose | −5.85 | Meropenem | −5.20 |
| 5 | Daphnetin | −5.78 | Penciclovir | −5.14 |
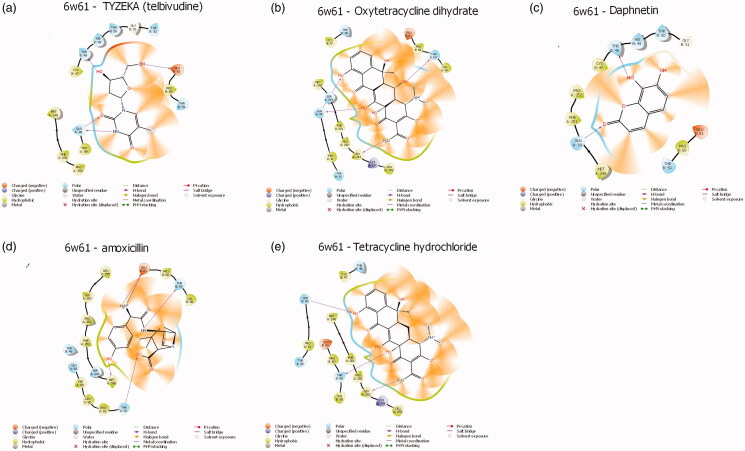
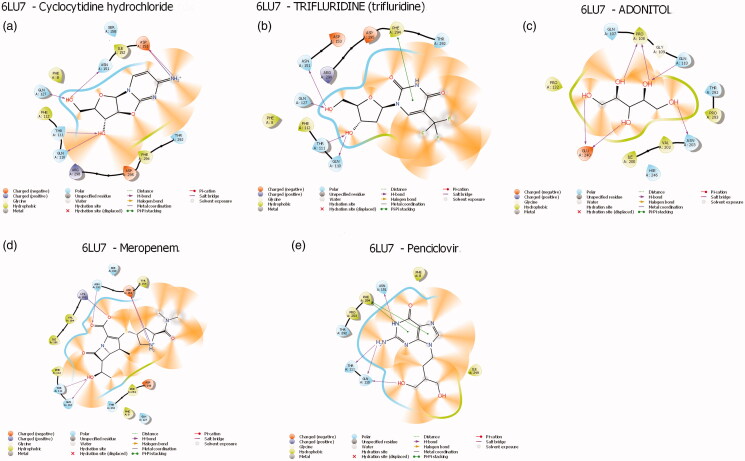
3.2. Interactions between the proteins and compounds
Protein-ligands interactions are generally highlighted by hydrophobic interactions and hydrogen bonds, which play a significant role in predicting the binding affinity of the ligand with proteins. All the selected compounds in the dataset were docked into the active site of the NSP10/NSP16 methyltransferase and the main Protease of SAR-CoV-2 (Figures 3 and 4) depicts the binding conformations of the selected compounds in the binding pocket of the NSP10/NSP16 methyltransferase and protease. The active site of the NSP10/NSP16 methyltransferase showed hydrophobic interaction with the help of Met248, Pro252, Phe251, Cys47, and Pro60 amino acids. The amino acids are present in the active site of Protease like Phe112, Phe8, and Phe294 are found to create hydrophobic interaction with top ligands. Besides Hydrophobic interactions, there are some hydrogen bonds present between amino acids and compounds. In the case of NSP10/NSP16 methyltransferase, mostly Gln88, Glu61, and thr48 formed hydrogen bonds with the atoms of ligands and in the active site of main protease complex Gln110, Thr111, Gln127, and Asn151 amino acids have been found to form a hydrogen bond with the atoms of ligands (Table 2).
Figure 3.
Protein-ligand interactions profile of Methyltransferase-stimulatory factor NSP16/NSP10 complex (PDB-ID: 6w61) with selected compounds. (a) Telbivudine- NSP10/NSP16 methyltransferase; (b) Oxytetracycycine dihydrate- NSP10/NSP16 methyltransferase; (c) Daphnetin- NSP10/NSP16 methyltransferase; (d) Amoxicillin- NSP10/NSP16 methyltransferase; (e) Tetracycline hydrochloride- NSP10/NSP16 methyltransferase.
Figure 4.
Protein-ligand interactions profile of main protease (PDB-ID: 6lu7) with selected compounds. (a) Cyclocytidine hydrochloride-protease; (b) Trifluridine-protease; (c) Adonitol-protease; (d) Meropenem-protease; (e) Penciclovir-protease.
Table 2.
Protein-Ligand interaction profile of the NSP10/NSP16 methyltransferase (PDB: 6w61) and Protease (PDB: 6lu7) with selected compounds.
| SN | Compounds | Hydrophobic interactions | Hydrogen bonds |
|---|---|---|---|
| NSP10/NSP16 methyltransferase | |||
| 1 | TYZEKA (telbivudine) |
|
|
| 2 | Oxytetracycline dihydrate |
|
|
| 3 | Methylgallate |
|
|
| 4 | 2-deoxy-D-glucose |
|
|
| 5 | Daphnetin |
|
|
| Main protease | |||
| 1 | Cyclocytidine hydrochloride | Phe112; Phe8; Ile152; Phe294 | Gln110; Thr111; Gln127; Asn151; Asp153 |
| 2 | Trifluridine | Phe8; Phe112; Phe294 | Gln110; Thr111; Gln127; Asn151 |
| 3 | Adonitol | Pro108; Pro132; Pro293; Ile200; Val202 | Pro108; Asn203; Glu240; Gln110 |
| 4 | Meropenem | Phe112; Ile106; Val104; Tyr154; Phe8; Phe294 | Lys102; asn151; Asp153; Thr111; Gln110 |
| 5 | Penciclovir | Phe8; Phe294; Pro293; Ile249 | Thr111; Gln110; Asn151 |
A good number of amino acids are found to appear in the hydrophobic interactions and hydrogen bonding. Moreover, the active site of the NSP10/NSP16 methyltransferase and main protease have mostly hydrophobic interactions with the ligands. Hydrophobic interaction successfully delineates specific functional groups that may be responsible for the hydrophobic generating effect of these compounds with strong binding affinity against target proteins and may play a highly influencing against SARS-CoV-2 infection
3.3. Estimation of binding-free energy
MM/GBSA calculation was performed to estimate the free binding energy of ligands with docked proteins. Docked top five compounds from ligands library with proteins showed the binding-free energy values. Compounds selected in virtual screening has been found to show the good result of binding free energy with their respective Proteins (Table 3). All the compounds showed good binding free energy with their respective proteins of SARS-CoV-2.
Table 3.
DG Binding free energy of the NSP10/NSP16 methyltransferase (PDB: 6w61) and main protease (PDB: 6lu7) with selected compounds.
| SN | Compounds (NSP10/NSP16 methyltransferase) |
DG binding free energy | Compounds (Main protease) |
DG binding free energy |
|---|---|---|---|---|
| 1 | Oxytetracycline dihydrate | −52.55 | Penciclovir | −42.90 |
| 2 | TYZEKA (Telbivudine) | −51.56 | Cyclocytidine hydrochloride | −33.61 |
| 3 | 2-deoxy-D-glucose | −41.18 | Meropenem | −30.36 |
| 4 | Daphnetin | −38.47 | Trifluridine | −30.18 |
| 5 | Methylgallate | −37.61 | Adonitol | −29.13 |
3.4. Adme/T properties of leads compounds
ADME/T properties of the lead compounds (Figure 4) were appraised by using the Qikprop application of Maestro 12.0). The most attractive aspect of compounds is their admirable QPlogo/w, QPlogS [17] molecular weight, H-bond donor and acceptor, QplogBB, and percentage of human oral value which satisfy the Lipinski rule of five [18] (Table 4). Moreover, polar surface area, high oral bioavailability, H-bond donors, and acceptors are being important criteria for the development of therapeutic agents. All these models unscrew the qualitative prediction and ranking of absorption, formulation effects on drug permeability, determining the mechanism(s) of permeability, and the potential for transporter-mediated drug-drug interactions. All the good scoring ligand has drug-likeness properties by Lipinski’s rule. It may also be predictable that the combination of these drugs/s may be a good idea to prevent COVID-19 infection.
Table 4.
Structural; physicochemical; biochemical; pharmacokinetics and toxicity properties of compound.
| Compound ID | Molecule name | MW | Logs | Logo/w | Accept H | Donor H | QPPCaco | QPlogBB | %Human OralAbso | QPlogHERG | SASA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C9H12CIN3O4 | Cyclocytidine hydrochloride | 225.204 | −1.46 | −0.83 | 8.1 | 3 | 165.556 | −1.174 | 2 | −3.383 | 405.11 |
| C10H11F3N2O5 | Trifluridine | 296.203 | −2.39 | −0.369 | 8.6 | 3 | 64.161 | −1.313 | 2 | −3.425 | 468.898 |
| C5H12O5 | Adonitol | 152.147 | −0.39 | −1.833 | 8.5 | 5 | 116.056 | −1.571 | 2 | −2.772 | 343.539 |
| C17H25N3O2O5S | Meropenem | 383.462 | −1.52 | −1.357 | 9.7 | 2 | 3.353 | −1.491 | 2 | −1.869 | 651.295 |
| C10H15N5O3 | Penciclovir | 253.26 | −1.76 | −1.443 | 8.9 | 5 | 20.823 | −2.494 | 2 | −4.113 | 488.808 |
| C10H14N2O5 | Telbivudine | 242.231 | −1.88 | −1.058 | 8.6 | 3 | 59.067 | −1.595 | 2 | −4.089 | 452.618 |
| C22H28N2O11 | Oxytetracycline dihydrate | 460.44 | −2.31 | −0.097 | 9.95 | 4 | 1.985 | −2.494 | 1 | −5.178 | 643.727 |
| C8H8O5 | Methylgallate | 184.148 | −1.44 | −0.187 | 4.25 | 3 | 111.267 | −1.421 | 3 | −3.787 | 391.928 |
| C6H12O5 | 2-deoxy-D-glucose | 164.158 | −0.83 | −1.687 | 8.5 | 4 | 138.202 | −1.268 | 2 | −2.771 | 349.196 |
| C9H6O4 | Daphnetin | 178.144 | −1.37 | 0.113 | 4 | 2 | 221.972 | −0.943 | 2 | −3.796 | 359.267 |
MW: Molecular weight; logs: Predicted aqueous solubility; logo/w: Predicted octanol/water partition coefficient; AccptH: Estimated number of hydrogen bonds that would be accepted by the solute from water molecules in an aqueous solution; DonorH: Estimated number of hydrogen bonds that would be donated by the solute to water molecules in an aqueous solution; QPPCaco: Predicted apparent Caco-2 cell permeability in nm/sec (Caco2 cells are a model for the gut-blood barrier); QPlogBB: Predicted brain/blood partition coefficient; % Human Oral Abs: Predicted human oral absorption on 0 to 100% scale; QPlog HERG: Predicted IC50 value for blockage of HERG K + channels; SASA: Solvent Accessible Surface Area.
4. Discussion
Drug discovery has become an important area in the field of COVID 19 spreading worldwide. So, the identification of drug targets is a crucial phase to modulate the expression of any targets. Many scientists are involved in searching for a cure for COVID 19. Most of the researchers are targeting protease complex and NSP10/NSP16 methyltransferase, which play a crucial role in inhibits the infection of the virus of COVID 19 [10,11]. It has been seen that various anti-viral, anti-malarial drugs target so many proteins of SARS CoV-2 with a good binding affinity [12,19]. Our selected compounds are already approved by the regulatory agencies, present in the market, and easy to purchase. It has been observed in this study that protease complex with Cyclocytidine hydrochloride (anti-viral and anti-cancer), Trifluridine (anti-viral), Adonitol (anti-bacterial), Meropenem (antibiotic and anti-bacterial), and Penciclovir (anti-viral) bound with a good docking score ranging from −6.8 to −5.1 (Kcal/mol).
Moreover, the NSP10/NSP16 methyltransferase complex was found to docked with Telbivudine, Oxytetracycline dihydrate (anti-viral), Methylgallate (anti-malarial), 2-deoxyglucose (anti-cancer), and Daphnetin (anti-inflammatory, and antitumor) from the docking score of −7.0 to −5.7 (Kcal/mol). Hydrophobic interactions and hydrogen bonding have been observed between the protein complexes and our selected compounds. These hydrophobic interactions and hydrogen bonding have been found to show a strong bonding between the atoms of compounds and protein molecules [20]. This ligand-protein interaction also shows good DG free binding energy and the strength of bonding between the protein and ligands. All the chosen compound also follows the criteria of Lipinski’s rule of five and show properties to have the physicochemical and pharmacokinetic parameters of drugability [18]. Our ligands probably inhibit protease and NSP10/NSP16 methyltransferase molecules involved in the COVID-19. Moreover, it may be possible that applying inhibitors in combination will exert a synergetic effect against the SARS CoV-2.
5. Conclusion
In this study, it has been shown that selected screened compounds have good docking score, high DG binding free energy as well as strong hydrophobic interactions, and follows the Lipinski rule of five. Thus, these compounds might have the potential to be utilized against the NSP10/NSP16 methyltransferase and main protease complexes. Here, in this study, we can conclude that the obtained compounds from this in silico study may be an excellent agent to combat SARS-CoV-2 infection either alone or in combination. However, it warrants further experimental investigations.
Funding Statement
SK acknowledges DST-SERB, India, for providing financial assistance as a Junior Research Fellowship. HRS is thankful to the UGC [Grant no. F.30-377/2017(BSR)] and DST-SERB [Grant no. EMR/20l7/001758], New Delhi, for providing financial help.
Acknowledgments
The authors acknowledge the Indian Institute of Information technology and Aligarh Muslim University for providing infrastructure facilities.
Disclosure statement
The authors declare no conflict of interest.
References
- 1.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya AE, Baker SC, Baric RS, et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020;395(10225):689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackenzie JS, Smith DW. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol Aust. 2020;41(1):45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020; 579(7798):265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zumla A, Chan JF, Azhar EI, et al. Coronaviruses – drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paules CI, Marston HD, Fauci AS. Coronavirus infections—more than just the common cold. Jama. 2020;323(8):707–708. [DOI] [PubMed] [Google Scholar]
- 8.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design . Febs J. 2014;281(18):4085–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand K, Ziebuhr J, Wadhwani P, et al. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Lin D, Kusov Y, et al. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. J Med Chem. 2020;63(9):4562–4578. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Sun Y, Wu A, et al. Coronavirus NSP10/NSP16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J Virol. 2015;89(16):8416–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YW, Yiu CP, Wong KY. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke M, Chen Y, Wu A, et al. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein NSP10 can suppress the 2'-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012;167(2):322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314(5801):997–1001. [DOI] [PubMed] [Google Scholar]
- 15.Kandeel M, Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020; 251:117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Abel R, Zhu K, et al. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins Struct Funct Bioinf. 2011;79(10):2794–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen WL, Duffy EM. Prediction of drug solubility from structure. Adv Drug Deliv Rev. 2002; 54(3):355–366. [DOI] [PubMed] [Google Scholar]
- 18.Lipinski CA, Lombardo F, Dominy BW, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivr Rev. 1997;23(1-3):3–25. [DOI] [PubMed] [Google Scholar]
- 19.Liu K, Tang M, Liu Q, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pushpendra S, Kushwaha PP, Shashank K. Novel potent inhibitors of Plasmodium vivax dihydrofolate reductase: an in silico anti-malarial drug discovery. IJPER. 2018;52(1):122–134. [Google Scholar]