Abstract
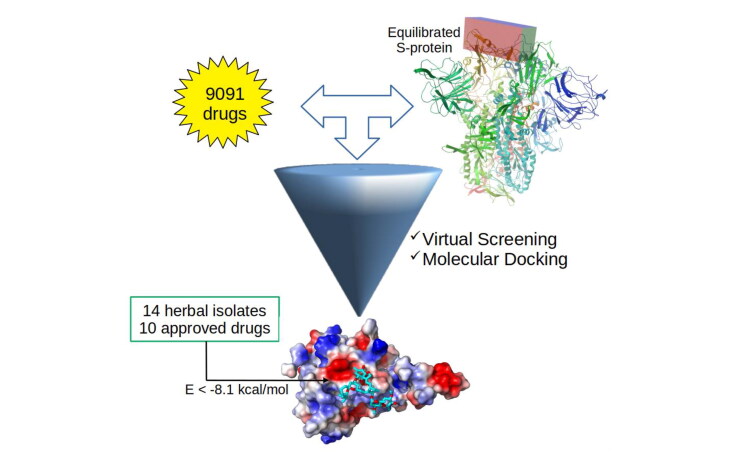
Herein, molecular modeling techniques were used with the main goal to obtain candidates from a drug database as potential targets to be used against SARS-CoV-2. This novel coronavirus, responsible by the COVID-19 outbreak since the end of 2019, became a challenge since there is not vaccine for this disease. The first step in this investigation was to solvate the isolated S-protein in water for molecular dynamics (MD) simulation, being observed a transition from “up” to “down” conformation of receptor-binding domain (RBD) of the S-protein with angle of 54.3 and 43.0 degrees, respectively. The RBD region was more exposed to the solvent and to the possible drugs due to its enhanced surface area. From the equilibrated MD structure, virtual screening by docking calculations were performed using a library contained 9091 FDA approved drugs. Among them, 24 best-scored ligands (14 traditional herbal isolate and 10 approved drugs) with the binding energy below –8.1 kcal/mol were selected as potential candidates to inhibit the SARS-CoV-2 S-protein, preventing the human cell infection and their replication. For instance, the ivermectin drug (present in our list of promise candidates) was recently used successful to control viral replication in vitro. MD simulations were performed for the three best ligands@S-protein complexes and the binding energies were calculated using the MM/PBSA approach. Overall, it is highlighted an important strategy, some key residues, and chemical groups which may be considered on clinical trials for COVID-19 outbreak.
Keywords: COVID-19, SARS-CoV-2 virus, Spike protein, drug design, molecular modeling
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
At the end of December 2019, it was reported an outbreak of many pneumonia cases in Wuhan, China (Li et al., 2020). Hereafter, according to the Chinese Center for Disease Control, this anomalous amount of cases was caused by a novel coronavirus. Initially, this coronavirus was called as 2019-nCoV (2019-novel coronavirus) by World Health Organization (WHO) in January 2020. Now, it has been officially renamed to SARS-CoV-2 (severe acute respiratory syndrome 2) (Guarner, 2020) and the disease caused by this coronavirus is known as COVID-19. Due to the COVID-19 outbreak rapidly spread worldwide, it was declared on 11 March 2020 as a pandemic disease by the WHO. To date at 14 March 2020, there are 216 countries affected by the COVID-19, which 4,307,287 cases and 295,101 deaths were confirmed (https://www.who.int/emergencies/diseases/novel-coronavirus-2019).
The symptoms of COVID-19 may appear between 2 and 14 days after exposure to the SARS-CoV-2 with main signs: cough, runny nose, fever, sore throat, headache and difficulty to breath (Wu et al., 2020). The SARS-CoV-2 is belong the SARS-CoV family as verified by Xu et al. (Xu et al., 2020) from comparison of its sequence with others coronavirus. Therefore, SARS-CoV-2 is composed by glycosylated spike protein (S-protein), membrane protein, envelope protein and nucleocapsid protein (Chen et al., 2006; Boopathi et al., 2020). Likewise, the SARS family, the SARS-CoV-2 infect the cell human using the angiotensin-converting enzyme 2 (ACE2) as receptor, which was confirmed by Ou et al. (Ou et al., 2020; Shang et al., 2020).Recent molecular modeling study developed by Xu et al. (2020) revealed that the S-protein from SARS-CoV-2 presents strong binding affinity with the ACE2 receptor.
The S-protein has the functional S1 and S2 subunits and they are responsible by the human cell attachment and membrane fusion, respectively (Boopathi et al., 2020). In the S1 subunit is located the receptor-binding domain (RBD) which binds to human ACE2 and mediates the viral fusion and cellular membranes (Song et al., 2018). However, the RBD from SARS-CoV-2 has highest affinity with the ACE2 than SARS-CoV as confirmed and evidenced by experimental (Wrapp et al., 2020; Wan et al., 2020), and theoretical data (Peng et al., 2020). In these studies, the authors suggest that the large spread from human to human can be attributed the higher affinity between the SARS-CoV-2 and human ACE2. Moreover, the optimized mutation in the RBD residues has evidenced high-affinity binding with ACE2 and suggest that the SARS-CoV-2 surged from a natural selection process instead of a human manipulation (Andersen et al., 2020).
Thus, S-protein emerges an interesting target to drug design for the SARS-CoV-2 inhibition, which blocking the RBD by a drug can prevent coupling between the S-protein and ACE2. Consequently, the initial step of fusion of the SARS-CoV-2 and human cell may be avoided inducing the virus death or at least no replication.
Although the S-protein is very important in the initial stage of infection, there is not experimental work reporting their use on drug design. On the other hand, there are various theoretical studies in the literature using the S-protein as target to provide its inhibition. For instance, Zhang et al. (Zhang et al., 2020) carried out docking calculations using Chinese medical herbs to S-protein inhibition, and they found 13 compounds with potential SARS-CoV-2 activity. In another study, the Smith et al. (Smith & Smith, 2020) used molecular dynamics simulation to generate an ensemble docking of the S-protein@ACE2 complex. In their docking calculations, it was found 19 potent candidates to bind at the S-protein:ACE2 interface and 30 top-scoring compounds were predicted to bind in the isolated S-protein. In same way, the Sandeep & McGregor (Sandeep & McGregor, 2020) used the S-protein:ACE2 interface to predict the binding of the hydroxychloroquine and azithromycin using docking calculations. Then Utomo et al. (Utomo et al., 2020) used the S-protein RBD domain as target in docking calculations using compounds presented in Curcuma sp., Citrus sp., Alpinia galanga, and Caesalpinia sappan. Likewise, herein we used virtual screening and molecular modeling to study the SARS-CoV-2 S-protein with the main insight to obtain potent candidates to inhibit the SARS-CoV-2.
Our approach adopted here differs from those studies in the following way: the isolated S-protein receptor for docking calculations will be obtained from molecular dynamics (MD) simulation, and not directly from crystal structure or S-protein@ACE2 complex. Where, in our understanding, the coupling of full structural flexibility accounted from MD technique and flexible docking calculations provide an efficient strategy to be used in virtual screening. In addition, the investigation of approved drugs is a straightforward way to repurpose quickly a good candidate against the virus. It is interesting to point that, up to date, there is not drug or vaccine against the COVID-19 and the social isolation is one way to control this outbreak as recommended by WHO.
2. Methodology
2.1. Molecular dynamics simulation
The initial structure of the SARS-CoV-2 viral S-protein was obtained by deleting the ACE2 enzyme from the S-protein@ACE2 complex elaborated by Shah et al. (Shah et al., 2020). The isolated S-protein was merged into a box with edge of 16.1 × 16.7 × 18.5 nm with 147,803 water molecules included. Six chloride ions were added to keep the system neutralized. The GROMOS54a7 force field (Schmid et al., 2011) was used to describe the structural and energetic parameters for the S-protein, and for water the SPC model (Berendsen et al., 1981) was considered. Periodic boundary conditions were considered within 1.0 nm cut-off for non-bonded interactions. The NpT ensemble was used to keep the constant pressure at 1 bar using the Berendsen barostat (Berendsen et al., 1984) (with coupling time of 5 ps) and the temperature at 310 K using the stochastic velocity rescaling method (Bussi et al., 2007) (with coupling time of 0.1 ps). Long-range electrostatic forces were taken into account using the smooth Particle-Mesh Ewald (PME) method (Berendsen, 2007, Deserno & Holm, 1998) with a real space interactions truncated at 1.0 nm cut-off. The system was minimized by the steepest descent method (Morse & Feshbach, 1953) to avoid unfavorable contacts between the atoms, and the convergence was archived at potential energy below 500 kJ/mol·nm. The water molecules were relaxed for 300 ps of MD simulation keeping S-protein rigid by means of a position restraint potential with a force constant of 1000 kJ/mol·nm. Afterwards, the system was equilibrated for 18 ns. All MD simulation steps were performed with the GROMACS package version 2019.2 (Berendsen et al., 1995; Hess et al., 2008). The total energy, temperature, pressure and the root mean square deviation (RMSD) of the S-protein were used to monitor the system equilibration. The equations of motions were integrated using the leap-frog algorithm (Berendsen, 2007) with an integration step of 2 fs. The covalent bonds to the hydrogen atoms were fixed using the P-LINCS method (Hess et al., 1997; Hess et al., 2008).
2.2. Docking calculations
MD equilibrated structure of the SARS-CoV-2 viral S-protein was used as input for docking calculations. The AutoDockTools software (ADT) (Morris et al., 2009) was employed to build the protein-ligand complexes. The AutoDock Vina method (Trott & Olson, 2010) was used in virtual screening calculations for docking the ligands into the S-protein RDB domain. The searching for ligands by docking calculations was done in a grid size of 1.8 × 4.8 × 2.2 nm, which was centered at 9.0226 × 9.3000 × 2.9897 nm in the RDB region. The S-protein and ligands structures were considered rigid and flexible, respectively, along the docking calculations. The ligands were retrieved from the SWEETLEAD library (Novick et al., 2013) and this was chosen because it was elaborated using approved drugs in USA, India, China, Brazil, WHO Essential Medicines List and others.
Moreover, the use of approved drug in virtual screening calculations is an efficient way to decrease the drug discovery costs and the time spend in research. Recently, Smith et al. (Smith & Smith, 2020) has been used this library to identify small molecules to inhibit the SARS-CoV-2 using a different approach than adopted here. Smith et al. (Smith & Smith, 2020) converted the ligands from SWEETLEAD library into the format (PDBQT) which is accepted in the Vina software. In this conversion, the authors have considered the same isomers of the respective ligands. Thus, here 9091 of these ligands and some their respectively isomers with PDBQT format were downloaded from the supplementary material available by Smith et al. (Smith & Smith, 2020) and used in docking calculations. For each ligand, 20 binding modes were generated. The visualization software PyMol (DeLano, 2009) and Discovery Studio Visualizer (Discovery Studio Visualizer Software, 2020) were used to prepare the figures and to analyze the results.
2.3. Binding free energy
MD simulations were carried out for the three best-scored ligands complexed to S-protein previously obtained from docking calculations. The ATB version 3.0 (http://compbio.chemistry.uq.edu.au/atb/) (Stroet et al., 2018) was used to generate the GROMOS force field for all ligands. Herein, we used the same protocol adopted in the Molecular Dynamics simulation section. Afterwards, the binding free energy (ΔGbind.) was calculated using the MM/PBSA method (Kollman et al., 2000) for each complex along 18 ns of equilibrated MD simulation. For each system, 60 snapshots were extracted at interval of 300 ps along the trajectory.
The ΔGbind is calculated with the following equation,
| (1) |
where Gcomplex is the Gibbs free-energy of the ligand@S-protein complex, while GS-protein and Gligand are the total G of S-protein and the ligands in the presence of the solvent, respectively. The individual G for complex, S-protein, and ligands was calculated with equation:
| (2) |
where 〈EMM〉 is the average of the potential energy obtained from the MD simulation in vacuum. The 〈Gsolv.〉 is the Gibbs free energy of solvation and its was calculated with the equation,
| (3) |
where Gpolar is the electrostatic contributions and it is calculated by solving the Poisson-Boltzmann equation (Honig & Nicholls, 1995; Srinivasan et al., 1998; Baker et al., 2001). The Gnonpolar is calculated from the solvent-acessible surface area (SASA). The APBS program (Baker et al., 2001) was used to calculate the energy components (Gpolar and Gnonpolar). The entropy contributions were not taken in account in the equation 3 due its high computational cost. The g_mmpbsa subroutine implemented in the GROMACS package by Kumari (Kumari et al., 2014) was used in the calculations.
3. Results and discussion
To our best knowledge, this work is shedding light on the potential candidates to inhibit the action of the SARS-CoV-2 S-protein on binding to ACE2 and then starting the replication in the human cell.
3.1. MD Simulation
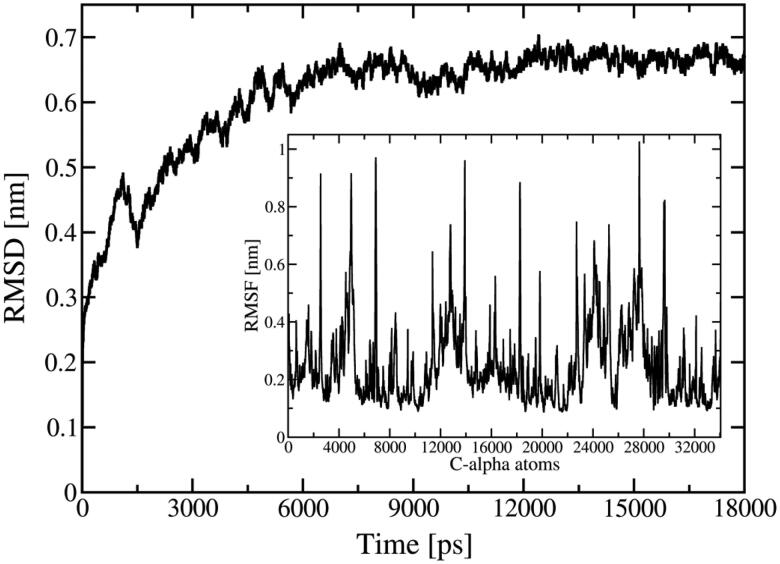
In this section, relevant results obtained from the MD trajectory are described. As shown in Figure S1, the potential energy, temperature and pressure were equilibrated. Figure 1 presents the RMSD of the C-alpha atoms of the S-Protein along the MD trajectory, showing that 10 ns was enough to reach a stable and well behaved structure.
Figure 1.
RMSD of the C-alpha atoms in the isolated S-protein as a function of the simulation time. Inset shows the RMSF of the C-alpha atoms.
RMSD values were calculated by the superposition of the S-protein structure along the trajectory with their initial structure (at 0 ps), increasing rapidly until 6 ns with a fluctuation around 0.65 nm and standard deviation of 0.015 nm at end of the simulation. Considering the large size of the protein, the RMSD value of 0.65 nm implies that the S-protein does not suffer significant structural change along the simulation. The inset in Figure 1 shows the RMSF (root mean square fluctuation) values, which indicate lowest fluctuation of the amino acids of the S-protein. The highest RMSF values are attributed the N- and C-terminals, and loop regions. For best comparison, Figure 2 shows the molecular representation of the initial and final snapshots obtained in the MD simulation.
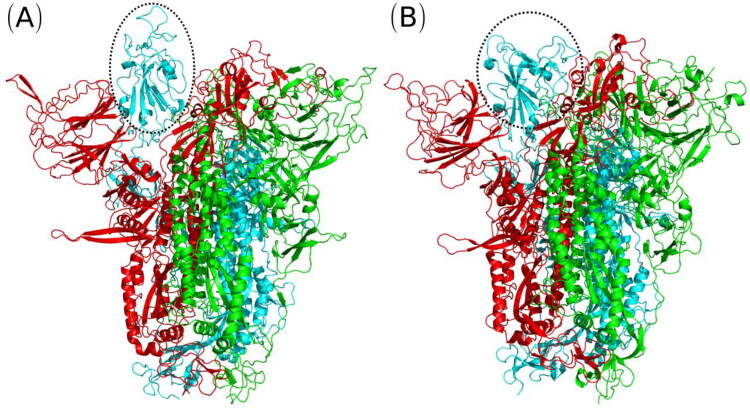
Figure 2.
Initial ((A), at 0 ps) and final ((B), at 18 ns) structures obtained from the molecular dynamics simulations. Chain A is in green, chain B is in red and chain C is in cyan colors. The RBD region is highlight by the dashed line.
Figure 2 reveals that the initial overall S-protein structure remains quite similar after 18 ns of MD simulation, in agreement with the low RMSD values (Figure 1). In general, the main difference observed is located in the loop regions, which is expected due to its high flexibility compared to helix motif.
From MD trajectory was observed an important change in RBD region (Figure 2, highlighted), which is dislocated along the simulation from the “up” to “down” conformation into direction of the S-protein center. Therefore, it indicates that when the RBD is in “down” configuration and it approximates to ACE2, a change is needed to the “up” conformation for an efficient interaction with the ACE2 receptor. In this transition, the angle formed by the C-alpha atoms of the D405, V622 and V991 residues decreases from 54.3 (“up” at 0 ns) to 43.0 degrees (“down” at 18 ns). These data are in agreement with the Peng et al. (Peng et al., 2020), which showed that the “up” angle between 84.8 and 52.2 degrees is accessible by ACE receptor and the “down” angle that ranges from 31.6 to 52.2 degrees is not accessible by the ACE2 human receptor. The RMSD value of the RBD region (C336-C525) is 0.33 nm (standard deviation of 0.02 nm) after 6 ns of simulation. In this region, the RMSF values are lowest than 0.32 nm, except for G482, V483, E484, G485 and F486, which remain the RMSF of 0.42, 0.64, 0.60 and 0.50 nm, respectively.
It is important to emphasize again that the initial S-protein model was obtained from the S-protein@ACE2 complex (Shah et al., 2020). Therefore, the results show that there is a significant structural difference between the RBD in isolated S-protein and that one when complexed with the human ACE2 receptor. It implies that RBD domain is very sensitive to the chemical environment, temperature and solvent effects. Thus, in drug design these differences must be considered in the search of compounds to inhibit the S-protein. Considering this, in the next section we used the final structure generated in the MD simulation to find good candidates for the SARS-CoV-2 S-protein inhibition using docking calculations.
3.2. Virtual screening
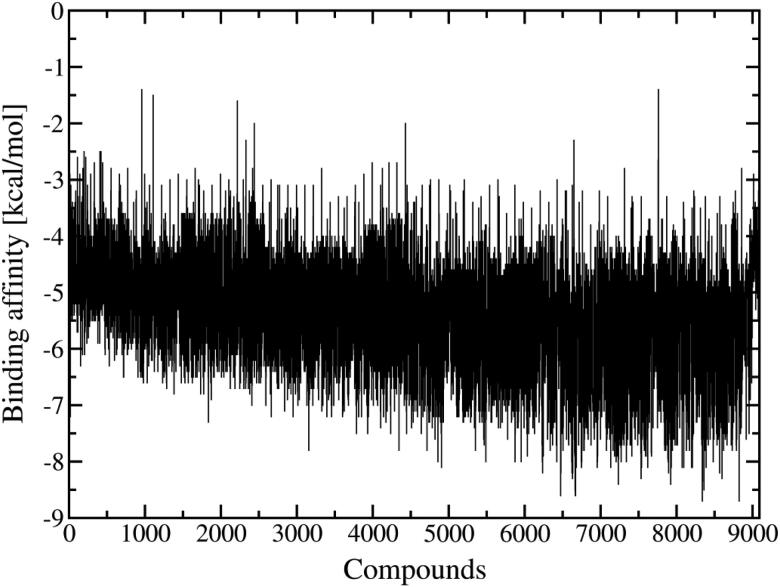
AutoDock Vina was used for docking calculations of 9091 approved drugs to obtain potential candidates for inhibition of the SARS-CoV-2 S-protein. Differently from other works (Zhang et al., 2020; Smith & Smith, 2020; Sandeep & McGregor, 2020; Utomo et al., 2020), the isolated S-protein receptor was previously equilibrated by MD simulation, providing a realistic structure when the virus infects humans. Then, the solvent and temperature effects are taking into account in the S-protein conformation. Figures 3 presents the binding affinity or energy for all ligands obtained in Docking calculations.
Figure 3.
Binding affinity between ligands and S-protein from Docking calculations.
Overall, all ligands docked in the RDB region has favorable energy. It is interesting to notice that the Smith et al. (Smith & Smith, 2020) has been used the present ligands library in docking calculation for S-protein. Therein the isolated protein receptor was retrieved from the S-protein@ACE2 complex previously obtained by their MD simulation. However, our MD results showed that the RBD region suffer significantly structural change from the S-protein@ACE2 complex to the isolated S-protein. For instance, the Smith et al. (Smith & Smith, 2020) identify only 30 compounds with binding affinity between –6.3 and –7.2 kcal/mol. However, as shown in Figure 3 it is possible to identify thousand compounds in this interval of energy, which indicates that our approach is more efficient to explore the S-protein RBD region than the strategy adopted by Smith et al. (Smith & Smith, 2020). There are 536 ranking docking poses of ligands (including isomers) with binding energy between –7.2 and –8.0 kcal/mol. However, the ligands with the lowest binding affinity is an important criterion in drug design. In this way, we selected 33 ligands with binding affinity bellow -8.1 kcal/mol, and among them, the isomers with highest energy were removed. In this process, 24 ligands were selected and analyzed here. In the Table 1, we present the results obtained from the docking calculations.
Table 1.
Ligands with lowest binding affinity (kcal/mol) obtained from docking calculations and the main interacting residues with the S-protein. T.H.I means Traditional Herbal Isolate.
| Compounds | Affinity | Residues |
|---|---|---|
| T.H.I: Theaflavin digallate (Lig8522) | –8.7 | R403, Y421, L455, F456, G476, Q493, G496, Y505 |
| WHO Essential Medicine: suramin sodium (Lig8970) | –8.7 | R403, R405, I418, Y449, L455, F456, Q493, G496, Q498, G504, Y505 |
| Indian Approved Name: 5-hydroxytrytophan (Lig6843) | –8.6 | R403, R405, R408, G496, Y505 |
| T.H.I: solamargine, beta-solamargine (Lig8541) | –8.5 | R403, Q493, Q498, Y505 |
| T.H.I: taraxanthin (Lig7527) | –8.4 | R403, K417, I418, L455, F456 |
| T.H.I: anthranil acid (Lig8148) | –8.4 | R403, I418, L455, Y505 |
| T.H.I: evomonoside (Lig6982) | –8.3 | R403, G496, S477 |
| NPC Approved Name: dihydroergocristine mesylate (Ergoloid) (Lig7798) | –8.3 | R403, R405, L455, Y505 |
| T.H.I: smilacin (Lig8757) | –8.3 | R403, R405, Y449, F490, G496, A475 |
| T.H.I: withaphysalin (Lig6629) | –8.2 | E406, Y453 |
| T.H.I: erysimosol (Lig8121) | –8.2 | R403, Y449, Y489, Q493, G496, Y505 |
| FDA Approved Drug: quinupristin (Lig8726) | –8.2 | R405, E406, I418, Y449, Q493, S494, Y505 |
| T.H.I: tigogenin; sarsasapogenin (Lig5429) | –8.1 | R405, Q498, Y505 |
| Australia Approved Name: Nilotinib; FDA Approved Drug: nilotinib hydrochloride monohydrate (Lig7028) | –8.1 | R403, G447, L455, Y449, F490, G496, F497, Q498, Y505 |
| T.H.I: deacetylo-leandrin (Lig7089) | –8.1 | R403, I418, Y505 |
| NPC Approved Name: dexamethasone-21-sulfobenzoate (Lig7435) | –8.1 | R403, Y449, Y453, S477, Q493, G496, |
| T.H.I: dauricinoline (Lig7801) | –8.1 | Y421, L355, F456, A475, S477, Y489, F490, L492 |
| NPC Approved Name: tirilazad (Lig7875) | –8.1 | L455, S477 |
| T.H.I: swertifrancheside (Lig8152) | –8.1 | R403, R405, Q409, Y449, Y505 |
| T.H.I: digitoxin; FDA Approved Drug: Digitoxin (Lig8283) | –8.1 | R403, Y449, L455, Q498 |
| NPC Approved Name: selamectin (Lig8301) | –8.1 | R403, L455, Y449, Q493, G496 |
| FDA Approved Drug: acetyldigitoxin (Lig8395) | –8.1 | R403, L455, L492, G496, Y505 |
| T.H.I: gitaloxin (Lig8402) | –8.1 | R403, Y453, F456, L455, Y489 |
| NPC Approved Name: doramectin (Lig8568) | –8.1 | R403, R405, L455, F456, A475 |
Recently, the azithromycin, hydroxychloroquine and chloroquine have been tested as possible treatment of the COVID-19 outbreak with some potential results (Juurlink, 2020; Gautret et al., 2020; Touret & de Lamballerie, 2020), but with undesirable side effects. However, our results shows high binding energy of the –6.8, −5.1 and –4.7 kcal/mol, respectively. Therefore, these values indicate that these drugs may act in the S-protein RDB region, but with lowest efficiency than our best-scored ligands in the same concentration. In Table 1, we can see that 14 compounds are derived from the traditional herbal isolate. The advantages of this type of therapeutics is due the cheap cost, little side effects, good availability and local cultural aspects. On the other hands, 10 approved drugs were selected from the docking calculations. Thus, the laboratories specialized can buy these drugs and test those to verify their efficiency against COVID-9 outbreak. The most scored-ligands presented in Table 1 are rich in hydroxyl group and eight (Lig6982, Lig6982, Lig7089, Lig8121, Lig8283, Lig8402, Lig8541 and Lig8757) has the presence at least one monosaccharides group attached in their structures.
This finding is very interesting because the S-protein is a glycoprotein, which has high affinity with oligosaccharides and various sugar chains are covalently linked. Therefore, we suggest that a drug against SARS-CoV-9 S-protein may have the presence of the oligosaccharide groups, which is important in recognition of S-protein. Analyzing the interacting residues with the S-protein (Table 1), we found that the R403, R405, Y449, L455, G496 and Y505 residues are common in the most ligands. Therefore, theses residues play an important role in the ligand-S-protein interactions. Thereby, we suggest that in drug design these residues can be used do draw a candidate to S-protein inhibition.
Although the lopinavir and emetine drugs does not appear in the Table 1, the binding energy obtained in our docking calculations were –7.0 and –6.5 kcal/mol, respectively, see supporting information. These results are interesting because recently the Choy et al. (Choy et al., 2020) showed that these drugs inhibit the SARS-CoV-2 replication with EC50 under 100 μM. On the other hands, Muralidharan et al. (Muralidharan et al., 2020) obtained from docking calculations a binding energy of –4.1 kcal/mol using the SARS-CoV-2 protease and lopinavir drug, respectively, as receptor and ligand. Thus, the lopinavir may act preferentially in the S-protein than protease, due the lowest binding energy (–7.0 kcal/mol), predicted using the S-protein as receptor.
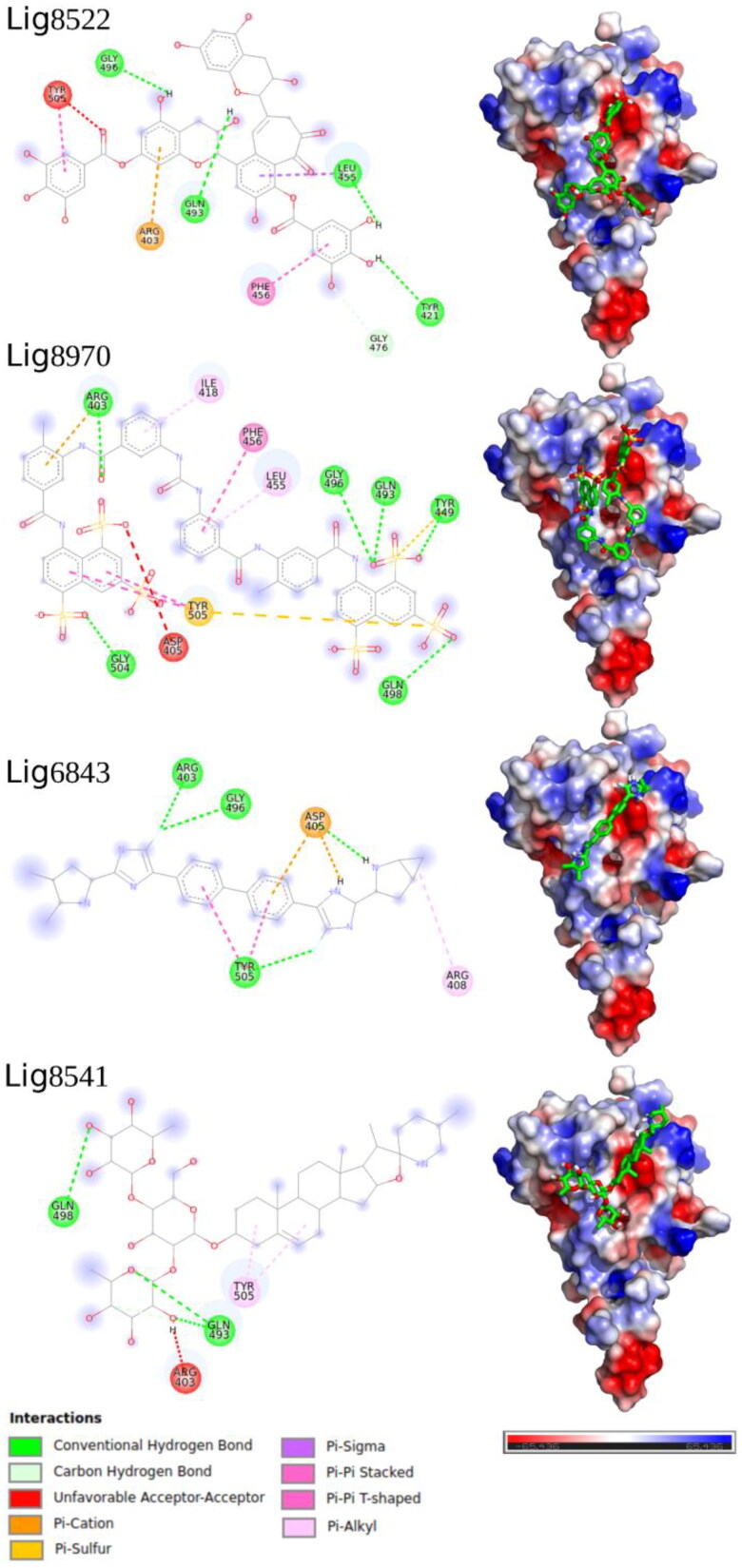
For best visualization of the interacting residues of the S-protein and the best-scored ligand, in the Figure 4 is presented the 2 D ligand interaction diagram and molecular electrostatic map (MEP) for the four ligand with lowest binding affinity. In the supporting information (Figure S2), we present the 2 D diagram for all compounds in the Table 1.
Figure 4.
2 D ligand interaction diagram and molecular electrostatic map (MEP) of four best docking poses.
Overall, in the 2 D diagram, the π-π stacking, π-cation and hydrogen bond are the most interactions observed in the ligands@S-protein RBD complexes. For best visualization, in Figure 2, the MEP was showed only for the RDB region and its position is in front compared with RDB highlighted in the Figure 1. In the MEP, it can see that all ligands interact with a negative (intense red color) of the RDB region. Contrary, the ligands bind with RBD in a region with partial positive (light blue color) charge.
Along the manuscript written, the ivermectin drug has been tested as anti-SARS-CoV-2 (Caly et al., 2020) and their results showed a reduction of ∼500 times the cell culture of the virus in 48 h. The authors also highlighted the anti-viral activity of the ivermectin against a broad range of viruses in vitro (Tay et al., 2013; Wagstaff et al., 2012). Thus, our docking calculation using this drug gives a binding energy of –8.1 kcal/mol. This finding is very promising since the second best-scored ligand also an anti-parasitic agent like the Lig8301 (selamectin) and Lig8568 (doramectin) drugs, Table 1. Moreover, the main interacting residues with the S-protein are: R403, I418, Y489 and F490. In this way, our calculations indicate that the ivermectin drug may bind in the RBD region, inhibiting the coupling of the SARS-CoV-2 S-protein with the human ACE2 receptor. Recently, Fan et al. (Fan et al., 2020) showed that the selamectin (lig8301) inhibits completely the spike protein of coronavirus GX_P2V, which shares 92.2% amino acid identity with the 2019-nCoV.
3.3. Binding free energy analysis
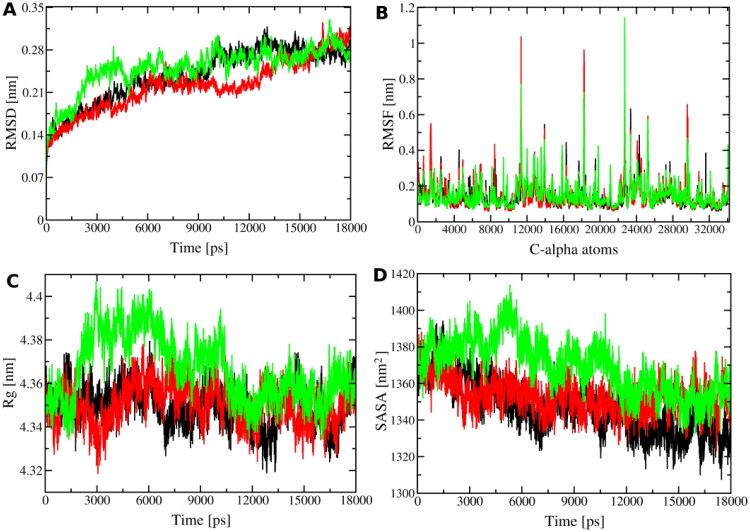
Although docking calculations are useful to predict the binding of a ligand into a protein, they are limited due to the lack of temperature and solvent effects, which have an important role in the stability and conformational flexibility of a system. Therefore, we carried out 18 ns of MD simulation for the three best-scored ligands complexed with the S-protein. These simulations were carried out using the Ohio Supercomputer Center (OSC, 1987). Figure 5 depicts some relevant properties obtained along the MD trajectories.
Figure 5.
RMSD (A), RMSF (B) and radius of gyration (Rg, C) of the C-alpha atoms, and solvent accessible surface area (SASA, D) of the S-Protein complexed with Lig8522 (black line), Lig8970 (red line) and Lig6843 (green line) ligands.
Overall, Figure 5 highlights that the S-protein is equilibrated at the last 5 ns of MD simulation for all system, as shown by RMSD (Figure 5(A)) of the S-protein, in which the values were ∼0.28 nm with a standard deviation of 0.02 nm for the three complexes. This imply that the presence of the ligands does not change significantly the S-protein structure along the MD simulation. Likewise, the RMSF per residues using their C-alpha atoms (Figure 5(B)) was calculated to determine and to identify the flexibility of the S-protein regions. Comparing the RMSF of the ligands@S-protein complexes with the apo S-protein (Figure 1), it is possible to see that the residues have lowest flexibility in the presence of the ligands.
However, it is important to empathize that we used an equilibrated S-protein as an initial configuration in the MD simulation of the complexes. In addition, all ligands are positioned in the S-protein RDB domain, which is composed by the atoms CA26100 (C336)–CA28081 (C525 residue). In this region is observed low residue fluctuations, keeping the RMSF values below ∼0.3 nm (Figure 5(B)). The compactness of the S-protein was achieved by the radius of gyration (Figure 5(C)). The S-Protein complexed with Lig8522 and Lig8970 ligands present a similar compactness along the entire MD simulation. For Lig6843, the Rg of the protein remains low fluctuation at the last 7 ns of the trajectory. Overall, the S-protein in all systems (apo e holo forms) is compact with Rg value of ∼4.35 nm. In general, the RMSD, RMSF and Rg analyzes show that the S-protein has low conformation change at the last 6 ns of the MD trajectories. Thus, as expected, the S-protein present a similar solvent accessible surface area (Figure 5(C)), retaining its accessibility in the last 6 ns of the MD simulations.
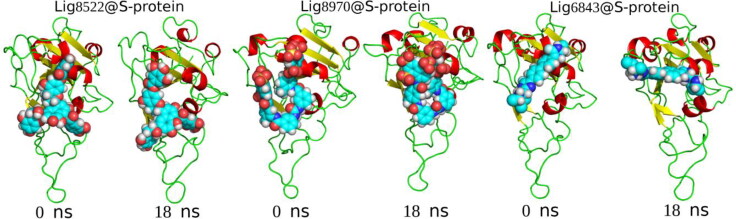
In Figure 6, we present the representation of the initial and final structures obtained in simulations.
Figure 6.
Snapshots obtained in the initial (0 ns) and final (18 ns) of MD simulations. Balls represent the ligands. For best visualization, only the S-protein RBD is shown.
As one can see in Figure 6, the ligands are attached on the S-protein RBD along the MD trajectory, in which only the Lig6843 differs from its initial configuration, but it remains in the RBD region.
Table 2 shows the binding energetic components obtained in the MM/PBSA calculations.
Table 2.
Binding energy components (kcal/mol) from MM/PBSA for the three complexes. The error for each one is in parenthesis.
| Energies | Lig8522 | Lig8970 | Lig6843 |
|---|---|---|---|
| van der Waal | –50.47 (± 0.81) | –51.07 (± 0.93) | –32.41 (± 0.42) |
| Electrostatic | –20.27 (± 0.97) | –145.80 (± 2.21) | 4.81 (± 0.12) |
| Polar solvation | 38.25 (± 1.37) | 163.24 (± 3.90) | 33.60 (± 1.45) |
| SASA | –6.01 (± 0.11) | –6.82 (± 0.08) | –3.35 (± 0.09) |
| Binding energy | –38.51 (± 1.59) | –40.43 (± 1.92) | 2.67 (± 1.18) |
Although the binding energy obtained from MM/PBSA method differs from the docking calculations, they follow the same energetic order. Lig8522 and Lig8970 present the same binding energy with similar van der Waals and apolar solvation (SASA) energies. However, the electrostatic and polar solvation energies are very different due to the large negatively charge (–6e) of the Lig8970 salt. The positive value of the binding energy for the Lig6843 was an unexpected result. However, this result is a consequence of the Lig6843 reorientation along the MD simulation, see Figure 6. In this process, the initial position of the Lig6843 (charge +2e) is favorable for interactions with two negative regions of the RBD (Figure 4). However, in the MD trajectory it is dislocated for an unfavorable positive region. Moreover, the large error in the binding energy of this ligand indicate that is necessary more time to be simulated in the MD simulation. Therefore, the Lig6841 remain binding in the RBD region by the van der Waals interactions along the 18 ns of the MD simulation. Nowadays, we are performing long MD simulations for the best-scored ligands presented in the Table 1. Forthcomingly, when in vitro activity were disposable in the literature, we will do receiver operating characteristic (ROC) curve analysis from machine learning to identify possible true positive ligands.
4. Conclusions
Currently, we are surviving at a global health crisis caused by the SARS-CoV-2 virus, which is responsible by the COVID-19 outbreak since the end of the 2019. To date, there is not a vaccine against COVID-19 and the social isolation is the main strategy adopted to avoid the spread of this novel coronavirus. Therefore, herein we used molecular dynamics (MD) simulation and docking calculations to study the SARS-CoV-2 S-protein with the main goal to obtain possible drugs candidates for repurposing them against to COVID-19. Our approach was study the SARS-CoV-2 S-protein before it bounded with the human angiotensin-converting enzyme 2 receptor preventing the human cell infection. Overall, the S-protein does not undergo significantly structural change along the 18 ns MD simulation equilibration in water solvent. The main change occurs in the receptor-binding domain (RBD) which was converted from “up” (angle of 54.3 degrees) to “down” (angle of 43.0 degrees) conformation. In this transition, the RBD increased surface area enhanced the possibility of interactions with the solvent and also with the possible drugs. In this way, we carried out virtual screening by docking calculations using the S-protein at 18 ns of the MD simulation as receptor and 9091 approved drugs as ligands. In these calculations, 14 traditional herbal isolate and 10 approved drugs were obtained as a potent candidates to inhibit the SARS-CoV-2 S-protein with binding affinity bellow –8.1 kcal/mol. In addition, our calculations were successful to predict the binding of the ivermectin in ACE2, which this drug was used recently with high efficiency to control viral replication in vitro. Overall, the R403, R405, Y449, L455, G496 and Y505 residues are the most interacting with ligands. MD simulations of the three ligands@complexes show that the ligands remain bounded in the RBD region along the simulation. Where the binding energies obtained from MM/MBSA calculations were –38.51 (± 1.59), −40.43 (± 1.92) and 2.67 (± 1.18) kcal/mol for Lig8522, Lig8970 and Lig6843, respectively. Finally, herein we highlight some important issues to be taken into account for repurposing known drugs for the COVID-19 outbreak.
Supplementary Material
Funding Statement
This work was financial supported by FAPESP (São Paulo Research Foundation) under process number of 2018/19844-8. LTC is thankful for CNPq award fellowship and FAPERJ for the financial support. Also, this study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) through the research project Bioinformática Estrutural de Proteínas: Modelos, Algoritmos e Aplicações Biotecnológicas (Edital Biologia Computacional 51/2013, No. AUXPE1375/2014 da CAPES). G.B.R. acknowledges support from the Brazilian National Council for Scientific and Technological Development (CNPq grant no. 309761/2017-4).
Acknowledgements
The authors also acknowledge the computing support from the Ohio Supercomputing Center for the binding free energy analysis calculations. We are grateful for the researcher Charly Empereur-mot for his valuable discussions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., & Garry R. F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. A., Sept D., Joseph S., Holst M. J., & McCammon J. A. (2001). Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America, 98(18), 10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., & Hermans J. (1981). In Intermolecular Forces (Pullman B., ed., p. 331). D. Reidel. [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., & Haak J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Berendsen H. J. C., van der Spoel D., & van Drunen R. (1995). GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications, 91(1–3), 43–56. 10.1016/0010-4655(95)00042-E [DOI] [Google Scholar]
- Berendsen H. J. C. (2007). Simulating the physical world: hierarchial modeling from quantum mechanics to fluid dynamics. Cambridge University Press. [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G., Donadio D., & Parrinello M. (2007). Canonical sampling through velocity rescaling. The Journal of Chemical Physics, 126(1), 014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- Caly L., Druce J. D., Catton M. G., Jans D. A., & Wagstaff K. M. (2020). The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178, 104787 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Luo H., Chen L., Chen J., Shen J., Zhu W., Chen K., Shen X., & Jiang H. (2006). An overall picture of SARS coronavirus (SARS-CoV) genome-encoded major proteins: Structures, functions and drug development . Curr. Pharm. Des, 12(35), 4539–4553. 10.2174/138161206779010459 [DOI] [PubMed] [Google Scholar]
- Choy K.-T., Wong A. Y.-L., Kaewpreedee P., Sia S. F., Chen D., Hui K. P. Y., Chu D. K. W., Chan M. C. W., Cheung P. P.-H., Huang X., Peiris M., & Yen H.-L. (2020). Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Research, 178, 104786 10.1016/j.antiviral.2020.104786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deserno M., & Holm C. (1998). How to mesh up Ewald sums. I. A theoretical and numerical comparison of various particle mesh routines. The Journal of Chemical Physics, 109(18), 7678–7693. 10.1063/1.477414 [DOI] [Google Scholar]
- Discovery Studio Visualizer Software. (2020). Version 4.0., http://www.accelrys.com
- Fan H.-H., Wang L.-Q., Liu W.-L., An X.-P., Liu Z.-D., He X.-Q., Song L.-H., & Tong Y.-G. (2020). Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus-related coronavirus model. Chinese Medical Journal, 133, 1051–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Hoang V. T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V. E., Dupont H. T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., & Raoult D. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open label non-randomized clinical trial. International Journal of Antimicrobial Agents, 105949 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guarner J. (2020). Three emerging coronaviruses in two decades . Am. J. Clin. Pathol, 153(4), 420–421. 10.1093/ajcp/aqaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H. J. C., & Fraaije J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [DOI] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., & Lindahl E. (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4(3), 435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Hess B. (2008). P-LINCS: A parallel linear constraint solver for molecular simulation. Journal of Chemical Theory and Computation, 4(1), 116–122. 10.1021/ct700200b [DOI] [PubMed] [Google Scholar]
- Honig B., & Nicholls A. (1995). Classical electrostatics in biology and chemistry. Science (New York, N.Y.), 268(5214), 1144–1149. 10.1126/science.7761829 [DOI] [PubMed] [Google Scholar]
- Juurlink D. N. (2020). Safety considerations with chloroquine, hydroxychloroquine and azithromycin in the management of SARS-CoV-2 infection. Cmaj : Canadian Medical Association Journal = Journal de L'association Medicale Canadienne, 192(17), E450–E453. 10.1503/cmaj.200528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman P. A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., Lee M., Lee T., Duan Y., Wang W., Donini O., Cieplak P., Srinivasan J., Case D. A., & Cheatham T. E. 3rd. (2000). Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts of Chemical Research, 33(12), 889–897. 10.1021/ar000033j [DOI] [PubMed] [Google Scholar]
- Kumari R., Kumar R., & Lynn A. (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations . Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Li Q.-H., Ma Y.-H., Wang N., Hu Y., & Liu Z.-Z. (2020). New coronavirus-infected pneumonia Engulfs Wuhan. Asian Toxicology Research, 1, 1–7. [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., & Olson A. J. (2009). Autodock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse P. M., & Feshbach H. (1953). Asymptotic series, method of steepest descent. Methods Theoretical Physics, Part I, 434–443. [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Novick P. A., Ortiz O. F., Poelman J., Abdulhay A. Y., & Pande V. S. (2013). SWEETLEAD: An in silico database of approved drugs, regulated chemicals, and herbal isolates for computer-aided drug discovery. Plos One, 8(11), e79568 10.1371/journal.pone.0079568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohio Supercomputer Center (OSC). (1987). Ohio supercomputer center. Ohio Supercomputer Center; http://osc.edu/ark:/19495/f5s1ph73. [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., & Ren L. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communication, 11, 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Zhu Z., Shi Y., Wang X., Mu K., Yang Y., Zhang X., Xu Z., & Zhu W. (2020). Exploring the binding mechanism and accessible angle of SARS-CoV-2 spike and ACE2 by molecular dynamics simulation and free energy calculation. ChemRxiv. 10.26434/chemrxiv.11877492.v1 [DOI] [Google Scholar]
- Sandeep S., & McGregor K. (2020). Energetics based modeling of hydroxychloroquine and azithromycin binding to the SARS-CoV-2 spike (S)Protein-ACE2 Complex. ChemRxiv. 10.26434/chemrxiv.12015792.v1 [DOI] [Google Scholar]
- Schmid N., Eichenberger A. P., Choutko A., Riniker S., Winger M., Mark A. E., & van Gunsteren W. F. (2011). Definition and testing of the GROMOS force-field versions 54A7 and 54B7. European Biophysics Journal, 40(7), 843–885. 10.1007/s00249-011-0700-9 [DOI] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., & Li F. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581(7807), 221–224. https://www.nature.com/articles/s41586-020-2179-y 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Ahmad B., Choi S., & Woo H. G. (2020). Sequence variation of SARS-CoV-2 spike protein may facilitate stronger interaction with ACE2 promoting high infectivity. Research Square, Preprint. 10.21203/rs.3.rs-16932/v1 [DOI] [Google Scholar]
- Smith M. D., & Smith J. (2020). Repurposing therapeutics for COVID-19: Supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface. ChemRxiv. 10.26434/chemrxiv.11871402.v4 [DOI] [Google Scholar]
- Song W., Gui M., Wang X., & Xiang Y. (2018). Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathogens, 14(8), e1007236 10.1371/journal.ppat.1007236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., Cheatham T. E., Cieplak P., Kollman P. A., & Case D. A. (1998). Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate - DNA helices. Journal of the American Chemical Society, 120(37), 9401–9409. 10.1021/ja981844+ [DOI] [Google Scholar]
- Stroet M., Caron B., Visscher K., Geerke D., Malde A. K., & Mark A. E. (2018). Automated topology builder version 3.0: Prediction of solvation free enthalpies in water and hexane. Journal of Chemical Theory and Computation, 14(11), 5834–5845. 10.1021/acs.jctc.8b00768 [DOI] [PubMed] [Google Scholar]
- Tay M. Y. F., Fraser J. E., Chan W. K. K., Moreland N. J., Rathore A. P., Wang C., Vasudevan S. G., & Jans D. A. (2013). Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Research, 99(3), 301–306. 10.1016/j.antiviral.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Touret F., & de Lamballerie X. (2020). Of chloroquine and COVID-19. Antiviral Research, 177, 104762. 10.1016/j.antiviral.2020.104762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W. L. (2009). The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific.
- Trott O., & Olson A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utomo R. Y., Ikawati M., & Meiyanto E. (2020). Revealing the potency of citrus and galangal constituents to halt SARS-CoV-2 infection. Preprints. 10.20944/preprints202003.0214.v1 [DOI] [Google Scholar]
- Wagstaff K. M., Sivakumaran H., Heaton S. M., Harrich D., & Jans D. A. (2012). Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus . The Biochemical Journal, 443(3), 851–856. 10.1042/BJ20120150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R. S., & Li F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology, 94(7), e00127–20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., & McLellan J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wu T., Liu Q., & Yang Z. (2020). The SARS-CoV-2 outbreak: What we know. International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases, 94, 44–48. https://www.sciencedirect.com/science/article/pii/S1201971220301235 10.1016/j.ijid.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., & Hao P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China. Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.-H., Wu K.-L., Zhang X., Deng S.-Q., & Peng B. (2020). In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. Journal of Integrative Medicine, 18(2), 152–158. 10.1016/j.joim.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.