Abstract
COVID-19, caused by SARS-CoV-2, is a virulent pneumonia, with >4,000,000 confirmed cases worldwide and >290,000 deaths as of May 15, 2020. It is critical that vaccines and therapeutics be developed very rapidly. Mice, the ideal animal for assessing such interventions, are resistant to SARS-CoV-2. Here, we overcome this difficulty by exogenous delivery of human ACE2 with a replication-deficient adenovirus (Ad5-hACE2). Ad5-hACE2-sensitized mice developed pneumonia characterized by weight loss, severe pulmonary pathology, and high-titer virus replication in lungs. Type I interferon, T cells, and, most importantly, signal transducer and activator of transcription 1 (STAT1) are critical for virus clearance and disease resolution in these mice. Ad5-hACE2-transduced mice enabled rapid assessments of a vaccine candidate, of human convalescent plasma, and of two antiviral therapies (poly I:C and remdesivir). In summary, we describe a murine model of broad and immediate utility to investigate COVID-19 pathogenesis and to evaluate new therapies and vaccines.
Keywords: COVID-19, SARS-CoV-2, mouse model, pathogenesis, therapeutics, vaccine
Graphical Abstract
Highlights
-
•
Mice are sensitized for SARS-CoV-2 infection by Ad5-hACE2 transduction
-
•
Genetically deficient strains can be directly assessed without additional breeding
-
•
Mice useful for determining host factors necessary for optimal virus clearance
-
•
Useful for assessing efficacy of vaccines and therapies such as convalescent plasma
An adenoviral transduction-based mouse model that can be infected with SARS-CoV-2 provides a tool to understand host factors involved in viral infection and clearance as well as potential therapeutic modalities.
Introduction
Severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), a betacoronavirus, emerged in China as the etiological agent of coronavirus disease 2019 (COVID-19), a severe pneumonia with systemic manifestations. COVID-19 has been classified as a pandemic by the WHO. COVID-19 has the transmissibility of coronaviruses (CoV) that cause the common cold and the virulence of two previously described zoonotic human highly pathogenic respiratory CoVs, SARS-CoV and MERS (Middle East respiratory syndrome)-CoV. These characteristics help explain the pathogenicity of COVID-19 and highlight the urgent need to develop broadly useful experimental animal models for additional studies. Although SARS-CoV-2, like SARS-CoV, uses ACE2 to enter cells, mouse ACE2 (mACE2) does not sensitize cells for infection (Zhou et al., 2020).
As the COVID-19 pandemic progresses, the need to understand mechanisms of cell and tissue injury, and to apply this knowledge to therapeutics, increases. Animal models of infection play important roles in such discoveries, with mice being the most widely used animal. Mice offer the convenience of small size and wide availability. Previous studies in laboratory mice facilitated our understanding of SARS and MERS. Mice infected with human SARS-CoV developed mild disease, but because SARS-CoV, unlike SARS-CoV-2, could infect mice, it was possible to develop a mouse-adapted virus that caused severe disease. Rodent-adapted SARS-CoV were isolated by several laboratories and used in a wide variety of studies (Nagata et al., 2008, Roberts et al., 2007). MERS-CoV, like SARS-CoV-2 does not naturally infect mice. However, we and others showed that by providing the human receptor (DPP4) by transduction with a replication-deficient adenovirus, or by transgenic or “knocked-in” human DPP4 expression, mice were sensitized for MERS-CoV infection (Cockrell et al., 2016, Li et al., 2016, Li et al., 2017, Pascal et al., 2015, Zhao et al., 2014). Further mouse adaptations resulted in isolation of highly pathogenic viruses that recapitulated the disease seen in human CoV infections (Cockrell et al., 2016, Li et al., 2017).
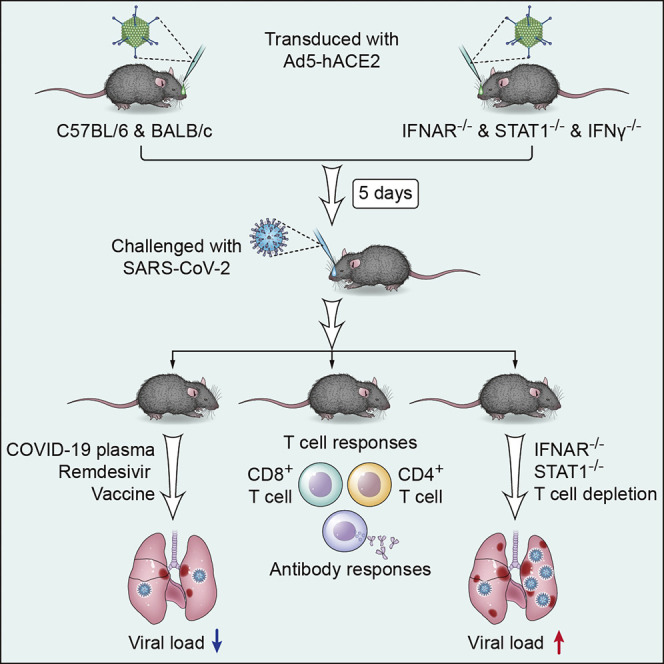
Prior to the isolation of rodent-adapted SARS-CoV, several transgenic mouse lines were developed for studies of SARS (McCray et al., 2007, Yang et al., 2007, Yoshikawa et al., 2009). Because SARS-CoV-2 also binds to the hACE2 receptor, these mice may have immediate applications. Indeed, Bao and colleagues recently re-purposed their hACE-2 transgenic mice for studies of SARS-CoV-2 (Bao et al., 2020). Although these mice are potentially useful, SARS-CoV-2 replication in these mice was suboptimal (less than 103 Log10 TCID50 per 100 μL of mouse lungs), and weight loss and lung pathological changes were minimal (Bao et al., 2020). Most importantly, many studies would also benefit from using genetically modified mice, which would require time-consuming backcrossing to meet the demand. Development of a murine infection system in which all mice were easily and rapidly sensitized to SARS-CoV-2 infection would circumvent this problem and would be very useful for these and other studies. Here, we show that providing hACE2 by adenovirus transduction sensitizes a broad range of immunocompetent and immunodeficient mice for SARS-CoV-2 infection, expediting studies of COVID-19 pathogenesis and the development of multiple interventions.
Results
Development of Mice Sensitized for SARS-CoV-2 Infection
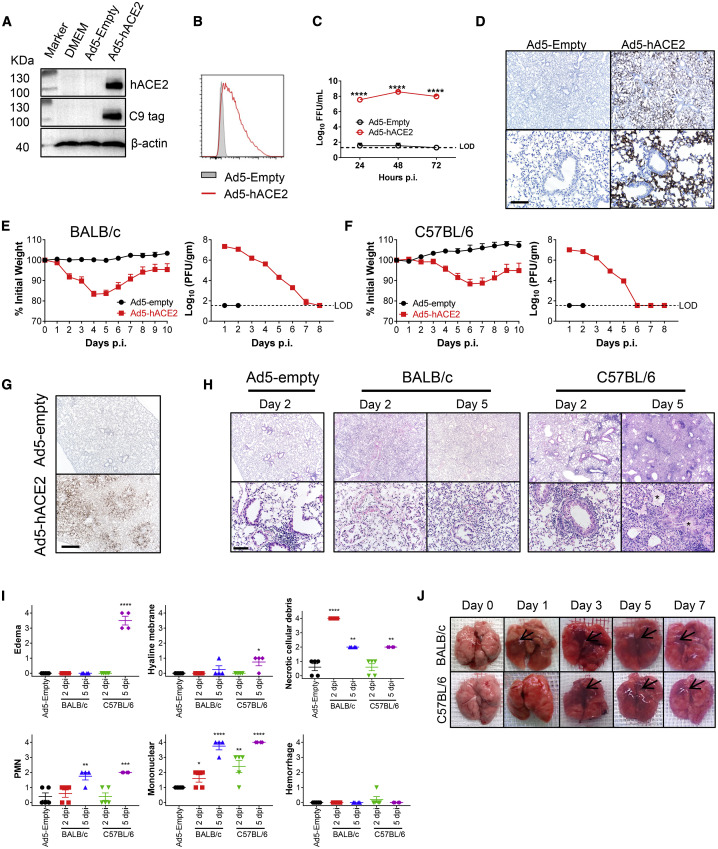
The adenoviral vector expressing hACE2 under the control of the CMV promoter was generated as previously described (Jia et al., 2005, McCray et al., 2007, Zhao et al., 2014). When we transduced mouse 17CL-1 cells with Ad5-hACE2, but not Ad5-empty (an adenoviral vector with no expression cassette) (MOI = 100), hACE2 expression was detected by immunoblot and flow cytometry (Figures 1A and 1B). High titers of SARS-CoV-2 were detected in the supernatants of 17CL-1 cells transduced with Ad5-hACE2, but not Ad5-empty (Figure 1C), after SARS-CoV-2 infection. Ad5 can transduce a large percentage of pulmonary epithelial cells and robustly express the encoded protein of interest (Crystal et al., 1994, Nabel, 2004). Consequently, we modified a previously described approach, to provide hACE2, by transducing mice with Ad5-hACE2 (Zhao et al., 2014b). When we transduced 6-to-8-week-old BALB/c mice intranasally with 2.5 × 108 PFU Ad5-hACE2, we observed hACE2 expression predominantly in the alveolar epithelium with occasional positive cells in the airway epithelium (Figure 1D). Five days later, mice received 1 × 105 PFU of SARS-CoV-2 and were monitored over a 10-day time course. Control mice received the Ad5-empty vector. Ad5-hACE2 transduced BALB/c mice infected with SARS-CoV-2 showed ruffled fur, hunching, and difficulty breathing beginning 2 days post infection (d.p.i.). The mice lost up to ∼20% of their body weight in the first 4–6 days of infection, and virus grew to high titers in lung tissue and gradually declined over the course of the infection (Figure 1E), similar to SARS-CoV infection of BALB/c mice (Roberts et al., 2007). Similarly treated C57BL/6 mice followed an almost identical course, with 10%–15% weight loss and highest virus titers at 1–2 d.p.i. (Figure 1F). As expected, robust viral antigen was detected in the lungs of mice transduced with Ad5-hACE2 but not Ad5-empty control (Figure 1G). Examination of lung tissues from both strains of mice demonstrated a variety of lesions including perivascular to interstitial inflammatory cell infiltrates, necrotic cell debris, and alveolar edema (Figures 1H and 1I). Consistent with the histological findings, examination of gross lung specimens from infected Ad5-hACE2-transduced mice revealed increased vascular congestion and hemorrhage, with the most severe changes observed at 5 d.p.i. (Figure 1J).
Figure 1.
Development of Mice Sensitized to SARS-CoV-2 Infection
(A and B) To assess hACE2 expression and surface localization, 17CL-1 cells were transduced with Ad5-hACE2 or Ad5-Empty at MOI of 100 at 37°C for 4 h. hACE2 expression was monitored by western blot assay (A) or flow cytometry (B).
(C) Ad5-hACE2 transduced 17CL-1 cells were infected with SARS-CoV-2 at MOI of 0.5 at 48 h post transduction, and virus titers were determined by foci forming assay (FFA) at 24, 48, and 72 hours post infection (h.p.i.).
(D) Five days after transduction with 2.5 × 108 FFU of Ad5-hACE2 or Ad5-Empty in 75 μL of DMEM intranasally, lungs were harvested from BALB/c mice, fixed in zinc formalin, and embedded in paraffin. Sections were stained with an anti-hACE2 antibody (brown color). hACE2 protein (brown color) was detected only in Ad-hACE2-treated mice and was predominantly localized to alveolar epithelial cells. Scale bars, 467 and 94 μm, top and bottom panels, respectively.
(E and F) Ad5-hACE2- or Ad5-Empty-transduced BALB/c or C57BL/6 mice were intranasally infected with 1 × 105 PFU of SARS-CoV-2 in 50 μL of DMEM. Weight changes in 6-to-8-week old BALB/c (E) and C57BL/6 (F) mice were monitored daily (n = 5 mice per group). To obtain virus kinetics in BALB/c (E) and C57BL/6 (F) mice, lungs were harvested and homogenized at the indicated time points, and virus was titered by plaque assay. Titers are expressed as PFU/g lung tissue (n = 3 mice per group per time point). Data are representative of two independent experiments.
(G) 2 d.p.i., lungs were harvested from BALB/c mice, fixed in zinc formalin, and embedded in paraffin. Sections were stained with anti-SARS-CoV-2 N protein. Scale bar, 476 μm.
(H) Representative Hematoxylin-eosin (HE) staining of lungs from BALB/c and C57BL/6 mice harvested at the indicated time points p.i. Scale bars, 443 and 88 μm, top and bottom panels, respectively. Asterisk, edema.
(I) Summary histology scores determined at the indicated time points (n = 4 to 5 mice per group). PMN, neutrophils.
(J) Photographs of lung specimens isolated from infected mice at indicated time points are shown. Arrowheads indicate regions with vascular congestion and hemorrhage.
Role of Innate Immune Signaling in SARS-CoV-2 Infections
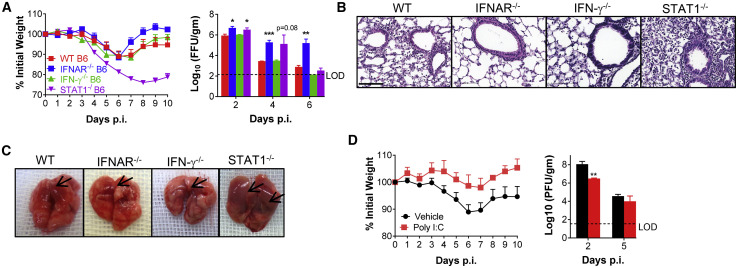
To investigate the possible contributions of interferon (IFN)-I signaling to COVID-19 lung disease, we transduced wild-type and IFNAR−/− C57BL/6 mice with Ad5-hACE2, then infected them with SARS-CoV-2. As shown in Figure 2 , IFNAR−/− C57BL/6 mice had delayed virus clearance and diminished inflammation, although this did not translate into a significant difference in weight loss, suggesting that IFN signaling might have a variety of roles in virus clearance, cellular infiltration, and clinical disease, as was recently postulated for STAT2 in a hamster model of SARS-CoV-2 infection (Boudewijns et al., 2020) (Figures 2A–2C). In comparison, the absence of type II IFN (IFN-γ) signaling had no effect on clinical disease, virus clearance, or pathological changes (Figures 2A–2C). In contrast, SARS-CoV-2-infected, Ad5-hACE2 transduced STAT1−/− C57BL/6 mice exhibited greater weight loss, enhanced inflammatory cell infiltration into the lungs, and delayed virus clearance (Figures 2A–2C). A key role for STAT1 in protection against SARS-CoV was also previously reported (Frieman et al., 2010). Although further studies are warranted, the disparity in phenotypes between IFNAR−/− and STAT1−/− C57BL/6 could reflect the protective effects of IFN-λ, an additional antiviral and immunomodulatory cytokine that signals through STAT1 (Lazear et al., 2019, Prokunina-Olsson et al., 2020). To further examine the role of IFN-I in protection against SARS-CoV-2, we treated mice intranasally with poly I:C, a potent inducer of IFN-I, 6 h prior to infection. Poly I:C treatment resulted in significantly diminished clinical disease and induced more rapid kinetics of initial virus clearance (Figure 2D). Together, our results suggest that IFNs have antiviral and immunomodulatory activity that affects SARS-CoV-2 disease outcome.
Figure 2.
The Role for IFN and STAT1 Signaling in SARS-CoV-2 Infection
(A) 5 days after transduction with 2.5 × 108 FFU of Ad5-hACE2, C57BL/6 mice were intranasally infected with 1 × 105 PFU of SARS-CoV-2. Weight changes were monitored daily (n = 5 mice per group), and virus titers in the lungs were measured at the indicated time points using FFA (n = 3–4 mice per group per time point). Titers are expressed as FFU/g tissue.
(B) Sections of paraffin embedded lungs from SARS-CoV-2-infected Ad5-hACE2-transduced, wild-type and genetically modified C57BL/6 mice at 4 d.p.i. were stained with hematoxylin/eosin. Scale bar, 100 μm.
(C) Photographs of gross pathological lung specimens isolated from infected C57BL/6 mice at 4 d.p.i. Arrowheads indicate regions with vascular congestion and hemorrhage.
(D) Ad5-hACE2-transduced C57BL/6 mice were treated with 80 μg of poly I:C in 50 μL of PBS 6 h before intranasal infection with SARS-CoV-2. Weight changes were monitored daily, and viral titers in lungs were measured at the indicated time points.
∗p values ≤ 0.05; ∗∗p values ≤ 0.005; ∗∗∗p values ≤ 0.0005; ∗∗∗∗p values ≤ 0.0001.
Differentially Expressed Genes in the Lungs of SARS-CoV-2-Infected Mice
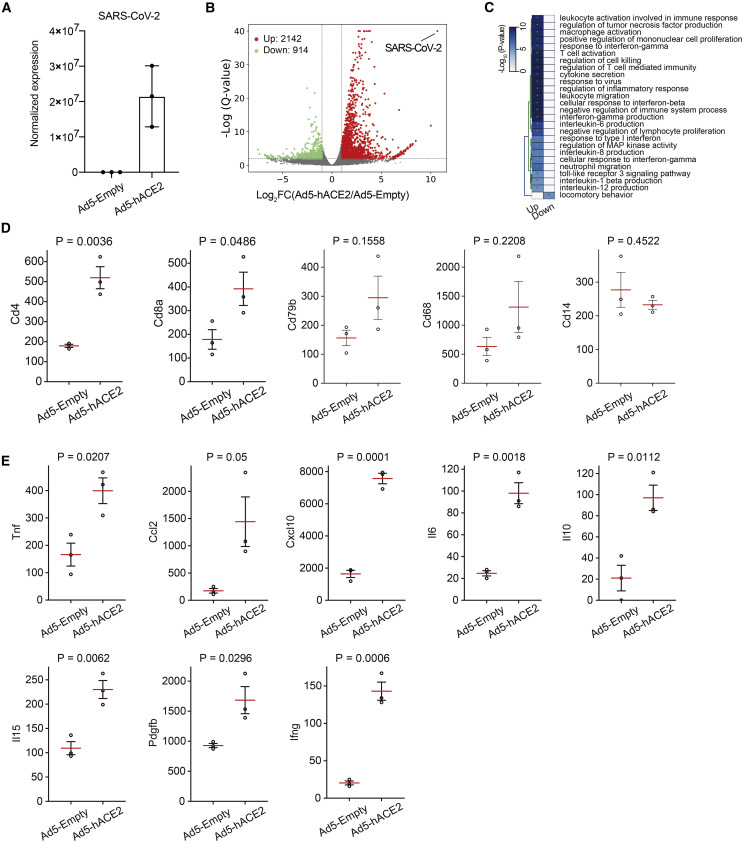
To further investigate the innate immune and host defense responses of mice to SARS-CoV-2 infection, we performed RNA sequencing (RNA-seq) using RNA extracted from lungs of Ad5-Empty and Ad5-ACE2 transduced BALB/c mice at 2 d.p.i. Abundant SARS-CoV-2 viral RNA was detected solely in Ad5-ACE2 transduced mice indicating robust viral replication in their lungs (Figure 3 A). In total, 3,056 genes were differentially expressed, with 2,142 genes upregulated and 914 genes downregulated in Ad5-ACE2-transduced mice compared with Ad5-Empty-transduced mice (Figure 3B). Most upregulated genes were associated with inflammation pathways and innate and adaptive immune response pathways, including ISGs, immune cell activation, and migration (Figure 3C). In particular, CD4 and CD8 transcript abundance were significantly upregulated, indicating T cell migration from periphery to the site of infection (Figure 3D). B cell and macrophage abundance as determined by CD79b and CD68 expression showed a trend toward increased migration. It is possible that since RNA-seq was performed at 2 d.p.i., B cell and macrophage infiltration could occur at later time points p.i. (Figure 3D). Several cytokines and chemokines, including tumor necrosis factor (TNF), IFN-γ, interleukin (IL)-10, IL-15, IL-6, CCL2, and CXCL10 were upregulated, consistent with observations in COVID-19 patients (Figure 3D) (Chen et al., 2020, Huang et al., 2020). In addition, we found that expression of platelet-derived growth factor subunit B (PDGFb) was upregulated in Ad5-ACE2 transduced mice. PDGFb is secreted by platelets during coagulation, and also by activated macrophages, fibroblasts, and endothelial cells during tissue damage and regeneration (Figure 3D) (Bennett and Schultz, 1993, Deuel et al., 1988, Wahl et al., 1989). In summary, these results demonstrate that Ad5-ACE2-transduced mice reproduced some key manifestations observed in COVID-19 patients.
Figure 3.
Differentially Expressed Genes in the Lungs of SARS-CoV-2-Infected Mice
(A) SARS-CoV-2 viral RNA detected by RNA-seq in Ad5-Empty- and Ad5-ACE2-transduced mouse lungs. Data are expressed as normalized read counts.
(B) Volcano plot showing differentially expressed genes in the lungs of Ad5-ACE2-transduced mice compared with Ad5-Empty-transduced mice. A total of 3,056 transcripts were differentially regulated.
(C) Gene ontology (GO) analysis showing the differentially expressed genes from (B).
(D) CD4+ (Cd4), CD8+ (Cd8a) T cell and B cell (Cd79b), macrophage (Cd68), and monocyte (Cd14) lineage marker expression. The red lines are the means of the three biological replicates, and the error bars are the standard error of the mean. Data are expressed as normalized read counts. p values are from a one-tailed Student’s t test.
(E) Selected cytokines and chemokines differentially regulated in the lungs of Ad5-Empty- and Ad5-ACE2-transduced BALB/c mice at 2 d.p.i., obtained from the RNA-seq data. The red lines are the means of the three biological replicates, and the error bars are the standard error of the mean. Data are expressed as normalized read counts. p values are from a one-tailed Student’s t test.
Requirement for Virus-Specific T Cells and Neutralizing Antibodies in SARS-CoV-2 Clearance from Infected Lungs
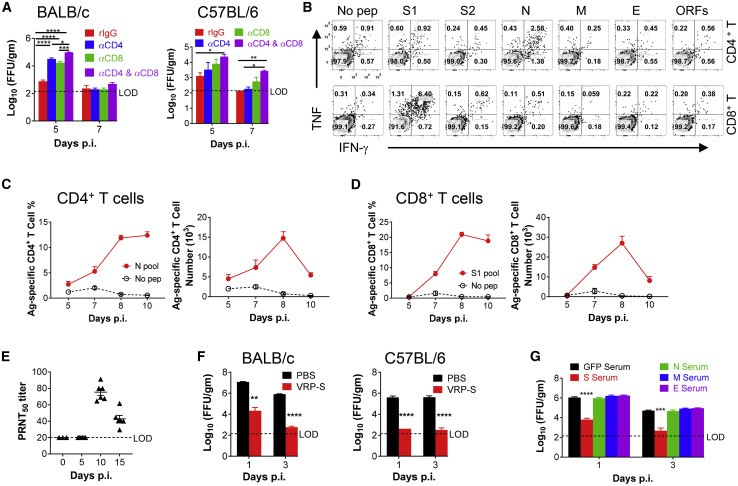
Virus-specific T cells are critical for SARS-CoV or MERS-CoV clearance (Zhao et al., 2016, Zhao et al., 2010). To examine the role of T cell responses in COVID-19, we depleted CD4+ and CD8+ T cells individually and together from Ad5-hACE2-sensitized BALB/c and C57BL/6 mice prior to SARS-CoV-2 infection. Our results indicate that optimal virus clearance required both CD4+ and CD8+ T cell responses in SARS-CoV-2 infected mice (Figure 4 A). To further characterize the T cell response in infected mice, peptide pools encompassing SARS-CoV-2 structural proteins and the accessory protein ORFs (3a, 6, 7a, 7b, 8, 10) were synthesized and used to stimulate cells harvested from infected mouse lungs. We found that CD4+ and CD8+ T cell epitopes were predominantly located in the N protein and the S1 region of the S protein, respectively, in BALB/c mice (Figure 4B). The virus-specific CD4+ and CD8+ T cell responses peaked at 8 d.p.i. in BALB/c mice, as expected (Figures 4C and 4D). In addition, Ad5-hACE2-sensitized mice also produced neutralizing antibodies in sera after infection as determined by a plaque reduction neutralizing test (PRNT50) (Figure 4E). To evaluate the utility of Ad5-hACE2-sensitized mice in vaccine evaluation, we developed Venezuelan equine encephalitis replicon particles (VRPs) expressing the SARS-CoV-2 spike (VRP-S), transmembrane (VRP-M), nucleocapsid (VRP-N), and envelope (VRP-E) proteins as previously described (Deming et al., 2006). Immunization with VRP-S reduced SARS-CoV-2 titers by greater than 3 logs in both BALB/c and C57BL/6 mice by 1 d.p.i. (Figure 4F). This protection was largely mediated by anti-S antibodies, which blocked virus attachment, as passive transfer of sera from VRP-S-immunized mice also accelerated the kinetics of virus clearance in naive mice (Figure 4G). In contrast, sera from mice immunized with VRP-M, VRP-N, or VRP-E failed to enhance the kinetics of virus clearance.
Figure 4.
Requirements for T Cells and Antibodies for SARS-CoV-2 Clearance and Protection from Subsequent Challenge
Ad5-hACE2-transduced mice were infected with 1 × 105 PFU of SARS-CoV-2.
(A) For systemic depletion of CD4+ or CD8+ T cells, mice were injected intraperitoneally (i.p.) with 0.5 mg anti-CD4 antibody (clone GK1.5) and/or 0.5 mg anti-CD8 antibody (clone 2.43) or 0.5 mg rat IgG at days −2 and 0 p.i. Virus titers in the lungs were measured at the indicated time points. Titers are expressed as FFU/g tissue (n = 4 mice per group per time point)
(B–D) To identify SARS-CoV-2 T cell responses, single-cell suspensions were prepared from the BALF of transduced/infected BALB/c mice and stimulated with 2 μM structural protein peptide pools for 5–6 h in the presence of brefeldin A. Flow plots (B, 7 d.p.i), and summary of frequencies and cell numbers of SARS-CoV-2-N pool specific CD4+ T cells (C) and S1 pool specific CD8+ T cells (D) (determined by IFN-γ intracellular staining) are shown (n = 3 to 4 mice per time point).
(E) PRNT50 titers in the sera of transduced/infected C57BL/6 mice at indicated time points p.i. are shown.
(F) BALB/c and C57BL/6 mice were immunized with 1 × 105 infectious units (IU) of VRP-S intranasally in 50 μL of PBS. Mice were transduced and infected with 1 × 105 PFU of SARS-CoV-2 3 weeks after vaccination. Virus titers in the lungs were measured at the indicated time points (n = 4 mice per group per time point).
(G) For adoptive transfer of serum, BALB/c mice were immunized with 1 × 105 IU of VRP in the footpad in 50 μL of PBS and boosted with the same dose 3 weeks later. Sera were obtained 1–2 weeks after VRP booster. Then, 150 μL of serum was transferred into transduced mice intravenously (i.v.) 1 day before SARS-CoV-2 infection (n = 3 mice per group per time point).
∗p values ≤ 0.05; ∗∗p values ≤ 0.005; ∗∗∗p values ≤ 0.0005; ∗∗∗∗p values ≤ 0.0001.
Ad5-hACE2-Sensitized Mice Are Useful for Evaluating SARS-CoV-2-Specific Therapies
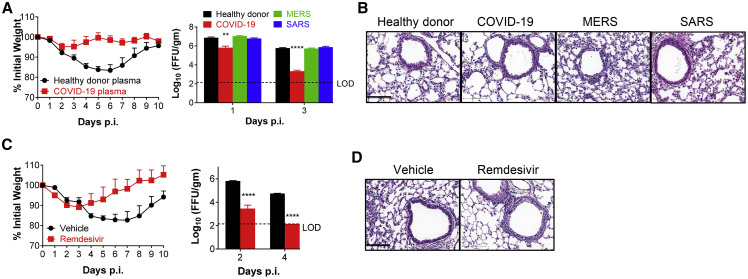
Several therapies have been proposed to treat COVID-19, including transfer of plasma from patients who have recovered from the infection. We obtained pooled plasma from 3 patients who recovered from SARS-CoV-2 infection (FRNT50 titer = 1:1,000) as well as plasma from a healthy donor, 3 SARS survivors (PRNT50 titer against SARS-CoV = 1:140), and 2 MERS convalescent patients (FRNT50 titer against MERS-CoV = 1:2,183). Administration of 150 μL of plasma one day prior to SARS-CoV-2 infection prevented weight loss and lung tissue histological changes and accelerated the rate of virus clearance (Figures 5A and 5B). More rapid clearance was not observed after treatment with pooled plasma from SARS survivors or MERS survivors.
Figure 5.
Convalescent Plasma from COVID-19 Patients and Remdesivir Protect Mice from SARS-CoV-2 Infection
(A and B) For plasma adoptive transfer, Ad5-hACE2-transduced mice were injected with 150 μL of plasma i.v. from a healthy donor or COVID-19, MERS, or SARS convalescent patients, at −1 d.p.i. Weight and virus titers in lung tissues were monitored (A) and expressed as FFU/g tissue (n = 4 mice per group per time point). Sections of paraffin embedded lungs from plasma adoptive transferred and infected mice were stained with HE at day 4 p.i. (B). Scale bar, 100 μm.
(C and D) For remdesivir treatment, Ad5-hACE2-transduced mice were treated with remdesivir (25 mg/kg, bid s.c.) or vehicle at −1 d.p.i. Weight loss of infected mice and virus titers in the lungs were monitored (C), and hematoxylin/eosin staining of sections of paraffin-embedded lungs is shown at 4 d.p.i. (D) (n = 4 mice per group per time point). Data are representative of two independent experiments. Scale bar, 100 μm.
∗p values ≤ 0.05; ∗∗p values ≤ 0.005; ∗∗∗p values ≤ 0.0005; ∗∗∗∗p values of ≤ 0.0001.
Remdesivir, an adenosine nucleotide triphosphate analog, was granted Emergency Use Approval by the U.S. Food and Drug Administration, based on recently published results (Grein et al., 2020, Wang et al., 2020, Yin et al., 2020). Treatment with remdesivir one day prior to infection and continued dosing of 25 mg/kg twice daily resulted in decreased weight loss, significantly accelerated virus clearance, and diminished cellular infiltration of lung tissue in infected Ad5-hACE2-transduced mice (Figures 5C and 5D).
Discussion
The COVID-19 pandemic, with associated high levels of morbidity and mortality and enormous economic losses, has made the discovery of vaccines and anti-viral treatments imperative. Here, we show that Ad5-hACE2-transduced mice develop pneumonia after infection with SARS-CoV-2, thus overcoming the natural resistance of mice to the infection. We show that the infection of transduced mice protocol is very reproducible. SARS-CoV-2 infection of these mice resulted in high levels of viral replication in the lungs. While the infection is non-lethal, Ad5-hACE2-sensitized mice are useful for evaluation of vaccines and anti-viral therapies. We used these mice to show critical roles for IFN-I and STAT1 signaling and virus-specific antibodies and T cells in the host response to SARS-CoV-2. We also illustrate the utility of the system by demonstrating the efficacy of patient-derived convalescent plasma, remdesivir, and poly I:C as therapeutic interventions and its use in vaccine evaluation.
While other animals including Syrian hamsters, ferrets, and rhesus monkeys can support virus replication (Chan et al., 2020, Kim et al., 2020, Shi et al., 2020, Yu et al., 2020), laboratory mice offer practical advantages including costs, multiple strains, and ease of genetic manipulation. Mice that are genetically modified to express hACE2 may be also useful. However, transgenic expression of hACE2 may not sensitize infection only in the correct organ, as shown in SARS and MERS transgenic mouse models (Agrawal et al., 2015, Li et al., 2017, McCray et al., 2007). Development of adenovirus vectors to express the SARS-CoV-2 receptor in the mouse lungs was efficient and rapid, resulting in the generation of an easily reproducible murine model for SARS-CoV-2 within 2–3 weeks. This compares favorably with the much longer time (several months to years) required to develop and breed sufficient numbers of hACE2-transgenic or hACE2-knockin mice for studies. Even longer time periods are required to cross or genetically modify these mice to study COVID-19 pathogenesis. Our strategy allows sensitization of all mouse strains and all genetically modified mice to SARS-CoV-2 infection. This facile and widely available approach helps address the current urgent need for small animal models for COVID-19.
Limitations of the Study
Mice transduced with Ad5-hACE2 do not develop severe disease. They also do not develop extrapulmonary manifestations of disease. Studies of pathogenesis, especially of Acute Respiratory Distress Syndrome (ARDS) will require development of infection models that include mild and severe COVID-19. This will likely involve development of mice “knocked in” for hACE2 expression and probably mouse adaptation of the virus by serial passage through mouse lungs.
Note Added as Our Study Was Finalized
Consistent with our studies, in a companion submission, Hassan et al. (2020) have analogously transduced mice with Ad5-hACE2 and successfully generated a SARS-CoV-2 model of lung infection and disease.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor® 488 anti-mouse CD8a Antibody | Biolegend | Clone 53-6.7 Cat.# 100723; AB_389304 |
| PerCP-Cyanine5.5 CD16/CD32 Monoclonal Antibody | eBioscience | Clone 93 Cat.# 45-0161-82; AB_996659 |
| eFluor 450 CD4 Monoclonal Antibody | eBioscience | Clone RM4-5 Cat.# 48-0042-82; AB_1272194 |
| PE TNF alpha Monoclonal Antibody | eBioscience | Clone MP6-XT22 Cat.# 12-7321-82; AB_466199 |
| APC IFN gamma Monoclonal Antibody | eBioscience | Clone XMG1.2 Cat.# 17-7311-82; AB_469504 |
| mouse anti-human hACE2 antibody | R&D Systems | Cat.# MAB933; AB_2223153 |
| mouse anti-β-actin | Genscript | Cat.# A01546; AB_1968817 |
| rabbit anti-SARS-CoV-2 nucleocapsid protein polyclonal antibody | Sino Biological | Cat.# 40143-T62 |
| HRP-labeled goat anti-rabbit secondary antibody | Jackson ImmunoResearch Laboratories | Cat.# 111-035-144; AB_2307391 |
| anti-CD4 antibody | Bio X Cell | Clone GK1.5 Cat.# BE0003-1; AB_1107636 |
| anti-CD8 antibody | Bio X Cell | Clone 2.43 Cat.# BE0061; AB_1125541 |
| Virus Strains | ||
| SARS-CoV-2/human/CHN/IQTC01/2020 (GenBank: MT123290.1) | N/A | |
| 2019-nCoV/USA-WA1/2020(GenBank: MN985325.1) | N/A | |
| Chemicals and Peptides | ||
| Remdesivir | MedChemExpress | Cat.# HY-104077 |
| Sulfobutylether-β-Cyclodextrin | MedChemExpress | Cat.# HY-17031 |
| Poly I:C | Invivogen | Cat.# 31852-29-6 |
| Agarose | RPI | Cat.# 9012-36-6 |
| Crystal violet | Solarbio | Cat.# C8470 |
| 4% Paraformaldehyde | Biosharp | Cat.# BL539A |
| Zinc formalin | Sigma | Cat.# Z2902 |
| Triton X-100 | Solarbio | Cat.# T8200 |
| GolgiPlug Protein Trnsp Inbb | BD Biosciences | Cat.# 555029 |
| Cytofix/Cytoperm Solution | BD Biosciences | Cat.# 554722 |
| Carboxymethylcellulose powder | Sigma | Cat.#C4888 |
| TrueBlue™ Peroxidase Substrate (KPL, Gaithersburg, MD) | SeraCare | Cat.# 510-0030 |
| Blocking reagent | Biocare Medical | Cat.# RBM961H |
| Secondary Kit (Mouse Envision) | Dako | Cat.# K00111-2 |
| DAB+ Substrate Chromogen System (Dako Omnis) | Dako | Cat.# GV825 |
| Streptavidin-horseradish peroxidase conjugate (Vectastain ABC Elite kit) | Vector Labs | Cat.# PK-8200 |
| Diaminobenzidine | Vector Labs | Cat.# SK-4100 |
| NovaRED | Vector Labs | Cat.# SK-4805 |
| RNA ScreenTape | Agilent Technologies | Cat.# 5067-5576 |
| RNA Reagent | Agilent Technologies | 5067-5577 |
| D1000 ScreenTape | Agilent Technologies | 5067-5582 |
| D1000 Reagent | Agilent Technologies | 5067-5583 |
| Qubit® dsDNA HS Assay Kit | Life Technologies | Q32851 |
| QIAseq FastSelect –rRNA HMR Kits | QIAGEN | 334387 |
| NEBNext® UltraTM RNA Library Prep Kit for Illumina | NEB | E7530L |
| SuperSignal West Pico Trial Kit | Thermo Scientific | N/A |
| Trizol | invitrogen | Cat.# 15596018 |
| Fetal Bovine Sera | GIBCO | Cat.# 10270-106 |
| Penicillin-Streptomycin | Invitrogen | Cat.# 60106-1 |
| L-GLUTAMINE,100X | Invitrogen | Cat.# 2503081 |
| DPBS | GIBCO | Cat.# 14190235 |
| RPMI1640 | Life | Cat.# C11875500BT |
| DMEM | Life | Cat.# C11965500BT |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: 150847 |
| Experimental Models: Cell Lines | ||
| Vero E6 cells | ATCC | CRL-1586 |
| Calu-3 cells | ATCC | HTB-55 |
| 17Cl-1 cells | ATCC | |
| Experimental Models: Organisms/Strains | ||
| Mouse: WT C57BL/6 mouse | Hunan SJA Laboratory Animal Co.(China) Charles River Laboratories (USA) |
N/A |
| Mouse: WT BALB/c mouse | Hunan SJA Laboratory Animal Co.(China) Charles River Laboratories (USA) |
N/A |
| Mouse: IFNARKO C57BL/6 mouse | Jackson Laboratories | JAX: 028288 |
| Mouse: B6.129S(Cg)-Stat1tm1Dlv/J | Jackson Laboratories | JAX: 012606 |
| Mouse: B6.129S7-Ifng tm1Ts J | Jackson Laboratories | JAX: 002287 |
| Software and Algorithms | ||
| FlowJo VX | N/A | |
| GraphPad Prism V7.04 | N/A | |
| Bowtie2 v2.2.5 | Langmead and Salzberg, 2012 | N/A |
| RSEM v1.2.22 | Li and Dewey, 2011 | N/A |
| EDASEQ v2.16 | Risso et al., 2011 | N/A |
| DESeq2 v1.22.1 | Love et al., 2014 | N/A |
| clusterProfiler v3.10.0 | Hutchins et al., 2014 | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jincun Zhao (zhaojincun@gird.cn)
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The accession number for the Next-gen RNA sequence data reported in this paper is Gene Expression Omnibus (GEO) database: GSE150847; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150847.
Experimental Model and Subject Details
Human subject approval
Written informed consent was obtained from all recovered COVID-19, SARS, and MERS patients and approval was obtained from the Institutional Review Boards of the First Affiliated Hospital of Guangzhou Medical University and the King Faisal Specialist Hospital, Jeddah, the two sites of patient enrollment. We also obtained Institutional Review Board approval from the Health Commission of Guangdong Province to use the anonymized healthy blood donor samples described in this study.
Mice, virus, and cells
Specific pathogen-free 6-10 week old male and female BALB/c and C57BL/6 mice and 5-6 month old C57BL/6 mice were purchased from Hunan SJA Laboratory Animal Co. (Hunan, China) or Charles River Laboratories and maintained in the Animal Care Facilities at the Guangzhou Medical University and University of Iowa. IFN-γ −/−, STAT1−/− and IFNAR−/− mice were purchased from Jackson Laboratories (Bar Harbor, Maine). All protocols were approved by the Institutional Animal Care and Use Committees of the Guangzhou Medical University and of the University of Iowa. The SARS-CoV-2 strains used in this research were isolated from COVID-19 patients in Guangzhou and in Washington state (Accession numbers: MT123290, MN985325.1), and passaged on Vero E6 and Calu-3 2B4 cells. African Green monkey kidney-derived Vero E6 cells and 17CL-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS). Calu-3 2B4 cells were grown in MEM (GIBCO, Grand Island, NY) supplemented with 20% FBS. The human serotype 5 adenoviral vector expressing human ACE2 under the control of the CMV promoter was previously described (Anderson et al., 2000, Jia et al., 2005).
Method Details
Chemicals, Cytokines and Peptides
Remdesivir (Cat. No. HY-104077) was purchased from MedChemExpress (Monmouth Junction, NJ). Poly I:C was purchased from Sigma (St. Louis, MO). A set of 20-mer peptides encompassing the four SARS-CoV-2 structural [S1, S2, N, and ME encompassing the N- and C-terminal portions of the spike (S) glycoprotein, the nucleocapsid (N) protein, and the transmembrane (M) and envelope (E) proteins], and 6 accessory proteins [ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10] and overlapping by 10 amino acids was synthesized by GL Biochem Ltd. (Shanghai, China), and used for stimulation of T cells.
Transduction and infection of 17Cl-1 cells and western blot analysis
Recombinant adenoviral vectors expressing hACE2 (Ad5-hACE2) with or without a C9 tag were prepared and used as previously described (Anderson et al., 2000, Jia et al., 2005). 17Cl-1 cells were transduced with Ad5-hACE2 or Ad5-Empty at a multiplicity of infection (MOI) = 100 for 4 h at 37°C. Extracts were prepared 48 h post transduction. Identical amounts of protein were separated on an 8% SDS/PAGE gel and transferred to PVDF membranes. Membranes were stained with a mouse anti-human hACE2 antibody (clone OTI2G7, Origene, Rockville, MD), a mouse anti-C9 antibody (clone Rho 1D4, EMD Millipore, Temecula, CA), or a mouse anti–β-actin (Cat: A01546, Genscript, Nanjing). Proteins were detected using a SuperSignal West Pico Trial Kit (Thermo Scientific). For infection, 17Cl-1 cells were transduced with Ad5-hACE2 or Ad5-Empty for 48 h at MOI = 100. The transduced cells were infected with SARS-CoV-2 at MOI = 0.5. Culture supernatants and cells were collected at the indicated time points and analyzed for infectious virus by a focus forming assay (see below). The Ad5-hACE2 or Ad5-Empty vectors are available from the University of Iowa Viral Vector Core (https://medicine.uiowa.edu/vectorcore/).
Transduction and infection of mice
Mice were lightly anesthetized with isoflurane and transduced intranasally with 2.5 × 108 FFU of Ad5-ACE2 or Ad5-Empty in 75 μL DMEM. Five days post transduction, mice were infected intranasally with SARS-CoV-2 (1 × 105 PFU) in a total volume of 50 μL DMEM. Mice were monitored weighted daily. All work with SARS-CoV-2 was conducted in the Biosafety Level 3 (BSL3) Laboratories of Guangzhou Customs District Technology Center or the University of Iowa.
SARS-CoV-2 plaque assay
Virus or lung homogenate supernatants were serially diluted in DMEM. Vero E6 cells in 12 well plates were inoculated at 37°C in 5% CO2 for 1 h with gentle rocking every 15 min. After removing the inocula, plates were overlaid with 1.2% agarose containing 4% FBS. After further incubation for 2 days, overlays were removed, and plaques were visualized by staining with 0.1% crystal violet. Viral titers were calculated as PFU per gram tissue.
Focus forming assay (FFA)
In some experiments, virus was titered using an FFA since it is high throughput as compared to the traditional plaque assay. Vero E6 cells were seeded in 96-well plates one day before infection. Virus cultures or lung homogenate were serially diluted and used to inoculate Vero E6 cells at 37°C for 1 h. Inocula were then removed before adding 125 μL 1.6% carboxymethylcellulose warmed to 37°C per well. After 24 h, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Cells were then incubated with a rabbit anti-SARS-CoV-2 nucleocapsid protein polyclonal antibody (Cat. No.: 40143-T62, Sino Biological, Inc. Beijing), followed by an HRP-labeled goat anti-rabbit secondary antibody (Cat. No.: 109-035-088, Jackson ImmunoResearch Laboratories, Inc. West Grove, PA). The foci were visualized by TrueBlue Peroxidase Substrate (KPL, Gaithersburg, MD), and counted with an ELISPOT reader (Cellular Technology Ltd. Cleveland, OH). Viral titers were calculated as FFU per ml or per gram tissue.
SARS-CoV-2 plaque reduction neutralization test (PRNT50)
Serum samples were serially diluted in DMEM and mixed with an equal volume of SARS-CoV-2 containing 80-100 PFU. After incubation at 37°C for 1 h, aliquots were added to Vero E6 cells in 12-well plates and incubated at 37°C in 5% CO2 for 1 h. After removing the inocula, plates were overlaid with 1.2% agarose containing 4% FBS. After further incubation for 2 days, overlays were removed, and plaques were visualized by staining with 0.1% crystal violet. The procedure of foci reduction neutralization test (FRNT50) is similar to PRNT, except that the plates were stained according to FFA above.
Venezuelan Equine Encephalitis Replicon particles (VRPs) and mouse immunization
VRPs expressing the SARS-CoV-2 spike protein (S), nucleocapsid protein (N), membrane protein (M), envelope protein (E) or GFP were constructed as previously described (Deming et al., 2006). Mice were primed and boosted (3 weeks after priming) with 1x105 Infectious Units (IU) of VRP-S, VRP-N, VRP-M, VRP-E, or VRP-GFP in the left footpad in 50 μL PBS or intranasally (i.n.) in 50 μL DMEM after light anesthesia with isoflurane. Sera were collected 1-2 weeks after booster. Mice were challenged with SARS-CoV-2 three weeks post VRP vaccination.
Antibody and poly I:C treatment
For depletion of CD4+ or CD8+ T cells, mice were injected intraperitoneally (i.p.) with 0.5 mg anti-CD4 antibody (clone GK1.5, Cat. No.: BE003-1, Bio X Cell, Lebanon, NH) or 0.5 mg anti-CD8 antibody (clone 2.43, Cat. No.: BE0061, Bio X Cell, Lebanon, NH), respectively at days −2 and 0 p.i. Poly I:C (80 μg in 50 μL of PBS, Cat. No. 31852-29-6, Invivogen, San Diego, CA) was administered intranasally to Ad5-hACE2 sensitized mice 6 h prior to infection.
Preparation of cells from bronchoalveolar lavage fluids (BALF)
Mice were sacrificed at the indicated time points. BALF was acquired by inflating lungs with 1 mL complete RPMI 1640 medium via cannulation of the trachea followed by lavaging four times. Cells in the BALF were collected by centrifugation.
Flow Cytometry
The following monoclonal antibodies were used: rat anti-mouse CD8α-Alexa 488 (clone 53-6.7, Cat. No.: 100723, Biolegend, San Diego, CA), anti-mouse CD16/32-PerCP-Cy5.5 (clone 93, Cat. No.: 45-0161-82, eBioscience, San Diego, CA), anti-mouse CD4-eFluor 450 (cloneRM4-5, Cat. No.: 48-0042-82 eBioscience, San Diego, CA), anti-mouse TNF-PE (cloneMP6-XT22, Cat. No.: 12-7321-82Bioscience, San Diego, CA), anti-mouse IFN-γ-APC (cloneXMG1.2, Cat. No.: 17-7311-82, eBioscience, San Diego, CA), and mouse anti-C9 antibody (clone Rho 1D4, Cat. No.: MAB5356, EMD Millipore). For intracellular cytokine staining (ICS), lymphocytes were cultured in 96-well dishes at 37°C for 5-6 h in the presence of 2 μM peptide pool and brefeldin A (BD Biosciences). Cells were then labeled for cell-surface markers, fixed/permeabilized with Cytofix/Cytoperm Solution (BD Biosciences), and labeled with anti-IFN-γ and anti-TNF antibody. All flow cytometry data were acquired on a BD FACSVerse and were analyzed using FlowJo software.
Histology and Immunohistochemistry
Animals were anesthetized and transcardially perfused with PBS followed by zinc formalin. Lungs were fixed in zinc formalin. For routine histology, tissue sections (∼4 μm each) were stained with hematoxylin and eosin. To detect virus antigen, sections were incubated with blocking reagent (Rodent Block-M, Biocare Medical, Pacheco, CA) incubated with a mouse monoclonal antibody to hACE2 (1:100 dilution, mouse anti-hACE2, MAB933,R&D Systems), then incubated with a secondary (polymer-based) kit (Mouse Envision, Dako, Carpinteria, CA), followed by incubation with DAB+ (Dako) or with an antibody to SARS-CoV-2 N protein (1:500 dilution, a monoclonal antibody to the SARS-N protein which also identifies the N-protein of SARS-CoV-2 provided by Professor John Nicholls, The University of Hong Kong), then incubated with Rabbit Envision (Dako) and diaminobenzidine (Dako). Tissues were examined and scored in a post-examination method of masking by a boarded experimental pathologist (Meyerholz and Beck, 2018). Ordinal scores for lesion parameters were assigned using the following tiers: 0 = within expected limits; 1 - uncommon, < 5%; 2 - detectable in 5%–33%; 3 - detectable in 34%–66% and 4 - detectable in > 66% of lung fields (200x objective magnification).
Human convalescent plasma
Plasma samples were collected from 3 patients 1 month following recovery from COVID-19 (FRNT50 = 1000). Plasma was collected from 3 patients who were 15 years post recovery from SARS-CoV infection (PRNT50 = 140). Plasma was collected from 2 patients who were 1-2 months post recovery from MERS (FRNT50 = 2183).
RNA sequencing and data analysis
Total RNA was extracted from infected lungs using Trizol (Invitrogen) according to the manufacturer’s protocol, and ribosomal RNA removed using QIAseq FastSelect-rRNA HMR Kits (QIAGEN, Germany). Fragmented RNA (average length approximately 200 bp) was subjected to first strand and second strand cDNA synthesis followed by adaptor ligation and enrichment with a low-cycle according to the instructions of NEBNext® UltraTM RNA Library Prep Kit for Illumina (NEB, USA). The purified library products were evaluated using the Agilent 2200 TapeStation and Qubit®2.0(Life Technologies, USA). The libraries were paired-end sequenced (PE150, Sequencing reads were 150 bp) at Guangzhou RiboBio Co., Ltd. (Guangzhou, China) using Illumina HiSeq 3000 platform.
Reads were aligned to the mouse reference genome mm10 with bowtie2 (Langmead and Salzberg, 2012), and RSEM (Li and Dewey, 2011) was used to quantify the reads mapped to each gene. Gene expression was normalized by EDASEQ (Risso et al., 2011). Differentially expressed genes were obtained using DESeq2 (version 1.10.1) (Love et al., 2014), a cutoff of Q-value < 0.05 and log2 (fold-change) > 1 was used for identify differentially expressed genes. All differentially expressed mRNAs were selected for GO analyses clusterProfiler (Yu et al., 2012). Other analysis was performed using glbase (Hutchins et al., 2014). Next-gen RNA sequence data supporting the findings in this study have been deposited in the Gene Expression Omnibus (GEO) database (GSE150847); https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150847).
Quantification and Statistical Analysis
ANOVA and Student’s t tests were used to analyze differences in mean values between groups using GraphPad Prism 7. All results are expressed as mean ± standard error of the mean (SEM) and were corrected for multiple comparisons. P values of < 0.05 were considered statistically significant. (∗, P values of ≤ 0.05. ∗∗, P values of ≤ 0.005. ∗∗∗, P values of ≤ 0.0005. ∗∗∗∗, P values of ≤ 0.0001).
Acknowledgments
This work is supported by the grants from The National Key Research and Development Program of China (2018YFC1200100) (J.Z.), National Science and Technology Major Project (2018ZX10301403) (J.Z.), the emergency grants for prevention and control of SARS-CoV-2 of Ministry of Science and Technology (2020YFC0841400) (J.Z.) and Guangdong province (2020B111108001, 2018B020207013) (J.Z.); the National Institutes of Health USA (NIH) (P01 AI060699 [S.P., P.B.M.] and RO1 AI129269 [S.P.]); and the Pathology Core, which is partially supported by the Center for Gene Therapy for Cystic Fibrosis (NIH grant P30 DK-54759), and the Cystic Fibrosis Foundation. P.B.M. is supported by the Roy J. Carver Charitable Trust. We thank Christine Wohlford-Lenane for technical assistance, Dr. Ralph Baric for providing VRP plasmids, and the patients who took part in this study.
Author Contributions
Conceptualization and Writing – Original Draft, P.B.M., S.P., and J.Z.; Data Curation, J.S., Z. Zhuang, J. Zheng, K.L., R.L.-Y.W., D.L., J. Huang, J. He, A.Z., Jingxian Zhao, X.L., Y.X., R.C., A.N.A., Z.C., Z. Zhang, C.C., X.H., F.L., X.L., D.C., L.W., J. Zhuo, Y.Z., Y.W., S.H., J.D., Y.S., K.Z., M.R.L., J.C., Y.L., and N.Z.; Formal Analysis, J.S., Z. Zhuang, J.Z., K.L., R.L.-Y.W., D.L., J. Huang, J. He, A.Z., Jingxian Zhao, X.L., D.K.M., P.B.M., S.P., and Jincun Zhao; Investigation, D.K.M., P.B.M., S.P., and Jincun Zhao; Visualization, D.K.M.; Resources, R.C., A.N.A., D.K.M., P.B.M., S.P., and Jincun Zhao; Writing – Review & Editing, J.S., Z. Zhuang, J. Zheng, P.B.M., S.P., and Jincun Zhao; Supervision, P.B.M., S.P., and Jincun Zhao; Project Administration, P.B.M., S.P., and Jincun Zhao; Funding Acquisition, P.B.M., S.P., and Jincun Zhao.
Declaration of Interests
The authors declare no competing interests.
Published: June 9, 2020
References
- Agrawal A.S., Garron T., Tao X., Peng B.H., Wakamiya M., Chan T.S., Couch R.B., Tseng C.T. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J. Virol. 2015;89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.D., Haskell R.E., Xia H., Roessler B.J., Davidson B.L. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7:1034–1038. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;2020:7. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bennett N.T., Schultz G.S. Growth factors and wound healing: Part II. Role in normal and chronic wound healing. Am. J. Surg. 1993;166:74–81. doi: 10.1016/s0002-9610(05)80589-6. [DOI] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., De Keyzer C., Sharma S., Jansen S., Weyenbergh J.V. STAT2 signaling as double-edged sword restricting viral dissemination but driving severe pneumonia in SARS-CoV-2 infected hamsters. bioRxiv. 2020 doi: 10.1101/2020.04.23.056838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020:ciaa325. doi: 10.1093/cid/ciaa325. Published online March 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S., Yount B.L., Scobey T., Jensen K., Douglas M., Beall A., Tang X.C., Marasco W.A., Heise M.T., Baric R.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016;2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal R.G., McElvaney N.G., Rosenfeld M.A., Chu C.S., Mastrangeli A., Hay J.G., Brody S.L., Jaffe H.A., Eissa N.T., Danel C. Administration of an adenovirus containing the human CFTR cDNA to the respiratory tract of individuals with cystic fibrosis. Nat. Genet. 1994;8:42–51. doi: 10.1038/ng0994-42. [DOI] [PubMed] [Google Scholar]
- Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3:e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuel T.F., Silverman N.J., Kawahara R.S. Platelet-derived growth factor: a multifunctional regulator of normal and abnormal cell growth. Biofactors. 1988;1:213–217. [PubMed] [Google Scholar]
- Frieman M.B., Chen J., Morrison T.E., Whitmore A., Funkhouser W., Ward J.M., Lamirande E.W., Roberts A., Heise M., Subbarao K., Baric R.S. SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism. PLoS Pathog. 2010;6:e1000849. doi: 10.1371/journal.ppat.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X. Compassionate Use of Remdesivir for Patients with Severe Covid-19. New Engl J Med. 2020 doi: 10.1056/NEJMoa2007016. Published online April 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Al Soussi W.B. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins A.P., Jauch R., Dyla M., Miranda-Saavedra D. glbase: a framework for combining, analyzing and displaying heterogeneous genomic and high-throughput sequencing data. Cell Regen. (Lond.) 2014;3:1. doi: 10.1186/2045-9769-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Kim E.J., Lee S., Casel M.A.B. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wohlford-Lenane C., Perlman S., Zhao J., Jewell A.K., Reznikov L.R., Gibson-Corley K.N., Meyerholz D.K., McCray P.B., Jr. Middle East Respiratory Syndrome Coronavirus Causes Multiple Organ Damage and Lethal Disease in Mice Transgenic for Human Dipeptidyl Peptidase 4. J. Infect. Dis. 2016;213:712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wohlford-Lenane C.L., Channappanavar R., Park J.E., Earnest J.T., Bair T.B., Bates A.M., Brogden K.A., Flaherty H.A., Gallagher T. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. USA. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz D.K., Beck A.P. Principles and approaches for reproducible scoring of tissue stains in research. Lab. Invest. 2018;98:844–855. doi: 10.1038/s41374-018-0057-0. [DOI] [PubMed] [Google Scholar]
- Nabel G.J. Genetic, cellular and immune approaches to disease therapy: past and future. Nat. Med. 2004;10:135–141. doi: 10.1038/nm990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N., Iwata N., Hasegawa H., Fukushi S., Harashima A., Sato Y., Saijo M., Taguchi F., Morikawa S., Sata T. Mouse-passaged severe acute respiratory syndrome-associated coronavirus leads to lethal pulmonary edema and diffuse alveolar damage in adult but not young mice. Am. J. Pathol. 2008;172:1625–1637. doi: 10.2353/ajpath.2008.071060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal K.E., Coleman C.M., Mujica A.O., Kamat V., Badithe A., Fairhurst J., Hunt C., Strein J., Berrebi A., Sisk J.M. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA. 2015;112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L., Alphonse N., Dickenson R.E., Durbin J.E., Glenn J.S., Hartmann R., Kotenko S.V., Lazear H.M., O’Brien T.R., Odendall C. COVID-19 and emerging viral infections: The case for interferon lambda. J. Exp. Med. 2020;217:e20200653. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D., Schwartz K., Sherlock G., Dudoit S. GC-content normalization for RNA-Seq data. BMC Bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S.M., Wong H., McCartney-Francis N. Role of growth factors in inflammation and repair. J. Cell. Biochem. 1989;40:193–199. doi: 10.1002/jcb.240400208. [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57:450–459. [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020:eabc1560. doi: 10.1126/science.abc1560. Published online May 1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa N., Yoshikawa T., Hill T., Huang C., Watts D.M., Makino S., Milligan G., Chan T., Peters C.J., Tseng C.T. Differential virological and immunological outcome of severe acute respiratory syndrome coronavirus infection in susceptible and resistant transgenic mice expressing human angiotensin-converting enzyme 2. J. Virol. 2009;83:5451–5465. doi: 10.1128/JVI.02272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., Bao L., Deng W., Gao H., Xiang Z. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr., Baric R.S., Enjuanes L., Gallagher T. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The accession number for the Next-gen RNA sequence data reported in this paper is Gene Expression Omnibus (GEO) database: GSE150847; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150847.