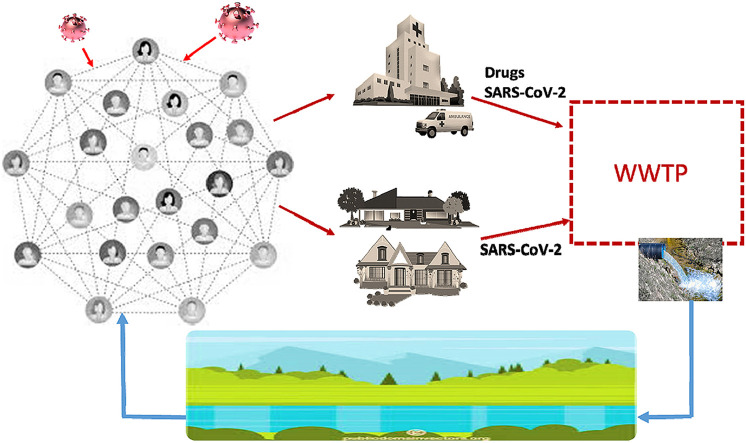
Abstract
The purpose of the present work is to provide a complete overview of possible direct/indirect implications on the quality of aquatic compartments due to the recent SARS-CoV-2 outbreak. With this aim, the environmental impacts are mainly related to i) the virus persistence in sewage and wastewaters, and ii) possible fate in aquatic compartments of drugs tested and administered to SARS-CoV-2 infected patients. Because SARS-CoV-2 spread is very recent, and there is a lack of specific studies on this strain, the virus persistence in wastewaters, the parameters influencing the persistence, as well as the detection methodologies are referenced to the general coronaviruses group. However, the present detailed report of up-to-date knowledge on this topic can provide a useful source for further studies focusing on more deepened investigations of SARS-CoV-2 behaviour in the environment. Such a perspective is significant not only for the control of virus diffusion but also represents a crucial point for the identification of produced alteration to the environmental quality.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Wastewater, Drugs persistence
Graphical abstract
Highlights
-
•
Current COVID-19 spread has important implications on the aquatic compartments.
-
•
Coronavirus persistence in wastewater can lead to further infection diffusion route.
-
•
Drugs for COVID-19 therapy in wastewater can cause negative environmental impact.
1. Introduction
At the end of December 2019, the first detection of SARS-CoV-2 virus occurred in Wuhan (Hubei Province, China) creating a big concern related to a possible outbreak (Zhu et al., 2020). The SARS-CoV-2 is a viral strain from a wide viruses group identified as coronaviruses in the sixties (Woo et al., 2010). The group includes strains such as MERS-CoV and SARS-CoV, which have been responsible for a widespread diffusion in the last 20 years (Lee and Hsueh, 2020; Lu et al., 2015). However, the current SARS-CoV-2 showed a wider and faster diffusion than the previous coronaviruses. In fact, its basic reproduction number (R0) is almost one order of magnitude higher compared to the others of the same group (R0-SARS-CoV-2>R0-SARS-CoV > R0-MERS-CoV) (Cao et al., 2020; Liu et al., 2020; WHO, 2003, 2019). Because of the SARS-CoV-2 worldwide diffusion, the World Health Organization (WHO) declared the state of pandemic on the last March 11, and each country all over the world adopted specific containment measures on the population and economic activities (Cucinotta and Vanelli, 2020; WHO, 2020a). It resulted in a global social restrictive action with no precedents. To date, the presence of the virus is confirmed in more than 200 countries with a number of confirmed infected individuals not far from 7.0 millions (WHO, 2020b). However, these numbers are likely to be much higher, as they do not include positive asymptomatic patients (Pan et al., 2020).
Because of their nature of respiratory pathogens, over the years the main concern about coronavirus strains transmission has been focused on aerosols and droplets, coming from infected individuals (Chen et al., 2020). Indeed aerosols and droplets control are primary means for the coronavirus spreading and play a key role for the prevention and containment of the contagion extension (Lai et al., 2020; Motta Zanin et al., 2020). Nonetheless, to fully control the outbreak, many researchers moved their attention on the potential correlation between the coronavirus spread and its survival in the environment outside the human host (Núñez-Delgado, 2020). In particular, different studies focused on the presence endurance of the virus in sewage and wastewaters identifying this environmental compartment as a potential mean of exposure to the contagion (Daughton, 2020; Reusken et al., 2020; Wigginton and Boehm, 2020). This concern is mainly due to the coronaviruses structure which is common to other viruses (such as H1N1 “Spanish flu”, avian influenza H5N1 and H7N9, SARS-CoV, MERS-CoV) detected in the stools and urine of infected individuals (Wigginton et al., 2015). For instance, Wang et al. (2005c) observed that SARS-CoV persistence in body fluids (stools and serum) was up to 96 h. Similarly some studies recently reported traces of the SARS-CoV-2 RNA in the wastewater of several European urban centres, during the current pandemic (Reusken et al., 2020; Wurtzer et al., 2020).
This is certainly the main aspect of the direct relationship existing between the current SARS-CoV-2 pandemic and the aquatic compartments (Venugopal et al., 2020; Zambrano-Monserrate et al., 2020). Nonetheless, there is an indirect relationship, which is worth highlighting too, related to the use of drugs administered to infectious individuals. In particular, several scientific studies are currently focusing on specific drugs already used for other medical diseases therapy in order to identify effective treatments, which could mitigate the dangerous effects of coronavirus infection (Guo et al., 2020). All these drugs are released, in massive amount, into the wastewater through the body fluids of the infected patients, potentially causing a dramatic alteration of the final aquatic receptor compartments and exposed biota (Richardson, 2012).
Therefore, the aim of the present manuscript is to provide a wide and detailed report about all the potential consequences of the current pandemic, which can be directly or indirectly exerted on the aquatic compartments. Accordingly, the manuscript focuses on three main sections analysing i) the correlations existing between the SARS-CoV-2 virus and the previously identified strains of the same coronavirus group; ii) the persistence characteristics of coronaviruses in wastewater, and the related methodologies for its detection; iii) the negative impact on water bodies, related to the release of therapeutic drugs for coronavirus infection treatments.
2. Genetics and virology
Coronaviruses are positive single stranded RNA viruses, having a diameter of about 60–140 nm (Bárcena et al., 2009; Cascella et al., 2020). They are named after their shape, observed at the electron microscope, which reminds a crown (in latin corona) because of the presence of spike proteins on the surface. Coronaviruses are common in many animal species and, in some cases, can be transmitted to humans, leading to zoonotic diseases (Su et al., 2016). Sequencing and phylogenetic analyses have shown that the novel SARS-CoV-2 is closely related (similarity rate of 88%) to a group of human SARS-like coronaviruses and bat SARS-related coronaviruses (bat-SL-CoVZC45, bat-SL-CoVZXC21).
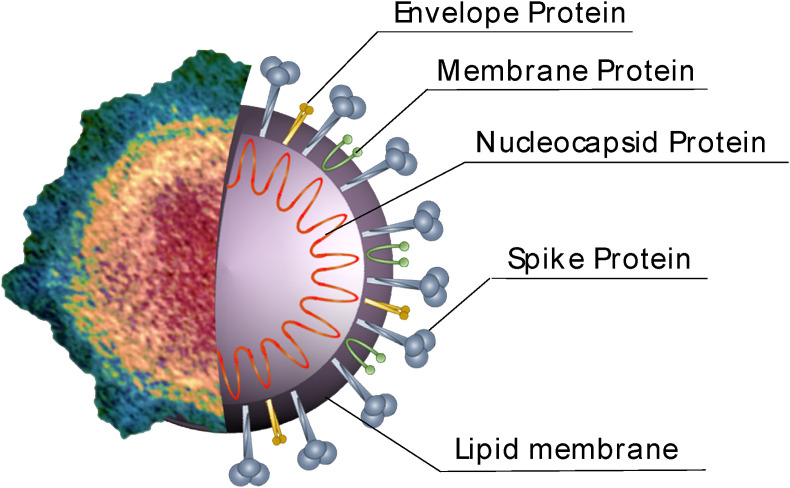
The viral composition of coronaviruses includes pericapsid envelope, which consists of a double layer of phospholipids and glycoproteins (Ashour et al., 2020). The several structural proteins (Fig. 1 ) are represented by the membrane (M), the envelope (E), the spike (S), and the nucleocapsid (N) one (Fehr and Perlman, 2015). The N protein is sited into the virus particle core and interacts with the virus RNA. The stabilization of this latter is due to the M protein, which is bound to the N protein. The structural M protein is the most abundant one and confers the shape to the virus (Nal et al., 2005). It forms the virion internal core, and together with the E protein, the mature viral envelopes. The amount of the E protein in the envelope is not high but, during the replication cycle, its presence inside the infected cells is reported to be remarkable. Moreover the E protein plays a fundamental role in the virus production (Siu et al., 2008). The S protein is responsible for the formation of homotrimeric spikes on the viral particle surface. It is a highly glycosylated protein, which mediates viral entry into the host cells. In certain coronaviruses, each S protein monomer can be present on viral particle as two subunits (i.e. S1 and S2). This is caused by the S protein separation occurring in the virus replication, due to the host furin-like proteases (Bosch et al., 2003; Izaguirre, 2019). In other strains, such as SARS-CoV, the S protein does not separate and forms the S1 and the S2 domains (Ashour et al., 2020; Xiao et al., 2003). In S1, the receptor-binding domain (RBD) is responsible for the mediation with the related host cell receptor while the S2 domain allows the fusion between the two cell membranes (host and viral) which is necessary for the entry of coronaviruses into the host cells (He et al., 2006).
Fig. 1.
Transmission electron microscope image (left side) and structural schematic representation (right side) of SARS-CoV-2.
It has been reported that some cellular receptors can be identified as coronaviruses receptor such as the case of angiotensin-converting enzyme 2 (ACE2) for the SARS-CoV (Wrapp et al., 2020). Moreover, Lu et al. (2020) observed similarities in the RBD between SARS-CoV and SARS-CoV-2. According to this, ACE2 could be a cellular receptor also for the new SARS-CoV-2 (Wrapp et al., 2020). ACE2 in humans is well expressed in the respiratory tract, so when the virus enters it is able to quickly replicate in the target cells, causing strong respiratory infections (Jia et al., 2006).
Also relevant, taking into account that cell entry of coronaviruses depends on binding of the viral S proteins to cellular receptors and on S protein priming by host cell proteases, Hoffmann et al. (2020) found that SARS-CoV-2 uses the SARS-CoV receptor ACE2 for entry and the serine protease TMPRSS2 for S protein priming.
3. Direct environmental impact on wastewater
3.1. Persistence of coronavirus in wastewaters
Because of the limited time span occurred from the beginning of the COVID-19 pandemic, very scarce research studies dealing with the presence of the SARS-CoV-2 in wastewaters are yet available (Lodder and de Roda Husman, 2020; Medema et al., 2020). However, the methodologies applied in the past for the detection of other strains of the same coronavirus group, such as virus RNA detection, Most Probable Number method, etc. can be considered applicable also for the present situation, and might turn out useful for monitoring the presence of the SARS-CoV-2 in wastewaters (Barcelo, 2020; C. C.G. Daughton, 2020; La Rosa et al., 2020; Orive et al., 2020).
Generally speaking, research studies concerning the fate of viruses in aquatic compartment, have been mainly focused on nonenveloped enteric viruses, as these viruses are characterized by higher resistance in various environmental conditions (Carducci et al., 2020). The number of studies concerning the fate of enveloped viruses in aquatic compartments, instead, is quite limited, as enveloped viruses are predisposed to deactivate in waters (Wigginton et al., 2015). Nonetheless, in the last 10–15 years, some authors have reported, the possible presence of enveloped viruses, such as coronaviruses in sewage systems (Cantalupo et al., 2011; Ikehata et al., 2009). Indeed, despite the faster inactivation rate of enveloped viruses compared to the nonenveloped ones, the survival time of enveloped viruses can be not negligible, according to the specific environmental conditions (Table 1 ). Main parameters affecting the potential survival of coronaviruses in wastewater are represented by temperature, relative humidity (Casanova et al., 2010; La Rosa et al., 2012) as well as suspended solids content and disinfection products in waters. For instance, Ye et al. (2016) observed a T90 (time for 90% virus titer decrease) equal to 7 h for enveloped Pseudomonas phage ɸ6 and equal to 13 h for murine hepatitis virus (MHV) in unpasteurized water at 25 °C. The value of T90 increased to 28 h for ɸ6 and to 36 h for MHV at 10 °C. On the contrary, Casanova and Weaver (2015) reported a faster inactivation of bacteriophage ɸ6 in sewage, increasing the temperature. In more details, the limit of virus detection occurred after 4 observation days at 30 °C, and after 10 observation days at 22 °C. In general, low temperatures and humidity can be favourable conditions for a prolonged persistence and more concerning viability of coronaviruses (Yeo et al., 2020). Comparing different waters (tap water filtered and unfiltered) or wastewaters (primary effluent filtered and unfiltered and secondary effluent), experimental results always indicated a decrease of coronavirus survival with increasing temperatures (Gundy et al., 2009) reporting a coronavirus T99.9 (time for 99.9% of virus titer decrease) growing from 10 to above 100 d with temperature decreasing from 23 to 4 °C in filtered tap water. Moreover, comparing filtered and unfiltered primary effluent from wastewater treatment plant at the same environmental temperature (23 °C), the same authors observed a higher T99.9 for the unfiltered wastewater than for the filtered one (Gundy et al., 2009). Such a result suggested that a higher solid content in water/wastewater samples can provide a higher protection to the virus, for its longer survival in the environment. This is expected considering the hydrophobicity of the coronavirus envelop, which leads to a lower virus solubility and to a potentially higher rate of virus adhesion onto solid particles (Gundy et al., 2009). Similar inactivation trend were observed for MHV and transmissible gastroenteritis virus (TGEV) in pasteurized settled sewage at two different temperatures (25 and 4 °C) (Casanova et al., 2009). A faster inactivation was observed for both viruses at 25 °C with a 99.99% of virus titer decrease (4log10 viral reduction kinetic) of 19 d for TGEV and 14 d for MHV. On the contrary, half of the time was required in order to achieve a 99% of virus titer decrease (2log10 viral reduction kinetic) for both viruses.
Table 1.
Information related to experimental conditions, investigated viruses, concentration/detection methods, and main results on virus persistence in wastewaters reported in literature studies.
| Experimental | Virus | Concentration method | Detection method | Virus persistence main results | Reference |
|---|---|---|---|---|---|
| 21 stool and urine samples collected from Xiao Tang Shan Hospital and 309th Hospital; sewage samples collected for 7 d before disinfection (2500 ml) and after disinfection (25,000-50000 ml) | SARS-CoV | Positively charged filter media particles | RT-PCR assay |
|
Wang et al. (2005a) |
| Sewage samples collected before disinfection (2500 ml) and after disinfection (25,000-50000 ml) from Xiao Tang Shan Hospital, 309th Hospital and housing estate | Bacteriophage f2 (as coronavirus model) and SARS-CoV | Positively charged filter media particles | RT-PCR assay |
|
Wang et al. (2005b) |
| Samples of stool (3) and urine (2) from Xiao Tang Shan Hospital; wastewater samples from 309th Hospital; sewage samples from housing estate; disinfection tests on wastewater with different chlorine (by dissolution of sodium hypochlorite) or chlorine dioxide concentration and disinfection time | Bacteriophage f2 (as coronavirus model) and SARS-CoV | – | RT-PCR assay |
|
Wang et al. (2005c) |
| Wastewater samples collected from wastewater treatment plant and pasteurized; comparison with reagent-grade and lake water; tests on temperature effect carried out at 23–25 °C and 4 °C | TGEV and MHV (as surrogates coronaviruses) | – | – |
|
Casanova et al. (2009) |
| Samples of unfiltered tap water tested at 23 °C and filtered tap water tested at 23 and 4 °C; samples of filtered and unfiltered primary effluent tested at 23 °C; samples of unfiltered secondary (activated sludge) effluent tested at 23 °C | Feline infectious peritonitis virus (FIPV), Human coronavirus 229 E (HCoV) and Poliovirus 1 LSc-2ab (PV-1) | – | Plaque assay or TCID50 |
|
Gundy et al. (2009) |
| Samples of wastewater collected and pasteurized at 70 °C for 3 h artificially spiked with enveloped virus surrogate; virus titer decrease tested at 22 and 30 °C | Bacteriophage ɸ6 (as surrogate of enveloped human viruses) | – | – |
|
Casanova and Weaver (2015) |
| Unpasteurized and pasteurized samples of wastewater from wastewater treatment plant artificially spiked and incubated at 10 and 25 °C for viruses survival tests; artificial spiking of untreated wastewater and centrifuged wastewater for solids removal incubated at 4 °C for viruses partitioning tests | MHV and Pseudomonas ɸ6 (as surrogates of enveloped human viruses); MS2 and T3 (as nonenveloped bacteriophages) | Polyethylene Glycol (PEG) precipitation method, ultracentrifugation method and ultrafiltration method | Plaque assay |
|
Ye et al. (2016) |
Further information concerning the survival of coronaviruses in aquatic environment can be found in the report of the World Health Organization (WHO, 2003), dealing with a significant SARS spreading occurred in a housing block of Hong Kong. According to a preliminarily investigation, in fact, the contagion was attributed to the contaminated air, flowing inside the apartments through the ventilation system located in the building bathrooms, having a flawed plumbing system. This hypothesis led to a pilot-scale investigation conducted with a model organism, Pseudomonas putida , aimed at assessing the potential virus transmission via building plumbing systems (Gormley et al., 2017). The experimental observations confirmed that the organisms spreading could have been favoured by the transportation through the ventilation system. Such an event allowed identifying the interconnectedness of a plumbing system and its conditions, as two fundamental factors to be monitored in order to prevent infection diffusion, especially in high risk location, such as hospitals (Gormley et al., 2020).
Strongly related to the high potential of sanitary structures to contribute to viruses spreading, are the two research studies conducted by Wang et al. (Wang et al., 2005a, 2005b). In these studies the RNA of SARS-CoV was isolated from patients’ stools, no RNA was found in their urine. Moreover, no live viruses were isolated from stools samples, suggesting the possibility of no infectious SARS-CoV excretion from digestive system of infected patients (Wang et al., 2005a). Also, the virus nucleic acid was mainly found in sewage samples collected before the disinfection treatment while less RNA occurrence was detected in disinfected sewage samples (Wang et al., 2005b). However, despite the RNA of SARS-CoV was detectable in sewage for 8 d, no active virus could be found (Wang et al., 2005b). Indeed, SARS-CoV virus showed a marked sensibility to inactivation due to disinfection products in wastewater. Experimental tests proved that both sodium hypochlorite and chlorine dioxide may exert an inactivation effect on the virus (Wang et al., 2005c). However, free chlorine from sodium hypochlorite dissolution resulted more efficient as inactivating agent, recommending its use as preferred disinfecting agent for hospital wastewater (Tsai and Lin, 1999; Wang et al., 2005c). It has to be highlighted that, besides chlorinated compounds, several other organic compounds, including alcohols (ethanol, and isopropanol), aldehydes (formaldehyde), and phenolic compounds (creosol soap) were found to be very efficient for some coronaviruses inactivation, such as MHV and canine coronavirus (Wolff et al., 2005). This generally highlights that an important approach to limit possible exposure to the virus infection could be represented by new disinfection technologies and upgrading of wastewater treatment plants for the remediation of wastewaters deriving from specific buildings (such as hospitals) (Naddeo and Liu, 2020).
3.2. Methodologies for detection of coronaviruses persistence in wastewater
Detection methods for viruses in the environment must be preceded by a suitable concentration technique especially in case of very low virus levels. Various concentration methods have been tested, each presenting some advantages and some disadvantages. Among others can be cited the methods based on adsorption-elution (negatively/positively charged filters, glass powder or fiber), precipitation (organic flocculation, ammonium sulphate precipitation), ultracentrifugation, ultrafiltration, and lyophilisation (Bosch et al., 2006).
Detection methods are often tested on artificially contaminated samples. A well-assessed practice includes the use of surrogate viruses. This practice is useful to overcome problem related to viruses which are not easily cultivable (Bosch et al., 2006). For instance, MHV and TGEV have been successfully used as surrogate viruses to identify the coronaviruses persistence at various ambient temperatures and in different water environments (Casanova et al., 2009).
Common traditional methods for virus determination are represented by plaque assay or, for viruses not forming plaques, by the 50% tissue culture infective dose (TCID50). In the plaque assay, it is possible to determine the viral titer in terms of plaque forming units (pfu). The assay is conducted using petri dishes, inoculating countable and statistically proper number of virus particles on a layer of immobilized cells (Cooper, 1962). The TCID50, in turns, allows to determine the sample dilution value corresponding to the occurrence of 50% cytopathic effect (CPE) (Gundy et al., 2009). Together with these traditional methods, nowadays more modern molecular techniques are frequently adopted. The most used ones are the polymerase chain reaction (PCR) and the reverse transcription polymerase chain reaction (RT-PCR). In particular, the RT-PCR is a technique used to transcript the RNA in a DNA chain through the reverse transcriptase enzyme and successively amplify the DNA fragment through the PCR technique in order to indirectly determine RNA species (Carter and Shieh, 2015). Real-time RT-PCR has been recently proposed as reliable technology to institute new diagnostic tests in the current pandemic emergency related to the SARS-CoV-2 (Corman et al., 2020). In this work, a validated diagnostic workflow has been suggested to detect the current coronavirus and methodology design/validation was allowed due to the genetic connection between the SARS-CoV-2 and the previous SARS-CoV.
Further methodology suggested for rapid and economic pathogens diagnosis is represented by paper-based devices (Magro et al., 2017). These devices are small and easily transportable analytical tools which can integrate various processes useful for tests on nucleic acid (from the extraction to the amplification and visual detection) (Mao et al., 2020). Paper-based devices could therefore potentially represent an useful tool for virus fast detection in wastewaters and fundamental monitoring system useable in emergency circumstances such as the current SARS-CoV-2 infection spreading.
4. Potential environmental impact of administered drugs
During an outbreak, the lack of information on effective antiviral drugs (AVs) or vaccines leads to nonspecific therapy for the minimization of the mortality rate. In the current pandemic, the existing drugs are being administered in much larger amount so representing an important threat to the quality of the receiving water bodies. Besides the treatment of disease due to coronaviruses, drugs are administered against a broad spectrum of viral infections such as HIV, herpes, hepatitis, Ebola and Malaria as well as autoimmune diseases such as lupus and rheumatoid arthritis (Babıć et al., 2017; Stebbing et al., 2020).
As part of clinical trials on drugs for the treatment of COVID-19 disease, the use of humanized monoclonal antibody like tocilizumab (TCZ) has been approved by FDA (Food and Drug Administration) for various therapies including those related to rheumatologic disease and lymphoproliferative disorder (Xu et al., 2020). TCZ is a recombinant monoclonal antibody against the interleukin-6 receptor (IL-6R) produced by recombinant DNA technology. IL-6R is a cytokine adopted in the development of immunological and inflammatory reactions. TCZ recognizes the IL-6 binding site on the cell membrane inhibiting the IL-6 transduction signalling (Kallen, 2002; Venkiteshwaran, 2009).
TCZ is considered a Protein and Peptide Therapeuticals (PPTs) not associated to environmental concern by the European Medicines Evaluation Agency (EMEA) guideline on environmental risk assessment (ERA) for human pharmaceuticals (EMEA, 2006). Although ecotoxicological data are not available, the half maximal effective concentration (EC50), as reported by the safety data sheet from the supplier, showed no TCZ adverse effects for concentrations higher than 100 ppm (Table 2 ). Biodegradability and acute ecotoxicity studies on TCZ report rapid biodegradability in sewage and surface waters as well as low ecotoxic characteristics (RCC Ltd, 2006a, 2006b; 2006c, 2006d; Straub, 2010).
Table 2.
Toxic effects of drugs used for the COVID-19 disease treatment on selected models and biomarkers.
| Compound | Organism | Species | Endpoint (exposure time) | EC50 (ppm) | Reference |
|---|---|---|---|---|---|
| Tocilizumab | Alga | Desmodesmus subspicatus | Growth rate inhibition (72 h) | >100 | Roche safety data sheet (2018) |
| Alga | Desmodesmus subspicatus | Biomass inhibition (72 h) | >100 | ||
| Crustacean | Daphnia magna | Immobility (48 h) | >100 | ||
| Fish | Danio rerio | Embryotoxicity (96 h) | >100 | ||
| Chloroquine | Bacteria | Aliivibrio fischeri | Bioluminescence Inhibition (24 h) | 132.1 | Zurita et al. (2005) |
| Alga | Chlorella vulgaris | Growth Inhibition (24 h) | 133.3 | ||
| Crustacean | Daphnia magna | Immobility (24 h) | 21.5 | ||
| Topminnow | PLHC-1 cell line | Protein content (24 h) | 158.3 | ||
| Basket willow | Salix viminalis | Relative transpiration (NRT) (117 h) (pH from 6 to 8) | 7–28 | Rendal et al. (2011) | |
| Crustacean | Daphnia magna | Immobility (48 h) (pH from 7 to 9) | 4–30 | ||
| Hydroxychloroquine | Alga | Raphidocelis subcapitata | Growth rate (72 h) | 3.1 | FASS safety data sheet (2019) |
| Crustacean | Daphnia magna | Immobility (48 h) | 14 |
The hydroxychloroquine (HCQ) and the chloroquine (CQ) represent another class of disease-modifying anti-rheumatic drug (DMARD) also added to the list of trial drugs in the guidelines for the diagnosis and treatment of COVID-19 (J. Liu et al., 2020). These drugs have also been used for years in antimalarial prevention. HCQ and CQ belong to the quinolone family and exert their action by blocking toll-like receptors (TLR) and reducing the activation of dendritic cells, with consequent mitigation of the inflammatory process.
The possible increasing release due to human excreta of HCQ and CQ in surface water through wastewater treatment plant effluents represents a concerning environmental issue. CQ and HCQ are highly soluble in water with partition coefficient octanol/water (log Kow) equal to 4.67 for HCQ and 3.03 for CQ. Moreover, available literature studies indicate that these drugs are only partially transformed inside the human body and can be almost completely excreted through urine and stools. It is reported that CQ is excreted unaltered at a percentage variable from 40 to 70% through kidney, and at a percentage variable from 5 to 10% through urine (Haładyj et al., 2018). Similarly, percentages of unaltered excreted HCQ range from 40 to 60% through kidney and from 8 to 25% through stools (Babıć et al., 2017).
CQ and HCQ can be also considered as persistent and/or bioaccumulative at high release extent in the environment potentially representing new emerging contaminants (Daughton, 2014; Howard and Muir, 2011; Zurita et al., 2005). Nevertheless, data on CQ and HCQ concentrations in the environment are very scarce. Chen et al. (2013) reported the CQ and HCQ detection in surface sediments near three rivers in southeast China. Similarly, Olaitan et al. (2017) found CQ in wastewater effluents in Nigeria (Olaitan et al., 2017). The general findings (Table 2) confirmed that CQ and HCQ compounds could be risky for the environment and should be classified as harmful to aquatic organisms (Ramesh et al., 2018; Zurita et al., 2005).
From an environmental perspective, different considerations should be made for the AVs used in COVID-19 disease therapies. The real concern related to AVs use and their environmental persistence, in analogy to the development of antibiotic resistance, is the potential formation of resistant strains through chronic exposure. This could consequently entail more adverse effects to human health than other classes of drugs (Jain et al., 2013). Moreover, further drawbacks are represented by AVs low biodegradability and their increasing use during pandemic outbreaks (Funke et al., 2016; Hill et al., 2014; Russo et al., 2017).
Most of the AVs are excreted as unchanged parent compounds with highly bioactive characteristics which are resistant to conventional treatments in wastewater treatment plants. Moreover, they can react with organic and inorganic constituents during wastewater treatment and can be transformed in additional molecules characterized by higher persistence (Funke et al., 2016; Jain et al., 2013). Despite no effective AV has been specifically approved for the treatment of COVID-19 disease, recent studies are focusing on the possible use of Lopinavir and Remdesivir (Grein et al., 2020). Regarding the Lopinavir, the environmental risk assessment in hospital effluents has been evaluated through the determination of the Predicted Environmental Concentration (PEC) and the Predicted No-Effect Concentration (PNEC) (Acree and Grubbs, 2012). The result from the risk assessment showed that PEC value of Lopinavir was higher than its PNEC value (0.26 and 0.05 ppb, respectively) therefore indicating a potential environmental harm. Moreover, the Lopinavir was listed among the top ten ranked active pharmaceutical ingredients (API), mainly due to its high bioaccumulation potential (log Kow>3.9) (Daouk et al., 2015).
The Remdesivir is a nucleoside analog, which incorporates into nascent viral RNA chains and inhibits viral RNA polymerases. This AV has broad-spectrum activity against members of the filoviruses, coronaviruses, and paramyxoviruses. The EMEA has recommended for compassionate use of the Remdesivir although information on the related environmental risk (ecotoxicity and degradability in the environment) are lacking. According to this, further researches are necessary to assess the magnitude of the environmental risk posed by the Remdesivir.
5. Conclusions
Since the beginning of globalization era, COVID-19 disease has been the first pandemic characterized by such a wide and significantly fast spread. This global emergency took all the world countries unawares. The occurrence of the infection spread also in poorly industrialized areas with limited resources for epidemic containment and healthcare systems represents an even more concerning issue.
In this context, the scientific community should not only pay close attention to the health aspect but also inevitably consider the environmental one. In fact, fundamental importance should be given to further deepened studies aimed at identifying accurate monitoring and analysis systems for prompt detection of potential viruses diffusion through aquatic media. An additional environmental element to be taken into account is related to the consumption of drugs for the coronavirus related disease therapy, which could lead to risky release of toxic substances in the receiving water bodies. To date, the removal efficiencies of these new contaminants from wastewaters through feasible treatments have been poorly investigated. Therefore, future researches should focus on the environmental fate of these contaminants, and should evaluate the effectiveness of tertiary treatments (such as advanced oxidation processes) on their removal.
Credit author statement
M. Race: Conceptualization, Supervision, Writing - original draft. A. Ferrraro: Conceptualization, Writing - original draft, Writing- Reviewing and Editing. E. Galdiero: Supervision, Writing - original draft. M. Guida: Supervision. A. Núñez-Delgado: Writing- Reviewing and Editing. F. Pirozzi: Funding acquisition, Supervision. A. Siciliano: Writing - original draft. M. Fabbricino: Project administration, Writing- Reviewing and Editing
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Dr. Ferraro would like to thank the Italian Ministry of Education, University and Research (MIUR), which provided financial support for a 12 months post-doctoral grant in the framework of the research project entitled “Dipartimenti di Eccellenza” per Ingegneria Civile, Edile e Ambientale – CUP E65D18000820006.
References
- Acree W.E., Grubbs L.M. Prediction of toxicity, sensory responses and biological responses with the Abraham model. Toxic. Drug Test. 2012:261–296. [Google Scholar]
- Ashour H.M., Elkhatib W.F., Rahman M., Elshabrawy H.A. Insights into the recent 2019 novel Coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens. 2020;9:186. doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babıć S., Dabıć D., Ćurkovıć L. 2017. Fate of Hydroxychloroquine in the Aquatic Environment. [Google Scholar]
- Barcelo D. An environmental and health perspective for COVID-19 outbreak: meteorology and air quality influence, sewage epidemiology indicator, hospitals disinfection, drug therapies and recommendations. J. Environ. Chem. Eng. 2020:104006. doi: 10.1016/j.jece.2020.104006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena M., Oostergetel G.T., Bartelink W., Faas F.G.A., Verkleij A., Rottier P.J.M., Koster A.J., Bosch B.J. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A., Pintó R.M., Abad F.X. Survival and transport of enteric viruses in the environment. In: Goyal S.M., editor. Viruses in Foods. Food Microbiology and Food Safety. Springer; Boston, MA: 2006. pp. 151–187. [DOI] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo P.G., Calgua B., Zhao G., Hundesa A., Wier A.D., Katz J.P., Grabe M., Hendrix R.W., Girones R., Wang D., Pipas J.M. Raw sewage harbors diverse viral populations. mBio. 2011;2 doi: 10.1128/mBio.00180-11. e00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Zhang Q., Lu X., Pfeiffer D., Jia Z., Song H., Zeng D.D. medRxiv; 2020. Estimating the Effective Reproduction Number of the 2019-nCoV in China. [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Verani M. Making Waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179:115907. doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M., Shieh J. Guide to Research Techniques in Neuroscience. Academic Press; 2015. Molecular cloning and recombinant DNA technology; pp. 219–237. [DOI] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Jeon S., Rutala W.A., Weber D.J., Sobsey M.D. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl. Environ. Microbiol. 2010;76:2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environ. Sci. Technol. Lett. 2015;2:76–78. doi: 10.1021/acs.estlett.5b00029. [DOI] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls [Internet] StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- Chen V., Ortube M.C., Nusinowitz S., Gorin M. Diagnostic disparities in testing for hydroxychloroquine (HCQ)-related ocular toxicity. Invest. Ophthalmol. Vis. Sci. 2013;54:1564. [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P.D. Advances in Virus Research. Academic Press; 1962. The plaque assay of animal viruses; pp. 319–378. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Bio-medica Atenei Parm. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daouk S., Chevre N., Vernaz N., Bonnabry P., Dayer P., Daali Y., Fleury-Souverain S. Prioritization methodology for the monitoring of active pharmaceutical ingredients in hospital effluents. J. Environ. Manag. 2015;160:324–332. doi: 10.1016/j.jenvman.2015.06.037. [DOI] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138149. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020:139631. doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. The Matthew Effect and widely prescribed pharmaceuticals lacking environmental monitoring: case study of an exposure-assessment vulnerability. Sci. Total Environ. 2014;466:315–325. doi: 10.1016/j.scitotenv.2013.06.111. [DOI] [PubMed] [Google Scholar]
- EMEA Note for guidance on environmental risk assessment of medicinal products for human use. Comm. Propr. Med. Prod. London. 2006 CPMP/SWP/4447/00 corr 1. [Google Scholar]
- FASS safety data sheet . 2019. Environmental Risk Assessment Summary Plaquenil. [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses. Springer; 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke J., Prasse C., Ternes T.A. Identification of transformation products of antiviral drugs formed during biological wastewater treatment and their occurrence in the urban water cycle. Water Res. 2016;98:75–83. doi: 10.1016/j.watres.2016.03.045. [DOI] [PubMed] [Google Scholar]
- Gormley M., Aspray T.J., Kelly D.A. COVID-19: mitigating transmission via wastewater plumbing systems. Lancet Glob. Heal. 2020;8 doi: 10.1016/S2214-109X(20)30112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley M., Aspray T.J., Kelly D.A., Rodriguez-Gil C. Pathogen cross-transmission via building sanitary plumbing systems in a full scale pilot test-rig. PloS One. 2017;12 doi: 10.1371/journal.pone.0171556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007016. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haładyj E., Sikora M., Felis-Giemza A., Olesińska M. Antimalarials–are they effective and safe in rheumatic diseases? Reumatologia. 2018;56:164. doi: 10.5114/reum.2018.76904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li J., Du L., Yan X., Hu G., Zhou Y., Jiang S. Identification and characterization of novel neutralizing epitopes in the receptor-binding domain of SARS-CoV spike protein: revealing the critical antigenic determinants in inactivated SARS-CoV vaccine. Vaccine. 2006;24:5498–5508. doi: 10.1016/j.vaccine.2006.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A., Khoo S., Fortunak J., Simmons B., Ford N. Minimum costs for producing hepatitis C direct-acting antivirals for use in large-scale treatment access programs in developing countries. Clin. Infect. Dis. 2014;58:928–936. doi: 10.1093/cid/ciu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard P.H., Muir D.C.G. Identifying new persistent and bioaccumulative organics among chemicals in commerce II: pharmaceuticals. Environ. Sci. Technol. 2011;45:6938–6946. doi: 10.1021/es201196x. [DOI] [PubMed] [Google Scholar]
- Ikehata K., Liu Y., Sun R. Health effects associated with wastewater treatment, reuse, and disposal. Water Environ. Res. 2009;81:2126–2146. doi: 10.2175/106143009X12445568400773. [DOI] [PubMed] [Google Scholar]
- Izaguirre G. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses. 2019;11:837. doi: 10.3390/v11090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Kumar P., Vyas R.K., Pandit P., Dalai A.K. Occurrence and removal of antiviral drugs in environment: a review. Water, Air. Soil Pollut. 2013;224:1410. [Google Scholar]
- Jia H.P., Look D.C., Hickey M., Shi L., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B. The Nidoviruses. Springer; 2006. Infection of human airway epithelia by SARS coronavirus is associated with ACE2 expression and localization; pp. 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen K.-J. The role of transsignalling via the agonistic soluble IL-6 receptor in human diseases. Biochim. Biophys. Acta Mol. Cell Res. 2002;1592:323–343. doi: 10.1016/s0167-4889(02)00325-7. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179:115899. doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Fratini M., della Libera S., Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super Sanita. 2012;48:397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.-I., Hsueh P.-R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020;53:365–367. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Trav. Med. 2020;27 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro L., Escadafal C., Garneret P., Jacquelin B., Kwasiborski A., Manuguerra J.-C., Monti F., Sakuntabhai A., Vanhomwegen J., Lafayee P., Tabeling P. Paper microfluidics for nucleic acid amplification testing (NAAT) of infectious diseases. Lab Chip. 2017;17:2347–2371. doi: 10.1039/C7LC00013H. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. MedRxiv. 2020 doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Motta Zanin G., Gentile E., Parisi A., Spasiano D. A preliminary evaluation of the public risk perception related to the COVID-19 health emergency in Italy. Int. J. Environ. Res. Publ. Health. 2020;17:3024. doi: 10.3390/ijerph17093024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6:1213–1216. doi: 10.1039/D0EW90015J. [DOI] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S. M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Núñez-Delgado A. What do we know about the SARS-CoV-2 coronavirus in the environment? Sci. Total Environ. 2020:138647. doi: 10.1016/j.scitotenv.2020.138647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan O.J., Okunuga Y.O., Kasim L.S., Chimezie A., Oderinde O. Determination of selected antimalarial pharmaceuticals in water from two hospital environments in abeokuta ogun state-Nigeria using SPE-LC. African J. Sci. Nat. 2017;3:50–56. [Google Scholar]
- Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020:139298. doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Chen D., Xia Y., Wu X., Li T., Ou X., Zhou L., Liu J. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect. Dis. 2020;20:410–411. doi: 10.1016/S1473-3099(20)30114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh M., Anitha S., Poopal R.K., Shobana C. Evaluation of acute and sublethal effects of chloroquine (C18H26CIN3) on certain enzymological and histopathological biomarker responses of a freshwater fish Cyprinus carpio. Toxicol. reports. 2018;5:18–27. doi: 10.1016/j.toxrep.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RCC Ltd . Ready biodegradability in a manometric respirometry test. RCC study no. A47327. In: Hoffmann F., editor. Formulated Actemra. La Roche Ltd; Basel, Switzerland: 2006. [Google Scholar]
- RCC Ltd . Toxicity to Scenedesmus subspicatus in a 72-hour algal growth inhibition test, limit test with 100 mg Tocilizumab/l. RCC study no. A47338. In: Hoffmann F., editor. Formulated Actemra. La Roche Ltd; Basel, Switzerland: 2006. [Google Scholar]
- RCC Ltd . Acute toxicity to Daphnia magna in a 48-hour immobilization test, limit test with 100 mg Tocilizumab/l. RCC study no. A47340. In: Hoffmann F., editor. Formulated Actemra. La Roche Ltd; Basel, Switzerland: 2006. [Google Scholar]
- RCC Ltd . Acute toxicity to zebra fish (Brachydanio rerio) in a 96-hour static test; limit test with 100 mg Tocilizumab/l. RCC study no. A47351. In: Hoffmann F., editor. Formulated Actemra. La Roche Ltd; Basel, Switzerland: 2006. [Google Scholar]
- Rendal C., Kusk K.O., Trapp S. The effect of pH on the uptake and toxicity of the bivalent weak base chloroquine tested on Salix viminalis and Daphnia magna. Environ. Toxicol. Chem. 2011;30:354–359. doi: 10.1002/etc.391. [DOI] [PubMed] [Google Scholar]
- Reusken C.B., Buiting A., Bleeker-Rovers C., Diederen B., Hooiveld M., Friesema I., Koopmans M., Kortbeek T., Lutgens S.P.M., Meijer A. Rapid assessment of regional SARS-CoV-2 community transmission through a convenience sample of healthcare workers, The Netherlands, March 2020. Euro Surveill. 2020;25:2000334. doi: 10.2807/1560-7917.ES.2020.25.12.2000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.D. Environmental mass spectrometry: emerging contaminants and current issues. Anal. Chem. 2012;84:747–778. doi: 10.1021/ac202903d. [DOI] [PubMed] [Google Scholar]
- Roche safety data sheet . In: Environmental Risk Assessment Summary Tocilizumab. Hoffmann F., editor. La Roche Ltd; 2018. [Google Scholar]
- Russo D., Siciliano A., Guida M., Galdiero E., Amoresano A., Andreozzi R., Reis N.M., Puma G.L., Marotta R. Photodegradation and ecotoxicology of acyclovir in water under UV254 and UV254/H2O2 processes. Water Res. 2017;122:591–602. doi: 10.1016/j.watres.2017.06.020. [DOI] [PubMed] [Google Scholar]
- Siu Y.L., Teoh K.T., Lo J., Chan C.M., Kien F., Escriou N., Tsao S.W., Nicholls J.M., Altmeyer R., Peiris J.S.M. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub J.O. Green and Sustainable Pharmacy. Springer; 2010. Protein and peptide therapeuticals: an example of “Benign by Nature” active pharmaceutical ingredients; pp. 127–133. [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.T., Lin S.T. Disinfection of hospital waste sludge using hypochlorite and chlorine dioxide. J. Appl. Microbiol. 1999;86:827–833. doi: 10.1046/j.1365-2672.1999.00732.x. [DOI] [PubMed] [Google Scholar]
- Venkiteshwaran A. MAbs. Taylor & Francis; 2009. Tocilizumab; pp. 432–438. [Google Scholar]
- Venugopal A., Ganesan H., Raja S.S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P.K.S.M., Gopalakrishnan A.V., Siama Z. Novel wastewater surveillance strategy for early detection of COVID–19 hotspots. Curr. Opin. Environ. Sci. Heal. 2020;17:8–13. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Guo T.-K., Zhen B., Kong Q.-X., Yi B., Li Z., Song N., Jin M., Wu X.-M., Xiao W.-J., Zhu X.-M., Gu C.-Q., Yin J., Wei W., Yao W., Liu C., Li J.-F., Ou G.-R., Wang M.-N., Fang T.-Y., Wang G.-J., Qiu Y.-H., Wu H.-H., Chao F.-H., Li J.-W. Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World J. Gastroenterol. 2005;11:4390–4395. doi: 10.3748/wjg.v11.i28.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Guo T.-K., Zhen B., Kong Q.-X., Yi B., Li Z., Song N., Jin M., Xiao W.-J., Zhu X.-M., Gu C.-Q., Yin J., Wei W., Yao W., Liu C., Li J.-F., Ou G.-R., Wang M.-N., Fang T.-Y., Wang G.-J., Qiu Y.-H., Wu H.-H., Chao F.-H., Li J.-W. Concentration and detection of SARS coronavirus in sewage from Xiao tang Shan hospital and the 309th hospital. J. Virol. Methods. 2005;128:156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.-J., Yin J., Wei W., Wang G.-J., Si B., Guo B.-Z., Liu C., Ou G.-R., Wang M.-N., Fang T.-Y., Chao F.-H., Li J.-W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. World Health Organization. WHO Announces COVID-19 Outbreak a Pandemic.http://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic [WWW Document] [Google Scholar]
- WHO . 2020. Coronavirus Disease 2019 (COVID-19) Situation Report – 87.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ [WWW Document] [Google Scholar]
- WHO . World Health Organization; 2019. WHO MERS Global Summary and Assessment of Risk, July 2019. [Google Scholar]
- WHO . World Health Organization; 2003. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS)https://apps.who.int/iris/handle/10665/70863 [Google Scholar]
- Wigginton K.R., Boehm A.B. Environmental engineers and scientists have important roles to play in stemming outbreaks and pandemics caused by enveloped viruses. Environ. Sci. Technol. 2020;54:3736–3739. doi: 10.1021/acs.est.0c01476. [DOI] [PubMed] [Google Scholar]
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1:735–746. doi: 10.1039/C5EW00125K. [DOI] [Google Scholar]
- Wolff M.H., Sattar S.A., Adegbunrin O., Tetro J. Environmental survival and microbicide inactivation of coronaviruses. In: Schmidt A., Weber O., Wolff M.H., editors. Coronaviruses with Special Emphasis on First Insights Concerning SARS. Birkhäuser Advances in Infectious Diseases BAID; Birkhäuser Basel: 2005. pp. 201–212. [DOI] [Google Scholar]
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80-. ) 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. medRxiv; 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics through Viral Genome Quantification in Paris Wastewaters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X. Effective treatment of severe COVID-19 patients with tocilizumab. China. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano-Monserrate M.A., Ruano M.A., Sanchez-Alcalde L. Indirect effects of COVID-19 on the environment. Sci. Total Environ. 2020:138813. doi: 10.1016/j.scitotenv.2020.138813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita J.L., Jos Á., del Peso A., Salguero M., López-Artíguez M., Repetto G. Ecotoxicological evaluation of the antimalarial drug chloroquine. Aquat. Toxicol. 2005;75:97–107. doi: 10.1016/j.aquatox.2005.07.009. [DOI] [PubMed] [Google Scholar]