Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a pandemic with millions of human infections. One limitation to the evaluation of potential therapies and vaccines to inhibit SARS-CoV-2 infection and ameliorate disease is the lack of susceptible small animals in large numbers. Commercially available laboratory strains of mice are not readily infected by SARS-CoV-2 because of species-specific differences in their angiotensin-converting enzyme 2 (ACE2) receptors. Here, we transduced replication-defective adenoviruses encoding human ACE2 via intranasal administration into BALB/c mice and established receptor expression in lung tissues. hACE2-transduced mice were productively infected with SARS-CoV-2, and this resulted in high viral titers in the lung, lung pathology, and weight loss. Passive transfer of a neutralizing monoclonal antibody reduced viral burden in the lung and mitigated inflammation and weight loss. The development of an accessible mouse model of SARS-CoV-2 infection and pathogenesis will expedite the testing and deployment of therapeutics and vaccines.
Keywords: SARS-CoV-2, coronavirus, COVID-19, pathogenesis, antibody, pneumonia, animal model, mice, inflammation
Graphical Abstract
Highlights
-
•
Adenovirus transduction of human ACE2 enables SARS-CoV-2 infection of BALB/c mice
-
•
High levels of viral RNA and infectious SARS-CoV-2 accumulate in lungs
-
•
Mice transduced with human ACE2 develop viral pneumonia after SARS-CoV-2 infection
-
•
Neutralizing mAbs protect from SARS-CoV-2-induced lung infection and inflammation
Laboratory mice transduced with adenoviruses encoding human ACE2 are permissive for SARS-CoV-2 and develop pneumonia. Passive transfer of a neutralizing monoclonal antibody reduces lung infection, inflammation, and disease.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense single-stranded RNA virus that was first isolated in Wuhan, China in December 2019 from a cluster of acute respiratory illness cases (Zhu et al., 2020). SARS-CoV-2 is related closely to other highly pathogenic betacoronoaviruses that emerged this century—SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV). SARS-CoV-2 is the cause of the coronavirus disease 2019 (COVID-19) pandemic that has infected millions of people worldwide, resulted in hundreds of thousands of deaths, and engendered severe and destabilizing global economic hardship. COVID-19 is characterized by fever, cough, and shortness of breath that can progress rapidly to respiratory and cardiac failure requiring mechanical ventilation (Guan et al., 2020). The elderly, immunocompromised, and those with co-morbid metabolic, pulmonary, and cardiac conditions are at greater risk of death from COVID-19 (Zhou et al., 2020). Most human infections occur after respiratory droplet exposure, with community transmission in asymptomatic or pauci-symptomatic individuals contributing to local and global spread (Day, 2020, Li et al., 2020).
Coronaviruses are spherical virions in which the viral spike (S) protein forms a characteristic crown on the virion surface. Three additional viral structural proteins (envelope [E], membrane [M], and nucleocapsid [N]) together with a lipid membrane and ∼25–31 kilobase viral genome comprise the virion. The S protein exists on virions as homotrimeric spikes that promote coronavirus entry into cells via attachment and membrane fusion. Both SARS-CoV and SARS-CoV-2 S proteins engage the human angiotensin-converting enzyme 2 (hACE2) as a cellular receptor for entry and infection (Hoffmann et al., 2020). After receptor engagement, SARS-CoV-2 S protein is enzymatically processed by a plasma membrane-associated serine protease, TMPRSS2, which is essential for efficient fusion and release of the virus contents into the host cell cytosol (Hoffmann et al., 2020, Matsuyama et al., 2020).
The rapid emergence of this highly pathogenic and readily transmissible SARS-CoV-2 has created an urgent need for the immediate development and deployment of antiviral agents and protective vaccines. Remdesivir, a direct-acting antiviral polymerase inhibitor with activity against coronaviruses in mice (Sheahan et al., 2017), was recently approved in the United States under an emergency order for human use against SARS-CoV-2 (Ledford, 2020), although an initial randomized, double-blinded trial in China did not show substantive benefit (Wang et al., 2020b). Several other repurposed drugs and antibody product-based agents are under evaluation in human studies (Sanders et al., 2020), Additionally, vaccine trials in humans with mRNA, DNA, and viral-vectored platforms have been initiated without published efficacy data in small or large animal models (Amanat and Krammer, 2020). The development of a high-throughput small animal model of SARS-CoV-2 infection could expedite testing of drugs and vaccines and enable down-selection before more costly evaluation in non-human primate and human clinical trials. Indeed, despite much anticipation (Jiang et al., 2020), neutralizing monoclonal antibodies against SARS-CoV-2 (Wang et al., 2020a) have not yet been demonstrated to have protective activity in any preclinical model.
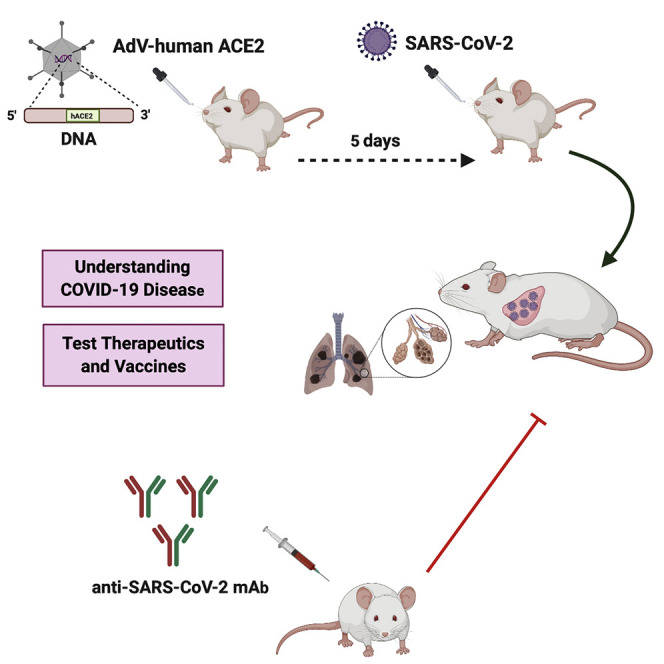
Reportedly, laboratory strains of mice are not readily infected by SARS-CoV-2 because hACE2 supports SARS-CoV-2 binding but mouse ACE2 does not (Letko et al., 2020, Wan et al., 2020). Although transgenic mice expressing hACE2 (hACE2-Tg) were developed for SARS-CoV research (McCray et al., 2007, Netland et al., 2008) and develop interstitial pneumonia after SARS-CoV-2 infection (Bao et al., 2020), they are not widely available for high-throughput drug and vaccine testing. Here, we transiently transduced a replication-defective adenovirus encoding hACE2 into the lungs of commercially available mice, which sensitized them to productive SARS-CoV-2 infection and pneumonia. In addition to creating an accessible model for studying SARS-CoV-2 pathogenesis in the lung, we established that systemic delivery of a neutralizing monoclonal antibody mitigates viral infection, inflammation, and disease.
Results and Discussion
Based on prior studies with the closely related SARS-CoV betacoronavirus (Hogan et al., 2004), we initially hypothesized that SARS-CoV-2 might infect and cause limited disease in immunocompromised laboratory strains of mice. To test this idea, we propagated SARS-CoV-2 (strain 2019n-CoV/USA_WA1/2020) in Vero CCL-81 cells and inoculated 3-to-4-week-old BALB/c, DBA/2J, Stat1 −/− C57BL/6, AG129 (type I and II interferon [IFN] receptor-deficient), and Rag1 −/− C57BL/6 (no mature B and T cells) mice with 105 focus-forming units (FFUs) of SARS-CoV-2 via an intranasal route. None of the mice tested had weight loss over the first week, and lungs harvested at 10 days post-infection (dpi) showed very low levels of viral RNA (Figures 1A and 1B). The observation that SARS-CoV-2 does not replicate efficiently in laboratory strains of mice is similar to that seen with MERS-CoV (Coleman et al., 2014), which was because of an inability to use the murine ortholog of the MERS-CoV receptor, dipeptidyl peptidase 4 (DPP4) (Raj et al., 2013). Given that human but not mouse ACE2 can act as a cellular receptor for SARS-CoV-2 in vitro (Hoffmann et al., 2020, Letko et al., 2020, Wan et al., 2020), our findings in conventional laboratory mice were anticipated and indeed recently reported by others (Bao et al., 2020).
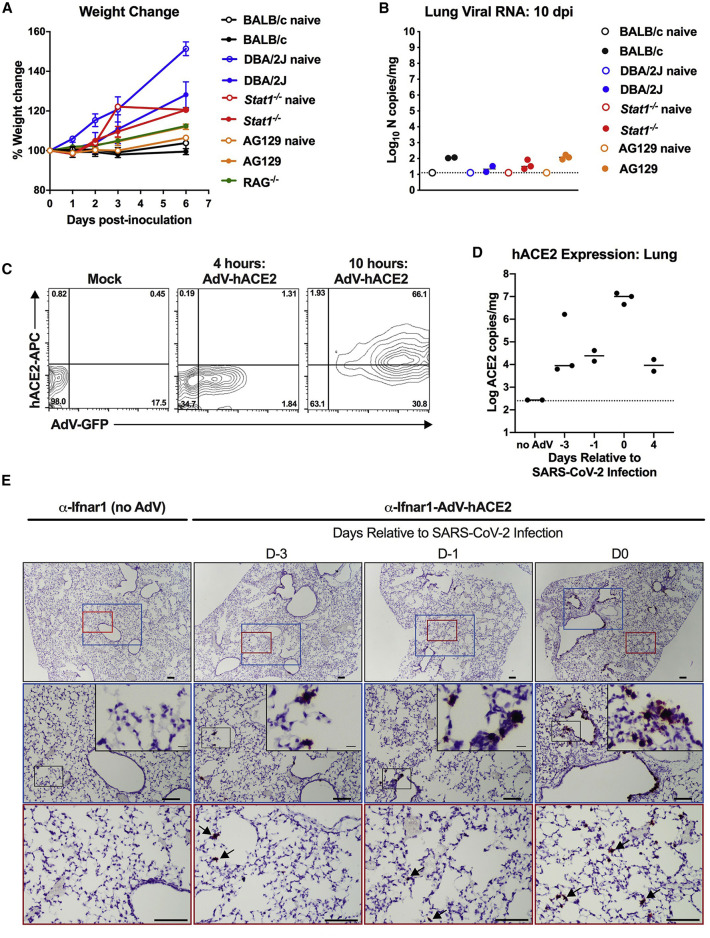
Figure 1.
SARS-CoV-2 Infection in Conventional Laboratory Strains of Mice and Expression of hACE2 after AdV Transduction
(A and B) 3-to-4-week-old BALB/c, DBA/2J, Stat1−/− C57BL/6, Rag1−/− C57BL/6, and AG129 mice were inoculated via combined intranasal and intravenous routes with 105 focus-forming units (FFU) of SARS-CoV-2. Weight change (A) was monitored, and viral burden (B) in the lung was determined at 10 dpi by RT-qPCR and expressed as copies of viral N gene per mg of tissue (n = 2 to 3 for each strain). Naive mice are shown as a control. Error bars indicate standard deviations (SD).
(C) HEK293 cells were transduced with AdV-hACE2-GFP. Flow cytometric analysis of hACE2 and GFP expression was analyzed at 4 and 10 h after AdV transduction. One of two experiments is shown.
(D) mRNA expression levels of hACE2 in the lungs of mice receiving the AdV-hACE2 at D-3, D-1, D0, and D4 relative to SARS-CoV-2 infection (2, 4, 5, and 8 days after AdV delivery) via intranasal route as measured by species-specific qRT-PCR of hACE2. Bars indicate median values.
(E) In situ hybridization using probes for hACE2 in lungs of control mice receiving anti-Ifnar1 mAb (left) or at D-3, D-1, and D0 relative to SARS-CoV-2 infection (2, 4, or 5 days post-transduction of mice receiving anti-Ifnar1 + AdV-hACE2) (right). Images show low-power (top; scale bars, 100 μm) and medium-power (middle [blue] and bottom [red]; scale bars, 100 μm) magnifications with an additional high-power magnification inset (scale bars, 10 μm). Arrows indicate hACE2-positive cells in medium-power magnification (representative images from n = 3 per group).
Mice transgenic for the expression of hACE2 are vulnerable to SARS-CoV, with infection of lungs observed after intranasal inoculation (McCray et al., 2007, Menachery et al., 2016). However, SARS-CoV also enters the brain in these hACE2-transgenic mice, and infection results in transneuronal spread and ultimately death due to central nervous system injury and dysfunction (Menachery et al., 2016, Netland et al., 2008). Because these hACE2-Tg mice are not yet widely available for drug and vaccine testing, we evaluated an alternative strategy in which hACE2 is transiently expressed following transduction with an adenoviral vector, akin to an approach that rendered mice susceptible to MERS-CoV infection by introducing DPP4 (Zhao et al., 2014). 9-week-old BALB/c mice were inoculated via an intranasal route with 2.5 × 108 plaque-forming units (PFUs) of a replication-defective adenovirus encoding for hACE2 (hACE2-expressing human Ad5, AdV-hACE2) (Figure 1C). Some of the animals received an anti-Ifnar1 monoclonal antibody (mAb MAR1-5A3, 2 mg via intraperitoneal injection) to transiently inhibit type I IFN signaling and possibly enhance SARS-CoV-2 infection. We detected hACE2 mRNA by qRT-PCR (Figure 1D) and by in situ hybridization in cells of the lungs including bronchiolar epithelial cells and pneumocytes (Figure 1E) with peak mRNA expression occurring about five days after administration.
In an attempt to maximize infectivity and pathogenesis, BALB/c mice were inoculated initially via intranasal and intravenous routes with 105 FFUs of SARS-CoV-2 5 days after AdV-hACE2 transduction (Figure 2 A). In contrast to mice transduced only with AdV-hACE2, animals administered AdV-hACE2 and inoculated with SARS-CoV-2 had weight loss during the first week (10%–25%, Figure 2B). More weight loss was observed in the SARS-CoV-2-infected mice that also received anti-Ifnar1 mAb treatment. High levels of SARS-CoV-2 infectious virus (by plaque assay) and viral RNA were detected in lung tissue homogenates at 4 dpi, whereas lower levels were present in other tissues (e.g., heart, spleen, and brain) and virtually none was measured in kidney, gastrointestinal tract tissues, or in serum (Figures 2C and 2D). These differences in tissue distribution of SARS-CoV-2 infection likely relate both to the delivery and expression of the AdV-hACE2 and the natural tropism of the virus. Virus inoculation by a systemic route was not required, as we did not observe substantive differences in lung infection when SARS-CoV-2 was administered via intranasal only versus combined intranasal and intravenous routes (Figure 2E).
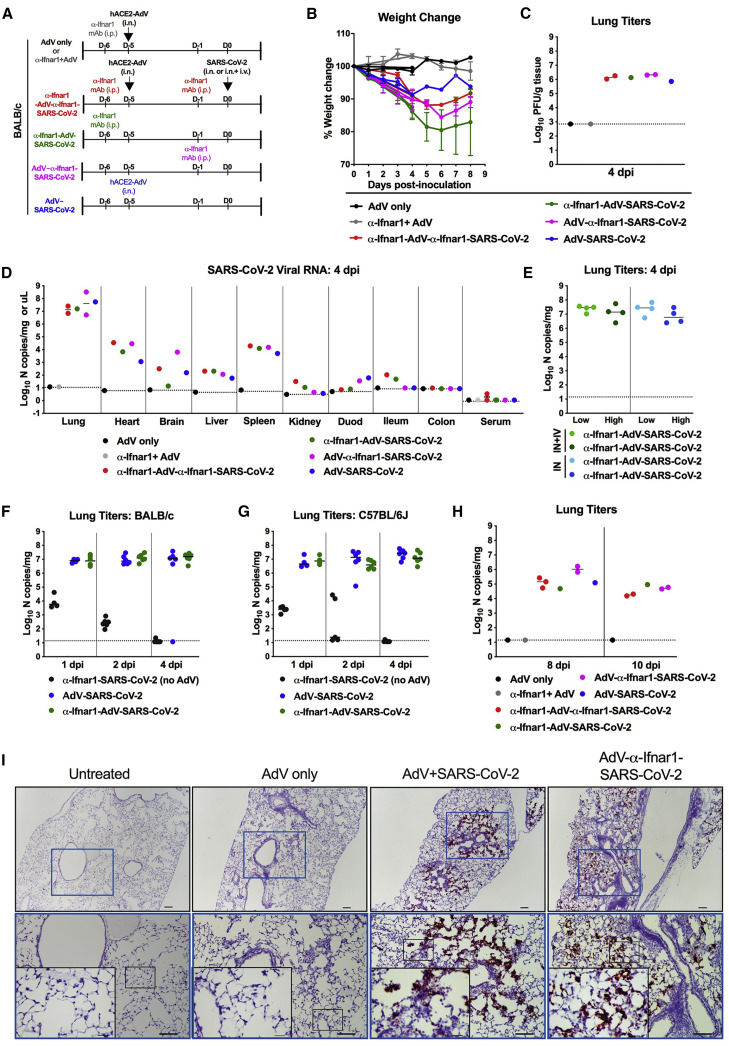
Figure 2.
SARS-CoV-2 Infection in AdV-hACE2-Transduced Mice
(A) 8-to-10-week-old male and female BALB/c mice received anti-Ifnar1 mAb (2 mg, intraperitoneal [i.p.] route; day −5), hACE2-AdV (2.5 × 108 PFU, intranasal [i.n.] route, day −5), or SARS-CoV-2 (105 FFU, i.n. + intravenous [i.v.] route, day 0) as indicated.
(B and C) Weight change was monitored (B) and viral burden in the lungs was analyzed at 4 dpi by plaque assay (C) (two experiments). Error bars indicate SD. The dashed line indicates the assay limit of detection.
(D–H) Viral RNA levels in tissues of BALB/c (D–F and H) or C57BL/6J (G) mice after AdV-hACE2 transduction (i.n.) and SARS-CoV-2 inoculation (i.n. only, E–G; i.n. + i.v., D–E and H, route) was measured by RT-qPCR after harvesting at indicated days. As indicated in each graph legend, in some experiments anti-Ifnar1 mAb was administered prior to hACE2-AdV transduction and/or SARS-CoV-2 inoculation. All symbols are color-coded to the indicated experimental conditional and represent data from individual mice. (E–H) Bars indicate median values.
(I) SARS-CoV-2 RNA in situ hybridization of lungs of naive mice or mice receiving AdV-hACE2 only, AdV-hACE2 + SARS-CoV-2, AdV-hACE2 + anti-Ifnar1 mAb + SARS-CoV-2 at 4 dpi. Images show low- (top; scale bars, 100 μm) and medium- (middle; scale bars, 100 μm) power magnification with a high-power magnification inset (scale bars, 10 μm; representative images from n = 3 per group).
To define the kinetics of infection, we monitored SARS-CoV-2 levels over time in AdV-hACE2-transduced and non-transduced mice after intranasal inoculation. While high and sustained levels of viral RNA were observed at early time points (days 1, 2, and 4) in AdV-hACE2-transduced, SARS-CoV-2-infected BALB/c and C57BL/6J mice, the input viral RNA rapidly decreased in animals receiving only SARS-CoV-2 in the absence of AdV-hACE2 (Figures 2F and 2G), consistent with the idea that hACE2 is required for robust productive infection. Although viral RNA levels waned by days 8 and 10 dpi (∼1,000-fold reduction), they were still readily detected in all cohorts of AdV-hACE2-transduced, SARS-CoV-2-infected mice (Figure 2H). The high levels of SARS-CoV-2 infection in the lung of AdV-hACE2 transduced mice at 4 dpi were confirmed by viral RNA in situ hybridization, with prominent staining observed in epithelial cells lining the bronchioles and alveoli (Figure 2I). Even in the absence of anti-Ifnar1 mAb treatment, SARS-CoV-2 RNA was readily detected in lung sections.
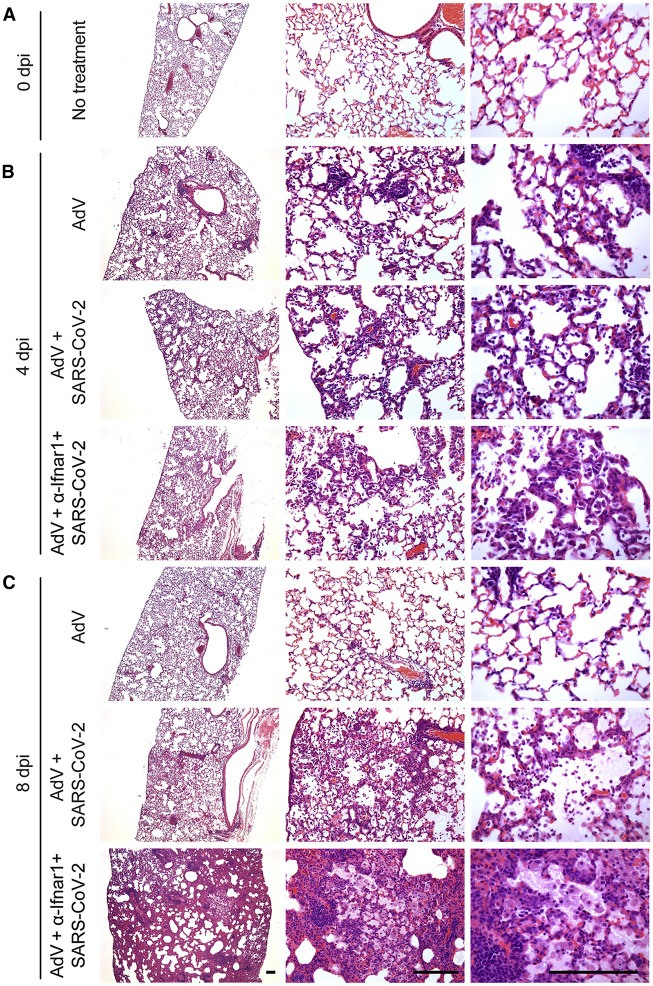
We evaluated the impact of SARS-CoV-2 infection on lung histopathology after staining with hematoxylin and eosin (H&E). At 4 dpi, mice transduced with recombinant AdV-hACE2 alone showed mild immune cell infiltration in peribronchiolar, perivascular, and alveolar sites compared with untreated mice (Figures 3A and 3B). At this early time point, these changes were similar in mice also infected with SARS-CoV-2 with or without additional treatment with anti-Ifnar1 antibody (Figure 3B). By 8 dpi, mice given AdV-hACE2 alone showed only focal perivascular inflammation that varied little from the untreated control mice (Figure 3C). However, at this later time point, AdV-hACE2-transduced, SARS-CoV-2-infected mice showed a substantial increase in immune cell infiltration featuring neutrophil accumulation in perivascular and alveolar locations and vascular congestion (Figure 3C). Moreover, AdV-hACE2-transduced, SARS-CoV-2-infected mice given anti-Ifnar1 mAb showed disease progression with a marked increase in immune cell infiltration in perivascular and alveolar sites along with parenchymal fluid, fibrin, and macrophage accumulation (Figure 3C). The findings seen after SARS-CoV-2 infection at 8 dpi are consistent with severe viral pneumonia found at autopsy in non-human primates with experimental SARS-CoV-2 infection (Munster et al., 2020) and humans with COVID-19 (Barton et al., 2020, Magro et al., 2020). Collectively, these studies establish that administration of an AdV encoding the hACE2 receptor to commercially available mice renders them susceptible to SARS-CoV-2 lung infection, clinical disease, and pathology.
Figure 3.
Histopathological Analysis of SARS-CoV-2 Infection in AdV-hACE2-Transduced BALB/c Mice
(A–C) Hematoxylin and eosin staining of lung sections from BALB/c mice after no treatment (A) or after intranasal administration of AdV-hACE2, AdV-hACE2 + SARS-CoV-2, or AdV-hACE2 + anti-Ifnar1 Ab + SARS-CoV-2 with tissue analysis at 4 dpi (B) or 8 dpi (C) using the protocol described in Figure 2. Scale bars, 500 μm. Each image is representative of a group of 3 mice.
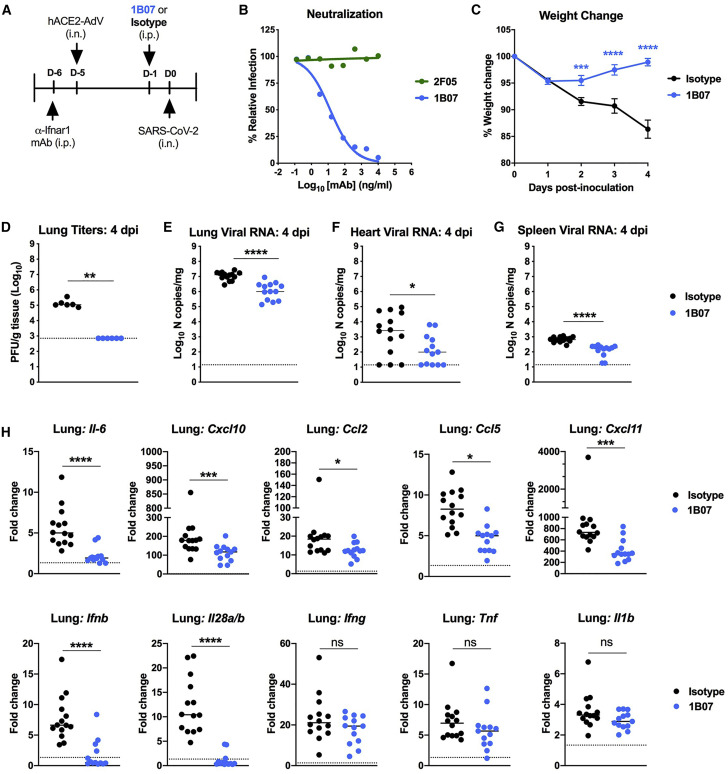
To assess the utility of this model for evaluating possible therapeutics, we passively transferred to anti-Ifnar1-antibody treated, AdV-hACE2-transduced BALB/c mice a single dose (10 mg/kg) of an anti-SARS-CoV-2 mAb 1B07 or isotype control mAb 1 day before intranasal SARS-CoV-2 inoculation (Figure 4 A). The 1B07 mAb is a chimeric mouse Fv-human Fc (IgG1) antibody that was generated after immunization of C57BL/6J mice with SARS-CoV-2 S protein, single B cell sorting with purified receptor-binding domain (RBD) and direct cloning and expression; this mAb recognizes the SARS-CoV-2 RBD as determined by ELISA (A. Ellebedy, unpublished data). 1B07 potently neutralized SARS-CoV-2 infection in Vero cells with an EC50 of 37 ng/mL whereas another anti-SARS-CoV-2 mAb (2F05) that recognized the viral S protein lacked inhibitory activity (Figure 4B). Prophylaxis with 1B07 prevented SARS-CoV-2-induced weight loss through the first 4 days of infection (Figure 4C) and markedly reduced infectious virus and viral RNA levels in the lung as determined by plaque assay, qRT-PCR assay, and viral RNA in situ hybridization (Figures 4D–4E and 5A). We also observed lower levels of SARS-CoV-2 RNA at distant tissue sites (e.g., heart and spleen) (Figures 4F and 4G). Finally, at 4 dpi, lower levels of several pro-inflammatory cytokines and chemokines (IL-6, CCL2, CCL5, CXCL10, CXCL11, IFN-λ, and IFN-β) were detected in bulk lung homogenates from mice treated with 1B07 compared with the isotype control mAb (Figure 4H), and this phenotype correlated with decreased immune cell infiltrates in the lungs at 6 dpi in animals treated with 1B07 (Figure 5B). These data suggest that neutralizing antibodies can protect against SARS-CoV-2-induced lung inflammation.
Figure 4.
Protective Effect of a Neutralizing mAb against SARS-CoV-2 Infection
(A) Experimental scheme. 8-week-old male BALB/c mice were treated with anti-Ifnar1 mAb and transduced with AdV-hACE2 via the i.n. route. Four days later, mice were treated via i.p. route with 200 ug of 1B07 (anti-SARS-CoV-2) or isotype control 2H09 (anti-influenza A virus) mAb. One day later, SARS-CoV-2 (4 × 105 FFU per mouse) was inoculated via an i.n. route.
(B) Anti-SARS-CoV-2 mAbs (1B07 and 2F05) were incubated with 102 FFU of SARS-CoV-2 for 1 h at 37°C followed by addition of mAb-virus mixture to Vero E6 cells. Virally infected foci were stained and counted. Wells containing mAb were compared with wells containing no mAb to determine the relative infection. One experiment of three is shown.
(C–G) In vivo outcomes. Weight change (C) was monitored (n = 15–16, two-way ANOVA with Sidak’s post-test: ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001), and viral burden at 4 dpi was determined in the lung (D and E), heart (F), and spleen (G) by plaque assay (D) or qRT-PCR (E–G) (D, n = 6; E–G, n = 13–16; Mann-Whitney test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001). The dashed line indicates the assay limit of detection.
(H) Fold change in gene expression of indicated cytokines and chemokines was determined by qRT-PCR, normalized to Gapdh, and compared with naive controls in lung homogenates at 4 dpi from 1B07 or isotype control mAb-treated mice (three experiments, n = 13-15 per group, Mann-Whitney test: ns, not significant, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Dotted line indicates the average level of cytokine or chemokine transcript in naive mice. Error bars indicate standard error of the mean (C), and bars represent median values (D–H).
Figure 5.
Pathological Analysis of SARS-CoV-2 Infection in Anti-SARS-CoV-2-mAb-Treated AdV-hACE2-Transduced BALB/c Mice
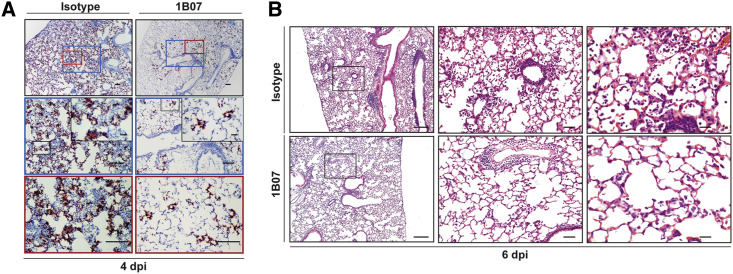
(A) SARS-CoV-2 RNA in situ hybridization at 4 dpi of lungs from 8-week-old BALB/c mice treated with 200 μg of 1B07 (anti-SARS-CoV-2) or isotype control 2H09 (anti-influenza A virus) that also received AdV-hACE2 + anti-Ifnar1 mAb + SARS-CoV-2 as described in Figure 4. Images show low- (top; scale bars, 100 μm) and medium- (middle; scale bars, 100 μm) power magnification with a high-power magnification inset (scale bars, 10 μm; representative images from n = 3 per group).
(B) Hematoxylin and eosin staining of lung sections from 8-week-old BALB/c mice at 6 dpi after treatment with 1B07 or isotype control mAbs. Mice were given an intranasal administration of AdV-hACE2 + anti-Ifnar1 Ab + SARS-CoV-2 as described in Figure 4. Images show low- (left; scale bars, 250 μm), medium- (middle; scale bars, 50 μm), and high-power magnification (scale bars, 25 μm). Each image is representative of a group of 3 mice.
In summary, we have developed a model of SARS-CoV-2 lung infection and disease in commonly available laboratory mice by rapidly sensitizing them via an intranasal delivery of replication-defective AdV encoding the hACE2 receptor. Although transient type I IFN blockade was not necessary or sufficient for SARS-CoV-2 infection in mice, we observed greater weight loss and lung pathology in the absence of intact IFN signaling. The basis for this remains uncertain but could be because of a lack of IFN-induced epithelial cell repair (Sun et al., 2015), sustained signaling of alternate pro-inflammatory pathways in the absence of IFN (Pinto et al., 2014), or a deficiency of protective IFN-λ responses in the lung (Galani et al., 2017, Klinkhammer et al., 2018) because IFN-λ itself is an IFN-stimulated gene (Lazear et al., 2019). Studies are planned to determine whether specific IFN subtypes mediate this protective effect against SARS-CoV-2 pathogenesis, as IFN-α and IFN-β can have disparate antiviral and immunomodulatory effects (Cook et al., 2019, Ng et al., 2016).
Further model refinement is planned using additional laboratory mouse strains (Boon et al., 2011), animals of different ages and sex, and mice with distinct underlying comorbidities (e.g., diabetes or obesity) that are risk factors for severe COVID-19 in humans (Zhou et al., 2020). Moreover, this AdV-hACE2 transduction system could be used immediately in the context of Collaborative Cross mice (Noll et al., 2019) or the large library of genetic knockout mice in academic, governmental, and commercial laboratories to identify host susceptibility and restriction factors for SARS-CoV-2 infection. Consistent with our studies, a separate group has analogously transduced hACE2 into C57BL/6 and BALB/c mice and successfully generated a SARS-CoV-2 pathogenesis model of lung infection and disease (Sun et al., 2020).
Limitations of Study
Our model has some limitations compared with fully transgenic hACE2 mice (Bao et al., 2020, McCray et al., 2007, Menachery et al., 2016) or other small animal models (e.g., hamsters and ferrets) of SARS-CoV-2 infection that are in development (Chan et al., 2020, Kim et al., 2020, Shi et al., 2020). These include potential mouse-to-mouse variation in hACE2 expression and tissue distribution and mild bronchial inflammation associated with AdV delivery. Notwithstanding these caveats, we consistently observed high levels of lung infection and pneumonia in our dual AdV-hACE2-transduced and SARS-CoV-2-infected mice. Moreover, viral infection and several inflammatory mediators in the SARS-CoV-2-infected lung were reduced by neutralizing antibody treatment. Of note, IL-6 levels in the lung were decreased after treatment with 1B07 anti-S protein mAb. In non-randomized trials in COVID-19 patients, treatment with tocilizumab, an IL-6 inhibitor, preliminarily was associated with less inflammation and clinical improvement (Alattar et al., 2020, Giamarellos-Bourboulis et al., 2020). Our mouse model provides definitive evidence that antibody-based treatments can mitigate SARS-CoV-2 infection in vivo, analogous to that described for SARS-CoV and MERS-CoV infections (Corti et al., 2015, Traggiai et al., 2004), and suggests a path forward for identifying therapeutic antibody candidates or combinations with even greater potency and protective activity. The availability of SARS-CoV-2 small animal models that are easily generated with commercially available mice can accelerate the pace of screening, identification, and development of countermeasures (drugs, vaccines, and antibody therapeutics) for advancement to pivotal non-human primate and human studies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 1B07, anti-SARS-CoV-2 mAb | This paper | N/A |
| 2F05, anti-SARS-CoV-2 mAb | This paper | N/A |
| 2H09, anti-influenza A virus mAb | This paper | N/A |
| Goat anti-human ACE2 antibody | R&D | AF933; RRID: AB_355722 |
| APC-conjugated donkey anti-goat IgG secondary antibody | Thermo Fisher | A32849; RRID: AB_2762840 |
| CR3022, anti-SARS-CoV-2 mAb | Yuan et al., 2020 | N/A |
| HRP-conjugated goat anti-human IgG | Thermo Fisher | 62-8420; RRID: AB_2533962 |
| MAR1-5A3, anti-Ifnar1 mAb | Leinco | I-401; RRID: AB_2491621 |
| Virus Strains | ||
| SARS-CoV-2 (strain 2019 n-CoV/USA_WA1/2020) | CDC/BEI Resources | NR52281 |
| AdV-hACE2-GFP | Jia et al., 2005 | N/A |
| AdV-hACE2 | Jia et al., 2005 | N/A |
| Experimental Models: Cell Lines | ||
| Vero CCL-81 | ATCC | CCL-81; RRID: CVCL_0059 |
| Vero E6 | ATCC | CRL-1586; RRID: CVCL_0574 |
| HEK293 | ATCC | CRL-1573; RRID: CVCL_0045 |
| HEK293T | ATCC | CRL-3216; RRID: CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | Cat#000664; RRID: IMSR_JAX:000664 |
| Mouse: BALB/c | Jackson Laboratory | Cat#000651; RRID: IMSR_JAX:000651 |
| Mouse: DBA/2J | Jackson Laboratory | Cat#000671; RRID: IMSR_JAX:000671 |
| Mouse: Rag1−/− C57BL/6 | Jackson Laboratory | Cat#002216; RRID: IMSR_JAX:002216 |
| Mouse: Stat1−/− C57BL/6 | Jackson Laboratory | Cat#012606; RRID: IMSR_JAX:012606 |
| AG129 | Marshall BioResources | AG129 |
| Oligonucleotides | ||
| SARS-CoV-2 N F: 5′-ATGCTGCAATCGTGCTACAA-3′ | This paper | N/A |
| SARS-CoV-2 N R: 5′-GACTGCCGCCTCTGCTC-3′ | This paper | N/A |
| SARS-CoV-2 N Probe: 5′-/56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/-3′ | This paper | N/A |
| SARS-CoV-2 RNA ISH probe (S gene) | Advanced Cell Diagnostics | Cat# 4848561 |
| Human ACE2 RNA ISH probe | Advanced Cell Diagnostics | Cat# 848151 |
| Gapdh TaqMan Primer/Probe set | IDT | Mm.PT.39a.1 |
| Human ACE2 TaqMan Primer/Probe set | IDT | Hs.PT.58.27645939 |
| Ifng TaqMan Primer/Probe set | IDT | Mm.PT.58.41769240 |
| Il6 TaqMan Primer/Probe set | IDT | Mm.PT.58.10005566 |
| Il1b TaqMan Primer/Probe set | IDT | Mm.PT.58.41616450 |
| Tnfa TaqMan Primer/Probe set | IDT | Mm.PT.58.12575861 |
| Cxcl10 TaqMan Primer/Probe set | IDT | Mm.PT.58.43575827 |
| Ccl2 TaqMan Primer/Probe set | IDT | Mm.PT.58.42151692 |
| Ccl5 TaqMan Primer/Probe set | IDT | Mm.PT.58.43548565 |
| Cxcl11 TaqMan Primer/Probe set | IDT | Mm.PT.58.10773148.g |
| Ifnb TaqMan Primer/Probe set | IDT | Mm.PT.58.30132453.g |
| Ifnl(2/3) TaqMan Primer/Probe set | Thermo Fisher | Mm04204156_gH |
| Software and Algorithms | ||
| BioRender | BioRender.com | N/A |
| FlowJo | FlowJo, LLC | v10 |
| GraphPad Prism | GraphPad | v 8.2.1 |
| Recombinant DNA | ||
| pABVec6W | Davis et al., 2019 | N/A |
| PCR-II topo | Invitrogen | Cat# 451245 |
Resource Availablity
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael S. Diamond (diamond@wusm.wustl.edu).
Materials Availability
All requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact author. This includes transgenic mice, antibodies, viruses, and proteins. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Data and Code Availability
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request.
Experimental Model and Subject Details
Cells and viruses
Vero E6 (CRL-1586, American Type Culture Collection (ATCC), Vero CCL81 (ATCC), HEK293 (ATCC), and HEK293T (ATCC) were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, 1 × non-essential amino acids, and 100 U/mL of penicillin–streptomycin.
SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 was obtained from the Centers for Disease Control and Prevention (gift of Natalie Thornburg). Virus was passaged in Vero CCL81 cells and titrated by focus-forming assay on Vero E6 cells. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and A-BSL3 facilities at Washington University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Monoclonal antibodies
1B07 and 2F05 are chimeric mouse Fv-human Fc (IgG1) mAbs that were generated after immunization of C57BL/6J mice with SARS-CoV-2 S protein, single B cell sorting and direct cloning into pABVec6W vectors (Davis et al., 2019). After transfection and expression in 293F cells, 1B07, 2F05, and a control anti-influenza A mAb (2H09) were purified by protein A chromatography.
Adenoviruses
The AdV-hACE2-GFP and AdV-hACE2 constructs and defective virus preparation was reported previously (Jia et al., 2005) and prepared initially by the University of Iowa Viral Vector Core and separately at Washington University. AdV-hACE2-GFP and AdV-hACE2 were propagated in 293T cells and purified using cesium chloride density-gradient ultracentrifugation, and the number of virus particles was determined using optical density (260 nm) measurement and plaque assay, as previously described (Mittereder et al., 1996).
Mouse experiments
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Mice were purchased from commercial vendors (BALB/c, C57BL/6J, and DBA2/J, Jackson Laboratories) or propagated at Washington University School of Medicine (Stat1 −/− C57BL/6; Rag1 −/− C57BL/6, and AG129). Animals were housed in groups and and fed standard chow diets. Mice of different ages and both sexes (three to four or eight to ten week-old, indicated in each Figure legend) were administered 2.5 × 108 PFU of AdV-hACE2 via intranasal administration. In some experiments, an intraperitoneal injection of 2 mg of anti-Ifnar1 mAb (MAR1-5A3 (Sheehan et al., 2006), Leinco) was given before (−24 h) or after (+ 4 days) AdV-hACE2 treatment. Five days after AdV transduction, mice were inoculated with SARS-CoV-2 via intranasal only or both intranasal and intravenous routes. Weights were monitored on a daily basis, and animals were sacrificed at 1, 2, 4, 8, or 10 dpi and tissues were harvested. 1B07 or isotype control 2H09-treated mice were euthanized and perfused with 20 mL of PBS after serum collection via cardiac puncture and before tissue harvest.
Method Details
Flow cytometry
HEK293 were transduced with AdV-hACE2-GFP. Cells were detached and stained with a primary human ACE2 antibody (R&D, 1:200) in PBS, 2% FBS, and 5% donkey serum for 1 h at 4°C and then stained with an APC-conjugated donkey anti-goat secondary antibody (Thermo Fisher, 1:1000) for 30 min at 4°C. Data was collected on a MACSQuant X Flow cytometer and analyzed using FlowJo V10.
Measurement of viral burden and hACE2 expression
Tissues were weighed and homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) in 1000 μL of DMEM media supplemented with 2% heat-inactivated FBS. Tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min and stored at −80°C. RNA was extracted using MagMax mirVana Total RNA isolation kit (Thermo Scientific) and a Kingfisher duo prime extraction machine (Thermo Scientific). For some samples, viral burden was determined by plaque assays on Vero E6 cells.
To identify highly conserved regions within coronaviruses, the originally published nucleotide sequence of SARS-CoV-2 (GenBank MN908947) was aligned with two related bat SARS-like CoV (GenBank MG772933.1 and MG772934.1) using MUSCLE (Edgar, 2004). We identified a ∼100 base pair nucleotide region in the nucleocapsid (N) gene coding sequence that had 100% sequence identity between the aligned coronaviruses. Primers were designed to target this conserved region using SARS-CoV-2 (MN908947) sequence as a guide (L Primer: ATGCTGCAATCGTGCTACAA; R primer: GACTGCCGCCTCTGCTC; probe: /56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/). To establish an RNA standard curve, we generated concatenated segments of the N gene in a gBlocks fragment (IDT) and cloned this into the PCR-II topo vector (Invitrogen). The vector was linearized and in vitro T7-DNA-dependent RNA transcription performed to generate materials for a quantitative standard curve. When used in combination with our generated primer-probe set and a known amount of the plasmid control, N gene copy-numbers could be determined down to 10 copies per reaction.
For hACE2 expression, RNA was DNase-treated (Thermo Scientific) following the manufacturer’s protocol. RNA levels were quantified as described above with the primer/probe set for hACE2 (IDT assay: Hs.PT.58.27645939), compared to an RNA standard curve, and normalized to mg of tissue.
Cytokine and chemokine mRNA measurements
RNA was isolated from lung homogenates as described above. cDNA was synthesized from DNase-treated RNA using the High-Capacity cDNA Reverse Transcription kit (Thermo Scientific) with the addition of RNase inhibitor following the manufacturer’s protocol. Cytokine and chemokine expression was determined using TaqMan Fast Universal PCR master mix (Thermo Scientific) with commercial primers/probe sets specific for IFN-γ (IDT: Mm.PT.58.41769240), IL-6 (Mm.PT.58.10005566), IL-1β (Mm.PT.58.41616450), TNF-α (Mm.PT.58.12575861), CXCL10 (Mm.PT.58.43575827), CCL2 (Mm.PT.58.42151692), CCL5 (Mm.PT.58.43548565), CXCL11 (Mm.PT.58.10773148.g), IFN-β (Mm.PT.58.30132453.g), and IFNλ−2/3 (Thermo Scientific Mm04204156_gH) and results were normalized to GAPDH (Mm.PT.39a.1) levels. Fold change was determined using the 2-ΔΔCt method comparing treated mice to naive controls.
Histology and RNA in situ hybridization
Animals were euthanized, and tissues were harvested before lung inflation and fixation. The left lung was tied off at the left main bronchus and collected for viral RNA analysis. The right lung was inflated with ∼1.2 mL of 10% neutral buffered formalin using a 3-mL syringe and catheter inserted into the trachea. For fixation after infection, inflated lungs were kept in a 40-mL suspension of neutral buffered formalin for 7 days before further processing. Tissues were embedded in paraffin, and sections were stained with hematoxylin and eosin. RNA in situ hybridization was performed using the RNAscope 2.5 HD Assay (Brown Kit) according to the manufacturer’s instructions (Advanced Cell Diagnostics). Briefly, sections were deparaffinized, treated with H2O2 and Protease Plus prior to probe hybridization. Probes specifically targeting hACE2 (cat no. 848151) or SARS-CoV-2 S sequence (cat no 848561) were hybridized followed by proprietary signal amplification and detection with 3,3′-Diaminobenzidine. Tissues were counterstained with Gill’s hematoxylin. An uninfected mouse was used as a negative control and stained in parallel. Tissue sections were visualized using a Nikon Eclipse microscope equipped with an Olympus DP71 color camera or a Leica DM6B microscope equipped with a Leica DFC7000T camera.
Neutralization assay
Serial dilutions of mAbs 1B07 or 2F05 were incubated with 102 focus-forming units (FFU) of SARS-CoV-2 for 1 h at 37°C. mAb-virus complexes were added to Vero E6 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were harvested 30 h later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with 1 μg/mL of CR3022 (Yuan et al., 2020) anti-S antibody and HRP-conjugated goat anti-human IgG in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies). Data were processed using Prism software (GraphPad Prism 8.0).
Quantification and Statistical Analysis
Statistical significance was assigned when P values were < 0.05 using Prism Version 8 (GraphPad) and are indicated in the relevant Figure legends. Analysis of weight change and viral burden in vivo were determined by two-way ANOVA and Mann-Whitney tests, respectively.
Acknowledgments
This study was supported by NIH contracts and grants (75N93019C00062, R01 AI127828, P01 AI60699, R21 AI139813, U01 AI141990, R01 AI130591, R35 HL145242, and HHSN272201400008C) and the Defense Advanced Research Projects Agency (HR001117S0019). B.T.M is supported by F32 AI138392 and J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. We thank Sean Whelan, Susan Cook, and Jennifer Philips for facilitating the high-containment studies with SARS-CoV-2 and Arthur Kim for purifying the CR3022 anti-S mAb.
Author Contributions
A.O.H. amplified the adenovirus, designed experiments, and performed intranasal inoculations of adenovirus. J.B.C. propagated the SARS-CoV-2 stocks; developed the focus forming, neutralization, and plaque assays; and performed intranasal inoculations of SARS-CoV-2. A.L.B. designed and constructed the viral qRT-PCR assays and performed intravenous inoculations of SARS-CoV-2. L.B.T., N.M.K., E.S.W., S.S., B.T.M., R.E.C., and J.M.F. performed clinical analysis, tissue harvests, histopathological studies, and viral burden analyses. E.S.W. optimized and performed the flow cytometry staining for hACE2. B.T.M. performed in situ hybridization. S.G, S.P.K., and M.J.H. analyzed the tissue sections for histopathology. S.P. and P.B.M. generated the hACE2 expressing adenovirus. W.B.A., J.S.T., A.J.S. T.L., and A.H.E. generated the anti-SARS-CoV-2 mAbs with reagents from D.H.F., and J.B.C. and R.E.C. performed neutralization assays. E.S.W., A.O.H., and M.S.D. wrote the initial draft, with the other authors providing editing comments.
Declaration of Interests
M.S.D. is a consultant for Inbios, Eli Lilly, Vir Biotechnology, NGM Biopharmaceuticals, and on the Scientific Advisory Board of Moderna. A.H.E. is a consultant for Inbios and Fimbrion Therapeutics. The Diamond laboratory has received unrelated funding under sponsored research agreements from Moderna and Emergent BioSolutions. The Ellebedy laboratory has received funding under a sponsored research agreement with Emergent BioSolutions. The Perlman laboratory has received research support from Eli Lilly and AbbVie.
Published: June 10, 2020
References
- Alattar R., Ibrahim T.B.H., Shaar S.H., Abdalla S., Shukri K., Daghfal J.N., Khatib M.Y., Aboukamar M., Abukhattab M., Alsoub H.A. Tocilizumab for the Treatment of Severe COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.25964. Published online May 5, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Published online May 7, 2020. [DOI] [PubMed] [Google Scholar]
- Barton L.M., Duval E.J., Stroberg E., Ghosh S., Mukhopadhyay S. COVID-19 Autopsies, Oklahoma, USA. Am. J. Clin. Pathol. 2020;153:725–733. doi: 10.1093/ajcp/aqaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon A.C., Finkelstein D., Zheng M., Liao G., Allard J., Klumpp K., Webster R., Peltz G., Webby R.J. H5N1 influenza virus pathogenesis in genetically diverse mice is mediated at the level of viral load. MBio. 2011;2 doi: 10.1128/mBio.00171-11. e00171–e00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa325. Published online March 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M., Matthews K.L., Goicochea L., Frieman M.B. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J. Gen. Virol. 2014;95:408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook L.E., Locke M.C., Young A.R., Monte K., Hedberg M.L., Shimak R.M., Sheehan K.C.F., Veis D.J., Diamond M.S., Lenschow D.J. Distinct Roles of Interferon Alpha and Beta in Controlling Chikungunya Virus Replication and Modulating Neutrophil-Mediated Inflammation. J. Virol. 2019;94 doi: 10.1128/JVI.00841-19. e00841–e00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.W., Jackson K.J.L., McElroy A.K., Halfmann P., Huang J., Chennareddy C., Piper A.E., Leung Y., Albarino C.G., Crozier I. Longitudinal Analysis of the Human B Cell Response to Ebola Virus Infection. Cell. 2019;177:1566–1582. doi: 10.1016/j.cell.2019.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M. Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ. 2020;368:m1165. doi: 10.1136/bmj.m1165. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.E., Triantafyllia V., Eleminiadou E.E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K., Andreakos E. Interferon-λ Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity. 2017;46:875–890. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.009. Published online April 17, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J., Kobinger G.P., Wivel N.A., Crystal R.G., Boyer J. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 2004;78:11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Kim E.J., Lee S., Casel M.A.B. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkhammer J., Schnepf D., Ye L., Schwaderlapp M., Gad H.H., Hartmann R., Garcin D., Mahlakõiv T., Staeheli P. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife. 2018;7:e33354. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Hopes rise for coronavirus drug remdesivir. Nature. 2020 doi: 10.1038/d41586-020-01295-8. Published online April 29, 2020. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020 doi: 10.1016/j.trsl.2020.04.007. Published online April 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittereder N., March K.L., Trapnell B.C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Perez-Perez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B. Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.21.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., Meyerholz D.K., Moore S., Cassell M., Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C.T., Mendoza J.L., Garcia K.C., Oldstone M.B. Alpha and Beta Type 1 Interferon Signaling: Passage for Diverse Biologic Outcomes. Cell. 2016;164:349–352. doi: 10.1016/j.cell.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll K.E., Ferris M.T., Heise M.T. The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell Host Microbe. 2019;25:484–498. doi: 10.1016/j.chom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto A.K., Ramos H.J., Wu X., Aggarwal S., Shrestha B., Gorman M., Kim K.Y., Suthar M.S., Atkinson J.P., Gale M., Jr., Diamond M.S. Deficient IFN signaling by myeloid cells leads to MAVS-dependent virus-induced sepsis. PLoS Pathog. 2014;10:e1004086. doi: 10.1371/journal.ppat.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020 doi: 10.1001/jama.2020.6019. Published online April 13, 2020. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan K.C., Lai K.S., Dunn G.P., Bruce A.T., Diamond M.S., Heutel J.D., Dungo-Arthur C., Carrero J.A., White J.M., Hertzog P.J., Schreiber R.D. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L.-Y., Liu D., Huang J., He J., Zhu A., Zhao J. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell. 2020;182:734–743. doi: 10.1016/j.cell.2020.06.010. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Miyoshi H., Origanti S., Nice T.J., Barger A.C., Manieri N.A., Fogel L.A., French A.R., Piwnica-Worms D., Piwnica-Worms H. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe. 2015;17:85–97. doi: 10.1016/j.chom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E., Becker S., Subbarao K., Kolesnikova L., Uematsu Y., Gismondo M.R., Murphy B.R., Rappuoli R., Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr., Baric R.S., Enjuanes L., Gallagher T. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request.