Table 1.
Pharmacokinetic and pharmacodinamic characteristics of natural compounds affecting PCSK9.
| Natural Compound | Chemical Structure | Bioavail. | Tmax (h) | Half-Life Time (h) | Metabolism | Mechanism of Action | Transport. | Level of Evidence on PCSK9 | Counteracts Statins | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Berberine |
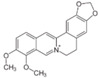
|
0.37% | 9.8 | 28.6 | Demethylation and glucuronide | Inhibits SREBP; HNF1α | P-gp and MRP1 | In vitro, in vivo and clinical | Yes | [23,24,28,30,33,35,36,37,188] |
| Sterol/Stanols and Vegetable Proteins | ||||||||||
| Lupin peptide | LILPKHSDAD | Poor (predicted) | Not known | Not known | Proteases | Inhibits interaction PCSK9-LDLR; Reduces HNF1α | No (predicted) | In vitro. Clinic (negative) | Not known | [61,64] |
| Polyphenols | ||||||||||
| Quercetin |
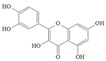
|
0.31% | 2.5 ÷ 3 | 2.1 | Liver: Sulfate and methyl glucuronide; Gut microbiota: free aglycone; 3,4-dihydroxyphenylacetic acid, 3-(3-hydroxyphenyl) propionic acid, 3,4-dihydroxybenzoic acid and 4-hydroxybenzoic acid | Inhibits secretion (sortilin) and SREBP | P-gp sand BCRP | In vitro and in vivo | Not known | [72,73] [78,80,81] |
| Epigallocatechin gallate |
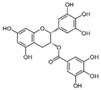
|
0.1% | 1 ÷ 2 | 3.4 | Liver: Methyl, sulfate, and glucuronide; Gut microbiota: phenylvalerolactones and phenylvaleric acids | Inhibits secretion and SREBP | MRP2 and OATP1B1 | In vitro | Yes | [85,91,92,95,96,97,98] |
| Resveratrol |
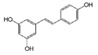
|
<1% | 3 | 9.2 | Liver: Sulfate and glucuronide; Gut microbiota: dihydroresveratrol | Inhibits SREBP1c and interaction PCSK9-LDLR | Not known | In vivo and in vivo | Not known | [100,101,102,105,106,107] |
| Curcumin |
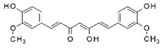
|
<1% | Not known | Not known | Liver: reduction and glucuronide, sulfate, and glutathione; Gut microbiota: tetrahydrocurcumin, demethylcurcumin, bisdemethylcurcumin etc. | Inhibits; HNF1α | Not known | In vitro and in vivo | Yes | [122,123,124,127,128,129] |
| Silibinin A |
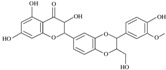
|
<1% | 1.4 | Not known | Sulfate and glucuronide | Inhibits transcription | P-gp inhibitor | In vitro | Yes | [108,109,110] |
| Naringin |
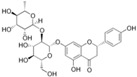
|
<1% | 0.5 | 9.5 | Liver: Hydrolysis and then glucuronide, sulfate, methylation; Gut microbiota: Phenolic derivatives | Inhibits SREBP | P-gp and OATP1A5 inhibitor | In vivo | Not known | [111,112] |
| Pinostrobin |
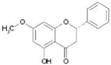
|
1.8% (S); 13.8% (R) | 6.0 | 38.1 | Glucuronide | Inhibits transcription and catalytic activity | Not known | In vitro | Not known | [113,114] |
| Eugenol |
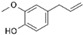
|
<1% | 2.1 | 14.0 | Phenol, glucuronide and sulphate | Direct interaction with PCSK9 and inhibits SREBP | Not known | In vitro | Not known | [115,117,118] |
| Nutrients | ||||||||||
| Kaempferol |
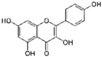
|
2.5% | Not known | Not known | Glucuronide and sulphate | Inhibits transcription | Not known | In vitro | Yes | [86,132] |
| p-Coumaric acid |
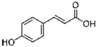
|
24% | 0.17 | 0.25 | Glucuronide and sulphate | Inhibits transcription | Not known | In vitro | Yes | [132] |
| Vitamin K7 |
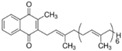
|
2% | 6.0 | 60 | Not known | Not known | Not known | In vitro and in vivo | Not known | [145,148,189] |
| Lycopene |
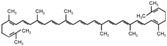
|
33.9% | 24 | 235 | Phase I, oxidation | Inhibits transcription and interaction PCSK9-LDLR | Not Known | In vitro and in vivo | Not Known | [151,152,154,190] |
| Other Inhibitors | ||||||||||
| Protodioscin |
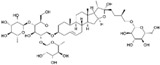
|
0.2% | 20 | 20 | Oxidation, deglycosylation and glucuronide | Inhibits transcription | Not known | In vitro and in vivo | Not known | [179,180] |
| Emodin |
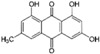
|
low | 0.13 | 8.6 | Glucuronide and sulphate | Inhibits SREBP; HNF1α | Not known | In vitro | Not known | [191,192] |
