Abstract
Small RNAs are important regulators of gene expression. They were first identified in Caenorhabditis elegans, but it is now apparent that the main small RNA silencing pathways are functionally conserved across diverse organisms. Availability of genome data for an increasing number of parasitic nematodes has enabled bioinformatic identification of small RNA sequences. Expression of these in different lifecycle stages is revealed by small RNA sequencing and microarray analysis. In this review we describe what is known of the three main small RNA classes in parasitic nematodes – microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs) and small interfering RNAs (siRNAs) – and their proposed functions. miRNAs regulate development in C. elegans and the temporal expression of parasitic nematode miRNAs suggest modulation of target gene levels as parasites develop within the host. miRNAs are also present in extracellular vesicles released by nematodes in vitro, and in plasma from infected hosts, suggesting potential regulation of host gene expression. Roles of piRNAs and siRNAs in suppressing target genes, including transposable elements, are also reviewed. Recent successes in RNAi-mediated gene silencing, and application of small RNA inhibitors and mimics will continue to advance understanding of small RNA functions within the parasite and at the host–parasite interface.
Key words: Extracellular vesicle, gene regulation, microRNA, nematode, parasite, Piwi-interacting RNA, small interfering RNA
Introduction
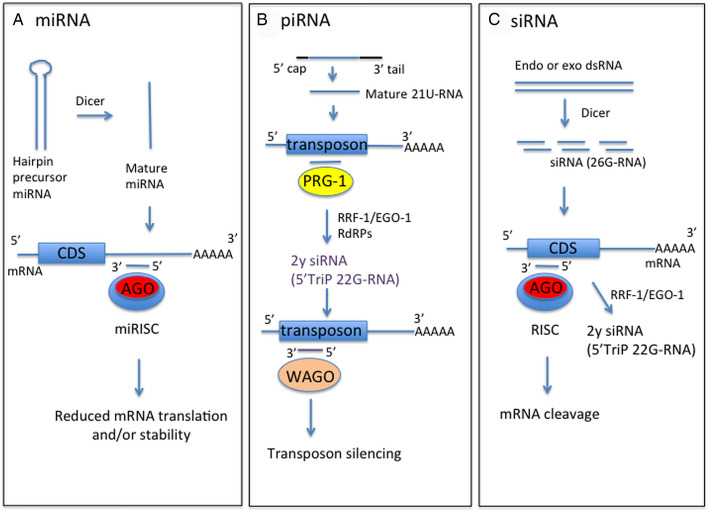
Knowledge of small RNA structure and function has increased greatly in the last decade. The free-living nematode Caenorhabditis elegans led the way, with the initial discovery of microRNAs (miRNAs) and small interfering RNAs (siRNAs) in this species (Lee et al., 1993; Reinhart et al., 2000; Fire et al., 1998). The subsequent identification of miRNAs in diverse organisms using experimental and/or bioinformatics approaches (Pasquinelli et al., 2000; Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001), and the discovery of siRNA gene silencing pathways in plants (Hamilton and Baulcombe, 1999), established the important roles that small RNAs play in suppressing gene expression. This review focuses on small RNAs in nematodes and what we know about these from genome, transcriptome and functional studies. Much progress has been made recently in characterizing nematode miRNAs and these are discussed in most detail. miRNAs were initially identified as regulators of nematode development (see below), however their presence in parasite excretory-secretory (ES) products has stimulated interest in these molecules as modulators of host–parasite interactions to promote parasite survival. The ability of miRNAs to alter levels of gene expression suggests they could have multiple, as yet undefined roles, in parasitic nematode biology. For example, altered expression of miRNAs may be associated with anthelmintic resistance (Devaney et al., 2010; Gillan et al., 2017), akin to changes in miRNA levels observed in drug-resistant tumour cells (reviewed in Ghasabi et al., 2019). This review will also discuss nematode Piwi-interacting RNAs (piRNA), required for silencing of transposable elements in the germline, but of which little is currently known in parasitic species. We also discuss the forms and functions of siRNAs, recent successes in RNA interference (RNAi)-mediated gene silencing in parasitic nematodes, and the application of this in progressing from genome to function. A diagram summarizing different small RNA classes discussed in this review is presented in Fig. 1.
Fig. 1.
Schematic of forms and functions of small RNA classes in nematodes, based on C. elegans information. (A) Mature miRNA strand, derived from precursor miRNA, is incorporated into the miRNA-induced silencing complex (miRISC) containing Argonaute protein (Ago). This complex directs binding to mRNA target sequences, commonly in the 3′-UTR. Binding specificity is determined by complementarity between the target sequence and miRNA seed sequence (nucleotides 2–7). (B) Mature piRNA (21U-RNA) are processed from a capped precursor and bind to Piwi Argonaute PRG-1 to recognize target sequences, often transposons, by imperfect complementary base-pairing. This initiates synthesis of secondary small inhibitory RNAs (siRNAs) with 5′ triphosphate (5′TriP 22G-RNAs) by RNA-dependent RNA polymerases (RdRPs) RRF-1 or EGO-1. 22G-RNAs associate with worm-specific Argonaute proteins (WAGOs) to mediate target silencing. (C) Endogenous or exogenous dsRNA is processed by dicer into siRNAs, which bind anti-sense to mRNA exonic sequence to mediate mRNA cleavage by RDE-1 Argonaute. siRNAs also act as primers for synthesis of 22G-RNAs by RRF-1 or EGO-1 to amplify the RNAi response, using target dsRNA as a template.
MicroRNAs
miRNA discovery
miRNAs regulate gene expression post-transcriptionally by binding with partial sequence complementary to the 3′-untranslated region (UTR) of their target mRNAs (Bartel, 2009, 2018). This interaction inhibits protein translation and results in miRNA degradation (Chekulaeva and Filipowicz, 2009). The first miRNAs, lin-4 and let-7, were discovered in C. elegans (Lee et al., 1993; Reinhart et al., 2000). The subsequent availability of genome data for a range of vertebrates and invertebrates revealed conservation of miRNA-mediated gene regulation (Pasquinelli et al., 2000; Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). Their conservation in diverse organisms has benefited parasitic nematode miRNA research through development of bioinformatics databases, such as miRBase (Release 22.1; http://www.mirbase.org/) (Griffiths-Jones et al., 2008), miRCarta (Version 1.1; https://mircarta.cs.uni-saarland.de/) (Backes et al., 2018), miRGeneDB (Version 2.0; http://mirgenedb.org/) (Fromm et al., 2015, 2020) and target prediction programmes [e.g. TargetScan (Lewis et al., 2005)], together with advancements in miRNA mimic and inhibitor chemistry. For example, mammalian mir-122, expressed in hepatocytes, is required for hepatitis C virus accumulation and a locked nucleic acid-modified oligonucleotide complementary to mir-122 can suppress viral load and is being evaluated as a therapeutic treatment (Titze-de-Almeida et al., 2017). In addition, specific miRNAs released into plasma or urine are being studied as potential diagnostic/prognostic biomarkers of diseases, including cancer (Wang et al., 2018), and filarial nematode infections (Buck et al., 2014; Tritten et al., 2014a, 2014b; Quintana et al., 2015).
miRNAs are derived from long primary transcripts that are processed to precursor miRNAs of approximately 70 nucleotides that fold into a hairpin structure (Kim et al., 2009; see Fig. 1). Mature miRNAs are 20–26 nucleotides in length (Fromm et al., 2015). Nucleotides 2–7 are referred to as the seed sequence and are important in determining the specificity of binding to target mRNAs. Novel miRNAs are most often identified by sequencing of small RNA libraries, mapping the sequences to the genome, where available, and testing that the region each miRNA is derived from folds into a hairpin structure, using programmes such as RNAfold (Hofacker et al., 1994). Discovery pipelines, such as miRDeep2 (Friedländer et al., 2012), incorporate RNAfold and other scoring criteria, and have been applied to miRNA discovery and validation in a range of parasitic nematodes, as summarized in Table 1.
Table 1.
Animal and human parasitic nematode species for which miRNA data are available from parasite extracts, EVs, ES supernatant or released into host serum/plasma. From small RNA sequencing data, unless indicated otherwise
| Nematode species (clade) | Developmental stage/host sample | Small RNA class | GEO Acc. No./in MiRBase | Reference |
|---|---|---|---|---|
| Angiostrongylus cantonensis (V) | Male and female adults | miRNA, siRNA | NA/No | Chen et al. (2011a) |
| Ascaris lumbricoides (III) | Female adults | miRNA | NA/No | Shao et al. (2014) |
| Ascaris suum (III) | Germline, zygote, embryo, L1–L3 | miRNA, siRNA | GSE 26956, GSE 26957/Yes | Wang et al. (2011) |
| Male and female adults | miRNA | NA/No | Xu et al. (2013) | |
| Female adults | miRNA | NA/No | Shao et al. (2014) | |
| Brugia malayi (III) | Male and female adults, mf | miRNA, siRNA | NA/Yes | Poole et al. (2010) |
| Male and female adults, mf | miRNA | NA/Yes | Poole et al. (2014) | |
| L3 EV | miRNA | SRA PRJNA 285132/No | Zamanian et al. (2015) | |
| Brugia pahangi (III) | L3, mixed sex adults | miRNA, siRNA | GSE 34539/Yes | Winter et al. (2012) |
| L3, L4, male and female adults | miRNA microarray | NA/No | Winter et al. (2015) | |
| Infected dog plasma | miRNA | NA/No | Tritten et al. (2014a) | |
| Dirofilaria immitis (III) | Mixed sex adult worms | miRNA | GSE 35646/No | Fu et al. (2013) |
| Infected dog plasma | miRNA | NA/No | Tritten et al. (2014a) | |
| Haemonchus contortus (V) | L3, mixed sex adults | miRNA, siRNA, piRNA | GSE 34539/Yes | Winter et al. (2012) |
| L3, L4, male and female adults, gut | miRNA microarray | GSE 101501/No | Marks et al. (2019) | |
| L4 EV and ES, adult EV and ES | miRNA | NA/No | Gu et al. (2017) | |
| Heligmosomoides polygyrus (V) | Egg, L3, adults, adult EV and ES | miRNA, siRNA, piRNA, YRNA | GSE 55941/Yes | Buck et al. (2014) |
| Litomosoides sigmodontis (III) | Infected mouse serum | miRNA | GSE 55978/No | Buck et al. (2014) |
| Loa loa (III) | Infected baboon plasma | miRNA | NA/No | Tritten et al. (2014b) |
| Onchocerca ochengi (III) | Infected cow plasma | miRNA | NA/No | Tritten et al. (2014b) |
| Infected cow nodule fluid | miRNA | GSE 63933/No | Quintana et al. (2015) | |
| Onchocerca volvulus (III) | Infected human serum | miRNA | NA/No | Tritten et al. (2014a) |
| Infected human serum | miRNA | GSE 63933/No | Quintana et al. (2015) | |
| Strongyloides ratti (IV) | Infective L3 and mixed stage | miRNA | GSE 41402/Yes | Ahmed et al. (2013) |
| Toxocara canis (III) | Male and female adults | miRNA | GSE 68710/No | Ma et al. (2016) |
| Trichinella spiralis (I) | Muscle stage larvae | miRNA | NA/No | Chen et al. (2011b) |
| Trichuris muris (I) | Mixed sex adult EV | miRNA | GSE 93667/No | Tritten et al. (2017) |
We applied a small RNA library sequencing approach to identify small RNAs from the clade V ovine gastrointestinal nematode (GIN) Haemonchus contortus and the clade III filarial parasite Brugia pahangi (Winter et al., 2012). Our data showed that while some miRNAs were conserved across diverse organisms or throughout the nematodes, many were unique to these species. As genome annotation improves for parasitic nematodes, it is likely that some of these unique miRNAs may be found to be clade or niche specific. However, it does suggest that miRNAs are evolving rapidly, perhaps reflecting roles in host–parasite interactions. In general, H. contortus novel miRNAs were found to be expressed at a low level compared to conserved miRNAs, based on small RNA sequencing (Winter et al., 2012) and microarray data (Marks et al., 2019). This is consistent with findings in other organisms (Liang and Li, 2009; Shen et al., 2011) and suggests there is conservation of the machinery regulating miRNA gene expression, of which little is currently known. More recent work has identified motifs within the upstream promoter region of C. elegans miRNA genes that may determine expression level (Jovelin et al., 2016).
miRNAs in parasitic nematode development
In C. elegans, miRNAs lin-4 and let-7 were discovered through genetic studies, based on their essential roles in regulating genes involved in development. lin-4 suppresses expression of heterochronic gene lin-14 to allow progression from larval stage L1 to L2 (Lee et al., 1993), while let-7 modulates gene expression to promote adult development (Reinhart et al., 2000). For parasitic species, sequencing or microarray analysis can identify miRNAs expressed in different lifecycle stages, to help determine their roles in regulating development. However, we currently know little of the functions of parasite miRNAs and the genes they modulate. As mentioned above, bioinformatics programmes are available to predict interactions between specific miRNA sequences and target mRNAs. Some of these employ experimental validation, such as mirWIP, which incorporates immunoprecipitation (IP) data, using antibodies to miRISC (RNA-induced silencing complex) proteins AIN-1 and AIN-2, to score miRNA target sites (reviewed in Ambros and Ruvkun, 2018). However, currently many programmes rely on custom databases designed for specific species, such human, mouse, C. elegans, zebrafish (e.g. TargetScan; http://www.targetscan.org/vert_72/), with only a few programmes available that allow input of a test miRNA and 3′-UTR sequence. In addition, current assembly and annotation of most parasitic nematode genomes is not of sufficient quality to allow reliable identification of 3′-UTR sequences to predict miRNA binding sites. We were able to generate 3′-UTR datasets for H. contortus (Gillan et al., 2017), due to the advanced nature of the genome data (Laing et al., 2013). With developments in technology, such PacBio long read sequencing of RNA libraries (Iso-seq), improved annotation of UTRs in genomes of other nematode species should be feasible.
By investigating stage-specific miRNAs using microarrays, we identified the potential roles of some miRNAs in H. contortus and B. pahangi development. Enrichment of specific miRNAs was found in pre- and post-infective larval stages and in adult male and female worms for both species (Winter et al., 2015; Marks et al., 2019). For H. contortus, we focused on two miRNAs that were significantly enriched in the arrested L3 stage. Target prediction and gene ontology analysis suggested that these miRNAs suppress metabolism to maintain an arrested state. Comparative functional studies using C. elegans mutants indicated that the two miRNAs may suppress development by synergizing with DAF-16 FOXO transcription factor (TF) activity (Marks et al., 2019). By combining these different approaches, we progressed from initial identification of miRNAs for determining their expression patterns, and predicting target genes and potential roles in regulating development. miRNAs often act to fine-tune gene expression and can regulate key switches in developmental pathways. Therefore, identifying the potential target genes and pathways regulated by miRNAs is important in improving understanding of parasitic nematode development and could prove useful in designing novel therapeutic interventions.
Microarray analysis was similarly informative in identifying enrichment of miRNAs in specific lifecycle stages of the filarial nematode B. pahangi, from mosquito-derived L3, through to L4 and male and female adult worms (Winter et al., 2015). We focused on B. pahangi mir-5364, which is significantly upregulated in the post-infective L3 within 24 h of infection of a mammalian host and is a novel member of the let-7 family. Target prediction programmes identified transcripts encoding several putative TFs, and interaction between bpa-mir-5364 and the 3′-UTR of these mRNAs were confirmed experimentally using dual luciferase reporter assays. Further analysis of differentially expressed B. pahangi and H. contortus miRNAs and their target genes will help reveal the roles that miRNAs play in regulating nematode development at key points in infection.
miRNAs in host–parasite interactions
In recent years there has been an explosion of interest in extracellular vesicles (EVs). These are small vesicles, between 50 and 200 nm in size, released by cells and are considered to be important for intercellular communication. EVs are released by a range of cell types, including tumours, and their uptake by recipient cells may alter cellular activity. Advances in protein and RNA sequencing technology, combined with genome data, have allowed detailed analysis of EV cargo from mammalian cells and from parasitic nematodes (Buck et al., 2014; Zamanian et al., 2015; Tzelos et al., 2016; Gu et al., 2017; Tritten et al., 2017, Hansen et al., 2019). miRNAs and to a lesser extent Y RNAs (small non-coding RNAs involved in RNA quality control and DNA replication), have been identified within parasitic nematode EVs (Buck et al., 2014). It is speculated that packaging of small RNAs within EV may protect them from degradation and facilitate their uptake by other cell types.
Buck et al. (2014) first identified miRNAs in the ES supernatant and within EV released by adult worms of the mouse GIN Heligmosomoides polygyrus during in vitro culture. Notably, within EV there was enrichment for miRNAs with identical seed sequences to mammalian miRNAs, including mir-100, let-7, lin-4 and bantam. Subsequent small RNA sequencing of ES supernatant and EV released in vitro by H. contortus L4 and adult worms (Gu et al., 2017), B. pahangi L3 (Zamanian et al., 2015) and Ascaris suum adults (Hansen et al., 2019) also found enrichment of some of the same miRNAs. This raises the interesting possibility that parasite miRNAs may be mimicking or hijacking host cell gene regulation, perhaps for their own benefit (Buck et al., 2014). Notably, the helminth-associated cytokine IL-13 was identified as a predicted target of A. suum lin-4 and let-7 (Hansen et al., 2019). From an evolutionary perspective, as the sequences of these abundant, secreted miRNAs are conserved between the parasite and the host, avoidance of this putative parasite manipulation of host genes through mutation is less likely to occur (Claycomb et al., 2017).
While presence within EV could reflect higher abundance of specific miRNAs, there does seem to be selectivity in what is loaded into EVs. Some miRNAs, abundant in somatic tissue, are not present in EVs and studies in mammalian cell systems also suggest that the EV profile is not a snapshot of total cellular miRNAs (Driedonks et al., 2018). Determining how selectivity is achieved and the mechanisms by which parasite miRNAs are loaded into EVs require further work. This is likely to be guided by data from mammalian cell culture systems, demonstrating that RNA binding proteins recognize specific motifs that dictate miRNA exosomal sorting (Villarroya-Beltri et al., 2013; Shurtleff et al., 2016). EV released by adult worms of both H. polygyrus and H. contortus show enrichment of miRNAs homologous to miRNAs expressed in C. elegans gut cells (e.g. mir-60 and mir-236; Martinez et al., 2008) and also identified in the gut of H. contortus (Marks et al., 2019) and of the pig GIN A. suum (Gao et al., 2017). This suggests that, at least for some nematodes, EV miRNAs may be derived from the gut, perhaps reflecting the metabolic activity of this tissue (Buck et al., 2014). In contrast, immunolocalization using an antibody to ALG-2 interacting protein X, suggests that for the microfilariae stage of Brugia malayi (which has no functional gut), EV are released from the excretory pore (Harischandra et al., 2018).
Uptake of parasitic nematode EV by mammalian cells was demonstrated for H. polygyrus (Buck et al., 2014) and for B. malayi (Zamanian et al., 2015) using labelled EV. Importantly, following exposure to H. polygyrus EV, alteration of gene expression in recipient cells was observed: treated cells showed down-regulation of immune-associated genes il-33r and Dusp1. In addition, specific miRNAs enriched within the H. polygyrus EV suppressed expression of a Dusp-3′-UTR reporter construct. This suggested, for the first time, that secreted parasitic nematode miRNAs may regulate host immune outcome. Studies using EV from H. polygyrus or Trichuris muris have also demonstrated the protective potential of parasitic nematode EV (Coakley et al., 2017; Shears et al., 2018). As EV may deliver small RNAs and proteins with immunoregulatory effects, vaccination has the potential to neutralize their functions and enhance immunity. Indeed, a reduction in worm burden of around 50% was observed following immunization of mice with EV purified from ES products of adult H. polygyrus or T. muris. Importantly, EV vaccination induced high levels of antibodies to EV and to ES supernatant, suggesting recognition of shared or cross-reactive epitopes that may limit infection. In addition, Coakley et al. (2017) detailed the suppressive effects of H. polygyrus EV on activation of both classical and alternatively stimulated macrophages. Importantly, they showed that exposure of macrophages to anti-EV antibodies abrogated this EV-mediated suppression. By tracking labelled EV within macrophage cells it was observed that, in the presence of anti-EV antibodies, EV localization was altered and led to accumulation within lysosomes, which reduced EV immunosuppressive effects. Whether the protective effect of EV vaccination may be mediated by neutralization of EV small RNA or protein function, or both, is currently unknown, however these studies are important in stimulating further investigation of the potential of EV to deliver parasite antigens for enhanced protection (Shears et al., 2018).
miRNAs released from GIN have not been identified in the serum or plasma of infected hosts (Buck et al., 2014; Britton et al., 2015), suggesting that they may act locally within the gastrointestinal tract. Consistent with this hypothesis, we were able to detect parasite-specific miRNAs in abomasal tissue and draining lymph nodes collected from H. contortus-infected, but not uninfected, sheep (Gu et al., 2017). Holz and Streit (2017) also failed to detect small RNAs of the GIN Strongyloides ratti in infected rat blood following infection. In contrast, parasite-derived miRNAs have been detected in the serum or plasma of hosts infected with tissue-dwelling parasitic nematodes, including the filariae Dirofilaria immitis (Tritten et al., 2014a), Litomosoides sigmodontis (Buck et al., 2014), Onchocerca volvulus and Onchocerca ochengi (Tritten et al., 2014a, 2104b; Quintana et al., 2015) or with the flatworm Schistosoma japonicum (He et al., 2013; Hoy et al., 2014). It was proposed that these released miRNAs may be involved in host–parasite interactions, although whether circulating parasite miRNAs have any effects on host gene expression are not yet known. Their release into the circulation has also stimulated interest in exploiting filarial and schistosome miRNAs as novel biomarkers of infection (reviewed in Cai et al., 2016 and Quintana et al., 2017). Parasite infection has also been shown to modulate expression of host miRNAs and many of these regulate genes involved in host innate and adaptive immune mechanisms. This has been detailed in previous reviews (e.g. Arora et al., 2017; Entwistle and Wilson, 2017). It is interesting to speculate on whether host miRNAs or other small RNAs could have any effect on the parasite. Uptake of labelled small RNAs by parasitic helminths maintained in vitro has been shown (e.g. Winter et al., 2014; Britton et al., 2016; Anandanarayanan et al., 2017). Whether sufficient levels of small RNAs could be transferred from the host to the parasite and whether these may be complexed to Ago or other RNA-binding proteins warrants further investigation to determine if host miRNAs could influence, for example, worm growth or reproduction.
Interestingly, recent work has shown that the widely-used anthelmintic drug ivermectin can reduce the release of EV from all life cycle stages of B. malayi (Mf, L3 and adult males and females) 24 h after exposure in vitro (Harischandra et al., 2018). The same approach showed a reduction in EV release from L3 of the related canine filarial nematode D. immitis, but not from an ivermectin resistant isolate, suggesting that the effect may be drug-specific (Harischandra et al., 2018). Ivermectin was previously shown to inhibit protein secretion by B. malayi microfilariae, leading to the hypothesis that one effect of ivermectin may be to reduce the release of immunosuppressive molecules from the excretory pore and thus enhance parasite clearance (Moreno et al., 2010). It is also possible that blocking the release and function of protein and/or small RNAs within EV may be responsible for this effect.
Piwi-interacting RNAs
Identification of piRNAs
While miRNAs repress mRNA translation by interacting with the Ago subfamily of Argonaute proteins, a different class of small RNA interacts with Piwi Argonautes (Seto et al., 2007). These are referred to as Piwi-interacting RNAs (piRNAs) of approximately 21–30 nucleotides in length that function mainly to silence mobile genetic elements, such as transposons. piRNAs have been identified from C. elegans and other animals, including Drosophila and mouse, and are required for fertility by protecting the germline from transposon insertion (Siomi et al., 2011).
The discovery that Piwi proteins interacted with small RNAs that were distinct from miRNA and siRNAs was first made in Drosophila. Sequencing of these piRNAs revealed that they mapped to retrotransposons and other repetitive sequences and were referred to as repeat-associated siRNAs (Saito et al., 2006). Parallel studies in mammals also identified Piwi-interacting small RNAs showing great complexity of sequence but with less identity to repetitive elements compared to those from flies (Lau et al., 2006). The diversity of piRNA sequences, both within and between species, makes their identification difficult, although in most species in which they have been identified, piRNAs are found clustered in the genome and are characterized by the presence of uracil as the 5′ nucleotide, with 5′ monophosphate and 2′-O-methyl 3′ termini (Brennecke et al., 2007).
While the function of piRNAs is conserved in different organisms, their biogenesis and mechanisms of silencing have diverged through evolution. In mouse and Drosophila, piRNAs are produced from long single-stranded precursors that are processed into multiple piRNA sequences (Brennecke et al., 2007). In contrast to miRNAs and siRNAs, processing does not depend in Dicer ribonuclease. Following the generation of initial primary piRNAs, sequences targeting active transposons are amplified by slicing of the target RNA to give rise to secondary piRNAs (referred to as the ping-pong cycle) (Gunawardane et al., 2007). It is thought that this amplification fine-tunes the piRNA response against active transposons (reviewed in Siomi et al., 2011).
Nematode piRNAs
Caenorhabditis elegans piRNAs differ in several aspects to those in other animals. In C. elegans, mature piRNAs are characterized as 21 nucleotides in length that are produced from short precursors of 26–30 nucleotides expressed from individual genetic loci (Weick et al., 2014). They have a bias for uracil as the 5′ nucleotide (referred to as 21U-RNAs), in common with mature piRNAs in other species, and possess an upstream promoter motif (GTTTC) (Batista et al., 2008). Rather than slicing their target RNA sequence, C. elegans piRNAs direct binding of the piRISC to the target sequence, which initiates the synthesis of siRNA molecules complementary to the target (see Fig. 1). These siRNA molecules, referred to as 22G-RNAs, also mediate target gene silencing in the RNAi pathway (see below). Importantly, piRNAs are present in parasitic nematodes but only those of clade V. Their absence from other nematode clades stimulated a study to discover alternative pathways involved in transposon silencing (Sarkies et al., 2015). This identified 22G-RNAs, mapping antisense to transposons, in nematodes of clades III and IV, while a different mechanism, involving Dicer-dependent RNA-directed DNA methylation, functions in nematode clades I and II. Currently, it is not known why Piwi and piRNAs are maintained in clade V species.
The above features of C. elegans piRNAs are conserved in the clade V parasitic nematodes in which they have been identified, including Pristionchus pacificus, H. contortus, H. polygyrus and Nippostrongylus brasiliensis, based on small RNA sequencing and genome data (de Wit et al., 2009; Winter et al., 2012; Buck et al., 2014; Sarkies et al., 2015), although their organization is different. Two large clusters of piRNAs are found in C. elegans on chromosome IV (Ruby et al., 2006), while clustering of piRNA loci is not observed in P. pacificus (de Wit et al., 2009) nor in H. contortus (Winter et al., 2012). Recent work by Beltran et al. (2019) also identified differences in the chromatin structure of domains encoding piRNAs between nematode species, suggesting different modes of regulation. Two types of genomic organization of piRNA genes were characterized: P-type (e.g. P. pacificus) in which piRNA loci are found within active chromatin (H3K36me3), and C-type (e.g. C. elegans) where piRNA genes are in regions of repressive chromatin, associated with H3K27me3. How and why these different control mechanisms have evolved and whether they can co-exist are areas of ongoing investigation.
From H. contortus small RNA libraries, we identified >1000 reads representing putative piRNA sequences in adult worms, but not in L3 stage, consistent with a role in germline development and maintenance (Winter et al., 2012). piRNAs were not identified in Brugia (Winter et al., 2012) nor A. suum (Wang et al., 2011). We, and others, speculated that piRNAs may be required for adaptation/survival of progeny in varying environmental conditions, such as differences in temperature (Wang et al., 2011; Winter et al., 2012). However, piRNAs are not found in Strongyloides nematodes (clade IV) that have environmental larval stages (Holz and Streit, 2017). Sarkies et al. (2015) suggested that there may be clade V-specific transposable elements that require piRNA for silencing, rather than other small RNA pathways. This hypothesis is consistent with the differences in transposable element loads across nematode clades identified by Szitenberg et al. (2016). Caenorhabditis elegans mutants of the piRNA Argonaute PRG-1 show fertility defects and become sterile over many generations (Simon et al., 2014), demonstrating the importance of piRNAs in protecting the germline against transposon-mediated mutations. Interestingly, in gonochorist species, that mate every generation, there are greater numbers of piRNAs than in androdioecious species (self-fertilize) (Shi et al., 2013). This suggests that additional piRNAs may be required to defend against paternal transposons. Of potential relevance to this, female worms of the clade V parasitic nematodes H. contortus and T. circumcincta are known to be polyandrous (mate with multiple males) (Redman et al., 2008; Doyle et al., 2017). It is interesting to speculate that the piRNA pathway may be required in these clade V parasites to maintain genome viability in the face of high levels of paternal transposon mixing.
Other functions of piRNAs
Recent small RNA sequencing has also identified differences in the number of piRNAs in different strains of H. contortus (Laing and Sarkies, unpublished data). Interestingly, this analysis compared H. contortus adult worms that are susceptible to anthelmintic drugs (MHco3.ISE) with a drug-resistant strain (MHco18.UGA). Whether these differences in piRNA levels have any consequence on drug sensitivity or may reflect different traits between the strains is not yet known.
While Piwi proteins and piRNAs localize predominantly to the germline, they have also been found in somatic stem cells. As germ cells and stem cells have the ability to replicate, the function of piRNAs in both may be to maintain genome integrity. The planarian flatworm Schmidtea mediterranea is used as model for stem cell proliferation and differentiation due to the presence of somatic stem cells (neoblasts) that allow tissue regeneration (Reddien and Sanchez Alvarado, 2004). Importantly, inhibition of S. mediterranea Piwi-encoding genes smedwi-2 or -3 resulted in lower abundance of piRNAs and failure of neoblast renewal and differentiation (Reddien et al., 2005). Studies in other flatworms and in cnidarians reported similar findings (De Mulder et al., 2009; Juliano et al., 2013). Piwi and piRNAs have been identified in lineage-restricted stem cells that do not give rise to germline cells, suggesting that they function to regulate gene expression and protect the genome in other cell types outside of the germline (reviewed in van Wolfswinkel, 2014). The progress being made in understanding the mechanisms and functions of piRNAs and Piwi proteins will help reveal new mechanisms of gene control.
Small interfering RNAs (siRNAs)
siRNAs are approximately 21–25 bases in length and, like miRNAs, are derived from longer double-stranded RNA (dsRNA) molecules processed by Dicer. The dsRNA precursor can be derived from infective pathogens, such as viruses, endogenous genes, or be introduced artificially into cells to induce RNAi-mediated gene silencing. siRNAs bind to siRISC, with the antisense strand guiding the active RISC to its target mRNA, which is cleaved by Argonaute protein within RISC (Hammond et al., 2000) (Fig. 1). In contrast to miRNAs, which can potentially interact with hundreds of target mRNAs through seed sequence interaction, siRNAs bind with sequence complementarity along their entire length to direct specific gene silencing. The RNAi pathway was first identified in C. elegans (Fire et al., 1998) and shown to be a natural defense against viral infection (Felix et al., 2011). The discovery that siRNAs could also silence genes in mammalian cells (Elbashir et al., 2001) (but not dsRNA >30 bp, which induces an interferon response) stimulated great interest in developing these small RNAs as potential therapeutics. A number of siRNAs targeting specific genes of viruses or cancer have been tested (reviewed in Haussecker, 2012 and Chakraborty et al., 2017), although ensuring specificity of silencing, with no off target effects, and delivery of siRNAs into cells, can be challenging.
RNAi in parasitic nematodes
Following the success of RNAi in C. elegans, a number of studies tested whether this approach could be applied as a functional genomics tool to identify novel drug and/or vaccine candidates for parasitic helminths (e.g. Hussein et al., 2002; Aboobaker and Blaxter, 2003; Skelly et al., 2003; Lustigman et al., 2004; Geldhof et al., 2006). While some parasite genes could be silenced, RNAi was not as effective as in C. elegans, particularly when dsRNA or siRNA was delivered by soaking (referred to as environmental RNAi). Our work in H. contortus showed that while some genes could be silenced using the soaking method, this was most effective for genes expressed in sites accessible to the environment, such as the gut, amphids and excretory cell. This suggested limited uptake and/or spreading of dsRNA in the non-feeding infective L3 stage used (Samarasinghe et al., 2011). This was supported by comparative genomic studies showing that while most genes required for siRNA-mediated gene silencing can be identified in parasitic nematodes (Dalzell et al., 2011; reviewed in Britton et al., 2016), a homologue of the transmembrane transporter SID-2, required for environmental RNAi in C. elegans (Winston et al., 2007), was absent in parasitic species.
More recent work has focused on delivering siRNA to the pre-infective L2 stage of H. contortus, which feed constitutively, in contrast to the more easily available, but non-feeding, infective L3 stage used previously. These recent studies by Blanchard et al. (2018) and Menez et al. (2019) were successful in determining a role for acetylcholine receptor subunit Hco-ACR-8 in mediating levamisole sensitivity and for Hco-NHR-8 in conferring tolerance to ivermectin, respectively. Their approach suggests that with improvements to the RNAi toolbox, including optimized delivery of siRNA, parasite stage examined, and phenotypic assays available, RNAi still holds promise for determining gene function. In addition, delivery by viral vectors may overcome the transient effect of exogenous siRNA or dsRNA. Effective gene silencing has been demonstrated following transfection of schistosome parasites with viral vectors expressing siRNAs (Hagen et al., 2014) and is being tested for the mouse GIN N. brasiliensis (Hagen and Selkirk, personal communication). Direct microinjection of siRNA or dsRNA can also be effective at gene silencing. The addition of lipofectamine to the microinjection mix used to deliver dsRNA was shown to enhance RNAi efficacy in a newly described nematode genus, Auanema, and to facilitate RNAi in an otherwise ‘resistant’ species, P. pacificus (Adams et al., 2019). These recent successes in determining function from RNAi phenotype, and development of new siRNA delivery methods, should stimulate further studies to progress from genome to function, in parallel with development of CRISPR/Cas9 gene editing for nematode species (Gang et al., 2017).
Forms and functions of nematode siRNAs
From sequencing studies in C. elegans, different types of endogenous siRNAs have been identified. These are: 26G siRNAs, 26 nucleotides long with a bias for guanosine monophosphate at the 5′ end, produced by RNA-dependent RNA-polymerase (RdRP) RRF-3; and 22G siRNAs, 22 nucleotides long with a 5′ bias for guanosine triphosphate, produced by RdRPs RRF-1 and EGO-1 (reviewed in Almeida et al., 2019). 22Gs have been identified as the secondary siRNAs that are produced downstream of exogenous dsRNA, 26G RNAs and piRNAs (Fig. 1). They map antisense to target transcripts and, in C. elegans, are responsible for amplification of the RNAi response (Sijen et al., 2001). Interestingly, while siRNAs can silence target gene sequences within nematodes, recent work from the Buck lab suggests that siRNAs may also be involved at the host–parasite interface. Much of the data on small RNAs present within EVs have focused on miRNAs and their potential targets within host cells. However, Chow et al. (2019) generated small RNA datasets from EVs that included only 5′ monophosphate RNAs (miRNAs, piRNAs, 26G siRNAs) or alternatively, all small RNAs. The latter included those with 5′ triphosphate, using 5′ polyphosphatase treatment, not carried out in previous studies of EV small RNAs. This identified enrichment of 23G triphosphate secondary siRNAs (equivalent to C. elegans 22G siRNAs) in EVs from adult Heligmosomoides bakeri (H. polygyrus) and these were bound to the worm-specific Argonaute WAGO. Interestingly, these siRNAs mapped to recently evolved regions and repeat sequences in the parasite genome. It is not yet known if or how these may be taken up by host cells and what roles they may play either within the host or within the parasite.
Conclusions and future directions
Genome data have enabled the identification of small RNA classes in parasitic nematodes. The challenge is to better understand the functions of these, both within the parasite and in host–parasite interactions.
With more advanced genome assembly and annotation, target identification based on miRNA–3′-UTR interaction, will improve. In addition, it is now feasible to sequence the transcripts expressed by each individual cell of an organism, including nematodes, using single cell RNA sequencing (Cao et al., 2017). Profiling of mRNAs and miRNAs in each cell can determine when and where these are expressed and, through inverse correlation analysis, identify potential regulatory functions (Wang et al., 2019). Microscopic or laser dissection of specific nematode tissues or cells, followed by mRNA and miRNA sequencing, is an alternative approach to reveal potential miRNA–mRNA networks. Dissection is possible with larger adult stage parasites and miRNA microarray profiling was successful for H. contortus gut tissue, isolated from adult female worms (Marks et al., 2019). However, confirmation of miRNA–mRNA interaction requires experimental verification, such as IP studies using antibodies to Argonaute proteins to isolate interacting complexes. While this has been achieved in C. elegans, only a few antibodies specific to parasite RNA binding proteins have been generated. These are available for ALG-2 interacting protein X (Harischandra et al., 2018) and extracellular WAGO, associated with EV of H. bakeri (Chow et al., 2019), but not yet for the major RISC complex Argonautes. IP studies, together with gene knockout approaches, would greatly advance knowledge of the specific interactions and functions of parasite small RNAs.
Little is currently known about how expression of small RNAs is regulated. Detailed studies in C. elegans and related species are beginning to reveal the mechanisms involved. Intergenic miRNAs have promoters similar to those of protein-coding genes and recent work has identified specific sequence motifs that determine expression pattern (Jovelin et al., 2016). As gene annotation improves, the same approach, using motif discovery tool MEME, can be applied to parasitic species. Interestingly, while miRNAs may be regulated by TFs, a number of miRNAs are known to target TFs, forming regulatory loops (Shalgi et al., 2007). Further details of the regulatory mechanisms and networks of miRNAs will help identify their control, function and evolution.
piRNAs target transposons and give rise to the 22G class of small RNAs. While 22G-RNAs are present in all nematode clades, piRNAs are restricted to clade V. It would be of interest to determine if this reflects a biological/genetic feature of this clade, such as mating behaviour, larger number of progeny, larger genome size for some species, and whether piRNAs are essential for fertility in parasitic species. It may be speculated that in sexually reproducing worms, and especially in polyandrous species, piRNAs may protect against male transposons to allow fertilization and development of progeny. Interestingly, Sargison et al. (2019) observed a reduced proportion of inter-strain hybrid F1 progeny developing following genetic mating of different H. contortus strains, relative to that observed following intra-strain mating. The underlying mechanisms of this post-zygotic incompatibility are as yet unknown but it would be important to determine if differences in expression of piRNAs or 22G-RNAs between strains could be responsible. As discussed above, approaches to enhance RNAi efficacy in parasitic nematodes are in progress; in the near future it may therefore be feasible to determine the roles of each class of small RNA, through knockout/knockdown of genes encoding Argonaute, RNA biogenesis or RNA binding proteins, such as PRG-1, required for piRNA function, as well as inhibition of specific small RNAs.
In addition to roles within parasitic nematodes, small RNAs, predominantly miRNAs, have been sequenced from EV released by parasites in vitro (Buck et al., 2014; Zamanian et al., 2015; Gu et al., 2017). As EV can be taken up by host cells, it is thought that these transported miRNAs play a role at the host–parasite interface. Development of tagged parasite miRNAs or knockout of miRNAs, combined with recipient host cell analysis, would be a useful approach to determine potential targets and functions within host cells. Studies to date have characterized EV released in vitro; whether EVs are released in vivo and transport the same RNAs and proteins is unknown. Evidence indicates release of miRNAs during infection with filarial (clade III) parasites and schistosomes, although it is not clear if these are released freely or may be derived from degraded EV or dying worms. Detailed analysis of small RNAs in EV of H. bakeri revealed, for the first time, the presence of 23G siRNAs (Chow et al., 2019). These are proposed to target repeat sequences and novel genomic regions of the parasite and were associated with Argonaute WAGO. While their abundance within EV may suggest host–parasite interaction, EV could also be a means of parasite–parasite communication. Determining the complete small RNA profile of EV from other parasitic nematode species, as well as C. elegans, and whether this changes following exposure to different environmental conditions may help determine if this is a novel and potentially important route of communication between worms.
In conclusion, recent studies have revealed novel mechanisms of small RNA regulation and packaging, helping advance our understanding of the diverse roles of these RNAs. Further efforts to effectively silence parasite genes by RNAi-mediated pathways will continue to reveal the importance of the different RNA classes in parasite development, communication, protection from invading genetic elements and in host–parasite interactions.
Acknowledgements
We acknowledge the input of Alan Winter, Neil Marks, Henry Gu, Victoria Gillan and Kirsty Maitland to some of the work described here.
Financial support
Funding for some of the work reviewed here was provided by The Wellcome Trust in project grants awarded to ED and CB (WT 086823/Z/08/Z and WT 094751) and by PhD studentships funded by UK Biotechnology and Biological Sciences Research Council (BBSRC; BB/J500732)/Knowledge Transfer Network (KTN)/Zoetis, and by EBLEX and the University of Glasgow. RL is supported by BBSRC BUG consortium LoLa grant (BB/M003949).
Conflict of interest
None.
Ethical standards
Not applicable.
References
- Aboobaker AA and Blaxter ML (2003) Use of RNA interference to investigate gene function in the human filarial nematode parasite Brugia malayi. Molecular and Biochemical Parasitology 129, 41–51. [DOI] [PubMed] [Google Scholar]
- Ahmed R, Chang Z, Younis AE, Langnick C, Li N, Chen W, Brattig N and Dieterich C (2013) Conserved miRNAs are candidate post-transcriptional regulators of developmental arrest in free-living and parasitic nematodes. Genome Biology and Evolution 5, 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams S, Pathak P, Shao H, Lok JB and Pires-daSilva A (2019) Liposome-based transfection enhances RNAi and CRISPR-mediated mutagenesis in non- model nematode systems. Scientific Reports 9, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MV, Andrade-Navarro MA and Ketting RF (2019) Function and evolution of nematode RNAi pathways. Non-coding RNA 5, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V and Ruvkun G (2018) Recent molecular genetic explorations of Caenorhabditis elegans microRNAs. Genetics 209, 651–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandanarayanan A, Raina OK, Lalrinkima H, Rialch A, Sankar M and Varghese A (2017) RNA interference in Fasciola gigantica: establishing and optimization of experimental RNAi in the newly excysted juveniles of the fluke. PLoS Neglected Tropical Diseases 11, e0006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora N, Tripathi S, Singh AK, Mondal P, Mishra A and Prasad A (2017) Micromanagement of immune system: role of miRNAs in helminthic infections. Frontiers Microbiology 8, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes C, Fehlmann T, Kern F, Kehl T, Lenhof H-P, Meese E and Keller A (2018) miRCarta: a central repository for collecting miRNA candidates. Nucleic Acids Research 46, D160–D167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018) Metazoan microRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D Jr, Luo S, Schroth GP, Carrington JC, Bartel DP and Mello CC (2008) PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Molecular Cell 31, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran T, Barroso C, Birkle TY, Stevens L, Schwartz HT, Sternberg PW, Fradin H, Gunsalus K, Piano F, Sharma G, Cerrato C, Ahringer J, Martınez-Perez E, Blaxter M and Sarkies P (2019) Comparative epigenomics reveals that RNA polymerase II pausing and chromatin domain organization control nematode piRNA biogenesis. Developmental Cell 48, 793–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard A, Guegnard F, Charvet CL, Crisford A, Courtot E, Sauve C, Harmache A, Duguet T, O'Connor V, Castagnone-Sereno P, Reaves B, Wolstenholme AJ, Beech RN, Holden-Dye L and Neveu C (2018) Deciphering the molecular determinants of cholinergic anthelmintic sensitivity in nematodes: when novel functional validation approaches highlight major differences between the model Caenorhabditis elegans and parasitic species. PLoS Pathogens 14, e1006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R and Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Britton C, Winter AD, Marks ND, Gu H, McNeilly TN, Gillan V and Devaney E (2015) Application of small RNA technology for improved control of parasitic helminths. Veterinary Parasitology 212, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton C, Marks ND and Roberts AB (2016) Functional genomics tools for Haemonchus contortus and lessons from other helminths. Advances in Parasitology 93, 599–623. [DOI] [PubMed] [Google Scholar]
- Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, Kumar S, Abreu-Goodger C, Lear M, Harcus Y, Ceroni A, Babayan SA, Blaxter M, Ivens A and Maizels RM (2014) Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nature Communications 5, 5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P, Gobert GN and McManus DP (2016) MicroRNAs in parasitic helminthiases: current status and future perspectives. Trends in Parasitology 32, 71–86. [DOI] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C and Shendure J (2017) Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Sharma G, Doss CGP and Lee S-S (2017) Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Molecular Therapy: Nucleic Acids 8, 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M and Filipowicz W (2009) Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Current Opinion in Cell Biology 21, 452–460. [DOI] [PubMed] [Google Scholar]
- Chen MX, Ai L, Xu MJ, Zhang RL, Chen SH, Zhang YN, Guo J, Cai YC, Tian LG, Zhang LL, Zhu XQ and Chen JX (2011a) Angiostrongylus cantonensis: identification and characterization of microRNAs in male and female adults. Experimental Parasitology 128, 116–120. [DOI] [PubMed] [Google Scholar]
- Chen MX, Ai L, Xu MJ, Chen SH, Zhang YN, Guo J, Cai YC, Tian LG, Zhang LL, Zhu XQ and Chen JX (2011b) Identification and characterization of microRNAs in Trichinella spiralis by comparison with Brugia malayi and Caenorhabditis elegans. Parasitology Research 109, 553–558. [DOI] [PubMed] [Google Scholar]
- Chow FW-N, Koutsovoulos G, Ovando-Vazquez C, Neophytou K, Bermudez-Barrientos JR, Laetsch DR, Robertson E, Kumar S, Claycomb JM, Blaxter M, Abreu-Goodger C and Buck AH (2019) Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Research 47, 3594–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J, Abreu-Goodger C and Buck AH (2017) RNA-mediated communication between helminths and their hosts: the missing links. RNA Biology 14, 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley G, McCaskill JL, Borger JG, Simbari F, Robertson E, Millar M, Harcus Y, McSorley HJ, Maizels RM and Buck AH (2017) Extracellular vesicles from a helminth parasite suppress macrophage activation and constitute an effective vaccine for protective immunity. Cell Reports 19, 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalzell JJ, McVeigh P, Warnock ND, Mitreva M, Bird DM, Abad P, Fleming CC, Day TA, Mousley A, Marks NJ and Maule AG (2011) RNAi effector diversity in nematodes. PLoS Neglected Tropical Diseases 5, e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mulder K, Pfister D, Kuales G, Egger B, Salvenmoser W, Willems M, Steger J, Fauster K, Micura R, Borgonie G and Ladurner P (2009) Stem cells are differentially regulated during development, regeneration and homeostasis in flatworms. Developmental Biology 334, 198–212. [DOI] [PubMed] [Google Scholar]
- Devaney E, Winter AD and Britton C (2010) MicroRNAs: a role in drug resistance in parasitic nematodes? Trends in Parasitology 26, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Linsen SEV, Cuppen E and Berezikov E (2009) Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Research 19, 2064–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SR, Laing R, Bartley DJ, Britton C, Chaudhry U, Gilleard JS, Holroyd N, Mable BK, Maitland K, Morrison AA, Tait A, Tracey A, Berriman M, Devaney E, Cotton JA and Sargison ND (2017) A genome resequencing-based genetic map reveals the recombination landscape of an outbred parasitic nematode in the presence of polyploidy and polyandry. Genome Biology and Evolution 10, 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driedonks TAP, van der Grein SG, Ariyurek Y, Buermans HPJ, Jekel H, Chow FWN, Wauben MHM, Buck AH, ’t Hoen PAC and Nolte-’t Hoen ENM (2018) Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cellular and Molecular Life Sciences 75, 3857–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K and Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Entwistle LJ and Wilson MS (2017) MicroRNA-mediated regulation of immune responses to intestinal helminth infections. Parasite Immunology 39, e12406. [DOI] [PubMed] [Google Scholar]
- Felix M-A, Ashe A, Piffaretti J, Wu G, Nuez I, Belicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA and Wang D (2011) Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biology 9, e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE and Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W and Rajewsky N (2012) MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Research 40, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E and Peterson KJ (2015) A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annual Review of Genetics 49, 213–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Domanska D, Høye E, Ovchinnikov V, Kang W, Aparicio-Puerta E, Johansen M, Flatmark K, Mathelier A, Hovig E, Hackenberg M, Friedländer MR and Peterson KJ (2020) MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Research 48, D132–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Lan J, Wu X, Yang D, Zhang Z, Nie H, Hou R, Zhang R, Zheng W, Xie Y, Yan N, Yang Z, Wang C, Luo L, Liu L, Gu X, Wang S, Peng X and Yang G (2013) Identification of Dirofilaria immitis miRNA using illumina deep sequencing. Veterinary Research 44, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang SS, Castelletto ML, Bryant AS, Yang E, Mancuso N, Lopez JB, Pellegrini M and Hallem EA (2017) Targeted mutagenesis in a human-parasitic nematode. PLoS Pathogens 13, e1006675. 10.1371/journal.ppat.1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tyagi R, Magrini V, Ly A, Jasmer DP and Mitreva M (2017) Compartmentalization of functions and predicted miRNA regulation among contiguous regions of the nematode intestine. RNA Biology 14, 1335–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldhof P, Murray L, Couthier A, Gilleard JS, McLauchlan G, Knox DP and Britton C (2006) Testing the efficacy of RNA interference in Haemonchus contortus. International Journal for Parasitology 36, 801e810. [DOI] [PubMed] [Google Scholar]
- Ghasabi M, Mansoori B, Mohammadi A, Duijf PH, Shomali N, Shirafkan N, Mokhtarzadeh A and Baradaran B (2019) MicroRNAs in cancer drug resistance: basic evidence and clinical applications. Journal of Cellular Physiology 234, 2152–2168. [DOI] [PubMed] [Google Scholar]
- Gillan V, Maitland K, Laing R, Gu H, Marks ND, Winter AD, Bartley D, Morrison A, Skuce PJ, Rezansoff AM, Gilleard JS, Martinelli A, Britton C and Devaney E (2017) Increased expression of a microRNA correlates with anthelmintic resistance in parasitic nematodes. Frontiers in Cellular and Infection Microbiology 7, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S and Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Research 36, D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu HY, Marks ND, Winter AD, Weir W, Tzelos T, McNeilly TN, Britton C and Devaney E (2017) Conservation of a microRNA cluster in parasitic nematodes and profiling of miRNAs in excretory-secretory products and microvesicles of Haemonchus contortus. PLoS Neglected Tropical Diseases 11, e0006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H and Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590. [DOI] [PubMed] [Google Scholar]
- Hagen J, Young ND, Every AL, Pagel CN, Schnoeller C, Scheerlinck J-PY, Gasser RB and Kalinna BH (2014) Omega-1 knockdown in Schistosoma mansoni eggs by lentivirus transduction reduces granuloma size in vivo. Nature Communications 5, 5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ and Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond S, Bernstein E, Beach D and Hannon G (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Hansen EP, Fromm B, Andersen SD, Marcilla A, Andersen KL, Borup A, Williams AR, Jex AR, Gasser RB, Young ND, Hall RS, Stensballe A, Ovchinnikov V, Yan Y, Fredholm M, Thamsborg SM and Nejsum P (2019) Exploration of extracellular vesicles from Ascaris suum provides evidence of parasite-host cross talk. Journal of Extracellular Vesicles 8, 1578116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harischandra H, Yuan W, Loghry HJ, Zamanian M and Kimber MJ (2018) Profiling extracellular vesicle release by the filarial nematode Brugia malayi reveals sex-specific differences in cargo and a sensitivity to ivermectin. PLoS Neglected Tropical Diseases 12, e0006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D (2012) The business of RNAi therapeutics in 2012. Molecular Therapy-Nucleic Acids 1, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Sai X, Chen C, Zhang Y, Xu X, Zhang D and Pan W (2013) Host serum miR-223 is a potential new biomarker for Schistosoma japonicum infection and the response to chemotherapy. Parasites & Vectors 6, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M and Schuster P (1994) Fast folding and comparison of RNA secondary structures. Chemical Monthly 125, 167–188. [Google Scholar]
- Holz A and Streit A (2017) Gain and loss of small RNA classes – characterization of small RNAs in the parasitic nematode family Strongyloididae. Genome Biology and Evolution 9, 2826–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy AM, Lundie RJ, Ivens A, Quintana JF, Nausch N, Forster T, Jones F, Kabatereine NB, Dunne DW, Mutapi F, MacDonald AS and Buck AH (2014) Parasite-derived microRNAs in host serum as novel biomarkers of helminth infection. PLoS Neglected Tropical Diseases 8, e2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein AS, Kichenin K and Selkirk ME (2002) Suppression of secreted acetylcholinesterase expression in Nippostrongylus brasiliensis by RNA interference. Molecular and Biochemical Parasitology 122, 91–94. [DOI] [PubMed] [Google Scholar]
- Jovelin R, Krizus A, Taghizada B, Gray JC, Phillips PC, Claycomb JM and Cutter AD (2016) Comparative genomic analysis of upstream miRNA regulatory motifs in Caenorhabditis. RNA 22, 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Reich A, Liu N, Gotzfried J, Zhong M, Uman S, Reenan RA, Wessel GM, Steele RE and Lin H (2013) PIWI proteins and Piwi-interacting RNAs function in Hydra somatic stem cells. Proceedings of the National Academy of Sciences USA 111, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J and Siomi MC (2009) Biogenesis of small RNAs in animals. Nature Reviews Molecular and Cellular Biology 10, 126–139. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W and Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. [DOI] [PubMed] [Google Scholar]
- Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, Holroyd N, Bartley DJ, Beasley H, Britton C, Curran D, Devaney E, Gilabert A, Hunt M, Jackson F, Johnston SL, Kryukov I, Li K, Morrison AA, Reid AJ, Sargison N, Saunders GI, Wasmuth JD, Wolstenholme A, Berriman M, Gilleard JS and Cotton JA (2013) The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biology 14, R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG and Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP and Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science 313, 363–367. [DOI] [PubMed] [Google Scholar]
- Lee RC and Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL and Ambros V (1993) The C. elegans Heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB and Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Liang H and Li WH (2009) Lowly expressed human microRNA genes evolve rapidly. Molecular Biology and Evolution 26, 1195–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustigman S, Zhang J, Liu J, Oksov Y and Hashmi S (2004) RNA interference targeting cathepsin L and cathepsin Z-like cysteine proteases of Onchocerca volvulus confirmed their essential function during L3 molting. Molecular and Biochemical Parasitology 138, 165–170. [DOI] [PubMed] [Google Scholar]
- Ma G, Luo Y, Zhu H, Luo Y, Korhonen PK, Young ND, Gasser RB and Zhou R (2016) MicroRNAs of Toxocara canis and their predicted functional roles. Parasites & Vectors 9, 229. doi: 10.1186/s13071-016-1508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks ND, Winter AD, Gu HY, Maitland K, Gillan V, Ambrož M, Martinelli A, Laing R, MacLellan R, Towne J, Roberts B, Hanks E, Devaney E and Britton C (2019) Profiling microRNAs through development of the parasitic nematode Haemonchus identifies nematode-specific miRNAs that suppress larval development. Scientific Reports 9, 17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR and Walhout AJ (2008) Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Research 18, 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menez C, Alberich M, Courtot E, Guegnard F, Blanchard A, Aguilaniu H and Lespine A (2019) The transcription factor NHR-8: a new target to increase ivermectin efficacy in nematodes. PLoS Pathogens 15, e1007598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Y, Nabhan JF, Solomon J, Mackenzie CD and Geary TG (2010) Ivermectin disrupts the function of the excretory-secretory apparatus in microfilariae of Brugia malayi. Proceedings of the National Academy of Sciences USA 107, 20120–20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI and Maller B (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. [DOI] [PubMed] [Google Scholar]
- Poole CB, Davis PJ, Jin J and McReynolds LA (2010) Cloning and bioinformatic identification of small RNAs in the filarial nematode, Brugia malayi. Molecular and Biochemical Parasitology 169, 87–94. [DOI] [PubMed] [Google Scholar]
- Poole CB, Gu W, Kumar S, Jin J, Davis PJ, Bauche D and McReynolds LA (2014) Diversity and expression of microRNAs in the filarial parasite, Brugia malayi. PLoS One 9, e96498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana JF, Makepeace BL, Babayan SA, Ivens A, Pfarr KM, Blaxter M, Debrah A, Wanji S, Ngangyung HF, Bah GS, Tanya VN, Taylor DW, Hoerauf A and Buck AH (2015) Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasites and Vectors 8, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana JF, Babayan SA and Buck AH (2017) Small RNAs and extracellular vesicles in filarial nematodes: from nematode development to diagnostics. Parasite Immunology 39, e12395. [DOI] [PubMed] [Google Scholar]
- Reddien PW and Sanchez Alvarado A (2004) Fundamentals of planarian regeneration. Annual Review of Cell and Developmental Biology 20, 725–757. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC and Sanchez Alvarado A (2005) SMEDWI-2 is a Piwi-like protein that regulates planarian stem cells. Science 310, 1327–1330. [DOI] [PubMed] [Google Scholar]
- Redman E, Grillo V, Saunders G, Packard E, Jackson F, Berriman M and Gilleard JS (2008) Genetics of mating and sex determination in the parasitic nematode Haemonchus contortus. Genetics 180, 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart J, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H and Bartel DP (2006) Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H and Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Development 20, 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasinghe B, Knox DP and Britton C (2011) Factors affecting susceptibility to RNA interference in Haemonchus contortus and in vivo silencing of an H11 aminopeptidase gene. International Journal for Parasitology 41, 51–59. [DOI] [PubMed] [Google Scholar]
- Sargison ND, Redman E, Morrison AA, Bartley DJ, Jackson F, Hoberg E and Gilleard JS (2019) Mating barriers between genetically divergent strains of the parasitic nematode Haemonchus contortus suggest incipient speciation. International Journal for Parasitology 49, 531–540. [DOI] [PubMed] [Google Scholar]
- Sarkies P, Selkirk ME, Jones JT, Blok V, Boothby T, Goldstein B, Hanelt B, Ardila-Garcia A, Fast NM, Schiffer PM, Kraus C, Taylor MJ, Koutsovoulos G, Blaxter ML and Miska EA (2015) Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS Biology 13, e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Kingston RE and Lau NC (2007) The coming of age for Piwi proteins. Molecular Cell 26, 603–609. [DOI] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M and Pilpel Y (2007) Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Computational Biology 3, e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao CC, Xu MJ, Alasaad S, Song HQ, Peng L, Tao JP and Zhu XQ (2014) Comparative analysis of microRNA profiles between adult Ascaris lumbricoides and Ascaris suum. BMC Veterinary Research 10, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears RK, Bancroft AJ, Hughes GW, Grencis RK and Thornton DJ (2018) Extracellular vesicles induce protective immunity against Trichuris muris. Parasite Immunology 40, e12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lv Y, Huang L, Liu W, Wen M, Tang T, Zhang R, Hungate E, Shi S and Wu CI (2011) Testing hypotheses on the rate of molecular evolution in relation to gene expression using microRNAs. Proceedings of the National Academy of Sciences USA 108, 15942–15947. 10.1073/pnas.1110098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Montgomery TA, Qi Y and Ruvkun G (2013) High- throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Research 23, 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S and Schekman R (2016) Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5, e19276. 10.7554/eLife.19276.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH and Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Simon M, Sarkies P, Ikegami K, Doebley A-L, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA and Ahmed S (2014) Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Reports 7, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D and Aravin AA (2011) PIWI-interacting small RNAs: the vanguard of genome defence. Nature Reviews. Molecular Cell Biology 12, 246–258. [DOI] [PubMed] [Google Scholar]
- Skelly PJ, Da'dara A and Harn DA (2003) Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. International Journal for Parasitology 33, 363–369. [DOI] [PubMed] [Google Scholar]
- Szitenberg A, Cha S, Opperman CH, Bird DM, Blaxter ML and Lunt DH (2016) Genetic drift, not life history or RNAi, determine long-term evolution of transposable elements. Genome Biology and Evolution 8, 2964–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titze-de-Almeida R, David C and Titze-de-Almeida SS (2017) The race of 10 synthetic RNAi-based drugs to the pharmaceutical market. Pharmacology Research 34, 1339–1363. [DOI] [PubMed] [Google Scholar]
- Tritten L, Burkman E, Moorhead A, Satti M, Geary J, Mackenzie C and Geary T (2014a) Detection of circulating parasite-derived microRNAs in filarial infections. PLoS Neglected Tropical Diseases 8, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritten L, O'Neill M, Nutting C, Wanji S, Njouendoui A, Fombad F, Kengne-Ouaffo J, Mackenzie C and Geary T (2014b) Loa loa and Onchocerca ochengi miRNAs detected in host circulation. Molecular and Biochemical Parasitology 198, 14–17. [DOI] [PubMed] [Google Scholar]
- Tritten L, Tam M, Vargas M, Jardim A, Stevenson MM, Keiser J and Geary TG (2017) Excretory/secretory products from the gastrointestinal nematode Trichuris muris. Experimental Parasitology 178, 30–36. [DOI] [PubMed] [Google Scholar]
- Tzelos T, Matthews JB, Buck AH, Simbari F, Frew D, Inglis NF, McLean K, Nisbet AJ, Whitelaw CB, Knox DP and McNeilly TN (2016) A preliminary proteomic characterisation of extracellular vesicles released by the ovine parasitic nematode, Teladorsagia circumcincta. Veterinary Parasitology 221, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC (2014) Piwi and potency: PIWI proteins in animal stem cells and regeneration. Integrative and Comparative Biology 54, 700–713. [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M and Sanchez-Madrid F (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ and Davis RE (2011) Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Research 21, 1462–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Peng R, Wang J, Qin Z and Xue L (2018) Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clinical Epigenetics 10, 59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zheng J, Chen Z, Liu Y, Dura B, Kwak M, Xavier-Ferrucio J, Lu Y-C, Zhang M, Roden C, Cheng J, Krause DS, Ding Y, Fan R and Lu J (2019) Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nature Communications 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick E-M, Sarkies P, Silva N, Chen RA, Moss SMM, Cording AC, Ahringer J, Martinez-Perez E and Miska EA (2014) PRDE-1 is a nuclear factor essential for the biogenesis of Ruby motif-dependent piRNAs in C. elegans. Genes & Development 28, 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Sutherlin M, Wright AJ, Feinberg EH and Hunter CP (2007) Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proceedings of the National Academy of Sciences USA 104, 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter AD, Weir W, Hunt M, Berriman M, Gilleard JS, Devaney E and Britton C (2012) Diversity in parasitic nematode genomes: the microRNAs of Brugia pahangi and Haemonchus contortus are largely novel. BMC Genomics 13, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter AD, Gillan V, Maitland K, Emes RD, Roberts B, McCormack G, Weir W, Protasio AV, Holroyd N, Berriman M, Britton C and Devaney E (2015) A novel member of the let-7 microRNA family is associated with developmental transitions in filarial nematode parasites. BMC Genomics 16, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Fu JH, Nisbet AJ, Huang SY, Zhou DH, Lin RQ, Song HQ and Zhu XQ (2013) Comparative profiling of microRNAs in male and female adults of Ascaris suum. Parasitology Research 112, 1189–1195. [DOI] [PubMed] [Google Scholar]
- Zamanian M, Fraser LM, Agbedanu PN, Harischandra H, Moorhead AR, Day TA, Bartholomay LC and Kimber MJ (2015) Release of small RNA-containing exosome-like vesicles from the human filarial parasite Brugia malayi. PLoS Neglected Tropical Diseases 9, e0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]