Abstract
Physical exercise represents one of the strongest physiological stimuli capable to induce functional and structural modifications in all biological systems. Indeed, beside the traditional genetic mechanisms, physical exercise can modulate gene expression through epigenetic modifications, namely DNA methylation, post-translational histone modification and non-coding RNA transcripts.
Initially considered as merely damaging molecules, it is now well recognized that both reactive oxygen (ROS) and nitrogen species (RNS) produced under voluntary exercise play an important role as regulatory mediators in signaling processes. While robust scientific evidences highlight the role of exercise-associated redox modifications in modulating gene expression through the genetic machinery, the understanding of their specific impact on epigenomic profile is still at an early stage. This review will provide an overview of the role of ROS and RNS in modulating the epigenetic landscape in the context of exercise-related adaptations.
Keywords: Physical activity, ROS, RNS, DNA methylation, hPTMs, ncRNAs
Graphical abstract
Highlights
-
•
Physical exercise can modulate gene expression through epigenetic modifications.
-
•
Epigenetic regulation of ROS/RNS generating, sensing and neutralizing enzymes can impact the cellular levels of ROS and RNS.
-
•
ROS might act as modulators of epigenetic machinery, interfering with DNA methylation, hPTMs and ncRNAs expression.
-
•
Redox homeostasis might hold a relevant role in the epigenetic landscape modulating exercise-related adaptations.
1. Introduction
It is well recognized that both reactive oxygen (ROS) and reactive nitrogen species (RNS) produced under voluntary exercise play an important role as regulatory mediators in signaling processes together with many other different stimuli, including metabolic disturbances, mechanical activation, increased temperature, hormonal environment, and inflammatory state [1,2]. Many of the ROS-mediated responses protect cells against oxidative stress and re-establish redox homeostasis [3]. Different tissues may potentially produce ROS and RNS during exercise, but in view of the complex physiological link among many organ systems, it is difficult to identify their specific contribution. Indeed, most of the reports assume that skeletal muscle provides the major source of ROS generation during exercise as part of the “stress response hormesis” [4,5], thus evoking specific adaptations, not only by increasing antioxidant/oxidative damage repairing enzyme activity and resistance to oxidative stress [6,7], but also contributing to the specific physiological adaptation of muscle function and plasticity, like mitochondrial biogenesis and angiogenesis [[8], [9], [10]], as well as adaptation in other biological systems [11,12].
Epigenetics can be shortly defined as “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” [13]. The historical definition of “epigenome” usually refers to the chemical changes to DNA and histone proteins (i.e., methylation and/or acetylation) in a cell, but the current definition of epigenetic modifications generally includes functional and inheritable changes of the genome driven by DNA methylation, histone post-translational modification and non-coding RNA (ncRNA) expression [14]. The epigenome is indeed highly dynamic and can be modified either in response to biological factors, such as development and aging processes [15], or under the influence of exogenous factors, such as nutrient availability and physical exercise [16,17]. The current literature on epigenetic modifications determined by exercise interventions points out biomarkers related to energy regulation, mitochondrial function and biosynthesis, as well as muscle regeneration, calcium signaling pathways and brain plasticity, that are consistent with the known exercise-induced redox signaling and/or ROS unbalance [18].
Although the global impact of exercise-associated redox modifications on epigenetics is far from being clarified, current knowledge suggests their pivotal role in DNA methylation, post-translational modification of histone residues (hPTMs) and ncRNA expression elicited by regular exercise [19]. This review aims at providing an overview of current evidence on exercise-related epigenetic modifications, with a specific emphasis on the epigenetic regulation of the ROS generating systems as well as antioxidant enzymes, and on the role of ROS as modulators of epigenetic machineries.
2. Exercise-induced epigenetic modifications
It is well known that exercise can induce deep effects on all physiological systems improving the performance and the health status through specific sub-cellular changes. In the past years, significant advances have been achieved in the understanding of the “cellular and molecular benefits of exercise” [[20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30]]. However, despite on-going investigations, the molecular mechanisms behind these changes/adaptations are yet to be fully elucidated. All processes involved are mediated by several signaling events, pre- and post-transcriptional regulation, translation and protein processing, which together orchestrate the integrated response to exercise practice.
Emerging evidences indicate that a large part of the positive effects of exercise are driven by epigenetic alterations, thus altering the expression level of various genes associated to physiological and pathological conditions [17,31] (Fig. 1). Moreover, several studies demonstrate the implication of epigenetic modifications as mediators of intergenerational transmission of exercise effects [32,33].
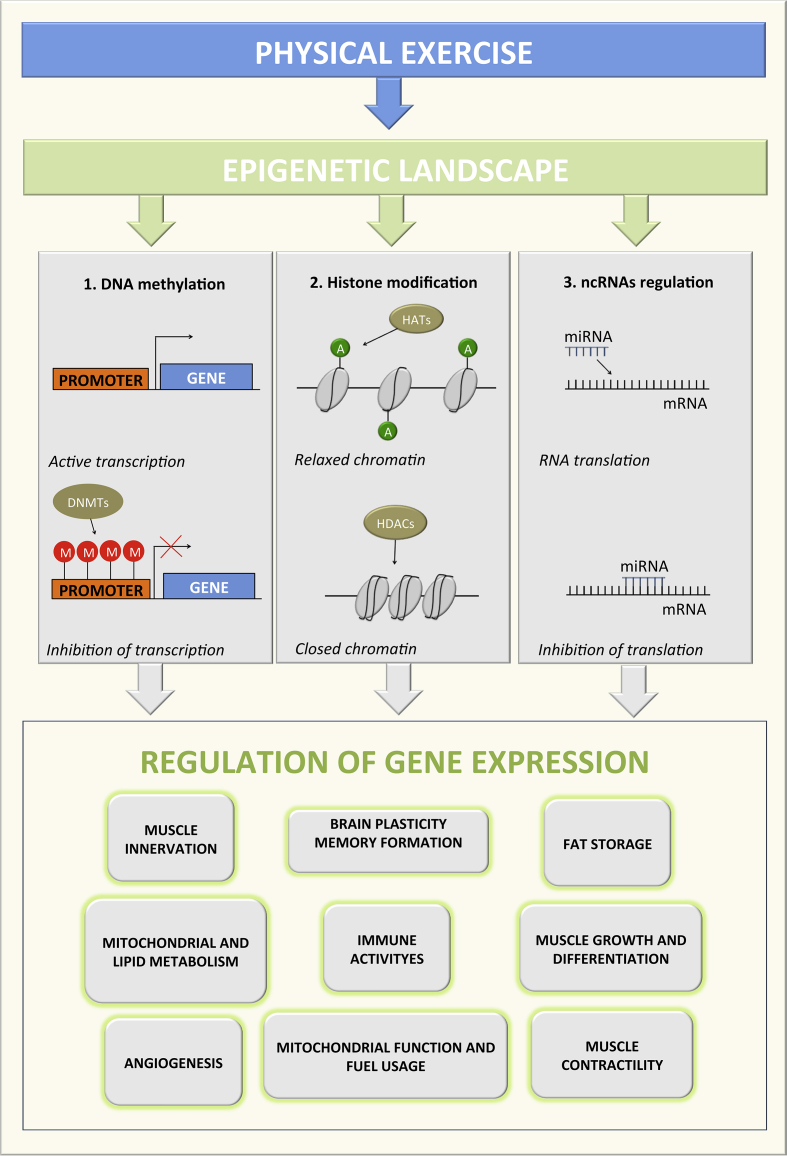
Fig. 1.
Schematic overview of epigenetic changes induced by physical exercise. DNA methylation, histone modification, and ncRNAs regulation are the main mechanisms related to changes in the expression of the relevant genes responsible for numerous changes/adaptation during physiological and pathological conditions. DNMTs, DNA methyltransferase; M, methylation; HATs, histone acetyiltransferases; HDACs, histone deacetyiltransferases; ncRNA, non-coding RNA; mRNA, messenger RNA.
The most intensely studied epigenetic modification induced by exercise is DNA methylation, although other common changes, such as histone acetylation and expression of different types of ncRNAs, are known to be relevant [[34], [35], [36], [37], [38]].
Many epigenetic studies presented here describe results deriving from animal or in vitro experiments and cannot be directly transferred into humans. However, they demonstrate that the epigenome has greater flexibility than previously thought, with a potentially high impact that extends to humans. In this section, we will offer an overview of the exercise-induced epigenetic modifications related to sport performance and health status, while their actual or putative connection with redox homeostasis alteration will be discussed in the following sections. For those interested into details of the molecular mechanism of epigenetic processes, the relevant literature is provided in the respective sub-sections.
2.1. DNA methylation
DNA methylation results from the addition of a methyl group to a cytosine by DNA methyltransferases (DNMTs) influencing transcription [39]. In vertebrates, the primary sites for DNA methylation are the 5′-positions of cytosine residues at cytosine-guanine dinucleotide (CpG) islands. Methylated sections of DNA are less accessible for DNA-binding proteins and RNA-polymerases and therefore downstream genes are generally decreased in their expression [40]. However, in some rare cases involving tumorigenesis and abnormal functions, the methylation can occur in non-CpG sites [41]. Finally, it seems that loss of DNA methylation (demethylation) is an active event mediated by Ten-eleven translocation (TET) enzymes [42].
A number of studies have sought to elucidate the relationship between exercise and DNA methylation, measured either across all the genome (i.e., global methylation) or at level of specific genes, showing an exercise-dependent and tissue specific fashion.
Skeletal muscle displays a remarkably high plasticity in its adaptive response to exercise. For instance, high-intensity acute exercise induces hypomethylation of genes involved in mitochondrial function and fuel usage, such as peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α), pyruvate dehydrogenase kinase, isozyme 4 (PDK4), mitochondrial transcription factor A (TFAM), peroxisome proliferator-activated receptor d (PPAR-δ), as well as citrate synthase (CS) and myocyte enhancer factor 2A (MEF2A) [43]. On the other hand, a chronic moderate-intensity exercise induces hypomethylation of genes involved in many aspects of muscle adaptation, such as mitochondrial and lipid metabolism (i.e., Stard10, Slc20a1, RUNX1, MEF2A, NDUFC2, ADIPOR1, ADIPOR2, and BDKRB2), glucose metabolism (Gfpt2, Cab39) [44], muscle growth and differentiation (Igfbp4, Plxna2) [45], as well as angiogenesis (Cds2) and muscle innervation (Dok7) [38,46].
Consequences of exercise on DNA methylation are also observable in adipose tissue. In particular, exercise training has been shown to produce positive changes in fat storage and reduce the chance of diabetes, increasing the DNA methylation level of thousands of genes, including RALBP1, HDAC4, NCOR2, as well as other candidate genes relevant for obesity and type 2 diabetes (e.g., TCF7L2, KCNQ1, HHEX, IGF2BP2 and JAZF1) [44,47].
In the peripheral blood, both acute and chronic exercise regulate the methylation level of genes involved in the inflammatory response and in inflammation-associated diseases (i.e., ASC, TNF, IL-10) [48,49].
Finally, Gomez-Penilla et al. [50] demonstrated that regular exercise promotes the stable increase in BDNF expression through the hypomethylation of CpG sites in BDNF promoter, thus suggesting one possible mechanism by which physical exercise can increase the hippocampal synaptic plasticity, learning and memory [51].
2.2. Histone post-translational modifications (hPTMs)
Histone modifications affect the accessibility of DNA through changes in chromatin structure by regulating histone binding to DNA. This association is governed by post-translational modifications of histones, including acetylation, methylation, phosphorylation and ubiquitination [52], and affects mainly lysine, arginine and serine residues to induce major changes on chromatin structure and, consequently, in gene expression [53].
A typical example of histone post-translational modification induced by exercise is represented by the regulation of the myocyte enhancer factor 2 (MEF2), a key transcription factor involved in muscle development [54]. The transcriptional activity of MEF2 is repressed by the class II histone deacetylases (HDACs), which do not possess HDAC activity by themselves, but rather recruit a repressive complex containing HDAC3 for this purpose [55]. Once formed, this molecular complex deacetylates the NH2-terminal tails of histone proteins, thus resulting in chromatin condensation and preventing the access of transcriptional coactivators. Indeed, following a single bout of endurance exercise, the phosphorylation of class II HDACs (i.e. HDAC4 and HDAC5) determines a reduced nuclear abundance of HDACs and MEF2-associated HDACs in skeletal muscle and an increase in the global H3 acetylation at lysine 36, changes that were associated with increased MEF2 DNA-binding activity and GLUT4 mRNA [34,35,56].
Another modification potentially involved in the exercise adaptations is the protein ubiquitination. Potthoff and colleagues [34] demonstrated that exercise-induced increase of MEF2 activity in slow oxidative soleus muscle was dependent upon the ubiquitin-mediated proteasomal degradation of HDACs 4, 5 and 7, leading to enhanced physical performance and resistance to fatigue. Other genes are regulated by the class II HDACs, including peroxisome proliferator activated receptor gamma coactivator1-α(PGC1α), carnitine palmitoyltransferase 1 (CPT-1), medium chain acyl-CoA dehydrogenase (MCAD), hexokinase II (HKII), glycogen phosphorylase, and ATP synthase β [57,58].
In addition to skeletal muscle, other tissues have shown this type of epigenetic changes induced by exercise: in 2011, Gomez-Pinilla et al. found that exercise-induced acetylation of histone H3 in the BDNF promoter IV, along with a reduction of HDAC5 levels, resulted in the transcription of BDNF gene and stable BDNF expression in hippocampus of rats [50].
Exercise can also alter the activity of hippocampus by changing the HAT/HDAC ratio. Experiments performed in mouse models document the reduction of HDAC activity and the increase in HAT activity induced by exercise in this specific brain region [59,60]. This allows the locals opening of condensed chromatin and thereby the transcription of specific genes, likely promoters of physical and mental health.
2.3. Modulation of non-coding RNAs expression
ncRNAs represent the majority of DNA transcripts and their role in mammalian biology is supported by the strong correlation between organismal complexity and size of the noncoding, rather than protein-coding, genome [61]. ncRNAs less than 200 nucleotides in length are generally classified as short, while all larger transcripts are regarded as long ncRNA (lncRNA). There are several subtypes of long and short ncRNA species, many of which are involved in the regulation of gene expression, and these can be further grouped according to their genomic origins and biogenic processes [62].
The best–characterized ncRNAs in exercise research are microRNAs (miRNAs) that are around 21 nucleotides in size and regulate gene expression mainly by base-pairing to the 3′-untranslated region (UTR) or 5′-UTR of target messenger RNAs (mRNA) [63]. To date, miRNAs have been suggested as key regulators of cardiac, skeletal muscle, immune and neuronal functions [[64], [65], [66], [67], [68]]. Only recently it has been highlighted the role of lncRNAs as epigenetic regulators during normal and diseased cellular function, however, their role in adaptive processes induced by physical exercise is just emerging and needs dedicated studies [69].
Many studies have demonstrated at both systemic and tissue levels the efficacy of exercise training in modulating the expression of several miRNAs, enough to be considered as essential mediators of processes associated with exercise in athletes and in general population [70], [71], [65], [68][65,68,70,71]. Changes in cardiac miRNA expression levels have been associated with the modalities of aerobic training regimes, which lead to physiological, non-pathological modification of heart. Indeed, both voluntary wheel and swimming exercise are recognized for its efficiency in inducing myocardial hypertrophy and ventricle compliance, cardiac angiogenesis, as well as cardiomyocyte survival, growth and proliferation through the modulation of several miRNAs (i.e. miR-1, miR-133a, miR-133b, etc.) influencing a broad spectrum of targets [70,72,73].
Similar to the cardiac tissue, miRNAs can mediate the exercise training-induced changes also in the skeletal muscle. In particular, exercise modifies the level of several small RNA molecules, including myomiRNAs (e.g., miR-1, miR-16, miR-21, miR-26a, miR-29a, miR-126, miR-133a, miR-133b, and miR-206, miR-210, miR-221, 328, miR-378, miR-451, and miR-494), which contribute to myocyte proliferation and differentiation, determination of muscle fiber types, and to muscle hypertrophy and atrophy [70,[74], [75], [76]].
As reported above, growing evidence suggests the importance of exercise for brain health [77]. Although the cellular and molecular mechanisms underlying the beneficial effects of exercise are still poorly understood, it is known that physical exercise is beneficial for brain functions through fine-tuning of sophisticated epigenetic mechanisms. In particular, exercise has demonstrated to be a potent modulator of miRNAs expression (i.e. miR-21, miR-15b, miR-199a, miR-28a, miR-98, miR-148b, etc.) that affects both growth-associated PTEN/mTOR and apoptosis-associated pathways. These pathways are critical regulators of protein synthesis, axon regeneration and plasticity in the central nervous system [68,78].
It is also known that physical exercise modulates several parameters of both natural and acquired immunity [79,80], reducing the number of infections and inhibiting tumorigenesis [81,82]. Several authors have highlighted as exercise affects the gene and the miRNA expression pattern (e.g., miR-21, miR-223, miR-126, miR-23b and miR-17, etc.) in different population of circulating cell lines such as monocytes, natural killer, and neutrophils [[83], [84], [85]].
Finally, and not in order of importance, it seems that exercise orchestrates a dynamic regulation of various circulating miRNAs (c-miRNAs) in the plasma (miR-21, miR-221, miR-20a, miR-146a, miR-133, and miR-222, etc.), which mediate many physiological processes such as angiogenesis, inflammation, skeletal and cardiac muscle contractility [70,86,87].
3. Epigenetic modulation of redox homeostasis components and response to exercise
3.1. Redox homeostasis and exercise
After the pioneering studies from Davies et al. [88] demonstrating that a single bout of exhaustive endurance exercise significantly increases ROS production and oxidative damage in rat skeletal muscle, the impact of different modalities of exercise in modifying the redox homeostasis have been extensively investigated [4,89]. At present, the dual role that exercise-induced ROS hold in cellular signaling and in the systemic or tissue specific adaptation to exercise training versus the exercise-induced oxidative stress and tissue damage is well recognized [4,5,7,18,90,91]. During moderate or customized intensive exercise, ROS signaling, possibly mediated by redox-sensitive thiols [92], plays a role in a broad range of biological processes, such as gene activation, detoxification, vasodilation, cell growth, proliferation and adaptation that, ultimately, explain the contribution of exercise in maintaining the healthy status in aging or in improving the prevention and treatment for several diseases [93]. In acute or exhaustive exercise, ROS production can overcome the cellular antioxidant capacity leading to biomolecules’ damage, impaired cellular function and apoptosis through a pro-inflammatory redox-sensitive response [65]. The present figure shall also take into account that contracting muscle produces nitric oxide (NO) mostly by nitric oxide synthase 1 (NOS1) and 2 (NOS2) 1 [94] and that NO, beside acting as signaling molecule per se, can react with superoxide producing peroxynitrite, determining a further depletion of thiol groups in cells [4].
In this context, any type of epigenetic modulator affecting the expression of genes related to ROS or RNS formation, neutralization and sensing has the potential to impact the outcomes of the redox perturbation during exercise acting on the upstream mediators, discussed in this section, while exercise-induced redox perturbation might act as downstream modulator of the epigenetic machinery, discussed in the next section. Due to space limitations, this chapter will highlight the main evidences directly or indirectly connected with exercise, recapitulated in Table 1 and Fig. 2. For an exhaustive analysis of the epigenetic landscape related to ROS formation and redox homeostasis, interested readers are directed to detailed reviews already available [[95], [96], [97], [98], [99], [100], [101], [102]].
Table 1.
Published articles on exercise-induced modulation of redox homeostasis components by epigenetic regulation.
| Subjects/Animals | Exercise intervention | Tissue | Target | Epigenetic modification | Outcome | Ref. |
|---|---|---|---|---|---|---|
| Human subjects, healthy sedentary | acute aerobic exercise (80% of maximal aerobic capacity) | skeletal muscle | PGC1α TFAM PPAR- δ PDK4 |
↓DNA methylation of genes' promoter | ↑Genes' expression | [43] |
| Rats, young (7 months) and old (34 months) | acute intramuscular induced electric stimulation at maximal tetanic force (10′ x 4 bouts) | skeletal muscle | PGC1α PDK4 |
↓DNA methylation of genes' promoter | ↑Genes' expression | [116] |
| Human subjects, sedentary with ≥2 CV risk factors | 12-week high-intensity interval training (HIIT) | blood cells | p66(Shc) | ↑DNA methylation of genes' promoter | ↓Gene expression ↓ 3-NT plasma concentration | [117] |
| C57BL mice, diet-induced hyperhomo-cysteinemia | Aerobic training (treadmill, 10 m/min x 60′), 5 days/week x 14 weeks | aortic endothelial cells | NADPH | ↑SIRT1-mediated hPTMs | ↓NADPH activity ↓ MDA plasma concentration | [132] |
| Male C57BL/6 mice | Aerobic training (voluntary wheel running x 8 weeks) | Skeletal muscle | Nrf1 PGC1α |
↓miR-494 ↓miR-696 | ↑Nrf1 mRNA = PGC1α mRNA ↑PGC1α protein |
[158] |
| Huma subjects, active female 66 ± 5 years | 5-month interval walking training (3′,70% + 3′, 40% peak aerobic capacity x 5 sets) plus low or high post-exercise protein intake | Whole blood | NFKB1 NFKB2 |
↑DNA methylation of genes' promoter | ↑Muscle strength | [188] |
| male C57/bl6 wild type and ATX mice | Aerobic training (voluntary wheel running x 12 weeks) | Whole blood Skeletal muscle |
Gpx1 | ↓DNA methylation of gene’ promoter | ↑Gpx1 mRNA | [197] |
| Human subjects, sedentary and lifelong active elderly | Structured exercise more than 3 times/week throughout life | Skeletal muscle |
MGST1 OXR1 SOD2 CAT |
↓DNA methylation of gene’ promoter | ↓ muscle protein carbonyls | [203] |
| Human subjects, type 2 diabetes | Aerobic exercise on treadmill (4 times/week x 10 weeks; 50–70% Vo2peak) | Skeletal muscle |
GSTM1 GSTM5 |
↓DNA methylation of gene’ promoter | ↑GSTM1 mRNA ↑GSTM5 mRNA |
[216] |
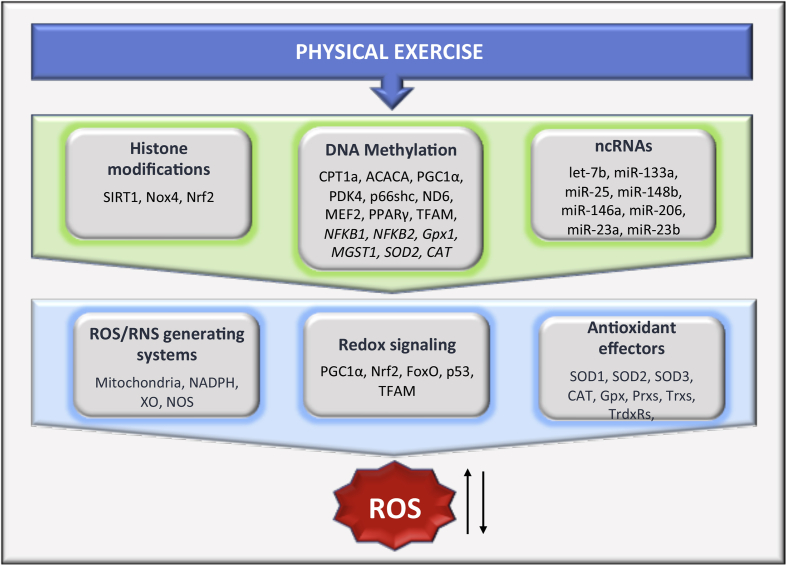
Fig. 2.
Schematic representation of putative overlapping between epigenetic modulation of redox homeostasis components and exercise-induced molecular response. In addition to genetic mechanism, physical activity can modulate ROS/RNS generating systems, redox-sensitive transcription factors and antioxidant enzymes through epigenetic mechanisms, including the methylation status or histone post-translational modifications (hPTMs) of relevant target genes, and modification of noncoding RNA (ncRNAs) expression. Within each category, only genes or ncRNAs reported to be involved in epigenetic regulation of redox homeostasis components and also known to be modulated by exercise have been included. ACACA, acetyl-CoA Carboxylase Alpha; CAT, catalase; CPT1a, carnitine palmitoyltransferase 1A; FoxO, forkhead box class O proteins; Gpx, glutathione peroxidases; Gpx1, glutathione peroxidase 1; MEF2, myocyte enhancer factor 2; MGST1, microsomal glutathione S-transferase 1; NADPH, NADPH oxidase family; ND6, NADH-ubiquinone oxidoreductase chain 6; NFKB1, Nuclear Factor Kappa B Subunit 1; NFKB2, Nuclear Factor Kappa B Subunit 2; NOS, nitric oxide synthase; Nox4, NADPH oxidase 4; Nrf2, nuclear respiratory factor 2; p53, tumor protein p53; p66shc, redox-regulating protein p66; PDK4, pyruvate dehydrogenase kinase, isozyme 4; PGC1α, PPAR Coactivator-1 α; PPARγ, peroxisome proliferator-activated receptor gamma; Prxs, peroxiredoxins; RNS, reactive nitrogen species; ROS, reactive oxygen species; SIRT1, NAD-dependent lysine deacetylases Sirtuin 1; SOD1, superoxide dismutases 1; SOD2, superoxide dismutases 2; SOD3, superoxide dismutases 3; TFAM, mitochondrial transcription factor A; Trxs, thioredoxins; TrdxRs, thioredoxin reductases; XO, xanthine oxidase.
3.2. Epigenetic modulation of ROS and RNS generating systems
The increased demand for ATP and the accelerated oxidation of food molecules during muscle voluntary contraction, together with additional molecular events, trigger also the primary or secondary generation of superoxide from the electron transport chains of mitochondria [103] and from other intracellular sources, such as sarcolemmal NADPH oxidase, 5-lipoxygenase, cyclooxygenase and xanthine oxidase (XO) [104].
Mitochondria represent the cell's powerhouse and their function depends upon the coordination of nuclear and mitochondrial genomes. Given the functional interconnection between mitochondria and environmental stimuli, such as nutritional fluctuations and stress conditions, mitochondria display a crucial role in the modulation of gene expression through the modification of the epigenome [105]. It is worthy to consider that acetyl-CoA and S-adenosyl methionine (SAM) are utilized for histone acetylation and for both histone and DNA methylation, respectively, and the levels of these metabolites are regulated by intermediate metabolism and mitochondrial function [106]. Indeed, available data confirm that epigenetic mechanisms are participating to the interplay between mitochondrial and nuclear interactions [107], [101][101,107]. DNA methylation is known to modulate many nuclear-encoded mitochondrial genes, such as the carnitine palmitoyltransferase 1A (CPT1a) and Acetyl-CoA Carboxylase Alpha (ACACA), two key enzymes of lipid oxidation [108,109], that have been recently shown being modulated by acute strength or endurance exercise in mice and in humans [[110], [111], [112]]. Also non-CpG methylation has been associated with mitochondrial function: the non-CpG hypermethylation at the PGC1α and PDK4 genes' promoter is associated with obesity in humans [113] and the methylation pattern of these genes can be restored by gastric bypass surgery, weight loss and exercise [[114], [115], [116]]. Recently, Streese et al. [117] demonstrated that 12 weeks of high-intensity interval training (HIIT) restored in blood mononuclear cells of ageing subjects with increased CV risk the methylation of the gene promoter for the redox-regulating protein p66(Shc), a mitochondrial adaptor that drives the ageing-induced reactive oxygen species [118]. They also demonstrate that HIIT-induced reduction of p66Shc gene expression is paralleled by a restored microvascular phenotype.
Although the biological significance of mitochondrial DNA (mtDNA) methylation is still a matter of debate [107,119], it has been reported that transcript variants of the human and mouse DNMT1 (mtDNMT1) translocate to the mitochondria, while DNMT3A and DNMT3B have been identified in mitochondria, too, with a possible role for GpC and not CpG methylation as regulator of mitochondrial gene expression [120]. Among the mtDNA genes, differently methylated in the experimental setting, a special attention should be given to NADH-ubiquinone oxidoreductase chain 6 (ND6) gene. In human colon carcinoma cells exposed to a pro-oxidant environment, ND6 displays inhibition via hypermethylation determined by NRF1 and PGC1α mediated up-regulation of the mtDNMT1 variant [121], opening the possibility that exercise-induced ROS production can contribute to mitochondrial adaptation via mtDNA methylation.
As described above, different levels of both acetylation and methylation of histones can modulate gene expression. Whereas mitochondria are involved in controlling histone acetylation and methylation [101], there is also regulation in the opposite direction. As example, the inhibition of the SET domain containing lysine methyltransferase 7 (SETD7, also called SET7/9) promotes mitochondrial biogenesis and activates an antioxidant response through PGC1α and NFE2L2, respectively [122].
A number of miRNAs, called mitomiRs, are encoded by the nuclear DNA and then translocate into the mitochondria targeting either mitochondrial or nuclear mRNAs. The role of exercise in modulating mitomiRs expression is still unknown but their species as well as cell type-specificity promises to identify a future role in training adaptation. Indeed, they represent a unique population of miRNAs and seem to be involved in the regulation of mitochondria-specific functions, in addition to the general cellular functions [123]. Bioinformatics analysis suggests that muscular mitomiRs let-7b has the potential to target mitochondrial transcripts, such as COX2, ND5, ATP6 and ATP8, while miR-133a was predicted to target ND1 [124]. Das et al. [125] demonstrated that overexpression of miR-181c-5p in rat myocytes results in a loss of mt-COX1 protein and imbalance among the mitochondria-encoded subunits in complex IV promoting ROS generation.
The NADPH oxidase (Nox) family of proteins generates a defined burst of superoxide or hydrogen peroxide as a controlled response to distinct stimuli, and thus are important contributors toward redox signaling [126]. Examples of epigenetic regulation of NADPH oxidases via DNA methylation of their promoter region or that of the genes required for their assembly [127], as well as throughout hPTMs and mi-RNA silencing have been reported [100]. Actually, beside the evidence that treatment of endothelial cells with inhibitors of HDAC reduces the transcriptional activity and expression of Nox4 [128], it has been clearly demonstrated that activation of SIRT1, a class IV HDAC, displays anti-aging and anti-oxidant effects partially by down-regulation of Nox-derived ROS production [129,130]. The role of exercise training in SIRT1 activation is well known, and it will be discussed in the next section [3,131,132]. Regarding the modulation of NOX transcripts by microRNA, it has been demonstrated that miR-25 directly targets the 3′UTR of the human NoxOX4 gene. Indeed, down-regulation of miR-25 expression leading to NOXox4 up-regulation and consequent ROS production has been described in some diseases, such as hypercholesterolemia [133], as well as after endurance training [134]. Other mi-RNAs (miR-106b, miR-148b, miR-204, miR-448-3p) targeting Nox2 in macrophages and heart of mice have been identified [135,136], with miR-148b also found to be negatively regulated in human skeletal muscle by transition from physical active to sedentary behavior [137].
Finally, in vitro and in vivo animal studies suggest that epigenetic changes can modulate NOS1, NOS2A and NOS3, affecting NO production or bioavailability [138,139]. Exercise training is effective in enhancing NOS activity and expression [140,141], but no data related to exercise-induced epigenetic regulation of NOS have been collected so far.
3.3. Epigenetic modulation of redox signaling pathway
The spontaneous or superoxide dismutases 1 and 2 (SOD1; SOD2) driven-dismutation of superoxide into hydrogen peroxide (H2O2) causes the ROS diffusion in the cytoplasm and the activation of redox-sensitive kinases such as AMP-activated protein kinase (AMPK) and p38 mitogen-activated protein kinase (MAPK). They activate redox-sensitive transcription factors that control both ROS generating systems and antioxidant enzymes, namely PGC1α, Nrf1, Nrf2, mitochondrial transcription factor A (TFAM), p53, NF-kB, the forkhead box class O proteins (FoxO) and others [142]. Considering the aim of this review, we will concentrate on PGC1α, Nrf2 and NF-kB, referring to the up-to-date available literature for an extended reading of the topic [95,[143], [144], [145], [146], [147]].
PGC1α, encoded by PPARGC1A gene, holds a fundamental role in mitochondrial biogenesis and antioxidants modulation, being activated by environmental stimuli via several signaling kinases (i.e., AMPK and p38) [95]. It targets several other transcription factors, such as Nrf1 and Nrf2, that regulate both ROS and RNS production and neutralization [148,149]. The key role of PGC1α in exercise adaptation is widely recognized [30,150] and its epigenetic modulation by exercise through the redox-sensitive NAD-dependent lysine deacetylases Sirtuin 1 (SIRT1) has been extensively reported [3,151]. In addition to the redox-sensitive post-translational modification of PGC1α exerted by SIRT1, PGC1α gene expression seems to be modulated through promoter hypermethylation [115,152], as well as via class I and class II histone deacetylases (HDACs) that, in addition to prevent the acetylation on PGC1α promoter area, also repress the myocyte enhancer factor 2 (MEF2), the main PGC1α upstream regulator [57]. In humans and in mice, Barrès et al. [43] demonstrated that acute exercise/contraction induces a decrease on whole genome methylation, as well as in the DNA methylation at PGC1α, MEF2, PPAR-δ and TFAM sequences with the subsequent increase in PGC1α expression. Conversely, it has been demonstrated that bed rest increases the DNA methylation of PGC1α gene together with other genes associated with insulin resistance [153]. Some results also suggest that exercise could inhibit HDAC5 thus increasing PGC1α expression in skeletal muscle, however, direct evidences about exercise-induced post-translational modifications of histones at the PGC1α promoter are missing [154]. Similar picture refers to the possible modulation of PGC1α by non-coding RNA: although it has been shown that PGC1α expression can be directly or indirectly downregulated by miRNAs (such as miR-22, miR-29, miR-27b), with critical consequences for skeletal and cardiac muscle cells [[155], [156], [157]], no direct evidences are available with respect exercise stimulus. In 2016, Sun et al. [158] demonstrated that 8 weeks of voluntary wheel running decrease the expression of miR-494 and miR-696 in mouse gastrocnemius muscle with an increase in Nrf1 mRNA and PGC1α protein, but not in PGC1α mRNA, suggesting that other factors might be involved in PGC1α modulation.
Nuclear factor erythroid 2-related factor 2 (Nrf2) transcription factor regulates about 250 genes, including antioxidant proteins, detoxifying enzymes and cytoprotective proteins and it is considered one of the key players in orchestrating the antioxidant response after acute and regular exercise [[159], [160], [161]]. In basal conditions, Nrf2 is associated with Keap1 (Kelch-like ECH-associated protein 1), a multi-domain protein rich in cysteines that, together with other factors, allows Nrf2 constant degradation through the proteasome. ROS increase leads to the oxidation of Keap1 critical cysteine residues and the release of Nrf2, preventing its degradation [162]. Although some recent results demonstrate the modulation of Nrf2 expression by epigenetic modulation of ROS production in muscle cells [163], at present no data have been collected regarding a direct epigenetic modulation of Nrf2 gene expression by exercise or muscle contraction. Both Nrf2 and Keap1 genes have a consistent number of CpG dinucleotides in their promoters, even though the epigenetic modulation through the CpG methylation level seems to occur preferentially in the Keap1 gene. In ageing, the loss of DNA methylation in the promoter of Keap1 gene decreases Nrf2-dependent antioxidant protection and results in a redox imbalance and decreased antioxidant protection in lens [164]. The downregulation of Nrf2 by hypermethylation of selected CpGs in its promoter has been described in prostate tumors of TRAMP (PB-Tag C57BL/6) mice, tumorigenic TRAMP-C1 cells and in tissue from patients with muscle-invasive urothelial carcinoma [165,166]. Moreover, the maintenance of Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation by bioactive nutrients has been also reported [167]. Regarding histone acetylation, histones H3 and H4 have been involved in epigenetic regulation of Nrf2 [168,169]. Ray et al. [170] demonstrated that acetylation of H4 in Lysine16 and Lysine 588 of the Neh1-promoter leads to the expression of Nrf2-dependent genes, whereas the induction of HDAC decreases H3 and H4 acetylation, resulting in lower Nrf2 expression in microglia [171]. Recently, in rat liver, Sun et al. [172] demonstrated that sodium butyrate up-regulates Nrf2 by down-regulating HDAC1 and increasing histone H3 acetyl K9 (H3K9Ac) modification in the Nrf2 promoter, thus protecting the animals from high-fat diet-induced oxidative stress. Different ncRNAs have been involved in the negative or positive regulation of Nrf2. The direct binding of miR-144, miR-153, miR-27a, and miR-142-5p to the 3′UTR end of Nrf2 messenger promotes Nrf2 down-regulation in lymphoblast cells [173] and/or neuronal cells [174], while bioinformatic analyses have shown that miR-200a can interact with the 3′UTR region of the Keap1 mRNA promoting its degradation and Nrf2 activation [158]. In recent years, various mi-RNAs have been identified to target the 3′UTR of Nrf2 and Keap1, as well as to alter the expression of other proteins in the Nrf2 “regulome” that would also alter ARE-mediated redox signaling [175].
The NF-κB family of redox sensitive transcription factors, consisting of five members [NF-κB1 (p50/p105), NF-κB2 (p52/p100), c-Rel, RelA (p65), RelB], is involved in the cell response to a wide spectrum of stress stimuli including oxidative stress. NF-kB activity is suppressed by an inhibitory protein (IκB) which, after phosphorylation by specific kinases (IKKα/β and NEMO), becomes ubiquitinylated and degraded via the ubiquitin/proteasome system, leading to the activation and translocation of NF-κB into the nucleus. NF-κB can be activated also through Tyr-181 nitration, which leads to dissociation from IκB. NF-κB has a crucial role in the regulation of inflammation and immunity, targeting genes such as NOX2, XOR, NOS1, NOS2, COX-2, LOX-5, LOX-12 (reviewed in Ref. [145]). NF-κB inhibition holds a prominent role in the early survival of muscle cells shifting the cellular function towards oxidative stress resistance and DNA repair, also via FOXO/SIRT1 pathway [176,177] and it is involved in the atrophy of skeletal muscle via protein degradation, particularly relevant in aging [178,179]. As for Nrf2, data on exercise-induced epigenetic modulation of NF-κB are scanty. Nevertheless, the epigenetic modification of NF-κB promoter related to oxidative stress improvement has been described in diseases such as type 2 diabetes [180], whose management via exercise intervention is linked, among others, to reduction in the pro-inflammatory response [181]. Indeed, in older women, a 5-months of combined intervention with interval walking training (IWT) and post-exercise milk product intake ameliorated muscle strength and induced NFKB1 and NFKB2 gene methylation [182]. In monocyte cells, Pinheiro et al. [183] demonstrated that in vitro treatment with the antioxidant resveratrol decreases the lipopolysaccharide-induced inflammatory response via histone deacetylation and methylation impacting the p65 NF-κB subunit, while different reports indicate a putative role of miRNAs (miR-146a, let-7a, miR-125b, miR-100, and miR-21) in regulating NF-κB and other pro-inflammatory signals in diabetic peripheral neuropathy [184] and periodontitis [185]. Actually, miR-21 and miR-146a have been often correlated with frailty [186], while decreased expression of miR-146a, NF-κB and inflammatory cytokines has been described in type-2 diabetic rats submitted to 12-week of swimming [187], as well as in plasma of obese human subjects or chronic kidney disease patients after 3 months of a physical activity program [188,189].
3.4. Epigenetic modulation of antioxidant effectors
As anticipated, in normal conditions the concentration of ROS is regulated by enzymatic and non‐enzymatic antioxidant defenses. The superoxide dismutases (SODs) family, including the mitochondrial MnSOD (SOD2) and the two isoforms of Cu/ZnSOD localized intracellularly (SOD1) or extracellularly (ECSOD, SOD3), disproportionate the superoxide anions into oxygen and hydrogen peroxide. The scavenging enzymes catalase (CAT) or glutathione peroxidases (GPx), as well as peroxiredoxins (Prxs), glutaredoxins (Grxs) and thioredoxins (Trxs), can detoxify H2O2 to oxygen and water [95]. Besides the indirect impact exerted by the epigenetic regulation of relevant transcription factors, such as Nrf2 and NF-κB, discussed in the previous section, the gene expression of antioxidant enzymes can be directly modulated by DNA methylation, hPTMs and microRNAs. For example, the epigenetic regulation of SOD2 gene expression can impact various regulatory sequences within the promoter, adjacent to the transcriptional start site, within the enhancer, in the second intron, and at numerous upstream regulatory elements [190]. Changes in both DNA methylation and hPTMs have been described in all these regions in cell types showing a very dynamic level of SOD2, especially during the carcinogenic progression [191]. Similar evidences have been accumulated for various members of the Prxs family (reviewed in Ref. [192]. Moreover, the modulation of DNA methylation and histone modification has been proven to effectively impact also the expression of CAT, GPx1, GPx4, SOD1, SOD3, GSTP1 (glutathione S- transferase pi 1), TXNRD2 (thioredoxin reductase 2) and HO-1 (heme oxygenase 1) genes in non-cancer cells [[193], [194], [195], [196]]. The exercise-associated dependence of the epigenetic regulation of antioxidant enzymes is still not well studied, but the available data suggest that regular exercise could alter the DNA methylation at regulatory DNA sequences of the relevant genes. In 2016, Nguyen et al. [197] demonstrated that 3 months of voluntary exercise in mice was able to transiently decrease the Gpx1 gene hypermethylation induced in skeletal muscle by severe dyslipidemia while, in a recent study, Sailani et al. [198] found that the promoter sequences of microsomal glutathione S-transferase 1 (MGST1), oxidation resistance protein 1 (OXR1), SOD2 and CAT genes show a reduced methylation in skeletal muscle of lifelong active elderly subjects when compared with age matched sedentary ones.
With respect the miRNAs contribution to the modulation of antioxidant genes’ expression, it is well established that “redoximiRs” are capable to regulate the endogeneous antioxidant defenses [98]. miR162, miR397, and miR408 have been suggested to potentiate the cellular antioxidant capacity by inhibiting the negative regulators of catalase gene expression [173]. Similarly, Kitscha et al. [199] demonstrated that modulation of miR‐320a, miR‐21‐3p and miR‐409‐5p decreases the oxidative damage by improving the expression of HO‐1 and increasing glutathione synthesis. In 2017, our group demonstrated that miR-23a and miR-23b bind directly to the 3′ UTR of thioredoxin reductase 1 (TrxR1) mRNA and are involved in the downregulation of TrxR1 expression during myogenesis [64]. Other miRNAs targeting mRNA for antioxidant enzymes and related proteins have been described, such as miR-146a, which has SOD2 as a target, miR-181a with GPX1, miR-30b with CAT and miR-210, which targets apoptosis–inducing factors, that, collectively, seem to potentiate ROS generation [200]. Also, miR-206, miR-335 and miR-34a, targeting SOD1 or SOD2 and Txnrd2, increase the ROS level [201,202].
Among the above mentioned miRNAs, the level of circulating miR-146a, but not miRNA-210, have been shown to be modulated by aerobic-exercise in a volume-related dependency [203], while primary miR-206 resulted up-regulated in skeletal muscle of young and old adults after an acute intervention combining resistance exercise plus amino acid supplementation (20-g leucine-enriched essential amino acid solution) [204]. Similarly, the expression of both miR-23a and miR-23b resulted upregulated in resting skeletal muscle of recreationally active adult males after 4 weeks of resistance exercise [205] or after concurrent, acute resistance and aerobic exercise, irrespective from post-exercise protein supplementation [206].
4. Redox control of the epigenetic machinery and response to exercise
Redox control of epigenetic mechanisms allows cells to adapt to changes in the environment. Adaptation to oxidative stress is obtained, at least in part, through a gene expression program achieved to protect cells from damage, to repair the damage, and to support cell survival. This involves cellular proteins responsive to redox changes, containing cysteine residues exhibiting nucleophilic thiols, which are promptly oxidized by ROS or alkylated by reactive carbonyl species (RCS) [207]. Reduction and oxidation are mechanisms of post-translational modification [208]. ROS arising from various origins, including exercise, can oxidize signaling proteins and transcription factors, but also components of the epigenetic machinery, including DNA and RNA molecules. For instance, thiol groups (R–SH) of specific cysteine residues can be reversibly oxidized in sulfenic acids (R–SOH) or irreversibly transformed in sulfinic (R–SO2−) or sulfonic (R–SO32−) acids [208]. These modifications cause conformational changes that affect enzymatic activity, protein-protein interaction, and protein subcellular localization [208,209].
Aerobic organisms have evolved defense systems to rapidly detoxify oxidants. However, hydrogen peroxide does not represent only a source of oxidative stress, but it also acts as a signaling molecule, which is rapidly generated and easily managed by antioxidant systems within the cell [210,211]. Several epigenetic regulators and their enzymatic activities can be affected by redox signaling, and RNA and DNA are often modified in this response. In this section we will briefly discuss the main modifications on epigenetic molecules achieved by redox perturbation (Fig. 3), pointing out available data on exercise-related modulation.
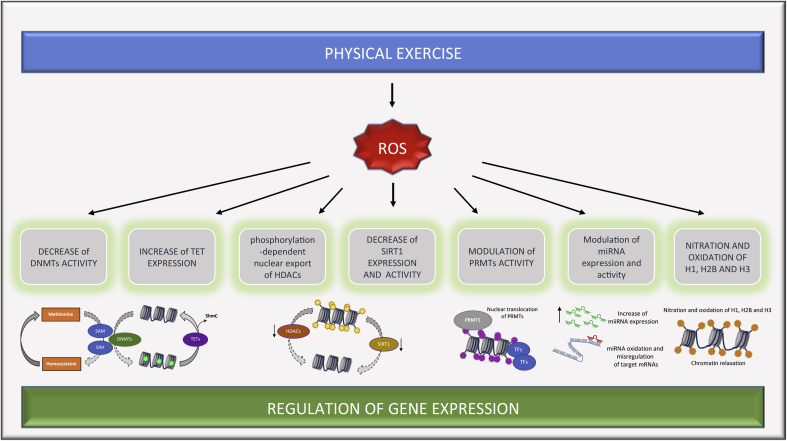
Fig. 3.
ROS affect the epigenetic machinery impacting gene expression programs. Schematic representation of ROS effect on the epigenetic machinery. On the left, ROS induce DNA demethylation by reducing the availability of SAM to DNMTs and increasing the expression of TET proteins, leading to global hypomethylation (Methylated CpG are indicated in green). Phosphorylation-dependent nuclear export of HDACs and decrease of SIRT1 expression and activity allow acetylation of chromatin (acetylated histones are indicated in yellow). Exposure to pro-oxidant environment can induce PRMTs translocation into the nucleus, histone methylation (in purple) and recruitment of transcription factors (TFs), thus ensuring the cellular antioxidant defense program. ROS also impacts expression of miRNAs and can induce their oxidation, thus causing misregulation of target mRNAs. On the right, direct oxidation and nitration of histones (in orange) can induce chromatin relaxation and recruitment of transcription factors. DNMTs, DNA methyltransferases; HDACs, histone deacetylases; PRMTs, protein arginine methyltransferases; ROS, reactive oxygen species; SAM, S-adenosyl methionine; SIRT1, NAD-dependent lysine deacetylases Sirtuin 1; TET, Ten Eleven Translocation family. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4.1. Redox control of DNA methylation
As already described, cytosine methylation is catalyzed by the DNA methyltransferases 1, 3A and 3B (DNMT1/3A/3B). DNMT enzymes use S-adenosyl methionine (SAM) as donor of the methyl group, which is then transferred to the 5-carbon of the cytosine ring, leading to the formation of S-adenosyl homocysteine [212]. ROS can affect DNA methylation by acting on the activity and expression of DNMTs. By reducing the availability of the cofactor SAM, ROS can hamper the activity of DNMTs thus causing hypomethylation. On the other hand, through the inhibition of methionine adenosyl-transferase ROS decrease the level of SAM synthesis, whereas by inhibiting methionine synthase, ROS impair methionine regeneration. In parallel, upon oxidative stress conditions, methionine residues are required to produce the antioxidant glutathione (GSH) through cysteine metabolism, thus decreasing SAM synthesis [96]. This critical link between the methylation intermediate SAM and the production of GSH highlights a role for the redox buffering capacity in influencing the epigenetic regulation via the availability of SAM pools, also important for the hPTMs by the HMTs [96,213]. Indeed, when cells are exposed to acute or prolonged conditions of oxidative stress, GSH pools can be significantly depleted. For example, during strenuous exercise GSH is oxidized, leading to the accumulation of disulfide glutathione (GSSG) and modification of the GSSG/GSH ratio [18,214], that is considered a redox central switch in the regulation of the cellular responses to oxidative stress induced by acute exercise or in the adaptation during regular exercise training [28,215]. Recently, Stephens et al. demonstrated through genome-wide DNA methylation analysis and RNA sequencing that in type 2 diabetic patients the efficacy of 10 weeks of aerobic training in improving insulin sensitivity and glycemic control is correlated to decreased promoter methylation for genes involved in mitochondrial function, insulin sensitivity, as well as glutathione metabolism, such as glutathione-S-transfrerase 1 and 5 (GSTM1, GSTM5) in skeletal muscle [216]. ROS can also induce DNA hypermethylation by increasing the expression of DNMTs [217]. In particular, it has been shown that human cardiac fibroblast cells exposed to prolonged hypoxia resulted in a pro-fibrotic state associated with global DNA hypermethylation and increased expression DNMT1 and DNMT3B by hypoxia-inducible factor (HIF)-1α [218]. Similarly, in isolated fetal rat hearts and cardiomyocytes, hypoxia was able to induce the hypermethylation of the PKC epsilon promoter, associated with cardiac dysfunction. Notably, this effect, which was not mediated by HIF, was attenuated by antioxidants [219].
Recent evidence suggests a role for the Ten Eleven Translocation (TET) family of Fe2+ and α-ketoglutarate-dependent 5mC dioxygenases in active DNA demethylation upon ROS exposure [220]. TET proteins catalyze the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), followed by deamination to 5-hydroxymethyluracil by cytidine deaminases [221]. Subsequent base excision repair replaces the 5-hydroxymethyluracil with unmethylated cytosine [168]. Cells exposed to ROS display significantly higher levels of 5hmC than controls, followed by a decrease in genomic methylation [220]. On the contrary, in hypoxic condition, which is frequently observed in tumors, the activity of TET enzymes is reduced, leading to increased hypermethylation at gene promoters in vitro [222].
Physical activity and lifestyle interventions widely contribute to these mechanisms, leading to a specific methylation signature, as mentioned above. In particular, it has been recently shown that acute aerobic exercise widely impacts the methylation status of natural killer cells, leading to a characteristic methylation pattern on specific genes [223]. Moreover, it has been shown that an acute bout of intense treadmill exercise is able to reduce DNMT3B nuclear accumulation in the plasma [224], possibly due to the downregulation of gene transcription and/or enhanced nuclear export, mediated by IL-6 accumulation and activation of JAK2/STAT3 and AKT kinases. In addition, increased Tet1 and decreased Dnmt3b mRNA expression were observed in the hippocampi of rats engaged in physical exercise, further documenting the epigenetic mechanisms underlying the impact of physical exercise.
4.2. Redox control and histone acetylation
After exercise, acetylation of the histone H3 at lysine 36 (K36) is significantly increased in promoter regions containing active RNA polymerase II (RNAPII) [35], due to the nuclear export of HDACs [225]. The dynamic nucleocytoplasmic shuttling has been proposed as a mechanism of regulation of HDACs activity [226]. For example, phosphorylation of HDACs at specific serine residues, occurring possibly by the AMP-activated protein kinase (AMPK) and the calcium–calmodulin-dependent protein kinase II (CaMKII) [35,227], can induce the interaction with 14-3-3, thus masking the nuclear localization signal (NLS) from importin and unmasking the nuclear export signal (NES) to CRM1 (exportin) [226]. Similarly, Trx1 regulate the nucleocytoplasmic shuttling of class II HDACs in a redox-dependent fashion. Increased ROS production causes thiol oxidation of cysteines 667 and 669 in HDAC4, unmasking the NES, which is then exposed to CRM1, inducing the nuclear export of HDAC4. Class II HDACs are thereby exported to the cytosol, where they can no longer suppress target transcription factors. In the cytosol, Trx1 reduces critical cysteines in HDAC4 by forming a multiprotein complex composed by DnaJb5, TBP-2, and importin, which returns HDAC4 to the nucleus [228]. Regarding endogenous thiols, it is worthy to mention that histone H3 can be S-glutathionylated in mammalian cells by GSH, inducing modification of the nucleosome structure [229]. Thus, the exercise-induced depletion of GSH or modification of the GSSG/GSH balance might represent an additional contribution to the post-translational modification of the histone code.
Likewise, the enzymatic activity of HDAC8 strongly depends on its redox state. The cysteines 102 and 153 are targeted by oxidation in the presence of ROS, leading to a switch from a reduced active to an oxidized inactive form of the enzyme [230]. Moreover, RCS generated by oxidative environment can induce covalent modification of two conserved cysteines in HDAC1, thus dampening histone deacetylase activity and promoting changes in histone H3 and H4 acetylation patterns [231].
Hence, physiological exposure to exercise-induced ROS can likely results in histone post translational modifications, including acetylation, methylation, and phosphorylation, thus fine-tuning gene expression programs and contributing to skeletal muscle plasticity in health and disease. In response to exercise, it has been observed specific activation of both AMPK and CaMKII, which in turn can induce phosphorylation-dependent nuclear export of class IIa HDAC [35,55]. Moreover, CaMKII activation during physical exercise can cause hyperacetylation of histones and increased MEF2 binding to cis elements in the promoter of the Glut4 gene, thus eliciting GLUT4 expression in skeletal muscle and improving glucose transport capacity and insulin sensitivity [232].
4.3. ROS impact on protein arginine methyltransferase
The activity of enzymes involved in the formation of asymmetric dimethylarginine (ADMA) precursors is regulated in a redox-sensitive fashion. This modification is catalyzed by the protein arginine methyltransferase (PRMT). The major type of protein arginine (R) methyltransferase (PRMT), type I, transfers the methyl group from S-adenosyl-l-methionine (AdoMet) to the guanidino group of arginines in protein substrates, resulting in mono-methylarginine and asymmetric dimethylarginine in substrate proteins [233]. It has been shown that oxidation at two cysteine residues destabilizes homodimerization of PRMT proteins, leading to diminished methyltransferase activity, whereas reduction can reverse the effects of oxidation, with sulfenic acid formation at both cysteine residues [234].
Arginine residues substrate of PRMTs are present in different kind of proteins, including nucleolar proteins, proteins involved in RNA processing and histones; all of them can be modified by the addition of one or two methyl groups. Mutagenesis studies revealed that homodimerization is essential for AdoMet binding and for arginine methyltransferase enzymatic activity [235]. In particular, removal of two cysteine residues implicated in PRMT1 dimerization and cofactor binding eliminates the redox-dependent control over PRMT1 methyltransferase activity [234].
Histone methylation by PRMTs can induce either activation or repression of gene expression. Arsenic exposure of human keratinocytes triggers PRMT1 and PRMT4 transient accumulation into the nucleus, leading to methylation of H4R3 and H3R17 in the ARE enhancer regions of ferritin transcription unit, recognized by Nrf2 [236]. In this way, ROS induced methylation of histones allow the activation of Nrf2-mediated antioxidant cellular defense program.
Remarkably, PRMT expression and activity do not change in response to acute exercise. However, subcellular fractionation experiments revealed that exercise elicited myonuclear translocation of PRMT1, correlating with increased PRMT1-methyltransferase activity, as indicated by the higher histone methylation levels of skeletal muscle after exercise [237]. These findings disclose PRMT-mediated histone arginine methylation as an essential player of the signaling driving skeletal muscle plasticity in response to pro-oxidant environment elicited by physical exercise.
4.4. ROS impact on sirtuins
The silencer information regulator (Sir) family of proteins is widely involved in metabolic homeostasis and aging [238]. Sirtuins are NAD-dependent deacetylases that catalyze deacetylation in specific lysine residues of histones and other proteins, including transcription factors and metabolic enzymes, thereby promoting chromatin silencing and transcriptional repression and regulating different biological processes, including cell cycle, differentiation, metabolism, stress resistance, senescence, and aging [239]. In addition to histone proteins, also other proteins can be deacetylated by sirtuins, with modulation of their activity [239]. SIRT1, -6, and -7 act directly on the transcription of genes involved in metabolism, whereas SIRT3-5 reside in the mitochondrial matrix and regulate enzymes involved in the tricarboxylic acid and urea cycles, oxidative phosphorylation, as well as ROS production [238].
The oxidative and metabolic stress induced by physical exercise greatly impacts sirtuins: exercise increases the SIRT3 protein content in skeletal muscle [240,241], whereas SIRT3 was shown to be downregulated in immobilized soleus muscle [241]. Importantly, SIRT3 dynamically responds to exercise modulating muscular energy homeostasis via AMPK and PGC1α [240].
Oxidative stress can affect sirtuins at different levels, thus causing a dysregulation in the normal Sir function. In particular, oxidative stress can induce or repress the expression of SIRT genes; it can trigger post-translational oxidative modifications of sirtuins, alter the SIRT-interactome and induce changes in intracellular NAD levels. In response to oxidative stress, SIRT1 was redistributed at the chromatin level causing deregulation of transcription [242]. Moreover, oxidative stress lowers SIRT1 mRNA and protein expression in human fibroblasts. Unlike the enhanced HuR affinity toward numerous target mRNAs in response to other stress agents, oxidant treatment was shown to dissociate HuR from the SIRT1 mRNA [243]. In particular, under oxidative stress and DNA damage, HuR interacts and is phosphorylated by the checkpoint kinase 2 (CHK2), leading to its dissociation from SIRT1 transcript, thus causing its destabilization [243]. In line with this observation, SIRT1 expression was decreased in atherosclerotic plaques of vascular smooth muscle cells, showing increased intracellular oxidants [244].
Notably, mild increase in ROS concentration induces the upregulation of SIRT1 protein, thus affecting SIRT1 targets, including p53 and FOXO3a, which achieve the antioxidant response through the expression of SOD2 and catalase [245]. In contrast, exposure to high levels of H2O2 led to sumoylation and proteasomal degradation of SIRT1 protein [246].
Oxidative stress can also affect sirtuin activity by altering its cellular interactors. The SIRT1 interactor DBC1 (deleted in breast cancer 1) [247] is phosphorylated by ATM/ATR during oxidative stress; this phosphorylation increases its affinity for SIRT1 and leads to inhibition of SIRT1 activity [248]. Similarly, obesity and aging impair SIRT1 activity both by promoting SIRT1 binding to DBC1 [249] and by declining the NAD levels [250].
Finally, it was recently shown that upon oxidative stress SIRT1 can be inactivated by sequestration into cytoplasmic caveolae, where it binds directly caveolin-1. In particular, caveolin-1-mediated inhibition of SIRT1 is sufficient to promote the acetylation, and activation, of the transcription factor p53, thus causing premature senescence in fibroblasts [251].
4.5. DNA and redox signaling
DNA itself is as a sensor of changes in the redox homeostasis and can be directly modified by ROS. For example, hydroxyl radicals promote the formation of 5-hydroxymethylcytosine (5hmC) from 5-methylcytosine (5mC) [252], thus causing indirect demethylation at CpG sites. These modifications interfere with the DNMT1 methylation signature [253]. Superoxide can also induce cytosine methylation by direct transfer of a methyl group from SAM ion [254]. Interestingly, ROS accumulate at G-rich regions, thus promoting oxidation of transcription factors at specific sites. Guanosine oxidation into 8-oxo-20-deoxyguanosine (8-oxodG) preferentially inhibits the methylation of adjacent cytosines, resulting in the hypomethylation and transcriptional activation [255]. In general, oxidative modifications of nuclear and mitochondrial DNA correlate with aging and age-associated diseases [256,257]. Remarkably, regular exercise can lower the oxidative damage of both nuclear and mitochondrial DNA thus delaying the aging process [[258], [259], [260]]. The observed effect was due, at least in part, to the exercise induced activity of the DNA damage repair enzymes 8-oxoguanine-DNA glycosilases (OGG) in the nucleus, and their exercise-training induced import into the mitochondrial matrix to repair oxidized guanosines [261]. On the contrary, a sedentary lifestyle impairs the transfer of OGG into the mitochondrial matrix [144,261]. These findings are in line with the beneficial effects of regular exercise with a moderate production of ROS, resulting in cellular adaptation and competence to face stronger oxidative insults. ROS can also modify histones, causing nitration and oxidation of H1, H2B and H3 [262]. Oxidation of basic residues, including arginine and lysine, in histone H3, promotes relaxation of chromatin and accumulation of transcription factors, thus activating gene expression programs [262]. Furthermore, the histone H3 is S-glutathionylated on Cys110 in response to redox changes producing structural transitions that affect nucleosomal occupancy, open the chromatin structure and activate transcription [229].
4.6. Noncoding RNA and redox signaling
Both lncRNAs and miRNAs can be regulated by ROS. Emerging evidences suggest a regulatory interplay between miRNAs and redox signaling: in fact, ROS profoundly impacts miRNA transcription, biogenesis and function, whereas miRNAs regulate the expression of redox sensors and components of the antioxidant machinery.
In general, miRNAs are induced by oxidative stress. ROS upregulates miR-146a, miR-181a, miR-30b and miR-874-3p, targeting respectively SOD2, glutathione peroxidase 1 (GPX1), Catalase, and Caspase 8 mRNAs [263]. Interestingly, miR200c is upregulated upon oxidative stress and targets SIRT1, FOXO1 and the endothelial nitric oxide synthase eNOS [264]. The upregulation of miR200c upon H2O2 exposure could explain, at least in part, the downregulation of SIRT1 expression and activity, as well as the hyperacetylation of the promoter of its target genes. However, how redox changes control miRNA expression at the mechanistic level is still largely unexplored.
ROS are involved in several steps of miRNA biogenesis [265]: they affect miRNA maturation by down-regulating Dicer and modifying the RISC complex [266]. MiRNA biogenesis is also redox-regulated via changes in the activity of the endonuclease Drosha, mainly through the redox-dependent enzyme glycogen synthase kinase that phosphorylates Drosha, thus promoting its translocation into the nucleus [267].
MiRNAs themselves can be directly oxidized by ROS, which alters their stability, structure, and ability to bind to target mRNAs. Moreover, miRNAs can be oxidatively modified by ROS. As mentioned above, ROS can hydroxylate guanine to produce 8-oxo-7,8-dihydro-2′-deoxyguanosine (8OHdG) in the DNA and 8-oxo-7,8-dihydroguanosine (8OHG) in the RNA [[268], [269], [270]]. The oxidized miRNAs mis-recognize target mRNAs, resulting in down-regulation in their protein synthesis native targets [271]. Oxidized miR-184 associates to the 3'untranslated region of Bcl-xL and Bcl-w that are not its native targets, thus causing the subsequent reduction in Bcl-xL and Bcl-w with the consequent apoptosis. Interestingly, not all miRNAs are oxidized in response to ROS, and the selective oxidation of specific miRNAs seems not related to the upregulation of miRNA expression induced by ROS [271].
As mentioned above, in response to oxidative stress, mammalian cells orchestrate a rapid and tightly regulated signaling response that results in the transcriptional induction of protein-coding genes, mostly involved in the regulation of the cellular redox-state [272], but it also involves transcriptional modulation of noncoding genes [273]. High throughput RNA sequencing revealed that thousands of lncRNA transcripts are transiently induced by oxidative exposure [273]. In particular, the sharp pausing of the RNA polymerase II at distinct promoters after oxidative exposure leads to a rapid and global induction of bidirectional transcription. This phenomenon can cause post-transcriptional processing defects leading to the downregulation of the mRNA levels of the host genes. Notably, these promoters associated noncoding transcripts (pancRNAs) are often capped and polyadenylated, and they can translocate from the nucleus to the cytoplasm to associate with polysomal fractions and inhibit translation [273]. However, the functional significance of these stress-induced lncRNAs needs to be further elucidated. Very little is known about the regulation of lncRNAs in response to physical exercise. It was recently reported that exercise can downregulate the lncRNA MALAT1, thus ameliorating the insulin resistance in type 2 diabetes mellitus by elevating miR-382-3p and reducing resistin, a protein hormone released by adipocytes, which contributes to insulin resistance phenotype [274]. MALAT1 acts as a competing endogenous RNA by sequestering miR-382-3p. Hence, the exercise-induced MALAT1 downregulation allows miR-382-3p to target resistin mRNA thus counteracting insulin resistance [274].
Collectively, these findings highlight miRNAs and lncRNAs as key molecules in the adaptive response of cellular systems to physical activity. MicroRNA levels can change depending on the training status and/or exercise protocol. Moreover, the differential expression of miRNAs is instrumental to achieve recovery and adaptation strategies to counteract exercise-induced ROS.
5. Conclusion and future perspectives
Growing body of evidence indicates ROS and oxidative stress as novel players in shaping the epigenetic landscape of the entire genome. Although we are only beginning to decipher the high complexity of this regulation, it seems clear that mitochondrial metabolism, various enzymes and signaling molecules, as well as antioxidant systems contribute to ROS balance in response to environmental stimuli. ROS, in turn, can impact the epigenetic landscape by modulating DNA and histone modifications, ncRNA transcripts and chromatin remodeling with important modification in the expression of genes related either to physiological or pathological adaptation. Indeed, increasing amount of data highlights the link between epigenetic regulation and oxidative stress-related pathways in aging, cancer and other pathologies, such as cardiovascular, neurological and metabolic diseases.
Physically active lifestyle and regular exercise contribute to improve health and prevent diseases, inducing both tissue specific and systemic adaption. Exercise has proven to be a potent environmental stimulus to induce epigenetic regulation of key regulatory, metabolic and myogenic genes, as well as genes that counteract in several points of disease. Although results on the direct connection between the modulation of redox homeostasis by exercise and epigenetic regulation are still very limited, the evidence that ROS and RNS produced under voluntary exercise represent important regulatory mediators in signaling processes suggests that exercise-induced ROS production is likely to be an important player in epigenetic events. Our current model for the role of exercise-induced ROS in the modification of the epigenome is shown in the graphical abstract, where exercise, by epigenetic modification of ROS producers, sensing and neutralization, can regulate ROS concentration. In turn, ROS can induce modifications in the epigenetic machinery, i.e. oxidative modification of DNA sequences, DNMTs, HDACs, ncRNAs, contributing to adaptation in the resulting phenotype. Of course, it should be considered that epigenetics represents a mechanism for cellular signal integration, thus the ideal models to study the causal link between exercise-induced ROS and epigenetic modifications are difficult to establish due to the extremely broad and interconnected nature of epigenetic control, as well as of redox homeostasis components. This represents indeed an area for future research, together with investigation of the metabolic changes and signaling events during exercise that may influence such interactions. As for most of the research questions in the field of exercise redox biology, the lack of precision tools to study the events that connect ROS production and redox signaling in the cell, especially in “in vivo” models, remains a major barrier also for advancement in understanding the role of ROS in exercise epigenomics. Moreover, further improvements in redox proteomics or in the detection of oxidative modifications to RNA promise to increase our knowledge on the exercise-induced post-translational, oxidative modification of relevant epigenetic components. Indeed, the main challenge for the future research is to move from a rather descriptive level of collected data to the mechanistic analysis of the causal involvement of exercise-induced redox modification in the epigenetic control of metabolic and physiological adaptations. The recent development of whole-genome epigenetic sequencing and new high-throughput technology, combined with new analytic methods in redox biology, promises to evaluate the global changes in epigenetic marks in relation to measurable changes in redox metabolism. Future studies should include well-defined exercise protocols in human subjects and animal models where key tissues (i.e., blood, skeletal muscle, adipose tissue) are broadly analyzed at multiple time points during both the exercise and recovery time, also matching the biochemical and molecular modifications with the functional physiological adjustment and/or adaptations.
Funding
This work was supported by grants from the University of Rome Foro Italico (Research Grant ID: CDR2.RIC182015) to DC and from the Associazione Italiana Ricerca sul Cancro AIRC; ID (IG21877) to MPP.
Declaration of competing interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101477.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ji L.L., Gomez-Cabrera Mc, Vina J. Exercise and hormesis: activation of cellular antioxidant signaling pathway. Ann. N. Y. Acad. Sci. 2006;1067:425–435. doi: 10.1196/annals.1354.061. [DOI] [PubMed] [Google Scholar]
- 2.Morales-Alamo D., Calbet J.A.L. AMPK signaling in skeletal muscle during exercise: role of reactive oxygen and nitrogen species. Free Radic. Biol. Med. 2016;98:68–77. doi: 10.1016/j.freeradbiomed.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Radak Z., Koltai E., Taylor A.W., Higuchi M., Kumagai S., Ohno H., Goto S., Boldogh I. Redox-regulating sirtuins in aging, caloric restriction, and exercise. Free Radic. Biol. Med. 2013 May;58:87–97. doi: 10.1016/j.freeradbiomed.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Powers S.K., Jackson M.J. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol. Rev. 2008;88(4):1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson MJ Control. Of reactive oxygen species production in contracting skeletal muscle. Antioxidants Redox Signal. 2011 Nov 1;15(9):2477–2486. doi: 10.1089/ars.2011.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vina J., Sanchis-Gomar F., Martinez-Bello V., Gomez-Cabrera M.C. Exercise acts as a drug; the pharmacological benefits of exercise. Br. J. Pharmacol. 2012;167(1):1–12. doi: 10.1111/j.1476-5381.2012.01970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimauro I., Mercatelli N., Caporossi D. Exercise-induced ROS in heat shock proteins response. Free Radic. Biol. Med. 2016;98:46–55. doi: 10.1016/j.freeradbiomed.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Keller P., Vollaard N.B., Gustafsson T., Gallagher I.J., Sundberg C.J., Rankinen T., Britton S.L., Bouchard C., Koch L.G., Timmons J.A. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J. Appl. Physiol. 1985;110(1):46–59. doi: 10.1152/japplphysiol.00634.2010. 2011 Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merry T.L., Ristow M. Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice. J. Physiol. 2016 Sep 15;594(18):5195–5207. doi: 10.1113/JP271957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shanmugam G., Challa A.K., Devarajan A. Exercise mediated Nrf2 signaling protects the myocardium from isoproterenol-induced pathological remodeling. Front Cardiovasc Med. 2019;6:68. doi: 10.3389/fcvm.2019.00068. Published 2019 Jun 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krüger K., Frost S., Most E., Völker K., Pallauf J., Mooren F.C. Exercise affects tissue lymphocyte apoptosis via redox-sensitive and Fas-dependent signaling pathways. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009 May;296(5):R1518–R1527. doi: 10.1152/ajpregu.90994.2008. [DOI] [PubMed] [Google Scholar]
- 12.Radak Z., Suzuki K., Higuchi M., Balogh L., Boldogh I., Koltai E. Physical exercise, reactive oxygen species and neuroprotection. Free Radic. Biol. Med. 2016 Sep;98:187–196. doi: 10.1016/j.freeradbiomed.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 13.Russo V.E.A., Martienssen R.A., Riggs A.D. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Press; 1996. Epigenetic Mechanisms of Gene Regulation. [Google Scholar]
- 14.Skvortsova K., Iovino N., Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018;19(12):774–790. doi: 10.1038/s41580-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 15.Gabbianelli R., Malavolta M. Epigenetics in ageing and development. Mech. Ageing Dev. 2018 Sep;174:1–2. doi: 10.1016/j.mad.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Meeran S.M., Ahmed A., Tollefsbol T.O. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin. Epigenet. 2010;1(3–4):101–116. doi: 10.1007/s13148-010-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grazioli E., Dimauro I., Mercatelli N., Wang G., Pitsiladis Y., Di Luigi L., Caporossi D. Physical activity in the prevention of human diseases: role of epigenetic modifications. BMC Genom. 2017 Nov 14;18(Suppl 8):802. doi: 10.1186/s12864-017-4193-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radak Z., Zhao Z., Koltai E., Ohno H., Atalay M. Oxygen consumption and usage during physical exercise: the balance between oxidative stress and ROS-dependent adaptive signaling. Antioxidants Redox Signal. 2013 Apr 1;18(10):1208–1246. doi: 10.1089/ars.2011.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powers S.K., Radak Z., Ji L.L. Exercise-induced oxidative stress: past, present and future. J. Physiol. 2016 Sep 15;594(18):5081–5092. doi: 10.1113/JP270646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes C.P.C., de Gonzalo-Calvo D., Toro R. Non-coding RNAs and exercise: pathophysiological role and clinical application in the cardiovascular system. Clin. Sci. (Lond.) 2018;132(9):925–942. doi: 10.1042/CS20171463. Published 2018 May 20. [DOI] [PubMed] [Google Scholar]
- 21.Hojman P., Gehl J., Christensen J.F., Pedersen B.K. Molecular mechanisms linking exercise to cancer prevention and treatment. Cell Metabol. 2018;27(1):10–21. doi: 10.1016/j.cmet.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Rebelo-Marques A., De Sousa Lages A., Andrade R. Aging hallmarks: the benefits of physical exercise. Front. Endocrinol. 2018;9:258. doi: 10.3389/fendo.2018.00258. Published 2018 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruegsegger G.N., Booth F.W. Health benefits of exercise. Cold Spring Harb Perspect Med. 2018;8(7):a029694. doi: 10.1101/cshperspect.a029694. Published 2018 Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimauro I., Sgura A., Pittaluga M. Regular exercise participation improves genomic stability in diabetic patients: an exploratory study to analyse telomere length and DNA damage. Sci. Rep. 2017;7(1):4137. doi: 10.1038/s41598-017-04448-4. Published 2017 Jun 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vega R.B., Konhilas J.P., Kelly D.P., Leinwand L.A. Molecular mechanisms underlying cardiac adaptation to exercise. Cell Metabol. 2017;25(5):1012–1026. doi: 10.1016/j.cmet.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimauro I., Scalabrin M., Fantini C. Resistance training and redox homeostasis: correlation with age-associated genomic changes. Redox Biol. 2016;10:34–44. doi: 10.1016/j.redox.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drake J.C., Wilson R.J., Yan Z. Molecular mechanisms for mitochondrial adaptation to exercise training in skeletal muscle. Faseb. J. 2016;30(1):13–22. doi: 10.1096/fj.15-276337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pittaluga M., Sgadari A., Dimauro I., Tavazzi B., Parisi P., Caporossi D. Physical exercise and redox balance in type 2 diabetics: effects of moderate training on biomarkers of oxidative stress and DNA damage evaluated through comet assay. Oxid Med Cell Longev. 2015;2015:981242. doi: 10.1155/2015/981242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmer P., Bloch W. Physical exercise and epigenetic adaptations of the cardiovascular system. Herz. 2015;40(3):353–360. doi: 10.1007/s00059-015-4213-7. [DOI] [PubMed] [Google Scholar]
- 30.Egan B., Zierath J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metabol. 2013 Feb 5;17(2):162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Sanchis-Gomar F., Garcia-Gimenez J.L., Perez-Quilis C., Gomez-Cabrera M.C., Pallardo F.V., Lippi G. Physical exercise as an epigenetic modulator: eustress, the "positive stress" as an effector of gene expression. J. Strength Condit Res. 2012;26(12):3469–3472. doi: 10.1519/JSC.0b013e31825bb594. [DOI] [PubMed] [Google Scholar]
- 32.Ingerslev L.R., Donkin I., Fabre O. Endurance training remodels sperm-borne small RNA expression and methylation at neurological gene hotspots. Clin. Epigenet. 2018;10:12. doi: 10.1186/s13148-018-0446-7. Jan 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laker R.C., Lillard T.S., Okutsu M. Exercise prevents maternal high-fat diet-induced hypermethylation of the Pgc-1α gene and age-dependent metabolic dysfunction in the offspring. Diabetes. 2014;63(5):1605–1611. doi: 10.2337/db13-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potthoff M.J., Wu H., Arnold M.A. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 2007;117(9):2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGee S.L., Fairlie E., Garnham A.P., Hargreaves M. Exercise-induced histone modifications in human skeletal muscle. J. Physiol. 2009;587(Pt 24):5951–5958. doi: 10.1113/jphysiol.2009.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen S., Scheele C., Yfanti C. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle [published correction appears in. J. Physiol. 2011 Mar 1;589(Pt 5):1239. doi: 10.1113/jphysiol.2010.189860. Laye, Matthew [corrected to Laye, Matthew J]] [published correction appears in J Physiol. 2015 Mar 1;593(5):1323]. J Physiol. 2010;588(Pt 20):4029–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidsen P.K., Gallagher I.J., Hartman J.W. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 1985;110(2):309–317. doi: 10.1152/japplphysiol.00901.2010. 2011. [DOI] [PubMed] [Google Scholar]
- 38.Voisin S., Eynon N., Yan X., Bishop D.J. Exercise training and DNA methylation in humans. Acta Physiol. 2015;213(1):39–59. doi: 10.1111/apha.12414. [DOI] [PubMed] [Google Scholar]
- 39.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 40.Rottach A., Leonhardt H., Spada F. DNA methylation-mediated epigenetic control. J. Cell. Biochem. 2009;108(1):43–51. doi: 10.1002/jcb.22253. [DOI] [PubMed] [Google Scholar]
- 41.Han H., Cortez C.C., Yang X., Nichols P.W., Jones P.A., Liang G. DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum. Mol. Genet. 2011;20(22):4299–4310. doi: 10.1093/hmg/ddr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guibert S., Weber M. Functions of DNA methylation and hydroxymethylation in mammalian development. Curr. Top. Dev. Biol. 2013;104:47–83. doi: 10.1016/B978-0-12-416027-9.00002-4. [DOI] [PubMed] [Google Scholar]
- 43.Barrès R., Yan J., Egan B. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metabol. 2012;15(3):405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Nitert M.D., Dayeh T., Volkov P. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61(12):3322–3332. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanzleiter T., Jähnert M., Schulze G. Exercise training alters DNA methylation patterns in genes related to muscle growth and differentiation in mice. Am. J. Physiol. Endocrinol. Metab. 2015;308(10):E912–E920. doi: 10.1152/ajpendo.00289.2014. [DOI] [PubMed] [Google Scholar]
- 46.Ntanasis-Stathopoulos J., Tzanninis J.G., Philippou A., Koutsilieris M. Epigenetic regulation on gene expression induced by physical exercise. J. Musculoskelet. Neuronal Interact. 2013;13(2):133–146. [PubMed] [Google Scholar]
- 47.Rönn T., Volkov P., Davegårdh C. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9(6) doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakajima K., Takeoka M., Mori M. Exercise effects on methylation of ASC gene. Int. J. Sports Med. 2010;31(9):671–675. doi: 10.1055/s-0029-1246140. [DOI] [PubMed] [Google Scholar]
- 49.Shaw B., Leung W.C., Tapp H.S., Fitzpatrick A.L., Saxton J.M., Belshaw N.J. A change in physical activity level affects leukocyte DNA methylation of genes implicated in cardiovascular disease in the elderly. Proc. Phys. Soc. 2014;31:C46. [Google Scholar]
- 50.Gomez-Pinilla F., Zhuang Y., Feng J., Ying Z., Fan G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur. J. Neurosci. 2011;33(3):383–390. doi: 10.1111/j.1460-9568.2010.07508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu P.Z., Nusslock R. Exercise-mediated neurogenesis in the Hippocampus via BDNF. Front. Neurosci. 2018;12:52. doi: 10.3389/fnins.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou V.W., Goren A., Bernstein B.E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 53.Wu H., Naya F.J., McKinsey T.A. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19(9):1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischle W., Dequiedt F., Hendzel M.J. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9(1):45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 55.McGee S.L., Hargreaves M. Epigenetics and exercise. Trends Endocrinol. Metabol. 2019;30(9):636–645. doi: 10.1016/j.tem.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 56.Egan B., Carson B.P., Garcia-Roves P.M. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signaling kinases in human skeletal muscle. J. Physiol. 2010;588(Pt 10):1779–1790. doi: 10.1113/jphysiol.2010.188011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czubryt M.P., McAnally J., Fishman G.I., Olson E.N. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. U. S. A. 2003;100(4):1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Collins A., Hill L.E., Chandramohan Y., Whitcomb D., Droste S.K., Reul J.M. Exercise improves cognitive responses to psychological stress through enhancement of epigenetic mechanisms and gene expression in the dentate gyrus. PloS One. 2009;4(1) doi: 10.1371/journal.pone.0004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elsner V.R., Lovatel G.A., Bertoldi K. Effect of different exercise protocols on histone acetyltransferases and histone deacetylases activities in rat hippocampus. Neuroscience. 2011;192:580–587. doi: 10.1016/j.neuroscience.2011.06.066. [DOI] [PubMed] [Google Scholar]
- 60.Liu G., Mattick J.S., Taft R.J. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle. 2013;12(13):2061–2072. doi: 10.4161/cc.25134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peschansky V.J., Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc. Natl. Acad. Sci. U. S. A. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation [published correction appears in Nat Rev Genet. 2004 Aug;5(8):631] Nat. Rev. Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 64.Mercatelli N., Fittipaldi S., De Paola E. MiR-23-TrxR1 as a novel molecular axis in skeletal muscle differentiation. Sci. Rep. 2017;7(1):7219. doi: 10.1038/s41598-017-07575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellison G.M., Waring C.D., Vicinanza C., Torella D. Physiological cardiac remodelling in response to endurance exercise training: cellular and molecular mechanisms. Heart. 2012;98(1):5–10. doi: 10.1136/heartjnl-2011-300639. [DOI] [PubMed] [Google Scholar]
- 66.Liu G., Detloff M.R., Miller K.N., Santi L., Houlé J.D. Exercise modulates microRNAs that affect the PTEN/mTOR pathway in rats after spinal cord injury. Exp. Neurol. 2012;233(1):447–456. doi: 10.1016/j.expneurol.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Güller I., Russell A.P. MicroRNAs in skeletal muscle: their role and regulation in development, disease and function. J. Physiol. 2010;588(Pt 21):4075–4087. doi: 10.1113/jphysiol.2010.194175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baltimore D., Boldin M.P., O'Connell R.M., Rao D.S., Taganov K.D. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 2008;9(8):839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 69.Shang J.L., Cheng Q., Duan S.J., Li L., Jia L.Y. Cognitive improvement following ischemia/reperfusion injury induced by voluntary running wheel exercise is associated with LncMALAT1mediated apoptosis inhibition. Int. J. Mol. Med. 2018;41(5):2715–2723. doi: 10.3892/ijmm.2018.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Denham J. Exercise and epigenetic inheritance of disease risk. Acta Physiol. 2018;222(1) doi: 10.1111/apha.12881. 10.1111/apha.12881. [DOI] [PubMed] [Google Scholar]
- 71.Ramasamy S., Velmurugan G., Shanmugha Rajan K., Ramprasath T., Kalpana K. MiRNAs with apoptosis regulating potential are differentially expressed in chronic exercise-induced physiologically hypertrophied hearts. PloS One. 2015;10(3) doi: 10.1371/journal.pone.0121401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fyfe J.J., Bishop D.J., Zacharewicz E., Russell A.P., Stepto N.K. Concurrent exercise incorporating high-intensity interval or continuous training modulates mTORC1 signaling and microRNA expression in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016;310(11):R1297–R1311. doi: 10.1152/ajpregu.00479.2015. [DOI] [PubMed] [Google Scholar]
- 73.Nie Y., Sato Y., Wang C., Yue F., Kuang S., Gavin T.P. Impaired exercise tolerance, mitochondrial biogenesis, and muscle fiber maintenance in miR-133a-deficient mice. Faseb. J. 2016;30(11):3745–3758. doi: 10.1096/fj.201600529R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ogasawara R., Akimoto T., Umeno T., Sawada S., Hamaoka T., Fujita S. MicroRNA expression profiling in skeletal muscle reveals different regulatory patterns in high and low responders to resistance training. Physiol. Genom. 2016;48(4):320–324. doi: 10.1152/physiolgenomics.00124.2015. [DOI] [PubMed] [Google Scholar]
- 75.Aoi W., Naito Y., Takanami Y. Oxidative stress and delayed-onset muscle damage after exercise. Free Radic. Biol. Med. 2004;37(4):480–487. doi: 10.1016/j.freeradbiomed.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 76.Nielsen S., Åkerström T., Rinnov A., Yfanti C., Scheele C., Pedersen B.K., Laye M.J. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PloS One. 2014 Feb 19;9(2) doi: 10.1371/journal.pone.0087308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Macpherson H., Teo W.P., Schneider L.A., Smith A.E. A life-long approach to physical activity for brain health. Front. Aging Neurosci. 2017;9:147. doi: 10.3389/fnagi.2017.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu G., Keeler B.E., Zhukareva V., Houlé J.D. Cycling exercise affects the expression of apoptosis-associated microRNAs after spinal cord injury in rats. Exp. Neurol. 2010;226(1):200–206. doi: 10.1016/j.expneurol.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walsh N.P., Gleeson M., Pyne D.B. Position statement. Part two: maintaining immune health. Exerc. Immunol. Rev. 2011;17:64–103. [PubMed] [Google Scholar]
- 80.Walsh N.P., Gleeson M., Shephard R.J. Position statement. Part one: immune function and exercise. Exerc. Immunol. Rev. 2011;17:6–63. [PubMed] [Google Scholar]
- 81.Navarro F., Bacurau A.V., Pereira G.B. Moderate exercise increases the metabolism and immune function of lymphocytes in rats. Eur. J. Appl. Physiol. 2013;113(5):1343–1352. doi: 10.1007/s00421-012-2554-y. [DOI] [PubMed] [Google Scholar]
- 82.Bacurau A.V., Belmonte M.A., Navarro F. Effect of a high-intensity exercise training on the metabolism and function of macrophages and lymphocytes of walker 256 tumor bearing rats. Exp. Biol. Med. 2007;232(10):1289–1299. doi: 10.3181/0704-RM-93. [DOI] [PubMed] [Google Scholar]
- 83.Radom-Aizik S., Zaldivar F., Haddad F., Cooper D.M. Impact of brief exercise on peripheral blood NK cell gene and microRNA expression in young adults. J. Appl. Physiol. 1985;114(5):628–636. doi: 10.1152/japplphysiol.01341.2012. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Radom-Aizik S., Zaldivar F., Jr., Leu S.Y., Adams G.R., Oliver S., Cooper D.M. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Clin Transl Sci. 2012;5(1):32–38. doi: 10.1111/j.1752-8062.2011.00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Radom-Aizik S., Zaldivar F., Jr., Oliver S., Galassetti P., Cooper D.M. Evidence for microRNA involvement in exercise-associated neutrophil gene expression changes. J. Appl. Physiol. 1985;109(1):252–261. doi: 10.1152/japplphysiol.01291.2009. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mooren F.C., Viereck J., Krüger K., Thum T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am. J. Physiol. Heart Circ. Physiol. 2014;306(4):H557–H563. doi: 10.1152/ajpheart.00711.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banzet S., Chennaoui M., Girard O. Changes in circulating microRNAs levels with exercise modality. J. Appl. Physiol. 1985;115(9):1237–1244. doi: 10.1152/japplphysiol.00075.2013. 2013. [DOI] [PubMed] [Google Scholar]
- 88.Davies K.J., Quintanilha A.T., Brooks G.A., Packer L. Free radicals and tissue damage produced by exercise. Biochem. Biophys. Res. Commun. 1982;107(4):1198–1205. doi: 10.1016/s0006-291x(82)80124-1. [DOI] [PubMed] [Google Scholar]
- 89.Sen C.K. Oxidants and antioxidants in exercise. J. Appl. Physiol. 1985;79(3):675–686. doi: 10.1152/jappl.1995.79.3.675. 1995. [DOI] [PubMed] [Google Scholar]
- 90.Jackson MJ Control. Of reactive oxygen species production in contracting skeletal muscle. Antioxidants Redox Signal. 2011 Nov 1;15(9):2477–2486. doi: 10.1089/ars.2011.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Radak Z., Chung H.Y., Goto S. Systemic adaptation to oxidative challenge induced by regular exercise. Free Radic. Biol. Med. 2008;44(2):153–159. doi: 10.1016/j.freeradbiomed.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 92.Jones D.P. Radical-free biology of oxidative stress. Am. J. Physiol. Cell Physiol. 2008;295(4):C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vina J., Rodriguez-Manas L., Salvador-Pascual A., Tarazona-Santabalbina F.J., Gomez-Cabrera M.C. Exercise: the lifelong supplement for healthy ageing and slowing down the onset of frailty. J. Physiol. 2016;594(8):1989–1999. doi: 10.1113/JP270536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moylan J.S., Reid M.B. Oxidative stress, chronic disease, and muscle wasting. Muscle Nerve. 2007;35(4):411–429. doi: 10.1002/mus.20743. [DOI] [PubMed] [Google Scholar]
- 95.Wang X., Hai C. Novel insights into redox system and the mechanism of redox regulation. Mol. Biol. Rep. 2016;43(7):607–628. doi: 10.1007/s11033-016-4022-y. [DOI] [PubMed] [Google Scholar]
- 96.Cyr A.R., Domann F.E. The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxidants Redox Signal. 2011 Jul 15;15(2):551–589. doi: 10.1089/ars.2010.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hayes P., Knaus U.G. Balancing reactive oxygen species in the epigenome: NADPH oxidases as target and perpetrator. Antioxidants Redox Signal. 2013 May 20;18(15):1937–1945. doi: 10.1089/ars.2012.4895. [DOI] [PubMed] [Google Scholar]
- 98.Gong Y.Y., Luo J.Y., Wang L., Huang Y. MicroRNAs regulating reactive oxygen species in cardiovascular diseases. Antioxidants Redox Signal. 2018 Oct 10;29(11):1092–1107. doi: 10.1089/ars.2017.7328. [DOI] [PubMed] [Google Scholar]
- 99.Kietzmann T., Petry A., Shvetsova A., Gerhold J.M., Görlach A. The epigenetic landscape related to reactive oxygen species formation in the cardiovascular system. Br. J. Pharmacol. 2017;174(12):1533–1554. doi: 10.1111/bph.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Manea S.A., Constantin A., Manda G., Sasson S., Manea A. Regulation of Nox enzymes expression in vascular pathophysiology: focusing on transcription factors and epigenetic mechanisms. Redox Biol. 2015 Aug;5:358–366. doi: 10.1016/j.redox.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matilainen O., Quirós P.M., Auwerx J. Mitochondria and epigenetics - crosstalk in homeostasis and stress. Trends Cell Biol. 2017 Jun;27(6):453–463. doi: 10.1016/j.tcb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 102.Silva-Palacios A., Ostolga-Chavarría M., Zazueta C., Königsberg M. Nrf2: molecular and epigenetic regulation during aging. Ageing Res. Rev. 2018;47:31–40. doi: 10.1016/j.arr.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 103.Barja G. Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J. Bioenerg. Biomembr. 1999;31(4):347–366. doi: 10.1023/a:1005427919188. [DOI] [PubMed] [Google Scholar]
- 104.Jackson 2015, Jackson M.J. Redox regulation of muscle adaptations to contractile activity and aging. J. Appl. Physiol. 1985;119(3):163–171. doi: 10.1152/japplphysiol.00760.2014. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mouchiroud L., Eichner L.J., Shaw R.J., Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metabol. 2014;20(1):26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Castegna A., Iacobazzi V., Infantino V. The mitochondrial side of epigenetics. Physiol. Genom. 2015 Aug;47(8):299–307. doi: 10.1152/physiolgenomics.00096.2014. [DOI] [PubMed] [Google Scholar]
- 108.Irvin M.R., Zhi D., Joehanes R. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study. Circulation. 2014;130(7):565–572. doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stimpfel M., Jancar N., Virant-Klun I. New challenge: mitochondrial epigenetics? Stem Cell Rev Rep. 2018;14(1):13–26. doi: 10.1007/s12015-017-9771-z. [DOI] [PubMed] [Google Scholar]
- 110.Dantas W.S., Murai I.H., Perandini L.A., Azevedo H., Moreira-Filho C.A., Camara N.O., Roschel H., Gualano B. Acute exercise elicits differential expression of insulin resistance genes in the skeletal muscle of patients with polycystic ovary syndrome. Clin. Endocrinol. 2017 May;86(5):688–697. doi: 10.1111/cen.13307. [DOI] [PubMed] [Google Scholar]
- 111.Zoladz J.A., Koziel A., Broniarek I. Effect of temperature on fatty acid metabolism in skeletal muscle mitochondria of untrained and endurance-trained rats. PloS One. 2017;12(12) doi: 10.1371/journal.pone.0189456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pereira R.M., Rodrigues K.C.D.C., Anaruma C.P., Sant'Ana M.R., de Campos T.D.P., Gaspar R.S., Canciglieri R.D.S., de Melo D.G., Mekary R.A., da Silva A.S.R., Cintra D.E., Ropelle E.R., Pauli J.R., de Moura L.P. Short-term strength training reduces gluconeogenesis and NAFLD in obese mice. J. Endocrinol. 2019 Apr 1;241(1):59–70. doi: 10.1530/JOE-18-0567. [DOI] [PubMed] [Google Scholar]
- 113.Cheng Z., Almeida F.A. Mitochondrial alteration in type 2 diabetes and obesity: an epigenetic link. Cell Cycle. 2014;13(6):890–897. doi: 10.4161/cc.28189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barres R., Kirchner H., Rasmussen M., Yan J., Kantor F.R., Krook A., Näslund E., Zierath J.R. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3(4):1020–1027. doi: 10.1016/j.celrep.2013.03.018. 2013 Apr 25. [DOI] [PubMed] [Google Scholar]
- 115.Barrès R., Osler M.E., Yan J. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metabol. 2009;10(3):189–198. doi: 10.1016/j.cmet.2009.07.01. [DOI] [PubMed] [Google Scholar]
- 116.Carter H.N., Pauly M., Tryon L.D., Hood D.A. Effect of contractile activity on PGC-1α transcription in young and aged skeletal muscle. J. Appl. Physiol. 1985;124(6):1605–1615. doi: 10.1152/japplphysiol.01110.2017. [DOI] [PubMed] [Google Scholar]
- 117.Streese L., Khan A.W., Deiseroth A. High-intensity interval training modulates retinal microvascular phenotype and DNA methylation of p66Shc gene: a randomized controlled trial (EXAMIN AGE) Eur. Heart J. 2019 doi: 10.1093/eurheartj/ehz196. ehz196. [DOI] [PubMed] [Google Scholar]
- 118.Bock F., Shahzad K., Wang H. Activated protein C ameliorates diabetic nephropathy by epigenetically inhibiting the redox enzyme p66Shc. Proc. Natl. Acad. Sci. U. S. A. 2013;110(2):648–653. doi: 10.1073/pnas.1218667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gao D., Zhu B., Sun H., Wang X. Mitochondrial DNA methylation and related disease. Adv. Exp. Med. Biol. 2017;1038:117–132. doi: 10.1007/978-981-10-6674-0_9. [DOI] [PubMed] [Google Scholar]
- 120.van der Wijst M.G., van Tilburg A.Y., Ruiters M.H., Rots M.G. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci. Rep. 2017;7(1):177. doi: 10.1038/s41598-017-00263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shock L.S., Thakkar P.V., Peterson E.J., Moran R.G., Taylor S.M. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2011;108(9):3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.He S., Owen D.R., Jelinsky S.A., Lin L.L. Lysine methyltransferase SETD7 (SET7/9) regulates ROS signaling through mitochondria and NFE2L2/ARE pathway. Sci. Rep. 2015;5:14368. doi: 10.1038/srep14368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Geiger J., Dalgaard L.T. Interplay of mitochondrial metabolism and microRNAs. Cell. Mol. Life Sci. 2017;74(4):631–646. doi: 10.1007/s00018-016-2342-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barrey E., Saint-Auret G., Bonnamy B., Damas D., Boyer O., Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PloS One. 2011;6(5) doi: 10.1371/journal.pone.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Das S., Ferlito M., Kent O.A., Fox-Talbot K., Wang R., Liu D., Raghavachari N., Yang Y., Wheelan S.J., Murphy E., Steenbergen C. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012 Jun 8;110(12):1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Krause K.H. Tissue distribution and putative physiological function of NOX family NADPH oxidases. Jpn. J. Infect. Dis. 2004;57(5):S28–S29. [PubMed] [Google Scholar]
- 127.Luxen S., Belinsky S.A., Knaus U.G. Silencing of DUOX NADPH oxidases by promoter hypermethylation in lung cancer. Canc. Res. 2008;68(4):1037–1045. doi: 10.1158/0008-5472.CAN-07-5782. [DOI] [PubMed] [Google Scholar]
- 128.Siuda D., Zechner U., El Hajj N. Transcriptional regulation of Nox4 by histone deacetylases in human endothelial cells. Basic Res. Cardiol. 2012;107(5):283. doi: 10.1007/s00395-012-0283-3. [DOI] [PubMed] [Google Scholar]
- 129.Zarzuelo M.J., López-Sepúlveda R., Sánchez M. SIRT1 inhibits NADPH oxidase activation and protects endothelial function in the rat aorta: implications for vascular aging. Biochem. Pharmacol. 2013;85(9):1288–1296. doi: 10.1016/j.bcp.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 130.Gano L.B., Donato A.J., Pasha H.M., Hearon C.M., Jr., Sindler A.L., Seals D.R. The SIRT1 activator SRT1720 reverses vascular endothelial dysfunction, excessive superoxide production, and inflammation with aging in mice. Am. J. Physiol. Heart Circ. Physiol. 2014;307(12):H1754–H1763. doi: 10.1152/ajpheart.00377.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sarga L., Hart N., Koch L.G. Aerobic endurance capacity affects spatial memory and SIRT1 is a potent modulator of 8-oxoguanine repair. Neuroscience. 2013;252:326–336. doi: 10.1016/j.neuroscience.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chan S.H., Hung C.H., Shih J.Y. Exercise intervention attenuates hyperhomocysteinemia-induced aortic endothelial oxidative injury by regulating SIRT1 through mitigating NADPH oxidase/LOX-1 signaling. Redox Biol. 2018;14:116–125. doi: 10.1016/j.redox.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Varga Z.V., Kupai K., Szűcs G. MicroRNA-25-dependent up-regulation of NADPH oxidase 4 (NOX4) mediates hypercholesterolemia-induced oxidative/nitrative stress and subsequent dysfunction in the heart. J. Mol. Cell. Cardiol. 2013;62:111–121. doi: 10.1016/j.yjmcc.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 134.Silva G.J.J., Bye A., El Azzouzi H., Wisløff U. MicroRNAs as important regulators of exercise adaptation. Prog. Cardiovasc. Dis. 2017;60(1):130–151. doi: 10.1016/j.pcad.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 135.Kyrychenko S., Kyrychenko V., Badr M.A., Ikeda Y., Sadoshima J., Shirokova N. Pivotal role of miR-448 in the development of ROS-induced cardiomyopathy. Cardiovasc. Res. 2015 Dec 1;108(3):324–334. doi: 10.1093/cvr/cvv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang J., Brown M.E., Zhang H. High-throughput screening identifies microRNAs that target Nox2 and improve function after acute myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 2017;312(5):H1002–H1012. doi: 10.1152/ajpheart.00685.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gastebois C, Chanon S, Rome S, Durand C, Pelascini E, Jalabert A, Euthine V, Pialoux V, Blanc S, Simon C, Lefai E. Transition from physical activity to inactivity increases skeletal muscle miR-148b content and triggers insulin resistance. Phys. Rep.. 4(17). pii: e12902. doi: 10.14814/phy2.12902. [DOI] [PMC free article] [PubMed]
- 138.Chan G.C., Fish J.E., Mawji I.A., Leung D.D., Rachlis A.C., Marsden P.A. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J. Immunol. 2005;175(6):3846–3861. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 139.Breton C.V., Park C., Siegmund K. NOS1 methylation and carotid artery intima-media thickness in children. Circ Cardiovasc Genet. 2014;7(2):116–122. doi: 10.1161/CIRCGENETICS.113.000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suhr F., Gehlert S., Grau M., Bloch W. Skeletal muscle function during exercise-fine-tuning of diverse subsystems by nitric oxide. Int. J. Mol. Sci. 2013;14(4):7109–7139. doi: 10.3390/ijms14047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fernandes J., Arida R.M., Gomez-Pinilla F. Physical exercise as an epigenetic modulator of brain plasticity and cognition. Neurosci. Biobehav. Rev. 2017 Sep;80:443–456. doi: 10.1016/j.neubiorev.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 143.Levine A.J., Berger S.L. The interplay between epigenetic changes and the p53 protein in stem cells. Genes Dev. 2017 Jun 15;31(12):1195–1201. doi: 10.1101/gad.298984.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nemes R., Koltai E., Taylor A.W., Suzuki K., Gyori F., Radak Z. Reactive oxygen and nitrogen species regulate key metabolic, anabolic, and catabolic pathways in skeletal muscle. Antioxidants. 2018;7(7):85. doi: 10.3390/antiox7070085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moldogazieva N.T., Mokhosoev I.M., Feldman N.B., Lutsenko S.V. ROS and RNS signaling: adaptive redox switches through oxidative/nitrosative protein modifications. Free Radic. Res. 2018 May;52(5):507–543. doi: 10.1080/10715762.2018.1457217. [DOI] [PubMed] [Google Scholar]
- 146.Hibler E, Huang L, Andrade J, Spring B. Impact of a diet and activity health promotion intervention on regional patterns of DNA methylation. Clin. Epigenet.;11(1):133. doi: 10.1186/s13148-019-0707-0. [DOI] [PMC free article] [PubMed]
- 147.Virág L., Jaén R.I., Regdon Z., Boscá L., Prieto P. Self-defense of macrophages against oxidative injury: fighting for their own survival. Redox Biol. 2019;26:101261. doi: 10.1016/j.redox.2019.101261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dinkova-Kostova A.T., Liby K.T., Stephenson K.K. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc. Natl. Acad. Sci. U. S. A. 2005;102(12):4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hayes J.D., McMahon M., Chowdhry S., Dinkova-Kostova A.T. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxidants Redox Signal. 2010;13(11):1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 150.Lira V.A., Brown D.L., Lira A.K. Nitric oxide and AMPK cooperatively regulate PGC-1 in skeletal muscle cells. J. Physiol. 2010;588(Pt 18):3551–3566. doi: 10.1113/jphysiol.2010.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Thirupathi A., de Souza C.T. Multi-regulatory network of ROS: the interconnection of ROS, PGC-1 alpha, and AMPK-SIRT1 during exercise. J. Physiol. Biochem. 2017;73(4):487–494. doi: 10.1007/s13105-017-0576-y. [DOI] [PubMed] [Google Scholar]
- 152.Garcia M.M., Guéant-Rodriguez R.M., Pooya S. Methyl donor deficiency induces cardiomyopathy through altered methylation/acetylation of PGC-1α by PRMT1 and SIRT1. J. Pathol. 2011;225(3):324–335. doi: 10.1002/path.2881. [DOI] [PubMed] [Google Scholar]
- 153.Alibegovic A.C., Sonne M.P., Højbjerre L. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am. J. Physiol. Endocrinol. Metab. 2010;299(5):E752–E763. doi: 10.1152/ajpendo.00590.2009. [DOI] [PubMed] [Google Scholar]
- 154.Santos J.M., Tewari S., Benite-Ribeiro S.A. The effect of exercise on epigenetic modifications of PGC1: the impact on type 2 diabetes. Med. Hypotheses. 2014;82(6):748–753. doi: 10.1016/j.mehy.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 155.Du J.K., Cong B.H., Yu Q., Wang H., Wang L., Wang C.N., Tang X.L., Lu J.Q., Zhu X.Y., Ni X. Upregulation of microRNA-22 contributes to myocardial ischemia-reperfusion injury by interfering with the mitochondrial function. Free Radic. Biol. Med. 2016 Jul;96:406–417. doi: 10.1016/j.freeradbiomed.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 156.Shen L., Chen L., Zhang S. MicroRNA-27b regulates mitochondria biogenesis in myocytes. PloS One. 2016;11(2) doi: 10.1371/journal.pone.0148532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Caravia X.M., Fanjul V., Oliver E. The microRNA-29/PGC1α regulatory axis is critical for metabolic control of cardiac function. PLoS Biol. 2018;16(10) doi: 10.1371/journal.pbio.2006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sun Y., Cui D., Zhang Z., Zhang Q., Ji L., Ding S. Voluntary wheel exercise alters the levels of miR-494 and miR-696 in the skeletal muscle of C57BL/6 mice. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016;202:16–22. doi: 10.1016/j.cbpb.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 159.Fittipaldi S., Mercatelli N., Dimauro I., Jackson M.J., Paronetto M.P., Caporossi D. Alpha B-crystallin induction in skeletal muscle cells under redox imbalance is mediated by a JNK-dependent regulatory mechanism. Free Radic. Biol. Med. 2015;86:331–342. doi: 10.1016/j.freeradbiomed.2015.05.035. [DOI] [PubMed] [Google Scholar]
- 160.O'Connell M.A., Hayes J.D. The Keap1/Nrf2 pathway in health and disease: from the bench to the clinic. Biochem. Soc. Trans. 2015;43(4):687–689. doi: 10.1042/BST20150069. [DOI] [PubMed] [Google Scholar]
- 161.Done A.J., Traustadóttir T. Nrf2 mediates redox adaptations to exercise. Redox Biol. 2016 Dec;10:191–199. doi: 10.1016/j.redox.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Niture S.K., Kaspar J.W., Shen J., Jaiswal A.K. Nrf2 signaling and cell survival. Toxicol. Appl. Pharmacol. 2010;244(1):37–42. doi: 10.1016/j.taap.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hu T., Schreiter F.C., Bagchi R.A., Tatman P.D., Hannink M., McKinsey T.A. HDAC5 catalytic activity suppresses cardiomyocyte oxidative stress and NRF2 target gene expression. J. Biol. Chem. 2019 May 24;294(21):8640–8652. doi: 10.1074/jbc.RA118.007006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Palsamy P., Ayaki M., Elanchezhian R., Shinohara T. Promoter demethylation of Keap1 gene in human diabetic cataractous lenses. Biochem. Biophys. Res. Commun. 2012 Jul 6;423(3):542–548. doi: 10.1016/j.bbrc.2012.05.164. [DOI] [PubMed] [Google Scholar]
- 165.Yu S., Khor T.O., Cheung K.L., Li W., Wu T.Y., Huang Y., Foster B.A., Kan Y.W., Kong A.N. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS One. 2010 Jan 5;5(1) doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sanford T., Meng M.V., Railkar R., Agarwal P.K., Porten S.P. Integrative analysis of the epigenetic basis of muscle-invasive urothelial carcinoma. Clin. Epigenet. 2018;10:19. doi: 10.1186/s13148-018-0451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Huang Y., Khor T.O., Shu L. A γ-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. J. Nutr. 2012;142(5):818–823. doi: 10.3945/jn.111.153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Guo J.U., Su Y., Zhong C., Ming G.L., Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hamm C.A., Costa F.F. Epigenomes as therapeutic targets. Pharmacol. Ther. 2015;151:72–86. doi: 10.1016/j.pharmthera.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 170.Ray P.D., Huang B.W., Tsuji Y. Coordinated regulation of Nrf2 and histone H3 serine 10 phosphorylation in arsenite-activated transcription of the human heme oxygenase-1 gene. Biochim. Biophys. Acta. 2015;1849(10):1277–1288. doi: 10.1016/j.bbagrm.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Correa F., Mallard C., Nilsson M., Sandberg M. Activated microglia decrease histone acetylation and Nrf2-inducible anti-oxidant defence in astrocytes: restoring effects of inhibitors of HDACs, p38 MAPK and GSK3β. Neurobiol. Dis. 2011;44(1):142–151. doi: 10.1016/j.nbd.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sun B., Jia Y., Yang S. Sodium butyrate protects against high-fat diet-induced oxidative stress in rat liver by promoting expression of nuclear factor E2-related factor 2. Br. J. Nutr. 2019;122(4):400–410. doi: 10.1017/S0007114519001399. [DOI] [PubMed] [Google Scholar]
- 173.Cheng X., Ku C.H., Siow R.C. Regulation of the Nrf2 antioxidant pathway by microRNAs: new players in micromanaging redox homeostasis. Free Radic. Biol. Med. 2013;64:4–11. doi: 10.1016/j.freeradbiomed.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 174.Narasimhan M., Patel D., Vedpathak D., Rathinam M., Henderson G., Mahimainathan L. Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PloS One. 2012;7(12) doi: 10.1371/journal.pone.0051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fabrizio F.P., Sparaneo A., Trombetta D., Muscarella L.A. Epigenetic versus genetic deregulation of the KEAP1/NRF2 Axis in solid tumors: focus on methylation and noncoding RNAs. Oxid Med Cell Longev. 2018 Mar 13;2018:2492063. doi: 10.1155/2018/2492063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Brunet A., Sweeney L.B., Sturgill J.F. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 177.Bakkar N., Wang J., Ladner K.J. IKK/NF-kappaB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J. Cell Biol. 2008;180(4):787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sandri M., Sandri C., Gilbert A. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jackson M.J., McArdle A. Age-related changes in skeletal muscle reactive oxygen species generation and adaptive responses to reactive oxygen species. J. Physiol. 2011;589(Pt 9):2139–2145. doi: 10.1113/jphysiol.2011.206623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Brasacchio D., Okabe J., Tikellis C. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58(5):1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Karstoft K., Pedersen B.K. Exercise and type 2 diabetes: focus on metabolism and inflammation. Immunol. Cell Biol. 2016 Feb;94(2):146–150. doi: 10.1038/icb.2015.101. [DOI] [PubMed] [Google Scholar]
- 182.Masuki S., Nishida K., Hashimoto S., Morikawa M., Takasugi S., Nagata M., Taniguchi S., Rokutan K., Nose H. Effects of milk product intake on thigh muscle strength and NFKB gene methylation during home-based interval walking training in older women: a randomized, controlled pilot study. PloS One. 2017 May 17;12(5) doi: 10.1371/journal.pone.0176757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pinheiro D.M.L., de Oliveira A.H.S., Coutinho L.G. Resveratrol decreases the expression of genes involved in inflammation through transcriptional regulation. Free Radic. Biol. Med. 2019;130:8–22. doi: 10.1016/j.freeradbiomed.2018.10.432. [DOI] [PubMed] [Google Scholar]
- 184.Wang L., Chopp M., Lu X. miR-146a mediates thymosin β4 induced neurovascular remodeling of diabetic peripheral neuropathy in type-II diabetic mice. Brain Res. 2019;1707:198–207. doi: 10.1016/j.brainres.2018.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Venugopal P., Koshy T., Lavu V. Differential expression of microRNAs let-7a, miR-125b, miR-100, and miR-21 and interaction with NF-kB pathway genes in periodontitis pathogenesis. J. Cell. Physiol. 2018;233(8):5877–5884. doi: 10.1002/jcp.26391. [DOI] [PubMed] [Google Scholar]
- 186.Rusanova I., Fernández-Martínez J., Fernández-Ortiz M. Involvement of plasma miRNAs, muscle miRNAs and mitochondrial miRNAs in the pathophysiology of frailty. Exp. Gerontol. 2019;124:110637. doi: 10.1016/j.exger.2019.110637. [DOI] [PubMed] [Google Scholar]
- 187.Alipour M.R., Yousefzade N., Bavil F.M., Naderi R., Ghiasi R. Swimming impacts on pancreatic inflammatory cytokines, miR-146a and NF-кB expression levels in type-2 diabetic rats. Curr. Diabetes Rev. 2019 doi: 10.2174/1573399815666191115154421. 10.2174/1573399815666191115154421. [DOI] [PubMed] [Google Scholar]
- 188.Russo A., Bartolini D., Mensà E. Physical activity modulates the overexpression of the inflammatory miR-146a-5p in obese patients. IUBMB Life. 2018;70(10):1012–1022. doi: 10.1002/iub.1926. [DOI] [PubMed] [Google Scholar]
- 189.Van Craenenbroeck A.H., Ledeganck K.J., Van Ackeren K., Jürgens A., Hoymans V.Y., Fransen E., Adams V., De Winter B.Y., Verpooten G.A., Vrints C.J., Couttenye M.M., Van Craenenbroeck E.M. Plasma levels of microRNA in chronic kidney disease: patterns in acute and chronic exercise. Am. J. Physiol. Heart Circ. Physiol. 2015 Dec 15;309(12):H2008–H2016. doi: 10.1152/ajpheart.00346.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cyr A.R., Hitchler M.J., Domann F.E. Regulation of SOD2 in cancer by histone modifications and CpG methylation: closing the loop between redox biology and epigenetics. Antioxidants Redox Signal. 2013 May 20;18(15):1946–1955. doi: 10.1089/ars.2012.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hitchler M.J., Oberley L.W., Domann F.E. Epigenetic silencing of SOD2 by histone modifications in human breast cancer cells. Free Radic. Biol. Med. 2008;45(11):1573–1580. doi: 10.1016/j.freeradbiomed.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ow S.H., Chua P.J., Bay B.H. Epigenetic regulation of peroxiredoxins: implications in the pathogenesis of cancer. Exp. Biol. Med. 2017 Jan;242(2):140–147. doi: 10.1177/1535370216669834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zelko I.N., Folz R.J. Regulation of oxidative stress in pulmonary artery endothelium. Modulation of extracellular superoxide dismutase and NOX4 expression using histone deacetylase class I inhibitors. Am. J. Respir. Cell Mol. Biol. 2015;53(4):513–524. doi: 10.1165/rcmb.2014-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tokarz P., Kaarniranta K., Blasiak J. Inhibition of DNA methyltransferase or histone deacetylase protects retinal pigment epithelial cells from DNA damage induced by oxidative stress by the stimulation of antioxidant enzymes. Eur. J. Pharmacol. 2016;776:167–175. doi: 10.1016/j.ejphar.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 195.Tran N.Q.V., Nguyen A.N., Takabe K., Yamagata Z., Miyake K. Pre-treatment with amitriptyline causes epigenetic up-regulation of neuroprotection-associated genes and has anti-apoptotic effects in mouse neuronal cells. Neurotoxicol. Teratol. 2017;62:1–12. doi: 10.1016/j.ntt.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 196.Zhu X., Li D., Du Y., He W., Lu Y. DNA hypermethylation-mediated downregulation of antioxidant genes contributes to the early onset of cataracts in highly myopic eyes. Redox Biol. 2018;19:179–189. doi: 10.1016/j.redox.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nguyen A., Duquette N., Mamarbachi M., Thorin E. Epigenetic regulatory effect of exercise on glutathione peroxidase 1 expression in the skeletal muscle of severely dyslipidemic mice. PloS One. 2016;11 doi: 10.1371/journal.pone.0151526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sailani M.R., Halling J.F., Møller H.D. Lifelong physical activity is associated with promoter hypomethylation of genes involved in metabolism, myogenesis, contractile properties and oxidative stress resistance in aged human skeletal muscle. Sci. Rep. 2019;9(1):3272. doi: 10.1038/s41598-018-37895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kitscha P., Mann G.E., Siow R.C. Mechanosensitive microRNAs in endothelial responses to shear stress and Nrf2‐mediated redox signaling. Free Radic. Biol. Med. 2017;108:S37. doi: 10.1016/j.freeradbiomed.2017.04.142. [DOI] [Google Scholar]
- 200.Leisegang M.S., Schröder K., Brandes R.P. Redox regulation and noncoding RNAs. Antioxidants Redox Signal. 2018 Sep 20;29(9):793–812. doi: 10.1089/ars.2017.7276. [DOI] [PubMed] [Google Scholar]
- 201.Bai X.Y., Ma Y., Ding R., Fu B., Shi S., Chen X.M. miR-335 and miR-34a Promote renal senescence by suppressing mitochondrial antioxidative enzymes. J. Am. Soc. Nephrol. 2011;22(7):1252–1261. doi: 10.1681/ASN.2010040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhang Y., Zheng S., Geng Y. MicroRNA profiling of atrial fibrillation in canines: miR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PloS One. 2015;10(3) doi: 10.1371/journal.pone.0122674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ramos A.E., Lo C., Estephan L.E. Specific circulating microRNAs display dose-dependent responses to variable intensity and duration of endurance exercise. Am. J. Physiol. Heart Circ. Physiol. 2018;315(2):H273–H283. doi: 10.1152/ajpheart.00741.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Drummond M.J., McCarthy J.J., Fry C.S., Esser K.A., Rasmussen B.B. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endocrinol. Metab. 2008;295(6):E1333–E1340. doi: 10.1152/ajpendo.90562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schwarz N.A., McKinley-Barnard S.K., Blahnik Z.J. Effect of Bang® Pre-Workout Master Blaster® combined with four weeks of resistance training on lean body mass, maximal strength, mircoRNA expression, and serum IGF-1 in men: a randomized, double-blind, placebo-controlled trial. J Int Soc Sports Nutr. 2019;16(1):54. doi: 10.1186/s12970-019-0310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Camera D.M., Ong J.N., Coffey V.G., Hawley J.A. Selective modulation of MicroRNA expression with protein ingestion following concurrent resistance and endurance exercise in human skeletal muscle. Front. Physiol. 2016;7:87. doi: 10.3389/fphys.2016.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Salsbury F.R., Jr., Knutson S.T., Poole L.B., Fetrow J.S. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17(2):299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Berndt C., Lillig C.H., Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 2007;292(3):H1227–H1236. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 209.Nakamura H., Nakamura K., Yodoi J. Redox regulation of cellular activation. Annu. Rev. Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 210.Paulsen C.E., Carroll K.S. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem. Biol. 2010;5(1):47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Spadaro D., Yun B.W., Spoel S.H., Chu C., Wang Y.Q., Loake G.J. The redox switch: dynamic regulation of protein function by cysteine modifications. Physiol. Plantarum. 2010;138(4):360–371. doi: 10.1111/j.1399-3054.2009.01307.x. [DOI] [PubMed] [Google Scholar]
- 212.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 213.García-Giménez J.L., Romá-Mateo C., Pérez-Machado G., Peiró-Chova L., Pallardó F.V. Role of glutathione in the regulation of epigenetic mechanisms in disease. Free Radic. Biol. Med. 2017 Nov;112:36–48. doi: 10.1016/j.freeradbiomed.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 214.Pittaluga M., Parisi P., Sabatini S., Ceci R., Caporossi D., Catani M.V., Savini I., Avigliano L. Cellular and biochemical parameters of exercise-induced oxidative stress: relationship with training levels. Free Radic. Res. 2006;40:607–614. doi: 10.1080/10715760600623015. [DOI] [PubMed] [Google Scholar]
- 215.Ceci R., Beltran Valls M.R., Duranti G., Dimauro I., Quaranta F., Pittaluga M., Sabatini S., Caserotti P., Parisi P., Parisi A., Caporossi D. Oxidative stress responses to a graded maximal exercise test in older adults following explosive-type resistance training. Redox Biol. 2014;2:65–72. doi: 10.1016/j.redox.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Stephens N.A., Brouwers B., Eroshkin A.M., Yi F., Cornnell H.H., Meyer C., Goodpaster B.H., Pratley R.E., Smith S.R., Sparks L.M. Exercise response variations in skeletal muscle PCr recovery rate and insulin sensitivity relate to muscle epigenomic profiles in individuals with type 2 diabetes. Diabetes Care. 2018;41:2245–2254. doi: 10.2337/dc18-0296. [DOI] [PubMed] [Google Scholar]
- 217.Kim G.H., Ryan J.J., Archer S.L. The role of redox signaling in epigenetics and cardiovascular disease. Antioxidants Redox Signal. 2013 May 20;18(15):1920–1936. doi: 10.1089/ars.2012.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Watson C.J., Collier P., Tea I. Hypoxia-induced epigenetic modifications are associated with cardiac tissue fibrosis and the development of a myofibroblast-like phenotype. Hum. Mol. Genet. 2014;23(8):2176–2188. doi: 10.1093/hmg/ddt614. [DOI] [PubMed] [Google Scholar]
- 219.Patterson A.J., Xiao D., Xiong F., Dixon B., Zhang L. Hypoxia-derived oxidative stress mediates epigenetic repression of PKCε gene in foetal rat hearts. Cardiovasc. Res. 2012;93(2):302–310. doi: 10.1093/cvr/cvr322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Coulter J.B., O'Driscoll C.M., Bressler J.P. Hydroquinone increases 5-hydroxymethylcytosine formation through ten eleven translocation 1 (TET1) 5-methylcytosine dioxygenase. J. Biol. Chem. 2013;288(40):28792–28800. doi: 10.1074/jbc.M113.491365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tahiliani M., Koh K.P., Shen Y. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Thienpont B., Steinbacher J., Zhao H. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537(7618):63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Schenk A., Koliamitra C., Bauer C.J., Schier R., Schweiger M.R., Bloch W., Zimmer P. Impact of acute aerobic exercise on genome-wide DNA-methylation in natural killer cells-A pilot study. Genes. 2019 May 19;10(5):E380. doi: 10.3390/genes10050380. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Horsburgh S., Todryk S., Toms C., Moran C.N., Ansley L. Exercise-conditioned plasma attenuates nuclear concentrations of DNA methyltransferase 3B in human peripheral blood mononuclear cells. Phys. Rep. 2015 Dec;3(12) doi: 10.14814/phy2.12621. pii: e12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.McKinsey T.A., Zhang C.L., Olson E.N. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell Biol. 2001;21(18):6312–6321. doi: 10.1128/mcb.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.McKinsey T.A., Zhang C.L., Lu J., Olson E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408(6808):106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Backs J., Backs T., Bezprozvannaya S., McKinsey T.A., Olson E.N. Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol. Cell Biol. 2008;28(10):3437–3445. doi: 10.1128/MCB.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ago T., Liu T., Zhai P. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133(6):978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 229.García-Giménez J.L., Òlaso G., Hake S.B., Bönisch C., Wiedemann S.M., Markovic J., Dasí F., Gimeno A., Pérez-Quilis C., Palacios O., Capdevila M., Viña J., Pallardó F.V. Histone h3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxidants Redox Signal. 2013 Oct 20;19(12):1305–1320. doi: 10.1089/ars.2012.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jänsch N., Meyners C., Muth M. The enzyme activity of histone deacetylase 8 is modulated by a redox-switch. Redox Biol. 2019;20:60–67. doi: 10.1016/j.redox.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Doyle K., Fitzpatrick F.A. Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J. Biol. Chem. 2010 Jun 4;285(23):17417–17424. doi: 10.1074/jbc.M109.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ojuka E.O., Goyaram V., Smith J.A. The role of CaMKII in regulating GLUT4 expression in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2012;303(3):E322–E331. doi: 10.1152/ajpendo.00091.2012. [DOI] [PubMed] [Google Scholar]
- 233.Gary J.D., Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 234.Morales Y., Nitzel D.V., Price O.M. Redox control of protein arginine methyltransferase 1 (PRMT1) activity. J. Biol. Chem. 2015;290(24):14915–14926. doi: 10.1074/jbc.M115.651380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhang X., Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11(5):509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Huang B.W., Ray P.D., Iwasaki K., Tsuji Y. Transcriptional regulation of the human ferritin gene by coordinated regulation of Nrf2 and protein arginine methyltransferases PRMT1 and PRMT4. Faseb. J. 2013;27(9):3763–3774. doi: 10.1096/fj.12-226043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Vanlieshout T.L., Stouth D.W., Tajik T., Ljubicic V. Exercise-induced protein arginine methyltransferase expression in skeletal muscle. Med. Sci. Sports Exerc. 2018;50(3):447–457. doi: 10.1249/MSS.0000000000001476. [DOI] [PubMed] [Google Scholar]
- 238.Abdellatif M. Sirtuins and pyridine nucleotides. Circ. Res. 2012;111(5):642–656. doi: 10.1161/CIRCRESAHA.111.246546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Vargas-Ortiz K., Pérez-Vázquez V., Macías-Cervantes M.H. Exercise and sirtuins: a way to mitochondrial health in skeletal muscle. Int. J. Mol. Sci. 2019;20(11):2717. doi: 10.3390/ijms20112717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Palacios O.M., Carmona J.J., Michan S. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging. 2009;1(9):771–783. doi: 10.18632/aging.100075. Published 2009 Aug 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hokari F., Kawasaki E., Sakai A., Koshinaka K., Sakuma K., Kawanaka K. Muscle contractile activity regulates Sirt3 protein expression in rat skeletal muscles. J. Appl. Physiol. 1985;109(2):332–340. doi: 10.1152/japplphysiol.00335.2009. 2010. [DOI] [PubMed] [Google Scholar]
- 242.Oberdoerffer P., Michan S., McVay M. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Abdelmohsen K., Pullmann R., Jr., Lal A. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Gorenne I., Kumar S., Gray K., Figg N., Yu H., Mercer J., Bennett M. Vascular smooth muscle cell sirtuin 1 protects against DNA damage and inhibits atherosclerosis. Circulation. 2013;127(3):386–396. doi: 10.1161/CIRCULATION.AHA.112.124404. [DOI] [PubMed] [Google Scholar]
- 245.Elliott P.J., Jirousek M. Sirtuins: novel targets for metabolic disease. Curr. Opin. Invest. Drugs. 2008;9(4):371–378. [PubMed] [Google Scholar]
- 246.Yang Y., Fu W., Chen J. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress [published correction appears in Nat Cell Biol. 2007 Dec;9(12):1442] Nat. Cell Biol. 2007;9(11):1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kim J.E., Chen J., Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451(7178):583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 248.Yuan J., Luo K., Liu T., Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 2012;26(8):791–796. doi: 10.1101/gad.18s8482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Escande C., Chini C.C., Nin V. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Invest. 2010;120(2):545–558. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yoshino J., Mills K.F., Yoon M.J., Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metabol. 2011;14(4):528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Volonte D., Zou H., Bartholomew J.N., Liu Z., Morel P.A., Galbiati F. Oxidative stress-induced inhibition of Sirt1 by caveolin-1 promotes p53-dependent premature senescence and stimulates the secretion of interleukin 6 (IL-6) J. Biol. Chem. 2015;290(7):4202–4214. doi: 10.1074/jbc.M114.598268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Madugundu G.S., Cadet J., Wagner J.R. Hydroxyl-radical-induced oxidation of 5-methylcytosine in isolated and cellular DNA. Nucleic Acids Res. 2014;42(11):7450–7460. doi: 10.1093/nar/gku334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Branco M.R., Ficz G., Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2011;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 254.Afanas'ev I. New nucleophilic mechanisms of ros-dependent epigenetic modifications: comparison of aging and cancer. Aging Dis. 2013;5(1):52–62. doi: 10.14336/AD.2014.050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Le D.D., Fujimori D.G. Protein and nucleic acid methylating enzymes: mechanisms and regulation. Curr. Opin. Chem. Biol. 2012;16(5–6):507–515. doi: 10.1016/j.cbpa.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ames B.N., Shigenaga M.K., Hagen T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. U. S. A. 1993 Sep 1;90(17):7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Hamilton M.L., Van Remmen H., Drake J.A., Yang H., Guo Z.M., Kewitt K., Walter C.A., Richardson A. Does oxidative damage to DNA increase with age? Proc. Natl. Acad. Sci. U. S. A. 2001 Aug 28;98(18):10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Nakamoto H., Kaneko T., Tahara S., Hayashi E., Naito H., Radak Z., Goto S. Regular exercise reduces 8-oxodG in the nuclear and mitochondrial DNA and modulates the DNA repair activity in the liver of old rats. Exp. Gerontol. 2007 Apr;42(4):287–295. doi: 10.1016/j.exger.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 259.Radák Z., Apor P., Pucsok J., Berkes I., Ogonovszky H., Pavlik G., Nakamoto H., Goto S. Marathon running alters the DNA base excision repair in human skeletal muscle. Life Sci. 2003 Feb 21;72(14):1627–1633. doi: 10.1016/s0024-3205(02)02476-1. [DOI] [PubMed] [Google Scholar]
- 260.Radak Z., Bori Z., Koltai E., Fatouros I.G., Jamurtas A.Z., Douroudos, Terzis G., Nikolaidis M.G., Chatzinikolaou A., Sovatzidis A., Kumagai S., Naito H., Boldogh I. Age-dependent changes in 8-oxoguanine-DNA glycosylase activity are modulated by adaptive responses to physical exercise in human skeletal muscle. Free Radic. Biol. Med. 2011 Jul 15;51(2):417–423. doi: 10.1016/j.freeradbiomed.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Radak Z., Atalay M., Jakus J., Boldogh I., Davies K., Goto S. Exercise improves import of 8-oxoguanine DNA glycosylase into the mitochondrial matrix of skeletal muscle and enhances the relative activity. Free Radic. Biol. Med. 2009 Jan 15;46(2):238–243. doi: 10.1016/j.freeradbiomed.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Khan M.A., Alam K., Dixit K., Rizvi M.M. Role of peroxynitrite induced structural changes on H2B histone by physicochemical method. Int. J. Biol. Macromol. 2016;82:31–38. doi: 10.1016/j.ijbiomac.2015.10.085. [DOI] [PubMed] [Google Scholar]
- 263.Kalinina E.V., Ivanova-Radkevich V.I., Chernov N.N. Role of MicroRNAs in the regulation of redox-dependent processes. Biochemistry (Mosc.) 2019;84(11):1233–1246. doi: 10.1134/S0006297919110026. [DOI] [PubMed] [Google Scholar]
- 264.Carlomosti F., D'Agostino M., Beji S., Torcinaro A., Rizzi R., Zaccagnini G., Maimone B., Di Stefano V., De Santa F., Cordisco S., Antonini A., Ciarapica R., Dellambra E., Martelli F., Avitabile D., Capogrossi M.C., Magenta A. Oxidative stress-induced miR-200c disrupts the regulatory loop among SIRT1, FOXO1, and eNOS. Antioxidants Redox Signal. 2017;27(6):328–344. doi: 10.1089/ars.2016.6643. [DOI] [PubMed] [Google Scholar]
- 265.He J., Jiang B.H. Interplay between reactive oxygen species and MicroRNAs in cancer. Curr Pharmacol Rep. 2016;2(2):82–90. doi: 10.1007/s40495-016-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Emde A., Hornstein E. miRNAs at the interface of cellular stress and disease. EMBO J. 2014;33(13):1428–1437. doi: 10.15252/embj.201488142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Tang X., Li M., Tucker L., Ramratnam B. Glycogen synthase kinase 3 beta (GSK3β) phosphorylates the RNAase III enzyme Drosha at S300 and S302. PloS One. 2011;6(6) doi: 10.1371/journal.pone.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kasai H., Chung M.H., Jones D.S. 8-Hydroxyguanine, a DNA adduct formed by oxygen radicals: its implication on oxygen radical-involved mutagenesis/carcinogenesis. J. Toxicol. Sci. 1991;16(Suppl 1):95–105. doi: 10.2131/jts.16.supplementi_95. [DOI] [PubMed] [Google Scholar]
- 269.Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355(6357):273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 270.Taddei F., Hayakawa H., Bouton M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278(5335):128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- 271.Wang J.X., Gao J., Ding S.L. Oxidative modification of miR-184 enables it to target Bcl-xL and Bcl-w. Mol. Cell. 2015;59(1):50–61. doi: 10.1016/j.molcel.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 272.Ma Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol. Ther. 2010;125(3):376–393. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 273.Giannakakis A., Zhang J., Jenjaroenpun P., Nama S., Zainolabidin N., Aau M.Y., Yarmishyn A.A., Vaz C., Ivshina A.V., Grinchuk O.V., Voorhoeve M., Vardy L.A., Sampath P., Kuznetsov V.A., Kurochkin I.V., Guccione E. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Sci. Rep. 2015;5:9737. doi: 10.1038/srep09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Liu S.X., Zheng F., Xie K.L., Xie M.R., Jiang L.J., Cai Y. Exercise reduces insulin resistance in type 2 diabetes mellitus via mediating the lncRNA MALAT1/MicroRNA-382-3p/resistin Axis. Mol. Ther. Nucleic Acids. 2019;18:34–44. doi: 10.1016/j.omtn.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.