Abstract
Mycobacterium bovis is the main pathogen of bovine, zoonotic, and wildlife tuberculosis. Despite the existence of programs for bovine tuberculosis (bTB) control in many regions, the disease remains a challenge for the veterinary and public health sectors, especially in developing countries and in high-income nations with wildlife reservoirs. Current bTB control programs are mostly based on test-and-slaughter, movement restrictions, and post-mortem inspection measures. In certain settings, contact tracing and surveillance has benefited from M. bovis genotyping techniques. More recently, whole-genome sequencing (WGS) has become the preferential technique to inform outbreak response through contact tracing and source identification for many infectious diseases. As the cost per genome decreases, the application of WGS to bTB control programs is inevitable moving forward. However, there are technical challenges in data analyses and interpretation that hinder the implementation of M. bovis WGS as a molecular epidemiology tool. Therefore, the aim of this review is to describe M. bovis genotyping techniques and discuss current standards and challenges of the use of M. bovis WGS for transmission investigation, surveillance, and global lineages distribution. We compiled a series of associated research gaps to be explored with the ultimate goal of implementing M. bovis WGS in a standardized manner in bTB control programs.
Keywords: bovine tuberculosis, Mycobacterium bovis, genomics, WGS, genotyping
1. Introduction
Tuberculosis (TB) is a transmissible disease of humans and animals accompanying societies for thousands of years [1]. Despite progress in its control and prevention, TB is a top cause of mortality by a single infectious agent in the world and has devastating effects on bovine livestock and wildlife populations. Ten million new cases and 1.2 million human deaths were reported in 2018, and the increasing incidence of multidrug resistant strains is a threat to public health [2]. In addition, bovine TB (bTB) is an OIE (World Organisation for Animal Health) notifiable disease and, of the 179 countries reporting disease status in 2015–2016, approximately 50% declared the presence of TB in animals, with higher prevalence in Africa and parts of Asia and the Americas [3]. Despite an effective global notification system, the actual impact of bTB in animals is not fairly quantified, especially in wildlife and in countries where disease control programs are not well-established [4]. TB in cattle has important socioeconomic consequences, as the loss of livestock severely affects producers in developing countries with poorly implemented disease control programs and in certain developed nations where specific wildlife reservoirs create pockets of infection [4,5,6,7,8,9,10].
bTB is also a major, but often neglected, public health concern [11]. The causative pathogen of the disease can be transmitted from cattle to humans through close contact or the consumption of unpasteurized milk [11,12]. It is estimated that zoonotic TB affects 143,000 people a year, killing approximately 12,300 individuals [2]. People with zoonotic TB face arduous challenges, as most strains of the bovine pathogen carry conferring resistance mutations to pyrazinamide [13,14,15], one of the first-line drugs in TB treatment, and a possible association with extra-pulmonary disease [16] delays diagnostics and treatment initiation [17].
The risk of zoonotic TB, economic losses in affected livestock, and benefits of a bTB-free status in international commerce, makes the eradication of bTB desirable in many places. Effective programs of bTB control and eradication are typically based on test-and-slaughter, movement restrictions, and post-mortem inspection measures [18]. When performed, active surveillance and contact investigation have played major roles in reducing or eliminating the disease, benefiting from effective bacterial genotyping systems used to guide targeted interventions [19,20,21,22]. These genotyping techniques applied for Mycobacterium bovis, the main causative pathogen of bTB, have been historically based on the evaluation of a limited set of genetic markers mainly through PCR-based assays [23,24,25,26,27,28,29]. Although these techniques have been useful for bTB control programs, M. bovis whole-genome sequencing (WGS) will likely replace some of these laborious assays as the cost per genome continuously decreases, while simultaneously allowing the investigation of outbreaks for which higher resolution is warranted [21,26,30].
Genomic approaches have been successfully applied to identify pathogens, study pathogen evolution and population structure, reconstruct transmission chains, detect sources of infection, calculate rates of geographical and temporal spread of disease, and determine antimicrobial resistance [31,32,33]. WGS has increasingly become the preferential technique for infectious disease epidemiology, moving from research settings to support public and veterinary health professionals in their decision-making process regarding treatment, outbreak response, and surveillance [21,34,35,36,37]. Accordingly, the World Health Organization (WHO) and the OIE have issued general and/or pathogen-specific technical standards for adopting WGS-based approaches in diagnostics, treatment guidance, and epidemiology studies [38,39,40,41]. More specifically for M. tuberculosis, the pathogen of human TB, WGS is being implemented in certain countries to direct patient treatment and improve surveillance systems [34,42]. In addition, the WHO launched a technical guide for routine genotypic drug susceptibility testing (DST) [41] to substitute traditional phenotypic assays, which will allow fast and accurate detection of resistant pathogens in the near future. As for M. bovis, certain developed countries have started to apply WGS in official bTB control programs over the past years [21]. Nevertheless, M. bovis specific guidelines for WGS data analysis are not yet available. Although it is possible that much of what is being developed for M. tuberculosis will be applicable to M. bovis, intrinsic genomic and disease dynamics differences of both pathogens will likely influence data analyses and interpretation moving forward.
Rapid, reliable, and interpretable notification of genomics-informed data from M. bovis outbreaks in the future is expected to improve source investigation and contact tracing [26]. By correctly identifying the source of M. bovis infection, as well as the transmission links that followed it, one can provide supportive evidence to delineate interventions to halt disease spread. An ideal WGS-based notification system would be able to detect M. bovis transmission links, store this information, and analyze and compare transmission networks in real-time and during selected time intervals. Over time disease surveillance and global dispersal of M. bovis lineages are also benefiting from whole-genome based data [21,43,44,45,46,47,48,49,50]. WGS has been a powerful tool to identify M. bovis lineages distributed worldwide [45] and also to provide the fine resolution needed to understand bTB introduction into countries, regions, and individual farms or wildlife populations over defined periods of time [21,45,48,50,51]. However, the widespread application of WGS and its resulting data faces technical challenges that need to be addressed. These challenges are dispersed from data collection to analysis and reporting to end-users, i.e., veterinarians and epidemiologists. As many stakeholders do not routinely work with WGS and phylogenetics, there is a need to analyze and present complex genomic data in a standardized, accurate, and succinct manner to inform outbreak response. Identifying and addressing challenges of M. bovis WGS analysis will pave the way towards the systematic application of such technology in bTB control and eradication programs. Therefore, the aim of this review is to describe M. bovis genotyping techniques and discuss current standards and challenges of M. bovis WGS data analysis and interpretation. The section on M. bovis WGS is focused on its applicability for pathogen transmission investigation, surveillance, and global lineages distribution, benefiting from transferable contributions of the rich literature surrounding M. tuberculosis WGS.
2. A Brief Background on MTBC Genomics
To precisely interpret genotyping and WGS data, it is necessary to understand the genetic make-up of M. bovis. This pathogen is part of the Mycobacterium tuberculosis complex (MTBC), a bacterial group composed of 11 species or ecotypes with variable host tropism and virulence [1,52]. Mycobacterium tuberculosis is the leading etiological agent of TB in humans, while M. bovis has a broader host range and is able to infect multiple host species, mainly cattle and including humans, with variable populational persistence [52]. The MTBC is a clonal group [1,53,54,55] that evolved from a common ancestor with the tuberculous Mycobacterium canettii thousands of years ago [56,57]. MTBC genomes are highly similar, with >99.95% identity over homologous nucleotide sequences, including the ribosomal RNA genes, while horizontal gene transfer and large recombination events are considered absent [1,54,55]. These pathogens have solely evolved through single nucleotide polymorphisms (SNPs), indels (small insertions and deletions), deletions of up to ≈26 Kb, insertion sequences (IS), and duplication of few paralogous gene families [1,54,58].
Some of these large deletions, called “regions of difference” (RD), were initially described through physical mapping and differential hybridization arrays amongst M. tuberculosis H37Rv, M. bovis BCG Pasteur, and M. bovis ATCC 19210 [59,60,61]. Fourteen evolutionarily stable regions of difference (RD1–14) were differentially present among these strains and ranged from 2 to 12.7 kb in size. The discovery of these RDs paved the way towards the molecular diagnosis and differentiation of MTBC species [62], and are considered the gold-standard to differentiate members of this complex. Accordingly, M. bovis can be accurately differentiated from other members of the MTBC by the deleted regions RD9 and RD4, and from M. bovis BCG by the absence of RD1BCG (which is deleted in BCG strains) [62].
The bovine tubercle bacillus was officially named M. bovis in 1970, albeit called this way since the beginning of the 20th century [63]. The type strain was defined as M. bovis ATCC 19210, still referenced in the most recent Bergey’s Manual of Systematic Bacteriology [64], along with CIP 105234 and NCTC 10772. For tuberculous mycobacteria, early taxonomic classification was based on specific phenotypic traits of the isolates, such as host of origin, virulence in animal models, and biochemical tests (e.g., pyrazinamide resistance, niacin accumulation, nitrate reduction, type of respiration, colony morphology) [64]. The high genetic relatedness between M. tuberculosis and M. bovis, as well as among other species of the MTBC, has always instigated discussions about their taxonomic classification, frequently suggesting to compile all members of the MTBC to a single species [65,66,67,68,69]. However, the biochemical differences and epidemiologic distinctions between infections, particularly regarding the bovine and human bacilli [63], emphasized the need for differentiating these organisms at some taxonomic level, which remains to be defined (e.g., species, subspecies, variant).
The average size of a virulent M. bovis genome is 4.3 Mb, containing approximately 4200 genes, including a single copy of each of the ribosomal RNA genes (5S, 16S, and 23S) and 45 tRNAs. As with other Actinobacteria [64], its genome has a high GC content (≈65%), which implies the use of appropriate sequencing reagents for library preparation in WGS [70]. MTBC genomes, including M. bovis, have a substantial number of repetitive elements, constituting one of the main challenges for WGS data analyses. These include, but are not restricted to, mobile elements (e.g., insertion sequences—IS), proline-glutamate (PE) or proline-proline-glutamate (PPE) family genes, integrases, two phage sequences, a CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), and the 13E12 repeat family genes. In particular, PE-PPE gene families account for approximately 10% of MTBC genomes, and have been associated with TB pathogenesis [71]. Repetitive elements are difficult to handle in genomic studies because the majority of and most commonly used sequencing platforms generate short reads, usually ranging from 50 to 300 bp, which are often shorter than the repeats themselves [72]. Some of these repetitive regions are the basis for the traditional genotyping techniques developed over the years (see next section).
3. Traditional Genotyping Techniques of M. bovis
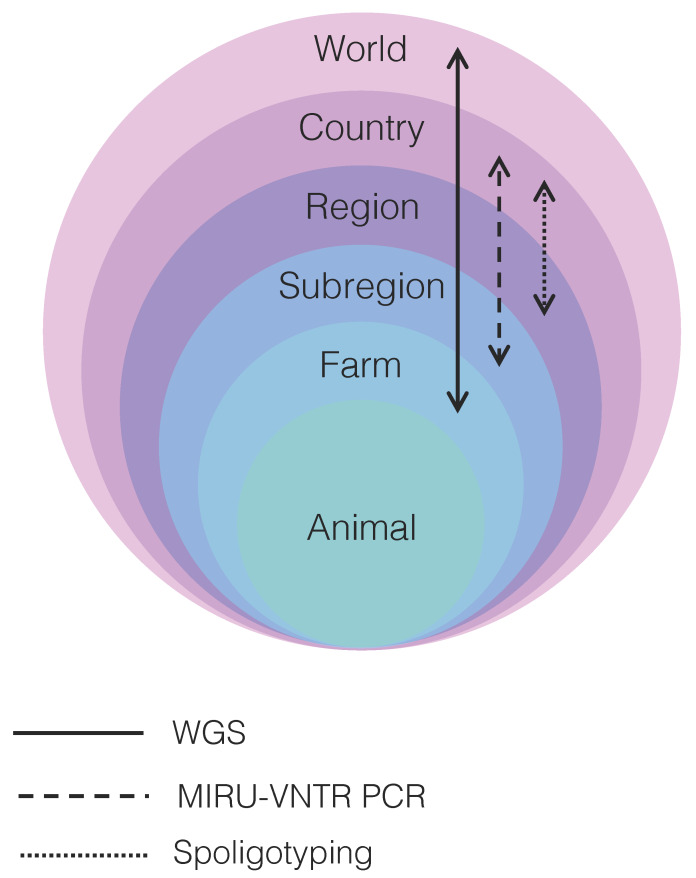
A number of reviews describe in detail traditional typing methods used for M. bovis outbreak investigations [23,24,25,26,27,28,29]. Nearly all techniques, briefly reviewed below, were first developed and applied for M. tuberculosis typing and later validated for M. bovis studies. Due to MTBC’s clonal nature, most polymorphisms in genotyping techniques originate from insertion sequences (e.g., IS6110) and other repeat regions (e.g., CRISPR, PE/PPE genes, PGRS genes). Evidence accumulated over the years indicates that each technique or a combination thereof presents distinct resolving power at the country, region, subregion, and farm levels [26] (Figure 1; Table S1).
Figure 1.
Resolution power of the main techniques used to resolve transmission clusters of Mycobacterium bovis depicted in relation to world, country, region, subregion, farm, and animal levels. WGS: whole-genome sequencing; MIRU-VNTR: mycobacterial interspersed repetitive unit-variable-number tandem repeat typing; PCR: polymerase chain reaction. Arrows indicate the level of resolution each technique is able to achieve. WGS provides fine resolution to discriminate between M. bovis strains distributed globally to the individual farm level, while MIRU-VNTR PCR and spoligotyping have more limited resolution, particularly at the individual farm level. WGS may be able to discriminate between different M. bovis strains infecting the same animal only if sampling is comprehensive, multiple isolate cultures are sequenced, and/or deep sequencing of the primary isolate is performed.
3.1. Restriction Endonuclease Analysis and Pulsed-Field Gel Electrophoresis
In 1985, Collins and de Lisle [73,74] developed the first intraspecific typing technique of M. bovis, the restriction endonuclease analysis (REA). REA consists of applying three different enzymes (BstEII, PvuII, and Bcll) to digest high amounts of total DNA extracted from M. bovis isolates, followed by band pattern visualization on agarose gels. Despite its use in molecular epidemiology studies in certain countries at the time [75,76,77], the assay soon proved to be technically demanding, with an excessive number of small DNA fragments difficult to resolve [78] (Table S2; Figure S1). Currently, its application is mostly restricted to a reference laboratory in New Zealand, in which it was developed, and was last used for routine typing of M. bovis in 2011 [26].
A pulsed-field gel electrophoresis (PFGE) [79] assay was later developed for M. tuberculosis and other MTBC strains and resulted in improved resolution of band patterns compared to REA (i.e., larger and fewer bands). However, PFGE had two main disadvantages: first, the MTBC’s lipid-rich cell wall inhibits the action of lytic enzymes used in PFGE, preventing the proper use of the PFGE’s agarose plugs [80,81,82]; and second, comparative studies developed in later years showed that PFGE of M. tuberculosis strains had a lower intra-specific discriminatory power compared to other genotyping techniques that were subsequently developed [83,84]. This low discriminatory power is associated with MTBC’s clonality; the low number of polymorphic positions between strains may result in undistinguishable band patterns [85] (Figure S1). PFGE has also some intrinsic disadvantages, such as being technically demanding and time consuming (Table S2). Given the drawbacks, there are only three published reports using this technique to type M. bovis strains [86,87,88].
As REA and PFGE proved insufficient to discriminate M. tuberculosis and M. bovis strains, the search for polymorphic and stable genetic markers allowed the elaboration of superior typing techniques. Currently, the most widely used genetic markers are the IS6110 (for M. tuberculosis), the direct repeat (DR) region (which is a mycobacterial CRISPR), the poly(GC) rich sequences (PGRS), and the variable number tandem repeats (VNTR) sequences. Each marker has its corresponding typing technique.
3.2. IS6110-RFLP
The 1358 bp IS6110 is MTBC’s specific [89] and differences in its location and copy numbers is what discriminate among isolates [90,91]. This repetitive element was first described in 1990, by screening a M. tuberculosis cosmid library constructed in pHC79 with labelled M. tuberculosis total DNA [92]. Presently, the most standardized and commonly used method to detect IS6110 in M. tuberculosis strains is the IS6110-RFLP (IS6110-Restriction Fragment Length Polymorphism) [93,94]. Briefly, the technique consists in extracting high amounts (2–3 µg) of total bacterial DNA, digesting it with PvuII endonuclease and subjecting the digested sample to standard electrophoresis on agarose gel. The agarose gel is then used to perform a Southern blot, in which the DNA fragments are transferred to a membrane, and probes complimentary to a portion of the 3′ end of the IS6110 sequence hybridize to reveal the number of IS elements and size of generated fragments through chemiluminescence (originally radiolabeling) [89,94,95]. IS6110-RFLP patterns can be compared and compiled using specific computer software. Mycobacterium tuberculosis isolates from individuals that are part of the same transmission link often display identical IS6110-RFLP patterns, constituting transmission clusters. IS6110 has also been shown to be stable over time (0.57–10.69 years to change, depending on the disease phase) [96], which means the technique can be used to study recent transmission or in long-term epidemiological studies.
A major drawback of IS6110-RFLP is the fact that nearly all M. bovis strains carries only 1–5 copies of the insertion element [91,97] and this technique has low discriminatory power in isolates containing five or less IS6110 copies [98] (Figure S1). In other words, many M. bovis isolates will have the same IS6110-RFLP pattern, making it impossible to distinguish among them. As with REA and PFGE, IS6110-RFLP also requires high amounts of DNA and is labor-intensive (Table S2). For these reasons, despite being commonly used for M. tuberculosis, IS6110-RFLP was of little use for M. bovis genotyping.
3.3. PGRS-RFLP
In 1991, a Southern blot-based RFLP was developed based on the digestion of M. tuberculosis DNA using enzymes of four-base recognition sites [99]. One of the detected DNA fragments showing high heterogeneity among isolates was cloned and sequenced, revealing a highly repetitive sequence, identified as PGRS [100]. This fragment served as a probe to identify the presence of up to 30 PGRS copies present in MTBC genomes. Owing to the poor applicability of IS6110 typing for M. bovis, PGRS-RFLP allowed significant improvement in M. bovis strain differentiation [98]. However, as with REA, the presence of multiple bands [101] makes it difficult to interpret [102] (Figure S1). As a Southern blot-RFLP based system, it also requires high amounts of DNA and is a laborious technique (Table S2).
3.4. Spoligotyping
As with many bacteria and archaea [103], MTBC organisms have a defense system against invading nucleic acids called type III-A CRISPR/Cas system. Even before much attention was given to bacterial CRISPR, this sequence in M. tuberculosis, known as DR locus, was described [104] and readily applied in genotyping [105]. Hermans et al. [104] originally described a genomic locus in M. bovis BCG containing a IS6110 element with many 36 bp direct repeats (DRs) interspersed by spacer sequences ranging from 35 to 41 bp in size. One DR and its neighboring spacer sequence is called a “direct variable repeat” (DVR). The order of the spacers is similar among MTBC strains, but DVRs can be deleted. Therefore, the difference between two isolates is given by the variable presence of spacers in the DR region. There is only one DR locus per MTBC genome (Figure 2 and Figure S1) and up to 43 unique spacers between DRs.
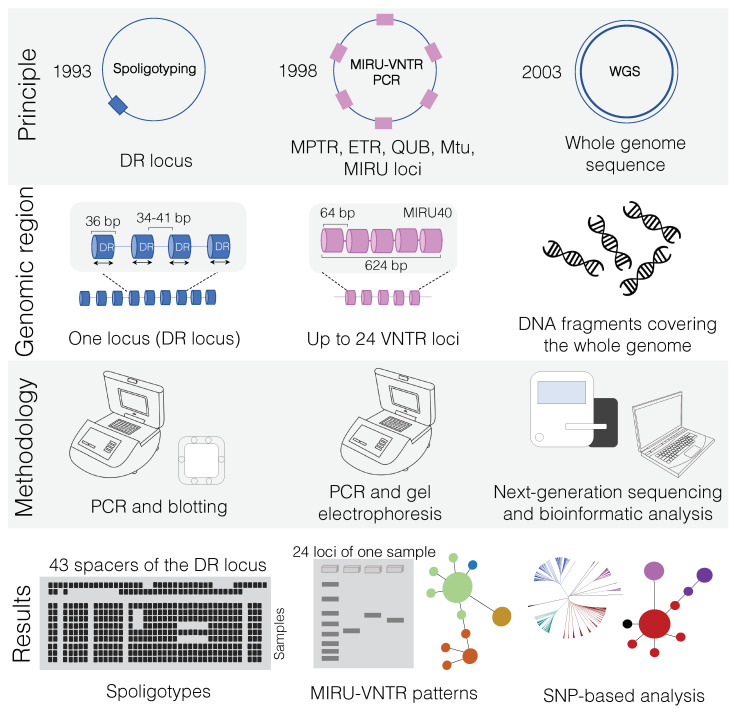
Figure 2.
Overview of main genotyping techniques (Spoligotyping and MIRU-VNTR) and whole-genome sequencing (WGS) used for transmission cluster investigation of Mycobacterium bovis. In “principle”, squares denote the quantity of specific genetic markers (i.e., DR locus and VNTR) on M. bovis genomes. While spoligotyping is based on a unique locus, MIRU-VNTR PCR amplifies genetic targets from multiple regions of the genome (up to 24 loci). In contrast, WGS uses information from the whole-genome sequence. Dates refer to the year in which each technique was developed. In “genomic region”, the MIRU40 locus is shown as an example of one of the 24 loci that can be used in MIRU-VNTR PCR. In “results”, the spoligotyping membrane is depicted accommodating several samples simultaneously, owing to the high-throughput capability of this technique (up to 45 samples can be simultaneously analyzed). In MIRU-VNTR PCR, although many samples can be amplified at once, each sample can occupy up to 24 wells in an agarose gel, so many electrophoresis runs may be needed depending on the laboratory. MIRU-VNTR databases can subsequently be used to generate a minimum spanning tree. WGS is a high-throughput technique that will lead to single nucleotide polymorphism (SNP)-based analysis. The same generated data can also be used to detect spoligotype and MIRU-VNTR patterns (see text). WGS results can be used to evaluate transmission clusters as well as phylogenetic relationships among the sequenced genomes. WGS: whole-genome sequencing; MIRU-VNTR: mycobacterial interspersed repetitive unit-variable-number tandem repeat typing; PCR: polymerase chain reaction; DR: direct repeat.
The first typing method for the DR locus was called DVR-polymerase chain reaction (DVR-PCR) [105], which was later substituted by spoligotyping (spacer oligotyping technique). Spoligotyping was developed in 1997 [106] and readily utilized to evaluate M. bovis strains [107]. This “reverse line blot hybridization technique” is PCR-based and detects the presence of the unique spacers in an MTBC isolate in two steps. First, the spacers between DRs are amplified using PCR. A single primer set complimentary to the two extremities of the DR sequences is used, but the reverse primer is biotin labelled, resulting in the synthesis of labelled reverse strands. Individual spacers are subsequently detected by hybridization of the biotin-labelled PCR product to a nylon membrane containing covalently linked oligonucleotides corresponding to 37 spacers of M. tuberculosis H37Rv and six spacers of M. bovis BCG. A mini-blotter is used for hybridization and up to 45 isolates can be simultaneously compared [28] (Figure 2). One advantage of this technique is that it can be applied directly to DNA extracted from infected tissue samples, not requiring bacterial isolation [108]. In the case of M. bovis, spacers 3, 9, 16, and 39–43 are lacking, allowing for species differentiation [106].
Further improvement and automatization of the technique led to the application of microbead-based detection systems, such as Luminex platforms [109,110,111], multiplexed primer extension-based spoligotyping assay using automated matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) [112], microarray [113,114,115,116], and ligation-based amplification and melting curve analysis [117].
The evolution of spoligotype patterns is given by the loss of spacer sequences, which cannot be restored by recombination and is, therefore, fixed in that population [118]. The problem of spoligotyping is the homoplasy, i.e., unrelated lineages can present identical spoligotype patterns because the loss of spacer sequences is a common event [119]. Thus, spoligotypes are not good indicators of phylogenetic relatedness. In addition, its resolving power has been frequently shown to be lower than REA and MIRU-VNTR PCR (mycobacterial interspersed repetitive unit-variable-number tandem repeat typing, polymerase chain reaction) [26] (Figure 1; Tables S1 and S2). Despite these factors, spoligotyping remains as one of the most applied genotyping techniques in M. bovis studies (Table S1).
3.5. Variable Number Tandem Repeat (VNTR)
VNTR is a locus in which a nucleotide sequence is arranged as tandem repeats, i.e., repeats clustered together and oriented in the same direction. The size (in bp) of this locus varies according to the number of times the nucleotide sequence is repeated. Each repeat can be added or removed through recombination or replication errors, resulting in alleles with different number of repeats. VNTR are present in eukaryotes and prokaryotes, and given its variability, it has been frequently used for DNA typing [120].
Compared to the single DR locus of spoligotyping, many VNTR loci exist in MTBC (Figure 2 and Figure S1) and they are detected using PCR. The sizes of resulting PCR products correspond to the number of repeats in each locus. Initially, 11 VNTR loci (five MPTR loci, with repeats of 15 bp, and six ETR loci, with repeats of 53–75 bp) were evaluated in MTBC strains [121,122,123]. Five ETR loci (ETR-A, -B, -C, -D, -E) showed more discriminatory power among strains [124]. However, these five loci did not provide higher resolution compared to the IS6110-RFLP in M. tuberculosis strains with high IS6110 copy numbers [121,124]. Since M. bovis has few IS6110 copies, ETR loci were indeed more discriminative than IS6110-RFLP [121,125], but spoligotyping continued to present higher resolution [124,126]. Thus, other loci were identified and tested, such as MIRU, QUB, and Mtu, and currently a 24-loci MIRU-VNTR PCR is commonly used [127]. MIRU-VNTR can also be evaluated along with spoligotyping to infer genotyping through the online platform MIRU-VNTRplus [128], providing a standardized manner of results delivery.
Among M. bovis studies, different sets of VNTR loci have been applied (Table S1). Each locus and combination thereof may present better or worse discriminatory power depending on the region and sample set (Table S1). It has been suggested that each region should define the best combination of loci for its reality [129], aiming also at decreasing the cost and time spent in running different PCR assays. It has also been shown that, in certain settings, the capacity of MIRU-VNTR PCR in detecting transmission clusters may be dependable on the M. tuberculosis lineage [130,131]. For instance, it has been described that standard 24-loci MIRU-VNTR PCR has low resolution power to precisely discriminate closely related isolates of the lineage 2, Beijing of M. tuberculosis [131]. It is unknown if this is also the case for M. bovis lineages or clonal complexes.
4. The Dawn of a New Era: WGS to Understand M. bovis Epidemiology and Ecology
The first complete genome sequence of M. bovis to become available originated from a strain denominated AF2122/97 isolated from a cow in England [132,133]. As this was the first M. bovis genome available, M. bovis AF2122/97 is now considered the reference genome of M. bovis in GenBank, as genomes of M. bovis type strains have never been sequenced. By December 2019, only 74 virulent M. bovis genomes (i.e., not BCG) are deposited as complete or draft forms in NCBI, compared to 6522 M. tuberculosis genomes. In SRA (Sequence Read Archive), the database for depositing raw reads, the number of sequenced M. bovis is in the thousands. The disparity in numbers between assembled complete or draft genomes and raw reads highlights that the majority of developed studies are based on SNP and/or indel detection using reads.
4.1. Current WGS Workflow
Overview
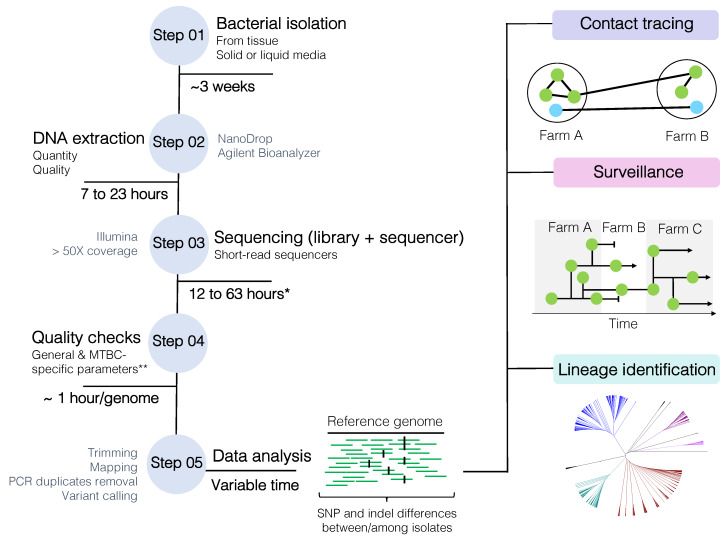
The current WGS workflow (Figure 3) begins with the isolation of M. bovis from de-contaminated tissue samples on solid (e.g., Stonebrink, 7H11-OADC) or liquid media (e.g., 7H9-OADC, MGIT—mycobacterial growth indicator tube), followed by the extraction of its DNA, library preparation, and WGS using short-read technologies (e.g., Illumina platforms). Special attention must be given to the quality of extracted DNA and the use of library kits that can accommodate high-GC content bacteria [70]. DNA extraction of mycobacteria is not trivial; the lipid-rich cell wall interferes with yield and DNA purity, which may affect library construction [70]. An optimized extraction protocol of non-tuberculous mycobacteria for long-read sequencing has been recently proposed [134]. Once DNA is successfully extracted and sequenced, generated reads need to undergo quality checks and are processed in specific data pipelines tailored to each need. For epidemiology purposes, WGS can be used to assess the genetic relatedness among isolates to address transmission investigation, surveillance, and/or lineage identification (Figure 3). Current methodologies are typically based on the identification of SNP and indel differences between or among isolates. Basically, the greater the SNP and indel difference between two isolates, the lower the probability they are related to each other. SNPs and indels are ultimately identified by mapping the quality-checked reads to a reference genome and calling the variants.
Figure 3.
Mycobacterium bovis whole-genome sequencing (WGS) workflow from bacterial isolation to data analysis. SNP: single nucleotide polymorphism. * Time is highly dependable on library kit and sequencing protocol. ** MTBC-specific and general parameters are described in detail in the text, but overall this includes FastQC parameters, minimum established sequencing coverage, contaminating reads, species confirmation, mixed-strain evaluation (depending on the purpose of the analysis), and homogeneous sequencing coverage (after reference genome mapping). MTBC: Mycobacterium tuberculosis complex.
5. Data Analyses Pipeline
5.1. Quality Assessment of Entry Data
The quality of WGS reads can dramatically impact the study outcome. Therefore, quality assessment is considered the first step in data analyses. Once laboratory-specific [135] and standard quality controls associated with the sequencer run are evaluated and errors originating from the sequencer itself are ruled out, generated FASTQ files normally undergo general and mycobacteria-specific quality assessments and processing. Accordingly, following adaptors removal with appropriate software [136,137,138,139], an overall quality evaluation of reads is typically performed using FastQC [140] or similar software [139,141,142,143]. Quality parameters are frequently evaluated rather manually, by analyzing the QC report of each FASTQ file or by using software that compile multiple-sample QC reports [141,142]. Nevertheless, these are important measures to ensure that high-quality sequencing data are used in downstream analyses. Based on this QC evaluation, FASTQ files are usually processed to remove low quality data.
Parameters such as anomalous GC content, duplicated sequences, per base and per sequence qualities, per base N content, sequence length distribution, among others can be addressed according to pre-established and/or default thresholds. The detection of anomalous GC content may indicate possible sample contamination, as any peak differing from the high value of mycobacteria (≈65%) are not expected. A high level of sequence duplication indicates errors or enrichment bias related to PCR amplification and sequencing that are not expected to occur in WGS [140]. Duplicated sequences (e.g., PCR duplicates) are normally removed downstream in the pipeline, after read mapping, using appropriate software [144]. In addition, reads are often trimmed and filtered out according to quality, with user-specified or default thresholds. Protocols of trimming with different stringency levels have been tested for eukaryotes, showing that variations of parameters may significantly affect end-results [145,146]. Basic FASTQ processing (e.g., adaptor removal, read trimming, read filtering, removal of duplicates, among others) can be performed by using a combination of different software or by using all-in-one tools [143]. Finally, QC results should be evaluated before and after file processing to guarantee that minimum quality standards have been reached with appropriate read length distribution.
Unfortunately, sequencing files can often contain contaminating reads, i.e., reads not originating from the target genome [147,148,149,150]. These contaminants may or may not result in discrepant GC content peaks. Their presence is sometimes inevitable and challenges for eliminating contaminant reads have been addressed previously [34]. If not evaluated beforehand, their presence may be detected only at read mapping or genome assembly, or go undetected and result in false positive or negative SNPs [151]. One way to check for contaminants is to use FastQ Screen [152] or Kraken [153], and if desired, filter out unwanted reads following a pre-established threshold of sample contamination acceptance [152]. One study using Kraken defined a threshold of at least 90% of the reads taxonomically assigned to MTBC for the sample to be included in the analyses [154]. However, because MTBC genomes are highly similar, it is difficult to control for cross-contamination when sequencing several MTBC isolates at once [155]. Heterozygous sites may occur, and the sample falsely considered a mixed-infection.
There are three mycobacteria-specific sequencing quality criteria that can be evaluated: (i) homogenous sequencing coverage; (ii) RD identification; and (iii) within-host genetic diversity. One of the advantages of MTBC clonality is that read-mapping coverage to a reference genome can be utilized as measure of homogeneous sequencing coverage of the target genome. When mapping high-quality reads to a MTBC genome, a high mapping coverage (>95%) of the reference genome is expected [156,157,158,159]. Percentage cut-offs may be established [45,160], because substantially low percentages are likely to indicate that the target genomes were not evenly sequenced. In addition, the presence of species-specific RDs in the target genome must be assessed. In our laboratory, we have identified a number of MTBC genomes deposited in public databases with mistakenly-assigned MTBC species [45,161]. As MTBC members have high genomic and phenotypic similarity, errors in species identification may occur. Therefore, even if the bacterial isolate was obtained from cattle tissue, M. bovis specific RD patterns should be confirmed. This confirmation can be performed using reads, by checking RD regions through reference genome-mapping [45], or by running the automated software RD analyzer [162].
Another major challenge of SNP-based approaches for MTBC WGS analysis is within-host genetic diversity [163]. The amount of MTBC genetic diversity an individual carry depends on the time between infection and development of active disease (within-host evolution, i.e., microevolution) and/or the number of strains this individual was exposed at single or multiple infection events through life (mixed-infection) [163]. Microevolution occurs during long-term co-existence between pathogen and host and is characterized by a single infection event leading to bacterial mutations over time. On the other hand, mixed-infection occurs when the individual is exposed to a single or repeated infection events through life of different strains, and is thus carrying distinct strains of MTBC [163]. If DNA is extracted from the primary isolate without bacterial propagation from a single, de-clumped colony, there may be simultaneous sequencing of more than one strain in a sample. Thus, when mapping reads to the reference genome and calling variants, heterozygous sites may arise. More details regarding this issue are given in the following sections.
5.2. Choice of Reference Genome for Read Mapping
A closed, complete genome must be chosen as reference for read mapping and variant calling. The choice of reference genome can dramatically alter the end-results [164,165] and it is still a controversial matter [34]. Lack of standardization of reference genomes halts comparisons between pipelines and laboratories. Ideally, the reference genome must have all DNA segments present in the bacterial population under study. If the reference genome has deleted regions compared to the genomes being tested, genetic diversity may be missed. The evolutionary distance between the microbial genomes under study and the reference genome should also be taken into account [165,166]. For instance, if M. tuberculosis genomes are used as reference for M. bovis studies, the number of detected SNPs increases dramatically [47,167], which may lead to errors in read mapping and variant calling [165], substantially increasing computer usage and time.
WGS-based studies of M. bovis often use as reference the genome of M. bovis AF2122/97 (Table S3). One recent study has proposed the use of an outbreak-matched M. bovis genome as reference in France [168]. Studies of M. tuberculosis have used the M. tuberculosis H37Rv genome, lineage- or outbreak-matched genomes, or an inferred ancestral MTBC genome, which have been reviewed elsewhere [34]. The use of a MTBC pan-genome as reference, i.e., a gene pool representing the whole diversity of MTBC genes, has also been suggested, but never evaluated [34]. Intergenic regions, however, should not be neglected in future technical validations of these approaches. Recently, a computational pan-genome of M. tuberculosis (in this case, a dataset of whole genome sequences, and not simply core and accessory genes) with 5,205,216 bp obtained from 146 M. tuberculosis genomes has been proposed as a reference genome for this species [169]. Considering that a M. bovis transmission cluster may be defined or ruled out by just few SNPs (see following sections), more comprehensive studies on the effect of the reference genome on SNP identification should be performed.
5.3. Reads Mapping and Variant Calling
Bacterial SNPs and indels, as well as structural variants (SVs—indels, duplications, inversions, and translocations >50 bp), can be identified through de novo genome assembly followed by comparison against a reference genome, or by mapping reads to a reference genome [170]. When using assembled genomes, failure to properly identify variant calls can occur due to assembly errors or misidentification of indels [170]. It is more appropriate and faster to use the complete information provided by the reads than relying on assemblers and consensus base callers [171] to detect variants. Thus, mapping reads to a reference genome is the preferred first step to detect high quality variants. Numerous short-read aligners have been developed (to cite a few [171,172,173,174,175,176,177,178,179,180,181]) and additional information about mapping principles have been reviewed [179].
Different approaches of read mapping and variant calling are described in M. bovis studies (Table S3). Among the most widely used short-read mapping tools are Bowtie/Bowtie2 [172] and BWA/BWA-SW [174]. Output alignment files are subsequently processed to call and generate a list of high-quality variants using tools available from toolkits or pipelines such as VarScan2 [182], SAMtools [173,183], and GATK (Genome Analysis Toolkit) [184,185]. PCR duplicates are frequently removed after read mapping with Picard (MarkDuplicates; https://broadinstitute.github.io/picard/) or SAMTools (rmdup) [173,183]; but the actual necessity for this step has not been systematically evaluated using MTBC genomes. VarScan2 combines a heuristic method coupled with statistical algorithm to detect mutations from read mapping, and integrates identification of SNPs, indels, or both (mpileup2snp, mpileup2indel, mpileup2cns, respectively). On the other hand, SAMtools and GATK are probabilistic methods, implementing Bayesian statistics. SAMtools contemplate the bcftools to call variants [173], while GATK version 4 uses HaplotypeCaller [184,185]. Unfortunately, variant callers have been mostly benchmarked with human genomes, which may lead to the report of false variants when analyzing microbial genomes [170]. More recent studies showed marked result differences among pipelines for variant detection in WGS studies of M. tuberculosis [160,165] and other bacteria [166,186].
One of the reasons for these discrepancies is that MTBC studies vary widely on the parameters adopted to map reads and call high-quality variants; no standards have been determined. The choice of parameters greatly influences variant detection (e.g., base call and mapping quality scores, tail distance, presence of variants on both strands for paired-end reads, read depth, minimum allele frequency, maximum number of SNP calls within 10–12 bp, local assembly or realignment around indels, and strand bias) [170]. Sequencing coverage, PCR duplicates, mapping artefacts around indels, SVs and repetitive or duplicated regions may also result in false positive (FP) and/or false negative (FN) calls [170]. As part of MTBC-specific measures, it is common practice to exclude SNPs and indels associated with repetitive DNA, such as PE/PPE family genes, phage genes, repetitive family 13E12 genes, transposases, and integrases, following their identification through annotation or genomic position, or by excluding selected genes from the reference genome [165,187]. However, comprehensive evaluation of the true probability of these calls being FP or FN are lacking. By using read simulation and comparison to long-read sequencing, a recent study has shown that SNPs detected in PE/PPE regions were highly unlikely to be FP calls when using BWA read mapping and Pilon variant caller [188] (which is a microbial variant caller). In contrast, another study has shown that both FP and FN calls are disproportionately present in PE/PPE regions in a multi-variant caller comparison [165]. This contradiction highlights the need for additional studies. Thus, significant challenges remain to be overcome in order to define the best parameters to call variants and how to handle low-quality variant calls.
5.4. Within-Host Genetic Diversity and Its Impact on Variant Calling
Two major technical challenges arise in variant calling when there is within-host genetic diversity: establishment of minimum allele frequency to identify a site as heterozygous, and the minimum number of heterozygous variants in a sample to be considered a “mixed-sample” condition. Importantly, the ability to uncover these parameters in M. tuberculosis studies is also directly affected by sequencing coverage [189]. The minimum allele frequency is a fixed threshold (≈75% to 95%) for the proportion of reads supporting a particular variant call. Sites with the percentage of reads falling below this threshold are considered heterozygous and hence used to support a mixed-infection. Unfortunately, there is no consensus on the percentage level that should be used.
Once heterozygous sites have been identified, two strategies are commonly applied to determine a mixed-sample condition: a cut-off proportion of heterozygous sites to total variants, or a minimum total number of heterozygous sites [21,45,51,190,191,192,193,194,195]. Certainly, the percentage threshold depends on the choice of the reference genome, e.g., M. bovis AF2122/97, M. tuberculosis H37Rv, or reconstructed MTBC ancestor. Currently, there are no established criteria or thresholds of what can be considered mixed-sample and what is variant calling error for M. bovis, especially in light of repetitive genomic regions and different parameters set for variant calling. Once a M. bovis mixed-sample is detected, researchers have either removed the sample from downstream analysis [21,45,51,192,194], consider these heterozygous sites in the context of the contact chain being analyzed to resolve transmission networks [21,194,196,197,198,199], or excluded heterozygous sites from downstream analysis [43,193,200,201,202].
5.5. Within-Host Genetic Diversity and Its Impact on Transmission Detection
Individual animals or humans carrying MTBC isolates with distinct SNP profiles have been described [21,48,51,156,163,194,195,196,197,198,203,204,205,206,207,208]. As explained above, such conditions occur when there is microevolution and/or mixed-infection. Both concepts, the manner in which within-host genetic diversity is detected, and their application to the definition of transmission clusters have been initially defined with M. tuberculosis and later applied to M. bovis WGS studies [21,45,47,51,194,195]. Owing to the low substitution rate of MTBC [46,190,196,202,209,210], the number of acquired SNPs by M. tuberculosis strains under a microevolution process (within-host evolution) has been estimated to be very low, usually in single digits [29,163,190,196,197,209,211]. On the other hand, mixed-infection is defined when two or more M. tuberculosis isolates obtained from an individual differ by a great number of SNPs [29,163,196].
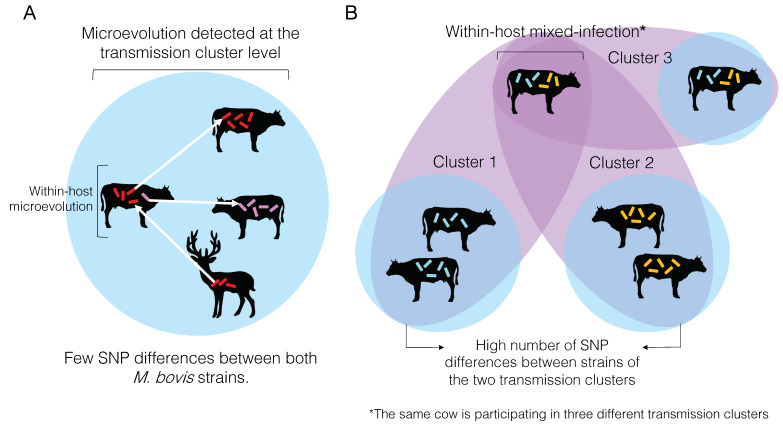
Microevolution can be detected at the individual or transmission cluster levels (Figure 4A). Very often, microevolution is only detected at the latter, because the whole extent of within-host genetic diversity is frequently missed due to insufficient individual sampling [163]. When these low SNP distances are inferred among individual samples in a cluster, they are used to define them as part of the same transmission cluster. In other words, the same M. bovis strain was transmitted from one animal to the other, and the amount of genetic changes accumulated is zero or just a reflection of within-host evolution (i.e., microevolution) represented by very few SNPs. Contrastingly, if the number of SNPs between two individual samples is too high, they are not considered part of the same transmission cluster (Figure 4B).
Figure 4.
Overview of microevolution and mixed-infection conditions and its relationship to and influence on the detection transmission clusters of bovine tuberculosis. (A) Microevolution condition. Microevolution is normally determined when two Mycobacterum bovis isolates obtained from the same host differ by a small number of SNPs (usually between 0 and 12 SNPs; see text). The same SNP threshold is used to define a transmission cluster, when two M. bovis isolates obtained from different hosts also differ by the same number of SNPs. (B) Mixed-infection condition. Mixed-infection is defined when two isolates obtained from the same host differ by a great number of SNPs (usually > 12 SNPs; see text). When a great SNP distance is found between two M. bovis isolates from different hosts, these animals are not considered part of the same transmission cluster. However, if an animal is infected with two strains differing by a great number of SNPs (i.e., mixed-infection), it may be identified as participating in two different transmission clusters (cluster 1 and cluster 2). If the within-host genomic diversity is not entirely captured, one of the transmission clusters may be missed. This animal with a mixed-infection may also transmit both strains to another animal (cluster 3), and if the diversity is entirely captured, both animals will be considered as part of the same cluster.
At the transmission cluster level, if this within-host genetic diversity is captured with adequate sampling, an individual may be considered part of two transmission clusters, representing different infection events that occurred over time (Figure 4B). More likely the within-host genetic diversity is not entirely captured, and pathogen transmission between or among individuals may be mistakenly discarded. In other words, if a cow is infected with two distinct strains of M. bovis differing by a great number of SNPs, and only one is sequenced but both are transmitted to other cows, one of the animals in the transmission chain will go undetected as part of that cluster. It is important to highlight that an individual may get re-infected with the same strain, which would be impossible to distinguish using current analytical methods. The actual impact of within-host genetic diversity on the transmission dynamics and pathogenesis of M. bovis remains to be comprehensively studied.
5.6. Where to Go after Detection of Variants?
5.6.1. SNP-Counting Method
Few approaches have been used to interpret variant calling data in M. tuberculosis and M. bovis studies. The simplest one is to use the absolute number of detected SNPs (indels are excluded) to infer relatedness based on predefined thresholds. This methodology has been often employed in M. bovis studies [21,47,51,210,212] (Table S3). Based on solid epidemiological links, SNP thresholds have been established to distinguish within-host microevolution from mixed-infection of M. tuberculosis [190,193,197,209]. Consequently, the same SNP thresholds were also applied to distinguish isolates belonging to a transmission cluster or not, helping define a transmission chain of M. tuberculosis or M. bovis and differentiate relapse from re-infection in M. tuberculosis infections [21,47,51,163,190,193,196,197,199,209,213,214,215,216,217] (Table S3). One of the first studies to define SNP thresholds evaluated M. tuberculosis isolates obtained from chronically infected patients, epidemiologically linked patients, and outbreaks with confirmed transmission chain observed in the UK (a low-burden country) from 1994 to 2011 [196]. A maximum of five SNPs was defined as the limit to infer a transmission cluster or microevolution. Similar thresholds have been confirmed in later studies performed at low and high burden settings [190,197,209,211], and is commonly accepted that five SNPs can be used as a stringent threshold and 10 or 12 as a more relaxed threshold [29,163]. Nevertheless, SNP thresholds described in the literature of M. tuberculosis vary [163]. It is known that these thresholds may be influenced by variant calling protocols, culture or sampling, read depth, and epidemiological links used to first define them, which makes them unlikely to be adequately transferred between settings and studies [163]. Thresholds have never been determined for M. bovis, which is likely subjected to different evolutionary pressures compared to M. tuberculosis. Moreover, owing to the possibility of false positives, indels are usually excluded from the analysis; just few studies of M. tuberculosis WGS have used this information to better resolve clusters [209,218].
Established SNP thresholds defining recent transmission events were calculated according to the evolutionary rate of M. tuberculosis, reported as 0.3–0.5 SNP per genome per year [190,196,209]. It is unknown if the same rate applies to M. bovis. Estimated substitution rates of M. bovis range from 0.15 to 0.53 substitutions per genome per year [46,202,210]. However, these studies either examined a limited number of isolates [210] or geographically restricted samples [46,202]. The correct estimation of M. bovis substitution rates has significant implications for the definition of the amount of genetic changes needed to define a transmission cluster, and for the temporal resolution WGS can provide to study disease dynamics in bTB [210]. Although it is possible that M. bovis-derived SNP thresholds are not very different from M. tuberculosis, the paucity of knowledge regarding M. bovis evolution and DNA repair mechanisms implies that more in-depth evaluation should be conducted. It is unknown, for instance, if the phenotype of broad host tropism [52] influences replication and substitutions rates of M. bovis over time.
5.6.2. Whole-Genome Based Multi-Locus Sequencing Typing
Traditional MLST (multi-locus sequencing typing) is based on the identification of mutations in a pre-established, limited number of bacterial genes. In order to incorporate the whole gene repertoire of a bacterial species and WGS technology, cgMLST or pgMLST (core genome or pan-genome MLST) schemes based on MTBC core or pan-genome genes (core genes plus some accessory genes), respectively, have been applied [219,220,221,222]. Briefly, the obtained list of SNPs (indels are normally excluded) is translated into a standardizable allele numbering system. SNPs are identified in a pre-defined allele dataset of selected MTBC species; any particular gene identified with a SNP is giving a number. Each sample is then given a sequence type (ST) determined by a combination of allele numbers. In other words, these schemes are based on the concept of allelic variation. STs generated from the bacterial population under study can then be used to generate minimum spanning trees to define transmission clusters [219,220,221,222]. One great advantage of these methods is the possibility of generating a nomenclature that can be readily compared between laboratories, which is vastly appreciated for disease control and eradication programs. However, by using these approaches, information from intergenic regions may not taken into account, unless intergenic loci are added to the initial reference dataset. In addition, there may be variation in the gene pool or gene annotation inconsistencies among different strains of M. bovis and MTBC [156,161,223,224] that may lead to errors when initially defining the gene repertoire to serve as alleles. This approach has been independently applied in M. bovis isolates from a Brazilian State approaching bTB eradication status, revealing recent transmission between farms and multiple M. bovis introductions within the same farm [225].
5.6.3. Phylogenetic Approaches
Most M. bovis WGS studies use phylogenetic methods to define potential clusters of pathogen transmission, to evaluate populational structure of M. bovis, and/or for surveillance purposes [21,43,45,46,47,48,49,50,51,161,167,194,202,210,212,217,226,227,228,229] (Table S3). In general, phylogenetic trees are constructed from alignments (i.e., matrices) of concatenated SNPs identified in each M. bovis genome under study. These trees are generated using different algorithms, such as maximum likelihood, maximum parsimony, neighbor-joining, or Bayesian inference. This approach provides clusters of associated M. bovis isolates, but additional analyses are normally performed to ascertain a transmission chain [21,44,47,50,51,202,210,212]. In phylogenetic trees, transmission pairs do not always appear phylogenetically related or associated; phylogenetic trees are not a complete substitute for a transmission network [163,230,231]. Nevertheless, Bayesian inference schemes have also been used to estimate temporal scales of bTB outbreaks by dating ancestries of the bacterial population under study [43,46,49]. When used to study M. bovis populational structure or evolutionary dynamics in countries or globally, phylogenetic reconstruction has always been the preferred method [21,45,46,48,50,167,194,226,227,228,229] (Table S3). However, it is important to understand that only core SNPs will be considered. All indels and variant sites that are not present in all strains are excluded from the analysis.
6. Errors Arising from Indels and Repetitive Regions
Genomic regions containing homopolymers or tandem repeats can lead to false reports of indels and/or SNPs due to sequencing errors or inaccurate read mapping. In addition, small and large indels are difficult to be accurately detected [154,170,232]. Therefore, current pipelines to infer M. tuberculosis or M. bovis transmission normally exclude indels or variants detected in repetitive, duplicated, and/or low-complexity regions [21,34]. Repetitive regions and duplicated genes are likely subjected to distinct evolutionary rate [233]. Thus, with the advent of more accurate variant callers and parameters, as well as long-read sequencing, the inclusion of such sites may provide further resolution for outbreaks as well as changes to the current SNP thresholds for definition of a transmission cluster in the future. Long-read sequencing technologies are powerful and promising tools that can uniquely identify the genomic origin of the read, helping resolve repeat regions, and determining large deletions or rearrangements [234]. However, a major drawback of such technologies is the low base calling accuracy when compared to short read technologies, which is detrimental for variant detection [234,235]. Hybrid systems, including the association of long- and short-read data, have been proposed to correct base calling errors [234,236]. In the future, it is expected that an increased accuracy in base calling of long-read technologies will revolutionize genome sequencing. Advantages and disadvantages of long-read sequencing compared to short-read sequencing have been recently reviewed elsewhere [234].
7. Software to Define Spoligotyping and MIRU-VNTR Profiles Using WGS Data
WGS does not eliminate the identification and reporting of spoligotypes and MIRU-VNTR patterns of samples under study. SpolPred [237] and SpoTyping [238] are two software developed to detect spoligotypes from short-read sequences in FASTQ format. SpoTyping also accepts assembled contigs in FASTA format as input and is reported to be 20–40 times faster than SpolPred [238]. Both software have reported identical spoligotypes in a dataset tested [238]. More recently, a methodology to reconstruct the whole CRISPR locus of MTBC strains have been proposed [239] and is awaiting further investigation for its applicability as a typing tool.
In contrast to spoligotyping, which is based on a single locus, the identification of MIRU-VNTR profiles using WGS data from short-read sequencing has been more challenging. An algorithm to assign 24-loci MIRU-VNTR profiles to isolates using draft and complete genomes have been described [240], provided that genomes meet a minimum-quality assembly. More recently, a software that uses long-read sequences obtained using Pacific Biosciences and Oxford Nanopore Technologies as input data has been developed [241], aiming to overcome the difficulties encountered with the long repeats of the MIRU-VNTR loci that may not be resolved with short-read sequencing.
8. Association of WGS with Epidemiological Data for Transmission Inference
Interpretation of genotyping and WGS data is challenging because the sampling of the population of interest is often partial and/or biased, and there is a variable interval between time of infection (i.e., when the transmission occurred) and time of sample collection [43,210]. In addition, transmission routes and intervals may be uncertain due to the slow evolving rates of the MTBC [46,190,196,202,209,210] and possible differences in substitution and replication rates between active replication state and latent state [43,210,242]. To circumvent some of these issues, the population of interest should be sampled in a manner that the epidemiological processes are captured [46,210].
Sole genetic data may not be sufficient to detect transmission in human or bovine TB outbreaks [26,163]. Identified transmission networks based solely on genetic data can be different from the network of actual transmission events if detailed field investigations are not performed [26,163]. In particular, highly clustered transmission networks can introduce uncertainty to the evaluation of transmission dynamics, especially when lower resolution genotyping is applied (e.g., MIRU-VNTR PCR and spoligotyping). Challenges associated with clustered networks have been reviewed elsewhere [26], but in general, clustering adds uncertainty to the identification of infection source and transmission patterns. To provide better resolution, the genetic data must be dense (i.e., well sampled) and complemented by good quality, collected epidemiological and demographic parameters. Accordingly, the association between WGS and network data have been elegantly applied to investigate bTB outbreaks at the local level [51,210]. The association between network, spatial-temporal mathematical models and WGS is the ideal situation to correctly describe the transmission dynamics of a particular outbreak [26,51,210] and propose targeted interventions. These are very powerful approaches to delineate disease control strategies in the long-term, particularly in a multi-host system; however, such refined analyses may not be easy to implement in bTB control programs requiring real-time transmission investigation.
9. Data Reporting in WGS Pipelines
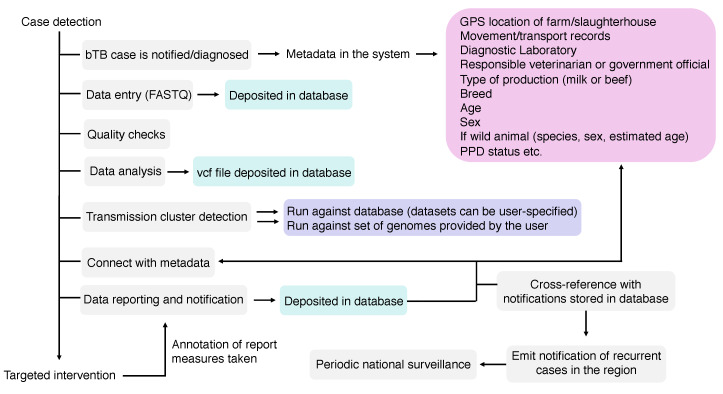
Once a transmission cluster, an infection source or a single infected-case or farm has been detected using WGS, such information needs to be communicated to end-users, e.g., veterinarians, epidemiologists, program officials, among others. Preferentially, reporting must be standardized and comparable among different veterinary services, connecting federal, state or province, and local stakeholders. Unfortunately, no standards exist on how these WGS reports must be, while capacity building is expected to play a crucial role in guaranteeing correct interpretation of the results. In other words, the improvement and acquirement of skills, knowledge, equipment, and general resources by personnel involved are vital for success of WGS-based programs. As bTB is an OIE-notifiable disease, M. bovis WGS-based surveillance systems can greatly benefit from general, robust disease reporting systems already in place in many countries [243]. An ideal system would be able to register: (i) an outbreak with genome-based transmission links that were detected using standardized data generation and bioinformatics pipelines, and (ii) individual cases or farms reporting M. bovis genomes that can be compared with a comprehensive database for the prospective identification of transmission links in a disease surveillance context, using the same standardized pipeline (Figure 5). A standardized bioinformatics pipeline, publicly available, has been developed by the National Veterinary Services Laboratories (NVSL) of the US Department of Agriculture (USDA), which implemented the use of M. bovis WGS in its official bTB program in 2013 [21]. Other pipelines (for detection of antibiotic resistance, strain typing, and/or transmission detection) have also been reported for M. tuberculosis and reviewed elsewhere [34]. Increasing efforts must be made to provide standardized end-to-end processes that are affordable and easily managed by non-experts.
Figure 5.
Components of a Mycobacterium bovis whole-genome sequencing (WGS) pipeline. In grey: components of the pipeline; in green: files that will compose the sequence and analysis databases (raw reads—FASTQ—and vcf file); in pink: metadata, which can also compose a metadata database linked to each FASTQ and vcf file; in purple: the possibilities of genome comparisons: (i) a user can choose to compare the genome against all genomes of the database or against a subset of genomes composing the database; or (ii) a user can input several genomes that can be compared against each other or with other genomes deposited in the database. An ideal pipeline would also allow periodic national surveillance reports, emitting alerts of newly detected clusters or outbreaks in certain regions that warrant further attention according to user-specified thresholds.
An important example of a standardized laboratory network for WGS reporting is GenomeTrakr [37] (US Food and Drug Administration), used for foodborne pathogens. Others exist for viral pathogens [244,245]. Reflecting GenomeTrakr structure, an effective integration between veterinary, public health, university, and industry laboratories would be of utmost interest to report M. bovis WGS data as part of national control programs. These laboratories can undergo proficiency tests to ensure quality control and standardization in generating and depositing data to a common database [246]. Once sequencing data is deposited in public databases, further comparison and identification can be fast and efficient, provided there is an effective bioinformatic pipeline established.
Report guidelines for animal health surveillance (AHSURED) were recently proposed, aiming at a systematic description of the means by which the output of surveillance has been generated for a particular disease [247]. Through a survey of experienced professionals working in animal surveillance for State Authorities, a consolidated checklist of items to be reported was generated. Although these guidelines are not specific to any bacterial typing technique or transmission cluster identification method, its applicability using WGS data remains to be tested. Other initiatives aimed at harmonizing the documentation of disease surveillance and reporting include the SANTERO (http://santero.fp7-risksur.eu/), HOTLINE (https://www.thehotlineproject.org/), and RISKSUR (http://www.fp7-risksur.eu/) projects. Guidelines for reporting cohort, case-control, and cross-sectional studies of veterinary diseases have also been proposed (STROBE-Vet Statement; https://meridian.cvm.iastate.edu/strobe). The inclusion of pathogen WGS for infection source identification and contact tracing in these projects has never been evaluated.
10. Resolution Power of WGS and Genotyping Techniques
Spoligotyping and MIRU-VNTR PCR have been often applied to resolve local clustering on larger scales [26] (Table S1). However, their power to discriminate within-cluster events or at the farm-to-farm scale is rather limited [26,29,202] (Figure 1). In such instances, WGS may provide the resolution to finely resolve transmission patterns happening at the individual herd level, in clusters of small spatial extent [48,210], or in countries where bTB prevalence is almost null and re-introduction outbreaks occur due to a single-sourced M. bovis strain [43]. Accordingly, many studies show that WGS is useful to differentiate M. tuberculosis strains with identical MIRU-VNTR genotypes, proving superior resolution [29,197,209,248,249]. Frequently, traditional typing methods of M. bovis depict the same or few genotypes distributed over relatively large local areas or encompassing a great proportion of the tested isolates under study [29,48,210,250,251,252]. Such lack of resolution is troublesome for the detection of M. bovis transmission between farms or between cattle and wildlife, especially in regions approaching free-status, with low bTB prevalence. WGS may provide an opportunity to solve this problem.
On the other hand, WGS studies evaluating transmission of M. bovis in high-burden countries or regions with high M. bovis genetic diversity are lacking. Sometimes, the genetic diversity given by MIRU-VNTR and spoligotyping is so high in the region and/or in the sample set being tested that an infection source cannot be accurately identified [199,253]. The applicability of M. bovis WGS in these instances remains to be elucidated.
The rate of genetic variation of a pathogen has implications for the scale at which the epidemiological events can be resolved using DNA typing data [254]. Accordingly, the use of WGS has been particularly advantageous to trace RNA virus outbreaks, owing to their high substitution rate. However, MTBC has a much lower evolutionary rate compared to these pathogens. As such, the resolution power of WGS for MTBC at the animal-to-animal or human-to-human level may be poor depending on the scenario [196,202,209,210,255]. In other words, zero or only very few SNPs between or among MTBC isolates are detected, leading to a failure in describing transmission links carrying meaningful information for prospective interventions. This is not a restraint of the WGS technology per se, yet a consequence of the low mutation rate of MTBC when compared to fast evolving pathogens, such as viruses. Regardless of this limitation, for both M. tuberculosis and M. bovis, it has been concluded that the epidemiology of outbreaks can greatly benefit from WGS data, providing better resolution than any other genotyping technique [26,34,202,249,256,257].
In a bacterial genome, repeat regions exhibit faster evolutionary rates compared to non-repeat regions [233]. MIRU-VNTR and spoligotype genomic regions have been successfully applied for genotyping because these are rapidly evolving regions of repetitive DNA. As explained above, the loss and gain of fragments within these regions drive the identification of genotyping patterns. Therefore, the genetic variation given by MIRU-VNTR PCR and spoligotyping is not depicted in current whole-genome data interpretation, which is based on SNP divergences. It also means that WGS is presently based on signals arising from the slowest evolving regions of the bacterial genome. The use of long-read technologies in the future may allow for more informative sites from repetitive regions to be included in the analysis, which may improve the applicability and resolution of WGS in epidemiology.
11. WGS Provides New Insights into the Global Distribution of M. bovis Lineages
In the past years, WGS has helped define MTBC lineages, particularly those adapted to humans (M. tuberculosis L1 through L4 and L7, and M. africanum L5 and L6) [258]. Mycobacterium tuberculosis and M. africanum global lineage distribution has been associated with geography and human populations, and later shown to have distinct profiles of virulence and drug resistance acquisition [258,259]. Similar attempts to classify M. bovis genetically have been made by using a limited set of markers, leading to the classification of clonal complexes (CCs). Accordingly, four M. bovis CCs have been described (African 1 and 2, European 1 and 2), and these are determined based on specific deletions ranging from 806 to 14,094 bp, few SNPs and spoligotypes [260,261,262,263]. Similarly to M. tuberculosis lineages, CCs appear to be geographically segregated, with African 1 and 2 restricted to Africa, European 2 usually found in the Iberian Peninsula, and European 1 distributed globally [118,260,261,262,263]. However, M. bovis WGS studies indicate that not all isolates can be classified into these complexes, indicating that CCs do not represent the whole genetic diversity of M. bovis [21,48,161]. More recently, a global collection of 1,969 M. bovis genomes from different countries has been analyzed using whole-genome based phylogenetics [45]. This study proposed the existence of at least four distinct global lineages of M. bovis (Lb1 to Lb4), geographically segregated and not fully represented by CCs. There were still few M. bovis genomes without CC markers that could not be classified in any of these lineages (unknown clusters 1, 2 and 3) [45]. Another study also described M. bovis isolates without CC classification in France and suggested that these might be country-specific lineages [228]. As these French M. bovis genomes have not been compared to global genome collections, their lineage classification remains to be unraveled. As more M. bovis genomes are sequenced in the future, particularly from Africa and Asia, a more complete picture of M. bovis lineages global distribution will be determined. The continuous investigation of M. bovis genomes at the global level will provide opportunities to understand differences in virulence and transmission profiles underlying the current disease distribution.
12. Other Pathogens Causing bTB
Mycobacterium caprae is a causative agent of TB in animals of the Bovidae family [264,265,266]. This pathogen has been mostly detected in the European continent, with few reports of M. caprae in animals outside of Europe and cases of zoonotic TB in European patients detected in other countries. Accordingly, one strain of M. caprae was isolated from cattle in Algeria but has been linked to a possible introduction from mainland Europe [267]. In Morocco, three isolates of animal MTBC with intact RD4 and M. caprae-associated spoligotype were obtained from cattle [268], and in Japan, one captive Borneo elephant was found infected with M. caprae [269]. With a similar generalist tropism for hosts compared to M. bovis, M. caprae has been isolated from humans, goats, sheep, cattle, pigs, red deer (Cervus elaphus), wild boars (Sus scrofa), foxes (Vulpes vulpes), European bisons (Bison bonasus), Borneo elephant, and captive dromedary camel (Camelus dromedarius) [264,265,266]. In Spain, the number of cattle farms from which M. caprae was isolated accounted for 0.85–6.67% of the total number of herds with bTB, a number that is increasing over years [264]. WGS has been successfully used for contact tracing of M. caprae in cattle herds from Germany, showing evidence of within and between farm transmission [44].
More recently, the possibility of M. orygis as a primary pathogen species causing bTB in South Asia has been raised due to the observation of TB caused by this species in people from the region [192,270]. However, very little is known about the true host range of M. orygis, as it has been isolated from cattle, oryxes, gazelles, deer, antelope, waterbucks, and non-human primates [271]. A single outbreak of M. orygis in a dairy farm of mixed-breed animals of Bos taurus (Friesian breed) and Bos taurus indicus (Sahiwal breed), with 18 affected animals was reported [272]. As similar outbreaks in alternative species are also described for M. tuberculosis (e.g., elephants) and M. bovis (e.g., dogs) [273,274,275,276], further studies should be conducted on the actual host range of M. orygis and if cattle is a reservoir for this bacterial species.
13. Conclusions and Perspectives
In this review, we outlined current standards and/or challenges that remain to be unraveled on genotyping and WGS of M. bovis as tools for epidemiologic investigations. One important step towards implementation of WGS in programs of bTB control and eradication is certainly the standardization of data analysis and reporting of M. bovis WGS outcome. Research gaps associated with these subjects have been identified and described throughout this review (Table 1). Although continuous efforts must be made to address these challenges, WGS ultimate implementation in bTB programs must also integrate systems administration, management of resulting databases, and maintenance of the pipeline. Another important aspect of standardizing data generation and analysis is to define sets of M. bovis isolates and genomes that can be used for validation of different approaches as well as between laboratories.
Table 1.
Proposed research gaps and areas that need further development and exploration.
| Pipeline Step | Areas in Need of Further Exploration |
|---|---|
| Bacterial isolation and sequencing | Methodologies to assess the possibility of cross-contamination with MTBC isolates |
| Quality assessment of entry data (FASTQ) |
Comparison of protocols with different parameters or stringency levels of read trimming and filtering, reference mapping, removal of PCR duplicates, minimum acceptable median read length, contaminants handling, etc. * |
| Read processing | Choice of reference genome Parameters of read mapping (e.g., realignment around indels) Parameters of variant calling How to handle low quality variant calls How to detect and handle variants within repetitive areas Methodologies for detection of mixed-sample (number of reads supporting an allele and number of acceptable heterozygous sites based on established parameters of variant calling) |
| Transmission cluster detection | Comparison and/or development of different approaches: SNP-count, cgMLST, pgMLST, phylogenetic inferences |
| Data reporting | Standardization of WGS data reporting to end-users |
| Validation and inter-laboratory quality control | Validation datasets (of bacterial isolates and genomes) Protocols for inter-laboratory standardization (from bacterial isolation to sequencing) |
* Technical validations should encompass the impact of choosing different parameters or stringency levels on the analysis output tailored for each need (contact investigation, surveillance, drug resistance detection), and also the relevance of these steps in the final outcome (are all these steps and parameters necessary to achieve the correct outcome?).
The field of bTB has unquestionably experienced many technique advancements for transmission investigation and surveillance, from genotyping to genome sequencing. Yet, the disease remains a significant challenge in numerous parts of the world. Many low-to-middle income countries have still to establish basic disease control and eradication programs, and they have not benefited from M. bovis genotyping in the past. Only few, developed countries, with well-established bTB control programs, have implemented M. bovis genotyping as an epidemiologic tool. In addition, many genotyping studies worldwide have been performed in a retrospective, research-oriented manner, frequently not providing real-time investigation to solve outbreaks. Nevertheless, these studies have been incredibly valuable to understand transmission dynamics at the local and country levels, providing important information for public policy implementations. Not surprisingly, the same developed countries with a tradition in applying genotyping techniques into their bTB programs have overcome barriers to apply M. bovis WGS in their transmission investigations or on a research-basis, such as the USA, Ireland, New Zealand, and France [21,43,46,50,51,202,210,212,228]. Data generated from these countries and beyond show that WGS provides superior resolution power when compared to traditional genotyping techniques. In addition, WGS provided the means to evaluate the global structure of M. bovis population, bringing valuable insights into the current disease distribution [45].
It is evident that the research community has proven the usefulness of genotyping techniques for M. bovis transmission detection and surveillance and is now accumulating evidence on the applicability of WGS for the same purposes. However, compared to genotyping, WGS will likely see a much slower pace of employment in bTB programs and research. The requirement for an articulate bTB control and eradication program, specialized personnel, laboratory and computing infra-structure, good internet connectivity, streamlined operational procedures and protocols for data generation, availability of reagents, bioinformatic pipelines, and integrated and effective veterinary services are obstacles for widespread M. bovis WGS implementation in many countries [26,34,277]. In addition, despite continuous drops in prices, WGS can still reach a high-cost per sample, especially if just a few isolates need to be sequenced [26]. Thus, successful implementation of M. bovis WGS depends on multiple factors and will be contingent on the veterinary service strength, country-specific willingness to eradicate and control bTB, and investments. Most importantly, current stakeholders have to understand the value of such tools in controlling the disease, and this requires continuous research in different scenarios showing its applicability to resolve outbreaks.
Acknowledgments
The authors are in debt to Carolina Bertelli de Souza Ferreira from the University of São Paulo, São Paulo, Brazil for invaluable technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/5/667/s1, Table S1: List of research articles comparing traditional DNA typing methods using Mycobacterium bovis isolates (1998 to mid-2019), Table S2: Characteristics of genotyping techniques and whole-genome sequencing (WGS) used in Mycobacterium bovis studies, Table S3: Characteristics of whole genome sequencing studies of Mycobacterium bovis. Figure S1: Schematic representation of the genomic regions involved in genotyping techniques used in Mycobacterium bovis studies.
Author Contributions
Researched and analyzed the data; wrote and approved the manuscript, A.M.S.G. and C.K.Z. All authors have read and agreed to the published version of the manuscript.
Funding
Fellowship for C.K.Z is provided by São Paulo Research Foundation (FAPESP; 2017/04617-3) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; 1721455). This study was financed in part by CAPES (Finance Code 001). Main research funding was made available through Morris Animal Foundation (grant number D17ZO-307) and FAPESP (2016/26108-0).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Galagan J.E. Genomic insights into tuberculosis. Nat. Rev. Genet. 2014;15:307–320. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) Global Tuberculosis Report 2019. World Health Organization; Geneve, Switzerland: 2019. [Google Scholar]
- 3.Cousing S. End TB Strategy. WHO; Geneva, Switzerland: 2018. pp. 82–83. [Google Scholar]
- 4.Ayele W.Y., Neill S.D., Zinsstag J., Weiss M.G., Pavlik I. Bovine tuberculosis: An old disease but a new threat to Africa. Int. J. Tuberc. Lung Dis. 2004;8:924–937. [PubMed] [Google Scholar]
- 5.Godfray H.C.J., Donnelly C.A., Kao R.R., Macdonald D.W., McDonald R.A., Petrokofsky G., Wood J.L.N., Woodroffe R., Young D.B., McLean A.R. A restatement of the natural science evidence base relevant to the control of bovine tuberculosis in Great Britain. Proc. R. Soc. B Biol. Sci. 2013;280:20131634. doi: 10.1098/rspb.2013.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller R.S., Sweeney S.J. Mycobacterium bovis (bovine tuberculosis) infection in North American wildlife: Current status and opportunities for mitigation of risks of further infection in wildlife populations. Epidemiol. Infect. 2013;141:1357–1370. doi: 10.1017/S0950268813000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nugent G., Buddle B.M., Knowles G. Epidemiology and control of Mycobacterium bovis infection in brushtail possums (Trichosurus vulpecula), the primary wildlife host of bovine tuberculosis in New Zealand. N. Z. Vet. J. 2015;63:28–41. doi: 10.1080/00480169.2014.963791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nugent G., Gortazar C., Knowles G. The epidemiology of Mycobacterium bovis in wild deer and feral pigs and their roles in the establishment and spread of bovine tuberculosis in New Zealand wildlife. N. Z. Vet. J. 2015;63:54–67. doi: 10.1080/00480169.2014.963792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer M.V. Mycobacterium bovis: Characteristics of Wildlife Reservoir Hosts. Transbound. Emerg. Dis. 2013;60:1–13. doi: 10.1111/tbed.12115. [DOI] [PubMed] [Google Scholar]
- 10.De Kantor I.N., Ritacco V. An update on bovine tuberculosis programmes in Latin American and Caribbean countries. Vet. Microbiol. 2006;112:111–118. doi: 10.1016/j.vetmic.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 11.Olea-Popelka F., Muwonge A., Perera A., Dean A.S., Mumford E., Erlacher-Vindel E., Forcella S., Silk B.J., Ditiu L., El Idrissi A., et al. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis—A call for action. Lancet Infect. Dis. 2017;17:e21–e25. doi: 10.1016/S1473-3099(16)30139-6. [DOI] [PubMed] [Google Scholar]
- 12.Cosivi O., Grange J.M., Daborn C.J., Raviglione M.C., Fujikura T., Cousins D., Robinson R.A., Huchzermeyer H.F.A.K., de Kantor I., Meslin F.X. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 1998;4:59–70. doi: 10.3201/eid0401.980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loiseau C., Brites D., Moser I., Coll F., Pourcel C., Robbe-Austerman S., Escuyer V., Musser K.A., Peacock S.J., Feuerriegel S., et al. Revised interpretation of the Hain Lifescience Genotype MTBC to differentiate Mycobacterium canettii and members of the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 2019;63:1–13. doi: 10.1128/AAC.00159-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scorpio A., Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 15.Konno K., Feldman F.M., McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am. Rev. Respir. Dis. 1967;95:461–469. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 16.Dürr S., Müller B., Alonso S., Hattendorf J., Laisse C.J.M., van Helden P.D., Zinsstag J. Differences in primary sites of infection between zoonotic and human tuberculosis: Results from a worldwide systematic review. PLoS Negl. Trop. Dis. 2013;7:e2399. doi: 10.1371/journal.pntd.0002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization (WHO) Food and Agriculture Organization of the United Nations (FAO) World Organisation for Animal Health (OIE) Roadmap for Zoonotic Tuberculosis. World Health Organization; Geneva, Switzerland: 2017. [Google Scholar]
- 18.OIE Bovine tuberculosis. Gen. Dis. Inf. Sheets. 2011:1–6. [Google Scholar]
- 19.Anderson D.P., Ramsey D.S.L., de Lisle G.W., Bosson M., Cross M.L., Nugent G. Development of integrated surveillance systems for the management of tuberculosis in New Zealand wildlife. N. Z. Vet. J. 2015;63:89–97. doi: 10.1080/00480169.2014.963830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skuce R.A., Mallon T.R., McCormick C.M., McBride S.H., Clarke G., Thompson A., Couzens C., Gordon A.W., McDowell S.W.J. Mycobacterium bovis genotypes in Northern Ireland: Herd-level surveillance (2003 to 2008) Vet. Rec. 2010;167:684–689. doi: 10.1136/vr.c5108. [DOI] [PubMed] [Google Scholar]
- 21.Orloski K., Robbe-austerman S., Stuber T., Hench B., Schoenbaum M. Whole genome sequencing of Mycobacterium bovis isolated from livestock in the United States, 1989–2018. Front. Vet. Sci. 2018;5:1–23. doi: 10.3389/fvets.2018.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Réveillaud É., Desvaux S., Boschiroli M.-L., Hars J., Faure É., Fediaevsky A., Cavalerie L., Chevalier F., Jabert P., Poliak S., et al. Infection of wildlife by Mycobacterium bovis in France assessment through a national surveillance system, Sylvatub. Front. Vet. Sci. 2018;5:262. doi: 10.3389/fvets.2018.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos D.F., Tavares L., da Silva P.E.A., Dellagostin O.A. Molecular typing of Mycobacterium bovis isolates: A review. Braz. J. Microbiol. 2014;45:365–372. doi: 10.1590/S1517-83822014005000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Sayed A., El-Shannat S., Kamel M., Castañeda-Vazquez M.A., Castañeda-Vazquez H. Molecular Epidemiology of Mycobacterium bovis in Humans and Cattle. Zoonoses Public Health. 2016;63:251–264. doi: 10.1111/zph.12242. [DOI] [PubMed] [Google Scholar]
- 25.Collins D.M. Advances in molecular diagnostics for Mycobacterium bovis. Vet. Microbiol. 2011;151:2–7. doi: 10.1016/j.vetmic.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Kao R.R., Price-Carter M., Robbe-Austerman S. Use of genomics to track bovine tuberculosis transmission. OIE Rev. Sci. Tech. 2016;35:241–258. doi: 10.20506/rst.35.1.2430. [DOI] [PubMed] [Google Scholar]
- 27.Durr P.A., Hewinson R.G., Clifton-Hadley R.S. Molecular epidemiology of bovine tuberculosis. I. Mycobacterium bovis genotyping. Rev. Sci. Tech. 2000;19:675–688. doi: 10.20506/rst.19.3.1241. [DOI] [PubMed] [Google Scholar]
- 28.Haddad N., Masselot M., Durand B. Molecular differentiation of Mycobacterium bovis isolates. Review of main techniques and applications. Res. Vet. Sci. 2004;76:1–18. doi: 10.1016/S0034-5288(03)00078-X. [DOI] [PubMed] [Google Scholar]
- 29.Merker M., Kohl T.A., Niemann S., Supply P. The evolution of strain typing in the Mycobacterium tuberculosis complex. Adv. Exp. Med. Biol. 2017;1019:79–93. doi: 10.1007/978-3-319-64371-7_3. [DOI] [PubMed] [Google Scholar]
- 30.Tsao K., Robbe-Austerman S., Miller R.S., Portacci K., Grear D.A., Webb C. Sources of bovine tuberculosis in the United States. Infect. Genet. Evol. 2014;28:137–143. doi: 10.1016/j.meegid.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 31.Gilchrist C.A., Turner S.D., Riley M.F., Petri W.A., Hewlett E.L. Whole-genome sequencing in outbreak analysis. Clin. Microbiol. Rev. 2015;28:541–563. doi: 10.1128/CMR.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryant J., Chewapreecha C., Bentley S.D. Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences. Future Microbiol. 2012;7:1283–1296. doi: 10.2217/fmb.12.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Didelot X., Bowden R., Wilson D.J., Peto T.E.A., Crook D.W. Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 2012;13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan C.J., Goig G.A., Kohl T.A., Verboven L., Dippenaar A., Ezewudo M., Farhat M.R., Guthrie J.L., Laukens K., Miotto P., et al. Whole genome sequencing of Mycobacterium tuberculosis: Current standards and open issues. Nat. Rev. Microbiol. 2019;17:533–545. doi: 10.1038/s41579-019-0214-5. [DOI] [PubMed] [Google Scholar]
- 35.Crisan A., Gardy J.L., Munzner T. A systematic method for surveying data visualizations and a resulting genomic epidemiology visualization typology: GEViT. Bioinformatics. 2019;35:1668–1676. doi: 10.1093/bioinformatics/bty832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belak S., Karlsson O.E., Leijon M., Granberg F. High-throughput sequencing in veterinary infection biology and diagnostics. Rev. Sci. Tech. l’OIE. 2013;32:893–915. doi: 10.20506/rst.32.2.2206. [DOI] [PubMed] [Google Scholar]
- 37.Allard M.W., Strain E., Melka D., Bunning K., Musser S.M., Brown E.W., Timme R. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 2016;54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.OIE (World Organisation for Animal Health) Chapter 1.1.7. Standards for High. Throughput Sequencing, Bioinformatics and Computational Genomics. 8th ed. OIE (World Organisation for Animal Health); Paris, France: 2018. [Google Scholar]
- 39.Van Bomr S., Wang J., Granberg F., Colling A. Next-generation sequencing workflows in veterinary infection biology: Towards validation and quality assurance. Rev. Sci. Tech. l’OIE. 2016;35:67–81. doi: 10.20506/rst.35.1.2418. [DOI] [PubMed] [Google Scholar]
- 40.World Health Organization (WHO) Whole Genome Sequencing for Foodborne Disease Surveillance: Landscape Paper. World Health Organization (WHO); Geneva, Switzerland: 2018. [Google Scholar]
- 41.World Health Organization (WHO) The Use of Next-Generation Sequencing Technologies for the Detection of Mutations Associated with Drug Resistance in Mycobacterium tuberculosis Complex.: Technical Guide. World Health Organization (WHO); Geneva, Switzerland: 2018. [Google Scholar]
- 42.Tagliani E., Cirillo D.M., Ködmön C., van der Werf M.J. EUSeqMyTB to set standards and build capacity for whole genome sequencing for tuberculosis in the EU. Lancet Infect. Dis. 2018;18:377. doi: 10.1016/S1473-3099(18)30132-4. [DOI] [PubMed] [Google Scholar]
- 43.Glaser L., Carstensen M., Shaw S., Robbe-Austerman S., Wunschmann A., Grear D., Stuber T., Thomsen B. Descriptive epidemiology and whole genome sequencing analysis for an outbreak of bovine tuberculosis in beef cattle and white-tailed deer in northwestern Minnesota. PLoS ONE. 2016;11:e0145735. doi: 10.1371/journal.pone.0145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broeckl S., Krebs S., Varadharajan A., Straubinger R.K., Blum H., Buettner M. Investigation of intra-herd spread of Mycobacterium caprae in cattle by generation and use of a whole-genome sequence. Vet. Res. Commun. 2017;41:113–128. doi: 10.1007/s11259-017-9679-8. [DOI] [PubMed] [Google Scholar]
- 45.Zimpel C.K., Patané J.S.L., Guedes A.C.P., Souza R.F., Pereira-Silva T.T., Soler Camargo N.C., de Souza Filho A.F., Ikuta C.Y., Soares Ferreira Neto J., Setubal J.C., et al. Global distribution and evolution of Mycobacterium bovis. Front. Microbiol. 2020;11:1–19. doi: 10.3389/fmicb.2020.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crispell J., Zadoks R.N., Harris S.R., Paterson B., Collins D.M., De-Lisle G.W., Livingstone P., Neill M.A., Biek R., Lycett S.J., et al. Using whole genome sequencing to investigate transmission in a multi-host system: Bovine tuberculosis in New Zealand. BMC Genom. 2017;18:180. doi: 10.1186/s12864-017-3569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohl T.A., Utpatel C., Niemann S., Moser I. Mycobacterium bovis persistence in two different captive wild animal populations in Germany: A longitudinal molecular epidemiological study revealing pathogen transmission by whole-genome sequencing. J. Clin. Microbiol. 2018;56:1–9. doi: 10.1128/JCM.00302-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghebremariam M.K., Hlokwe T., Rutten V.P.M.G., Allepuz A., Cadmus S., Muwonge A., Robbe-Austerman S., Michel A.L. Genetic profiling of Mycobacterium bovis strains from slaughtered cattle in Eritrea. PLoS Negl. Trop. Dis. 2018;12:e0006406. doi: 10.1371/journal.pntd.0006406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdelaal H.F.M., Spalink D., Amer A., Steinberg H., Hashish E.A., Nasr E.A., Talaat A.M. Genomic Polymorphism Associated with the Emergence of Virulent Isolates of Mycobacterium bovis in the Nile Delta. Sci. Rep. 2019;9:11657. doi: 10.1038/s41598-019-48106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salvador L.C.M., O’Brien D.J., Cosgrove M.K., Stuber T.P., Schooley A.M., Crispell J., Church S.V., Gröhn Y.T., Robbe-Austerman S., Kao R.R. Disease management at the wildlife-livestock interface: Using whole-genome sequencing to study the role of elk in Mycobacterium bovis transmission in Michigan, USA. Mol. Ecol. 2019;28:2192–2205. doi: 10.1111/mec.15061. [DOI] [PubMed] [Google Scholar]
- 51.Crispell J., Benton C.H., Balaz D., De Maio N., Akhmetova A., Allen A., Biek R., Presho E.L., Dale J., Hewinson G., et al. Combining genomics and epidemiology to analyse bi-directional transmission of Mycobacterium bovis in a multi-host system. Elife. 2019;8:e45833. doi: 10.7554/eLife.45833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordon S. Strain Variation in the Mycobacterium Tuberculosis Complex: Its Role in Biology, Epidemiology and Control. Volume 1019 Springer; Berlin/Heidelberg, Germany: 2017. [Google Scholar]
- 53.Supply P., Warren R.M., Bañuls A.-L., Lesjean S., Van Der Spuy G.D., Lewis L.-A., Tibayrenc M., Van Helden P.D., Locht C. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 2003;47:529–538. doi: 10.1046/j.1365-2958.2003.03315.x. [DOI] [PubMed] [Google Scholar]
- 54.Gagneux S., Small P.M. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 55.Hirsh A.E., Tsolaki A.G., DeRiemer K., Feldman M.W., Small P.M. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comas I., Coscolla M., Luo T., Borrell S., Holt K.E., Kato-Maeda M., Parkhill J., Malla B., Berg S., Thwaites G., et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bos K.I., Harkins K.M., Herbig A., Coscolla M., Weber N., Comas I., Forrest S.A., Bryant J.M., Harris S.R., Schuenemann V.J., et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514:494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brosch R., Gordon S.V., Marmiesse M., Brodin P., Buchrieser C., Eiglmeier K., Garnier T., Gutierrez C., Hewinson G., Kremer K., et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gordon S.V., Brosch R., Billault A., Garnier T., Eiglmeier K., Cole S.T. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 60.Brosch R., Gordon S.V., Billault A., Garnier T., Eiglmeier K., Soravito C., Barrell B.G., Cole S.T. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect. Immun. 1998;66:2221–2229. doi: 10.1128/IAI.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Philipp W.J., Nair S., Guglielmi G., Lagrande M., Gicquelv B., Cole S.T. Physical mapping of Mycobacterium bovis BCG Pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology. 2019;142:3135–3145. doi: 10.1099/13500872-142-11-3135. [DOI] [PubMed] [Google Scholar]
- 62.Warren R.M., Van Pittius N.C.G., Barnard M., Hesseling A., Engelke E., De Kock M., Gutierrez M.C., Chege G.K., Victor T.C., Hoal E.G., et al. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int. J. Tuberc. Lung Dis. 2006;10:818–822. [PubMed] [Google Scholar]
- 63.Karlson A.G. Mycobacterium bovis nom.nov. Int. J. Syst. Bacteriol. 1970;20:273–282. doi: 10.1099/00207713-20-3-273. [DOI] [Google Scholar]
- 64.Goodfellow M., Kämpfer P., Busse H.J., Trujillo M.E., Suzuki K.I., Ludwig W.W. Bergey’s Manual of Systematic Bacteriology. Springer; New York, NY, USA: 2012. Genus I. Streptomyces. [Google Scholar]
- 65.Wayne L., Kubica G. The Mycobacteria: A Sourcebook. Marcel Dekker Inc.; New York, NY, USA: 1984. pp. 1436–1457. [Google Scholar]
- 66.Shinnick T.M., Good R.C. Mycobacterial taxonomy. Eur. J. Clin. Microbiol. Infect. Dis. 1994;13:884–901. doi: 10.1007/BF02111489. [DOI] [PubMed] [Google Scholar]
- 67.Baess I. Deoxyribonucleic acid relatedness among species of slowly-growing mycobacteria. Acta Pathol. Microbiol. Scand. B. 1979;87:221–226. doi: 10.1111/j.1699-0463.1979.tb02430.x. [DOI] [PubMed] [Google Scholar]
- 68.Smith N., Kremer K., Inwald J., Dale J., Driscoll J., Gordon S., van Soolingen D., Hewinson R., Smith J. Ecotypes of the Mycobacterium tuberculosis complex. J. Theor. Biol. 2006;239:220–225. doi: 10.1016/j.jtbi.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 69.Riojas M.A., McGough K.J., Rider-Riojas C.J., Rastogi N., Hazbón M.H. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycob. Int. J. Syst. Evol. Microbiol. 2018;68:324–332. doi: 10.1099/ijsem.0.002507. [DOI] [PubMed] [Google Scholar]
- 70.Tyler A.D., Christianson S., Knox N.C., Mabon P., Wolfe J., Van Domselaar G., Graham M.R., Sharma M.K. Comparison of sample preparation methods used for the next-generation sequencing of Mycobacterium tuberculosis. PLoS ONE. 2016;11:e0148676. doi: 10.1371/journal.pone.0148676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delogu G., Brennan M.J., Manganelli R. PE and PPE genes: A tale of conservation and diversity. Adv. Exp. Med. Biol. 2017;1019:191–207. doi: 10.1007/978-3-319-64371-7_10. [DOI] [PubMed] [Google Scholar]
- 72.Tørresen O.K., Star B., Mier P., Andrade-navarro M.A., Bateman A., Jarnot P., Gruca A., Grynberg M., Kajava V., Promponas V.J., et al. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019;47:10994–11006. doi: 10.1093/nar/gkz841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collins D.M., de Lisle G.W. DNA Restriction Endonuclease Analysis of Mycobacterium tuberculosis and Mycobacterium bovis. BCG. 1984;130:1019–1021. doi: 10.1099/00221287-130-4-1019. [DOI] [PubMed] [Google Scholar]
- 74.Collins D.M., De Lisle G.W. DNA Restriction Endonuclease Analysis of Mycobacterium bovis and other members of the Tuberculosis complex. Microbiology. 1985;21:526–564. doi: 10.1128/JCM.21.4.562-564.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collins D.M., De Lisle G.W., Gabric D.M. Geographic distribution of restriction types of Mycobacterium bovis isolates from brush-tailed possums ( Trichosurus vulpecula) in New Zealand. J. Hyg. 1986;96:431–438. doi: 10.1017/S0022172400066201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Collins D.M., de Lisle G.W., Collins J.D. DNA restriction fragment typing of Mycobacterium bovis isolates from cattle and badgers in Ireland. Vet. Rec. 1994;134:681–682. doi: 10.1136/vr.134.26.681. [DOI] [PubMed] [Google Scholar]
- 77.Collins D.M., Gabric D.M., de Lisle G.W. Typing of Mycobacterium bovis isolates from cattle and other animals in the same locality. N. Z. Vet. J. 1988;36:45–46. doi: 10.1080/00480169.1988.35476. [DOI] [PubMed] [Google Scholar]
- 78.Collins D.M., Erasmuson S.K., Stephens D.M., Yates G.F., De Lisle G.W. DNA fingerprinting of Mycobacterium bovis strains by restriction fragment analysis and hybridization with insertion elements IS1081 and IS6110. J. Clin. Microbiol. 1993;31:1143–1147. doi: 10.1128/JCM.31.5.1143-1147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Mazurek G.H., Cave M.D., Eisenach K.D., Pang Y., Murphy D.T., Wallace R.J. DNA polymorphisms in strains of Mycobacterium tuberculosis analyzed by pulsed-field gel electrophoresis: A tool for epidemiology. J. Clin. Microbiol. 1992;30:1551–1556. doi: 10.1128/JCM.30.6.1551-1556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hughes V.M., Stevenson K., Sharp J.M. Improved preparation of high molecular weight DNA for pulsed-field gel electrophoresis from mycobacteria. J. Microbiol. Methods. 2001;44:209–215. doi: 10.1016/S0167-7012(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 81.Ghodousi A., Arash G., Vatani S., Darban-Sarokhalil D., Omrani M., Fooladi A., Khosaravi A., Feizabadi M.M. Development of a new DNA extraction protocol for PFGE typing of Mycobacterium tuberculosis complex. Iran. J. Microbiol. 2012;4:44–46. [PMC free article] [PubMed] [Google Scholar]
- 82.Choe Y.-K., Huh Y.-J., Park J.-H., Kim J.-R., Park J.-S., Song J.-C., Ko J.-H., Lee Y.-C., Nashiru O., Kim J.-K., et al. Improved Isolation of Genomic DNA from Mycobacteria in Agarose Plugs by Rapid Lysis with a Combination of N-Acetylglucosaminidase and Lysozyme. Biotechniques. 1996;20:547–552. doi: 10.2144/19962004547. [DOI] [PubMed] [Google Scholar]
- 83.Ravansalar H., Tadayon K., Ghazvini K. Molecular typing methods used in studies of Mycobacterium tuberculosis in Iran: A systematic review. Iran. J. Microbiol. 2016;8:338–346. [PMC free article] [PubMed] [Google Scholar]
- 84.Jeon S., Lim N., Park S., Park M., Kim S. Comparison of PFGE, IS6110-RFLP, and 24-Locus MIRU-VNTR for molecular epidemiologic typing of Mycobacterium tuberculosis isolates with known epidemic connections. J. Microbiol. Biotechnol. 2018;28:338–346. doi: 10.4014/jmb.1704.04042. [DOI] [PubMed] [Google Scholar]
- 85.Suffys P.N., De Araujo M.E.I., Degrave W.M. The changing face of the epidemiology of tuberculosis due to molecular strain typing: A review. Mem. Inst. Oswaldo Cruz. 1997;92:297–316. doi: 10.1590/S0074-02761997000300001. [DOI] [PubMed] [Google Scholar]
- 86.Njanpop-Lafourcade B.M., Inwald J., Ostyn A., Durand B., Hughes S., Thorel M.F., Hewinson G., Haddad N. Molecular typing of Mycobacterium bovis isolates from Cameroon. J. Clin. Microbiol. 2001;39:222–227. doi: 10.1128/JCM.39.1.222-227.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feizabadi M., Robertson I., Edwards R., Cousins D.V., Hampson D. Genetic differentiation of Australian isolates of Mycobacterium tuberculosis by pulsed-field gel electrophoresis. J. Med. Microbiol. 1997;46:501–505. doi: 10.1099/00222615-46-6-501. [DOI] [PubMed] [Google Scholar]
- 88.Hughes V.M., Skuce R., Doig C., Stevenson K., Sharp J.M., Watt B. Analysis of multidrug-resistant Mycobacterium bovis from three clinical samples from Scotland. Int. J. Tuberc. Lung Dis. 2003;7:1191–1198. [PubMed] [Google Scholar]
- 89.Thierry D., Brisson N.A., Vincent L.F., Nguyen S., Guesdon J.L., Gicquel B. Characterization of Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J. Clin. Microbiol. 1999;28:2668–2673. doi: 10.1128/JCM.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cave M.D., Eisenach K.D., McDermott P.F., Bates J.H., Crawford J.T. IS6110: Conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol. Cell. Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-Q. [DOI] [PubMed] [Google Scholar]
- 91.Gonzalo-Asensio J., Pérez I., Aguiló N., Uranga S., Picó A., Lampreave C., Cebollada A., Otal I., Samper S., Martín C. New insights into the transposition mechanisms of IS6110 and its dynamic distribution between Mycobacterium tuberculosis Complex lineages. PLoS Genet. 2018;14:e1007282. doi: 10.1371/journal.pgen.1007282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thierry D., Cave M.D., Eisenach K.D., Crawford J.T., Bates J.H., Gicquel B., Guesdon J.L., Froides S., Microbiologique D.G., Pasteur I. IS6110, an IS-like element of Mycobacterium tuberculosis. Nucleic Acids Res. 1990;18:6110. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hermans P.W.M., Van Soolingen D., Dale J.W., Schuitema A.R.J., McAdam R.A., Catty D., Van Embden J.D.A. Insertion element IS986 from Mycobacterium tuberculosis: A useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol. 1990;28:2051–2058. doi: 10.1128/JCM.28.9.2051-2058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Otal I., Martin C., Vincent-Levy-Frebault V., Thierry D., Gicquel B. Restriction fragment length polymorphism analysis using IS6110 as an epidemiological marker in tuberculosis. J. Clin. Microbiol. 1991;29:1252–1254. doi: 10.1128/JCM.29.6.1252-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Embden J.D., Cave M.D., Crawford J.T., Dale J.W., Eisenach K.D., Gicquel B., Hermans P., Martin C., McAdam R., Shinnick T.M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: Recommendations for a standardized methodology. J. Clin. Microbiol. 1993;31:406–409. doi: 10.1128/JCM.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Warren R.M., Van Der Spuy G.D., Richardson M., Beyers N., Borgdorff M.W., Behr M.A., Helden P.D. Van Calculation of the stability of the IS6110 banding pattern in patients with persistent Mycobacterium tuberculosis disease. J. Clin. Microbiol. 2002;40:1705–1708. doi: 10.1128/JCM.40.5.1705-1708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Soolingen D., Hermans P.W.M., De Haas P.E.W., Soll D.R., Van Embden J.D.A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: Evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 1991;29:2578–2586. doi: 10.1128/JCM.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cousins D.V., Williams S.N., Ross B.C., Ellis T.M. Use of a repetitive element isolated from Mycobacterium tuberculosis in hybridization studies with Mycobacterium bovis: A new tool for epidemiological studies of bovine tuberculosis. Vet. Microbiol. 1993;37:1–17. doi: 10.1016/0378-1135(93)90178-A. [DOI] [PubMed] [Google Scholar]
- 99.Ross B.C., Raios K., Jackson K., Sievers A., Dwyer B. Differentiation of Mycobacterium tuberculosis strains by use of a nonradioactive Southern Blot hybridization method. J. Infect. Dis. 1991;163:904–907. doi: 10.1093/infdis/163.4.904. [DOI] [PubMed] [Google Scholar]
- 100.Ross B.C., Raios K., Jackson K., Dwyer B. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J. Clin. Microbiol. 1992;30:942–946. doi: 10.1128/JCM.30.4.942-946.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van Soolingen D., De Haas P.E.W., Hermans P.W.M., Groenen P.M.A., Van Embden J.D.A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol. 1993;31:1987–1995. doi: 10.1128/JCM.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang Z.H., Ijaz K., Bates J.H., Eisenach K.D., Cave M.D. Spoligotyping and polymorphic GC-rich repetitive sequence fingerprinting of Mycobacterium tuberculosis strains having few copies of IS6110. J. Clin. Microbiol. 2000;38:3572–3576. doi: 10.1128/JCM.38.10.3572-3576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sorek R., Lawrence C.M., Wiedenheft B. CRISPR-Mediated Adaptive Immune Systems in Bacteria and Archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 104.Hermans P.W.M., Van Soolingen D., Bik E.M., De Haas P.E.W., Dale J.W., Van Embden J.D.A. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 1991;59:2695–2705. doi: 10.1128/IAI.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Groenen P.M.A., Bunschoten A.E., van Soolingen D., Errtbden J.D.A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 106.Kamerbeek J., Schouls L.E.O., Kolk A., Kuijper S., Bunschoten A., Molhuizen H., Shaw R., Goyal M. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/JCM.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aranaz A., Liébana E., Mateos A., Dominguez L., Vidal D., Domingo M., Gonzolez O., Rodriguez-Ferri E.F., Bunschoten A.E., Van Embden J.D.A., et al. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: A tool for studying epidemiology of tuberculosis. J. Clin. Microbiol. 1996;34:2734–2740. doi: 10.1128/JCM.34.11.2734-2740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Van Der Zanden A.G.M., Hoentjen A.H., Heilmann F.G.C., Weltevreden E.F., Schouls L.M., Van Embden J.D.A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. J. Clin. Pathol. Mol. Pathol. 1998;51:209–214. doi: 10.1136/mp.51.4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cowan L.S., Diem L., Brake M.C., Crawford J.T. Transfer of a Mycobacterium tuberculosis genotyping method, Spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 2004;42:474–477. doi: 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ocheretina O., Merveille Y.M., Mabou M.M., Escuyer V.E., Dunbar S.A., Johnson W.D., Pape J.W., Fitzgerald D.W. Use of Luminex MagPlex magnetic microspheres for high-throughput spoligotyping of Mycobacterium tuberculosis isolates in Port-au-Prince, Haiti. J. Clin. Microbiol. 2013;51:2232–2237. doi: 10.1128/JCM.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang J., Abadia E., Refregier G., Tafaj S., Boschiroli M.L., Guillard B., Andremont A., Ruimy R., Sola C. Mycobacterium tuberculosis complex CRISPR genotyping: Improving efficiency, throughput and discriminative power of “spoligotyping” with new spacers and a microbead-based hybridization assay. J. Med. Microbiol. 2010;59:285–294. doi: 10.1099/jmm.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 112.Honisch C., Mosko M., Arnold C., Gharbia S.E., Diel R., Niemann S. Replacing reverse line blot hybridization spoligotyping of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 2010;48:1520–1526. doi: 10.1128/JCM.02299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Song E.J., Jeong H.J., Lee S.M., Kim C.M., Song E.S., Park Y.K., Bai G.H., Lee E.Y., Chang C.L. A DNA chip-based spoligotyping method for the strain identification of Mycobacterium tuberculosis isolates. J. Microbiol. Methods. 2007;68:430–433. doi: 10.1016/j.mimet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 114.Ruettger A., Nieter J., Skrypnyk A., Engelmann I., Ziegler A., Moser I., Monecke S., Ehricht R., Sachse K. Rapid spoligotyping of Mycobacterium tuberculosis complex bacteria by use of a microarray system with automatic data processing and assignment. J. Clin. Microbiol. 2012;50:2492–2495. doi: 10.1128/JCM.00442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bespyatykh J.A., Zimenkov D.V., Shitikov E.A., Kulagina E.V., Lapa S.A., Gryadunov D.A., Ilina E.N., Govorun V.M. Spoligotyping of Mycobacterium tuberculosis complex isolates using hydrogel oligonucleotide microarrays. Infect. Genet. Evol. 2014;26:41–46. doi: 10.1016/j.meegid.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 116.Cabibbe A.M., Miotto P., Moure R., Alcaide F., Feuerriegel S., Pozzi G., Nikolayevskyy V., Drobniewski F., Niemann S., Reither K., et al. Lab-on-chip-based platform for fast molecular diagnosis of multidrug-resistant tuberculosis. J. Clin. Microbiol. 2015;53:3876–3880. doi: 10.1128/JCM.01824-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zeng X., Li H., Zheng R., Kurepina N., Kreiswirth B.N., Zhao X., Xu Y., Li Q., Diagnostic M. Spoligotyping of Mycobacterium tuberculosis complex isolates using ligation-based amplification and melting curve analysis. J. Clin. Microbiol. 2016;54:1–14. doi: 10.1128/JCM.00857-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smith N.H. The global distribution and phylogeography of Mycobacterium bovis clonal complexes. Infect. Genet. Evol. 2012;12:857–865. doi: 10.1016/j.meegid.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 119.Warren R.M., Streicher E.M., Sampson S.L., van der Spuy G.D., Richardson M., Nguyen D., Behr M.A., Victor T.C., van Helden P.D. Microevolution of the Direct Repeat Region of Mycobacterium tuberculosis: Implications for Interpretation of Spoligotyping Data. J. Clin. Microbiol. 2002;40:4457–4465. doi: 10.1128/JCM.40.12.4457-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ramazanzadeh R., McNerney R. Variable number of tandem repeats (VNTR) and its application in bacterial epidemiology. Pak. J. Biol. Sci. 2007;10:2612–2621. doi: 10.3923/pjbs.2007.2612.2621. [DOI] [PubMed] [Google Scholar]
- 121.Frothingham R., Meeker-O’Connell W.A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 122.Frothingham R. Differentiation of strains in Mycobacterium tuberculosis complex by DNA sequence polymorphisms, including rapid identification of M. bovis BCG. J. Clin. Microbiol. 1995;33:840–844. doi: 10.1128/JCM.33.4.840-844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Supply P., Magdalena J., Himpens S., Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 124.Kremer K., Van Soolingen D., Frothingham R., Haas W.H., Hermans P.W.M., Martín C., Palittapongarnpim P., Plikaytis B.B., Riley L.W., Yakrus M.A., et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: Interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 1999;37:2607–2618. doi: 10.1128/JCM.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barlow R.E.L., Gascoyne-binzi D.M., Gillespie S.H., Dickens A., Qamer S., Hawkey P.M. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 2001;39:2453–2457. doi: 10.1128/JCM.39.7.2453-2457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Filliol I., Ferdinand S., Negroni L., Sola C., Rastogi N. Molecular typing of Mycobacterium tuberculosis based on variable number of tandem DNA repeats used alone and in association with spoligotyping. J. Clin. Microbiol. 2000;38:2520–2524. doi: 10.1128/JCM.38.7.2520-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rusch-Gerdes S., Willery E., Savine E., de Haas P., van Deutekom H., Roring S., et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weniger T., Krawczyk J., Supply P., Niemann S., Harmsen D. MIRU-VNTRplus: A web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326–W331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hauer A., Michelet L., De Cruz K., Cochard T., Branger M., Karoui C., Henault S., Biet F., Boschiroli M.L. MIRU-VNTR allelic variability depends on Mycobacterium bovis clonal group identity. Infect. Genet. Evol. 2016;45:165–169. doi: 10.1016/j.meegid.2016.08.038. [DOI] [PubMed] [Google Scholar]
- 130.Wyllie D.H., Davidson J.A., Grace Smith E., Rathod P., Crook D.W., Peto T.E.A., Robinson E., Walker T., Campbell C. A quantitative evaluation of MIRU-VNTR typing against whole-genome sequencing for identifying Mycobacterium tuberculosis transmission: A prospective observational cohort study. EBioMedicine. 2018;34:122–130. doi: 10.1016/j.ebiom.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Allix-Béguec C., Wahl C., Hanekom M., Nikolayevskyy V., Drobniewski F., Maeda S., Campos-Herrero I., Mokrousov I., Niemann S., Kontsevaya I., et al. Proposal of a consensus set of hypervariable mycobacterial interspersed repetitive-unit-variable-number tandem-repeat loci for subtyping of Mycobacterium tuberculosis Beijing isolates. J. Clin. Microbiol. 2014;52:164–172. doi: 10.1128/JCM.02519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Garnier T., Eiglmeier K., Camus J.-C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., et al. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Malone K.M., Farrell D., Stuber T.P., Schubert O.T. Updated reference genome sequence and annotation of Mycobacterium bovis AF2122/97. Genome Announc. 2017;5:17–18. doi: 10.1128/genomeA.00157-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bouso J.M., Planet P.J. Complete nontuberculous mycobacteria whole genomes using an optimized DNA extraction protocol for long-read sequencing. BMC Genom. 2019;20:793. doi: 10.1186/s12864-019-6134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arnold C., Edwards K., Desai M., Platt S., Green J., Conway D. Setup, validation, and quality control of a centralized whole- genome-sequencing laboratory: Lessons learned. J. Clin. Microbiol. 2018;56:e-00261-18. doi: 10.1128/JCM.00261-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang M., Sun H., Fei Z., Zhan F., Gong X., Gao S. Fastq-clean: An optimized pipeline to clean the Illumina sequencing data with quality control; Proceedings of the 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); Belfast, UK. 2–5 November 2014; pp. 44–48. [Google Scholar]
- 137.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hannon G.J. [(accessed on 15 December 2019)];FASTX-Toolkit. Available online: http://hannonlab.cshl.edu/fastx_toolkit/
- 139.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Andrews S. FastQC: A Quality Control for High Throughput Sequence Data. [(accessed on 10 December 2019)]; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 141.Okonechnikov K., Conesa A., García-Alcalde F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:884–890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ebbert M.T.W., Wadsworth M.E., Staley L.A., Hoyt K.L., Pickett B., Miller J., Duce J., Kauwe J.S.K., Ridge P.G. Evaluating the necessity of PCR duplicate removal from next-generation sequencing data and a comparison of approaches. BMC Bioinform. 2016;17:239. doi: 10.1186/s12859-016-1097-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chiara M., Pavesi G. Evaluation of quality assessment protocols for high throughput genome resequencing data. Front. Genet. 2017;8:94. doi: 10.3389/fgene.2017.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Del Fabbro C., Scalabrin S., Morgante M., Giorgi F.M. An extensive evaluation of read trimming effects on illumina NGS data analysis. PLoS ONE. 2013;8:e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lusk R.W. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS ONE. 2014;9:e110808. doi: 10.1371/journal.pone.0110808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F., Turner P., Parkhill J., Loman N.J., Walker A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Breitwieser F.P., Pertea M., Zimin A.V., Salzberg S.L. Human contamination in bacterial genomes has created thousands of spurious proteins. Genome Res. 2019;29:954–960. doi: 10.1101/gr.245373.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Merchant S., Wood D.E., Salzberg S.L. Unexpected cross-species contamination in genome sequencing projects. Peer J. 2014;2:e675. doi: 10.7717/peerj.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Goig G.A., Garcia-basteiro A.L., Cambeve B. Contaminant DNA in bacterial sequencing experiments is a major source of false genetic variability. BMC Biol. 2020;18:748. doi: 10.1186/s12915-020-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wingett S.W., Andrews S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research. 2018;7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wood D.E., Salzberg S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ezewudo M., Borens A., Chiner-Oms Á., Miotto P., Chindelevitch L., Starks A.M., Hanna D., Liwski R., Zignol M., Gilpin C., et al. Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase. Sci. Rep. 2018;8:15382. doi: 10.1038/s41598-018-33731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wyllie D.H., Robinson E., Peto T., Crook D.W., Ajileye A., Rathod P., Allen R., Jarrett L., Smith E.G., Walker A.S. Identifying mixed Mycobacterium tuberculosis infection and laboratory cross-contamination during mycobacterial sequencing programs. J. Clin. Microbiol. 2018;56:e00923-18. doi: 10.1128/JCM.00923-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Silva-Pereira T.T., Ikuta C.Y., Zimpel C.K., Camargo N.C.S., Filho A.F.D.S., Neto J.S.F., Heinemann M.B., Guimarães A.M.S. Genome sequencing of Mycobacterium pinnipedii strains: Genetic characterization and evidence of superinfection in a South American sea lion (Otaria flavescens) BMC Genom. 2019:1030. doi: 10.1186/s12864-019-6407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Coscolla M., Lewin A., Metzger S., Maetz-Rennsing K., Calvignac-Spencer S., Nitsche A., Dabrowski P.W., Radonic A., Niemann S., Parkhill J., et al. Novel Mycobacterium tuberculosis Complex Isolate from a Wild Chimpanzee. Emerg. Infect. Dis. 2013;19:969–976. doi: 10.3201/eid1906.121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dou H., Lin C., Ch Y., Yang S., Chang J. Lineage-specific SNPs for genotyping of Mycobacterium tuberculosis clinical isolates. Sci. Rep. 2017;7:1425. doi: 10.1038/s41598-017-01580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bainomugisa A., Lavu E., Hiashiri S., Majumdar S., Honjepari A., Moke R., Dakulala P., Hill-cawthorne G.A., Pandey S., Marais B.J., et al. Multi-clonal evolution of multi-drug-resistant/extensively drug- resistant Mycobacterium tuberculosis in a high-prevalence setting of Papua New Guinea for over three decades. Microb. Genom. 2018;4:1–11. doi: 10.1099/mgen.0.000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jajou R., Kohl T.A., Walker T., Norman A., Cirillo D.M., Tagliani E., Niemann S., De Neeling A., Lillebaek T., Anthony R.M., et al. Towards standardisation: Comparison of five whole genome sequencing (WGS) analysis pipelines for detection of epidemiologically linked tuberculosis cases. Eurosurveillance. 2019;24 doi: 10.2807/1560-7917.ES.2019.24.50.1900130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zimpel C.K., Brandão P.E., de Souza Filho A.F., de Souza R.F., Ikuta C.Y., Ferreira Neto J.S., Camargo N.C.S., Heinemann M.B., Guimarães A.M.S. Complete Genome Sequencing of Mycobacterium bovis SP38 and Comparative Genomics of Mycobacterium bovis and M. tuberculosis Strains. Front. Microbiol. 2017;8:2389. doi: 10.3389/fmicb.2017.02389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Faksri K., Xia E., Tan J.H., Teo Y.Y., Ong R.T.H. In silico region of difference (RD) analysis of Mycobacterium tuberculosis complex from sequence reads using RD-Analyzer. BMC Genom. 2016;17:847. doi: 10.1186/s12864-016-3213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hatherell H.A., Colijn C., Stagg H.R., Jackson C., Winter J.R., Abubakar I. Interpreting whole genome sequencing for investigating tuberculosis transmission: A systematic review. BMC Med. 2016;14:21. doi: 10.1186/s12916-016-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee R.S., Behr M.A. Does choice matter? Reference-based alignment for molecular epidemiology of tuberculosis. J. Clin. Microbiol. 2016;54:1891–1895. doi: 10.1128/JCM.00364-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Walter K.S., Colijn C., Cohen T., Mathema B., Liu Q., Bowers J., Engelthaler D.M., Narechania A., Croda J., Andrews J.R. Genomic variant identification methods alter Mycobacterium tuberculosis transmission inference. bioRxiv. 2019:733642. doi: 10.1101/733642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bush S.J., Foster D., Eyre D.W., Clark E.L., De Maio N., Shaw L.P., Stoesser N., Peto T.E.A., Crook D.W., Walker A.S. Genomic diversity affects the accuracy of bacterial SNP calling pipelines. Gigascience. 2020;9 doi: 10.1093/gigascience/giaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dippenaar A., Parsons S.D.C., Miller M.A., Hlokwe T., Gey van Pittius N.C., Adroub S.A., Abdallah A.M., Pain A., Warren R.M., Michel A.L., et al. Progenitor strain introduction of Mycobacterium bovis at the wildlife-livestock interface can lead to clonal expansion of the disease in a single ecosystem. Infect. Genet. Evol. 2017;51:235–238. doi: 10.1016/j.meegid.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 168.Branger M., Loux V., Cochard T., Boschiroli M.L., Biet F., Michelet L. The complete genome sequence of Mycobacterium bovis Mb3601, a SB0120 spoligotype strain representative of a new clonal group. Infect. Genet. Evol. 2020;82:104309. doi: 10.1016/j.meegid.2020.104309. [DOI] [PubMed] [Google Scholar]
- 169.Jandrasits C., Kröger S., Haas W., Renard B.Y. Computational pan-genome mapping and pairwise SNP-distance improve detection of Mycobacterium tuberculosis transmission clusters. bioRxiv. 2019;15:1–20. doi: 10.1371/journal.pcbi.1007527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Olson N.D., Lund S.P., Colman R.E., Foster J.T., Sahl J.W., Schupp J.M., Keim P., Morrow J.B., Salit M.L., Zook J.M. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front. Genet. 2015;6:235. doi: 10.3389/fgene.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Inouye M., Dashnow H., Raven L.A., Schultz M.B., Pope B.J., Tomita T., Zobel J., Holt K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:1–16. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li H., Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Raczy C., Petrovski R., Saunders C.T., Chorny I., Kruglyak S., Margulies E.H., Chuang H.Y., Källberg M., Kumar S.A., Liao A., et al. Isaac: Ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 2013;29:2041–2043. doi: 10.1093/bioinformatics/btt314. [DOI] [PubMed] [Google Scholar]
- 176.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 177.Lunter G., Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Canzar S., Salzberg S.L. Short read mapping: An algorithmic tour. Proc. IEEE Inst. Electr. Electron. Eng. 2017;105:436–458. doi: 10.1109/JPROC.2015.2455551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koboldt D.C., Chen K., Wylie T., Larson D.E., McLellan M.D., Mardis E.R., Weinstock G.M., Wilson R.K., Ding L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv Prepr. 20121207.3907 [Google Scholar]
- 182.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yoshimura D., Kajitani R., Gotoh Y., Katahira K., Okuno M., Ogura Y., Hayashi T., Itoh T. Evaluation of SNP calling methods for closely related bacterial isolates and a novel high-accuracy pipeline: BactSNP. Microb. Genom. 2019;5:1–8. doi: 10.1099/mgen.0.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pouseele H., Supply P. Accurate whole-genome sequencing-based epidemiological surveillance of Mycobacterium tuberculosis. In: Sails A., Tang Y.-W., editors. Current and Emerging Technologies for the Diagnosis of Microbial Infections. Elsevier Ltd; Oxford, UK: 2015. pp. 359–386. [Google Scholar]
- 188.Vargas R., Jr., Freschi L., Marin M., Epperson L.E., Smith M., Oussenko I., Durbin D., Strong M., Salfinger M., Farhat M.R. In-host population dynamics of M. tuberculosis during treatment failure. bioRxiv. 2019;6:726430. doi: 10.7554/eLife.61805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lee R.S., Proulx J.-F., McIntosh F., Behr M.A., Hanage W.P. Previously undetected super-spreading of Mycobacterium tuberculosis revealed by deep sequencing. Elife. 2020;9:e53245. doi: 10.7554/eLife.53245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bryant J.M., Harris S.R., Parkhill J., Dawson R., Diacon A.H., van Helden P., Pym A., Mahayiddin A.A., Chuchottaworn C., Sanne I.M., et al. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: A retrospective observational study. Lancet Respir. Med. 2013;1:786–792. doi: 10.1016/S2213-2600(13)70231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sobkowiak B., Glynn J.R., Houben R.M.G.J., Mallard K., Phelan J.E., Guerra-Assunção J.A., Banda L., Mzembe T., Viveiros M., McNerney R., et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genom. 2018;19:613. doi: 10.1186/s12864-018-4988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Brites D., Loiseau C., Menardo F., Borrell S., Boniotti M.B., Warren R., Dippenaar A., Parsons S.D.C., Beisel C., Behr M.A., et al. A new phylogenetic framework for the animal-adapted Mycobacterium tuberculosis complex. Front. Microbiol. 2018;9:2820. doi: 10.3389/fmicb.2018.02820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Guerra-Assunção J.A., Houben R.M.G.J., Crampin A.C., Mzembe T., Mallard K., Coll F., Khan P., Banda L., Chiwaya A., Pereira R.P.A., et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: A whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J. Infect. Dis. 2015;211:1154–1163. doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sandoval-Azuara S.E., Muñiz-Salazar R., Perea-Jacobo R., Robbe-Austerman S., Perera-Ortiz A., López-Valencia G., Bravo D.M., Sanchez-Flores A., Miranda-Guzmán D., Flores-López C.A., et al. Whole genome sequencing of Mycobacterium bovis to obtain molecular fingerprints in human and cattle isolates from Baja California, Mexico. Int. J. Infect. Dis. 2017;63:48–56. doi: 10.1016/j.ijid.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 195.Bruning-Fann C., Robbe-Austerman S., Kaneene J., Thomsen B., Tilden J.D., Jr., Ray J., Smith R., Fitzgerald S., Bolin S., O’Brien D., et al. Use of whole-genome sequencing and evaluation of the apparent sensitivity and specificity of antemortem tuberculosis tests in the investigation of an unusual outbreak of Mycobacterium bovis infection in a Michigan dairy herd. J. Am. Vet. Med. Assoc. 2017;251:206–216. doi: 10.2460/javma.251.2.206. [DOI] [PubMed] [Google Scholar]
- 196.Walker T.M., Ip C.L., Harrell R.H., Evans J.T., Kapatai G., Dedicoat M.J., Eyre D.W., Wilson D.J., Hawkey P.M., Crook D.W., et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: A retrospective observational study. Lancet Infect. Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kato-Maeda M., Ho C., Passarelli B., Banaei N., Grinsdale J., Flores L., Anderson J., Murray M., Rose G., Kawamura L.M., et al. Use of whole genome sequencing to determine the microevolution of Mycobacterium tuberculosis during an outbreak. PLoS ONE. 2013;8:e58235. doi: 10.1371/journal.pone.0058235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pérez-Lago L., Comas I., Navarro Y., González-Candelas F., Herranz M., Bouza E., García-De-Viedma D. Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: Potential impact on the inference of tuberculosis transmission. J. Infect. Dis. 2014;209:98–108. doi: 10.1093/infdis/jit439. [DOI] [PubMed] [Google Scholar]
- 199.Luo T., Yang C., Peng Y., Lu L., Sun G., Wu J., Jin X., Hong J., Li F., Mei J., et al. Whole-genome sequencing to detect recent transmission of Mycobacterium tuberculosis in settings with a high burden of tuberculosis. Tuberculosis. 2014;94:434–440. doi: 10.1016/j.tube.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Comas I., Hailu E., Kiros T., Bekele S., Mekonnen W., Gumi B., Tschopp R., Ameni G., Hewinson R.G., Robertson B.D., et al. Population Genomics of Mycobacterium tuberculosis in Ethiopia Contradicts the Virgin Soil Hypothesis for Human Tuberculosis in Sub-Saharan Africa. Curr. Biol. 2015;25:3260–3266. doi: 10.1016/j.cub.2015.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Faksri K., Xia E., Ong R.T.-H., Tan J.H., Nonghanphithak D., Makhao N., Thamnongdee N., Thanormchat A., Phurattanakornkul A., Rattanarangsee S., et al. Comparative whole-genome sequence analysis of Mycobacterium tuberculosis isolated from tuberculous meningitis and pulmonary tuberculosis patients. Sci. Rep. 2018;8:4910. doi: 10.1038/s41598-018-23337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Trewby H., Wright D., Breadon E.L., Lycett S.J., Mallon T.R., McCormick C., Johnson P., Orton R.J., Allen A.R., Galbraith J., et al. Use of bacterial whole-genome sequencing to investigate local persistence and spread in bovine tuberculosis. Epidemics. 2016;14:26–35. doi: 10.1016/j.epidem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cohen T., van Helden P.D., Wilson D., Colijn C., McLaughlin M.M., Abubakar I., Warren R.M. Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin. Microbiol. Rev. 2012;25:708–719. doi: 10.1128/CMR.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Egbe N.F., Muwonge A., Ndip L., Kelly R.F., Sander M., Tanya V., Ngwa V.N., Handel I.G., Novak A., Ngandalo R., et al. Molecular epidemiology of Mycobacterium bovis in Cameroon. Sci. Rep. 2017;7:4652. doi: 10.1038/s41598-017-04230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ghielmetti G., Coscolla M., Ruetten M., Friedel U., Loiseau C., Feldmann J., Steinmetz H.W., Stucki D., Gagneux S. Tuberculosis in Swiss captive Asian elephants: Microevolution of Mycobacterium tuberculosis characterized by multilocus variable-number tandem-repeat analysis and whole-genome sequencing. Sci. Rep. 2017;7:14647. doi: 10.1038/s41598-017-15278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu Q., Via L.E., Luo T., Liang L., Liu X., Wu S., Shen Q., Wei W., Ruan X., Yuan X., et al. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci. Rep. 2015;5:17507. doi: 10.1038/srep17507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ssengooba W., de Jong B.C., Joloba M.L., Cobelens F.G., Meehan C.J. Whole genome sequencing reveals mycobacterial microevolution among concurrent isolates from sputum and blood in HIV infected TB patients. BMC Infect. Dis. 2016;16:1–7. doi: 10.1186/s12879-016-1737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lieberman T.D., Wilson D., Misra R., Xiong L.L., Moodley P., Cohen T., Kishony R., Author N.M. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated M. tuberculosis HHS Public Access Author manuscript. Nat. Med. 2016;22:1470–1474. doi: 10.1038/nm.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Roetzer A., Diel R., Kohl T.A., Rückert C., Nübel U., Blom J., Wirth T., Jaenicke S., Schuback S., Rüsch-Gerdes S., et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: A longitudinal molecular epidemiological study. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Biek R., O’Hare A., Wright D., Mallon T., McCormick C., Orton R.J., McDowell S., Trewby H., Skuce R.A., Kao R.R. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathog. 2012;8:e1003008. doi: 10.1371/journal.ppat.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Guerra-Assunção J.A., Crampin A.C., Houben R.M.G.J., Mzembe T., Mallard K., Coll F., Khan P., Banda L., Chiwaya A., Pereira R.P.A., et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015;2015:e05166. doi: 10.7554/eLife.05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Michelet L., Conde C., Branger M., Cochard T., Biet F., Boschiroli M.L. Transmission Network of Deer-Borne Mycobacterium bovis Infection Revealed by a WGS Approach. Microorganisms. 2019:687. doi: 10.3390/microorganisms7120687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Clark T.G., Mallard K., Coll F., Preston M., Assefa S., Harris D., Ogwang S., Mumbowa F., Kirenga B., O’Sullivan D.M., et al. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLoS ONE. 2013;8:e83012. doi: 10.1371/journal.pone.0083012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lee R.S., Radomski N., Proulx J.F., Manry J., McIntosh F., Desjardins F., Soualhine H., Domenech P., Reed M.B., Menzies D., et al. Reemergence and amplification of tuberculosis in the Canadian Arctic. J. Infect. Dis. 2015;211:1905–1914. doi: 10.1093/infdis/jiv011. [DOI] [PubMed] [Google Scholar]
- 215.Walker T.M., Lalor M.K., Broda A., Ortega L.S., Parker L., Churchill S., Bennett K., Golubchik T., Giess A.P., Del C., et al. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007—2012, with whole pathogen genome sequences: An observational study. Lancet Infect. Dis. 2015;2:285–292. doi: 10.1016/S2213-2600(14)70027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Witney A.A., Gould K.A., Arnold A., Coleman D., Delgado R., Dhillon J., Pond M.J., Pope C.F., Planche T.D., Stoker N.G., et al. Clinical application of whole-genome sequencing to inform treatment for multidrug-resistant tuberculosis cases. J. Clin. Microbiol. 2015;53:1473–1483. doi: 10.1128/JCM.02993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Acosta F., Chernyaeva E., Mendoza L., Sambrano D., Correa R., Rotkevich M., Tarté M., Hernández H., Velazco B., de Escobar C., et al. Mycobacterium bovis in Panama, 2013. Emerg. Infect. Dis. 2015;21:1059–1061. doi: 10.3201/eid2106.141821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Outhred A.C., Holmes N., Sadsad R., Martinez E., Jelfs P., Hill-Cawthorne G.A., Gilbert G.L., Marais B.J., Sintchenko V. Identifying likely transmission pathways within a 10-year community outbreak of tuberculosis by high-depth whole genome sequencing. PLoS ONE. 2016;11:e0150550. doi: 10.1371/journal.pone.0150550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kohl T.A., Diel R., Harmsen D., Rothgänger J., Meywald Walter K., Merker M., Weniger T., Niemann S. Whole-genome-based Mycobacterium tuberculosis surveillance: A standardized, portable, and expandable approach. J. Clin. Microbiol. 2014;52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kohl T.A., Harmsen D., Rothgänger J., Walker T., Diel R., Niemann S. Harmonized genome wide typing of tubercle bacilli using a web-based gene-by-gene nomenclature system. EBioMedicine. 2018;34:131–138. doi: 10.1016/j.ebiom.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ridom SeqSphere+ Software. [(accessed on 8 January 2020)]; Available online: https://www.ridom.de/seqsphere/
- 222.Bionumerics Bionumerics for Whole Genome Multi Locus Sequence Typing. [(accessed on 8 January 2020)]; Available online: http://www.applied-maths.com/applications/wgmlst.
- 223.Kavvas E.S., Catoiu E., Mih N., Yurkovich J.T., Seif Y., Dillon N., Heckmann D., Anand A., Yang L., Nizet V., et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat. Commun. 2018;9:4306. doi: 10.1038/s41467-018-06634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Periwal V., Patowary A., Vellarikkal S.K., Gupta A., Singh M., Mittal A., Jeyapaul S., Chauhan R.K., Singh A.V., Singh P.K., et al. Comparative Whole-Genome Analysis of Clinical Isolates Reveals Characteristic Architecture of Mycobacterium tuberculosis Pangenome. PLoS ONE. 2015;10:e0122979. doi: 10.1371/journal.pone.0122979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Anzai E.K. Sequenciamento do Genoma Completo do Mycobacterium bovis Como Instrumento de Sistema de Vigilância no Estado de Santa Catarina. University of São Paulo; Sao Paulo, Brazil: 2019. [Google Scholar]
- 226.Lasserre M., Fresia P., Greif G., Iraola G., Castro-Ramos M., Juambeltz A., Nuñez Á., Naya H., Robello C., Berná L. Whole genome sequencing of the monomorphic pathogen Mycobacterium bovis reveals local differentiation of cattle clinical isolates. BMC Genom. 2018;19:2. doi: 10.1186/s12864-017-4249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Otchere I.D., van Tonder A.J., Asante-Poku A., Sánchez-Busó L., Coscollá M., Osei-Wusu S., Asare P., Aboagye S.Y., Ekuban S.A., Yahayah A.I., et al. Molecular epidemiology and whole genome sequencing analysis of clinical Mycobacterium bovis from Ghana. PLoS ONE. 2019;14:e0209395. doi: 10.1371/journal.pone.0209395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hauer A., Michelet L., Cochard T., Branger M., Nunez J., Boschiroli M.-L., Biet F. Accurate phylogenetic relationships among Mycobacterium bovis strains circulating in France based on whole genome sequencing and single nucleotide polymorphism analysis. Front. Microbiol. 2019;10:955. doi: 10.3389/fmicb.2019.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Patané J.S.L., Martins J., Castelão A.B., Nishibe C., Montera L., Bigi F., Zumárraga M.J., Cataldi A.A., Junior A.F., Roxo E., et al. Patterns and processes of Mycobacterium bovis evolution revealed by phylogenomic analyses. Genome Biol. Evol. 2017;9:521–535. doi: 10.1093/gbe/evx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ypma R.J.F., van Ballegooijen W.M., Wallinga J. Relating phylogenetic trees to transmission trees of infectious disease outbreaks. Genetics. 2013;195:1055–1062. doi: 10.1534/genetics.113.154856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Jombart T., Eggo R.M., Dodd P.J., Balloux F. Reconstructing disease outbreaks from genetic data: A graph approach. Heredity. 2011;106:383–390. doi: 10.1038/hdy.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zojer M., Schuster L.N., Schulz F., Pfundner A., Horn M., Rattei T. Variant profiling of evolving prokaryotic populations. Peer J. 2017;5:e2997. doi: 10.7717/peerj.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Persi E., Wolf Y.I., Koonin E.V. Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins. Nat. Commun. 2016;7:13570. doi: 10.1038/ncomms13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:1–16. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Laehnemann D., Borkhardt A., McHardy A.C. Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief. Bioinform. 2016;17:154–179. doi: 10.1093/bib/bbv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Fu S., Wang A., Au K.F. A comparative evaluation of hybrid error correction methods for error-prone long reads. Genome Biol. 2019;20:1–17. doi: 10.1186/s13059-018-1605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Coll F., Mallard K., Preston M.D., Bentley S., Parkhill J., McNerney R., Martin N., Clark T.G. SpolPred: Rapid and accurate prediction of Mycobacterium tuberculosis spoligotypes from short genomic sequences. Bioinformatics. 2012;28:2991–2993. doi: 10.1093/bioinformatics/bts544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Xia E., Teo Y.-Y., Ong R.T.-H. SpoTyping: Fast and accurate in silico Mycobacterium spoligotyping from sequence reads. Genome Med. 2016;8:19. doi: 10.1186/s13073-016-0270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Guyeux C., Sola C., Refrégier G. Exhaustive reconstruction of the CRISPR locus in Mycobacterium tuberculosis complex using short reads. bioRxiv. 2019 doi: 10.1101/844746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rajwani R., Shehzad S., Siu G.K.H. MIRU-profiler: A rapid tool for determination of 24-loci MIRU-VNTR profiles from assembled genomes of Mycobacterium tuberculosis. Peer J. 2018;6:e5090. doi: 10.7717/peerj.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tang C.Y., Ong R.T.H. MIRUReader: MIRU-VNTR typing directly from long sequencing reads. Bioinformatics. 2020;36:1625–1626. doi: 10.1093/bioinformatics/btz771. [DOI] [PubMed] [Google Scholar]
- 242.Ford C.B., Lin P.L., Chase M.R., Shah R.R., Iartchouk O., Galagan J., Mohaideen N., Ioerger T.R., Sacchettini J.C., Lipsitch M., et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat. Genet. 2011;43:482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Food and Agriculture Organization of the United Nations (FAO) Challanges of Animal Health Information Systems And Surveillance for Animal Diseases And Zoonoses. FAO; Rome, Italy: 2011. pp. 1–124. FAO Animal Profuction and Health Proceedings. [Google Scholar]
- 244.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pickett B.E., Greer D.S., Zhang Y., Stewart L., Zhou L., Sun G., Gu Z., Kumar S., Zaremba S., Larsen C.N., et al. Virus Pathogen Database and Analysis Resource (ViPR): A Comprehensive Bioinformatics Database and Analysis Resource for the Coronavirus Research Community. Viruses. 2012;4:3209–3226. doi: 10.3390/v4113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Timme R.E., Rand H., Leon M.S., Hoffmann M., Strain E., Allard M., Roberson D., Baugher J.D. GenomeTrakr proficiency testing for foodborne pathogen surveillance: An exercise from 2015. Microb. Genom. 2018;4 doi: 10.1099/mgen.0.000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Comin A., Grewar J., van Schaik G., Schwermer H., Paré J., El Allaki F., Drewe J., Lopes Antunes A.C., Estberg L., Horan M., et al. Development of reporting guidelines for animal health surveillance—AHSURED. Front. Vet. Sci. 2019;6:426. doi: 10.3389/fvets.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Gardy J.L., Johnston J.C., Sui S.J.H., Cook V.J., Shah L., Brodkin E., Rempel S., Moore R., Zhao Y., Holt R., et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 249.Jajou R., de Neeling A., van Hunen R., de Vries G., Schimmel H., Mulder A., Anthony R., van der Hoek W., van Soolingen D. Epidemiological links between tuberculosis cases identified twice as efficiently by whole genome sequencing than conventional molecular typing. PLoS ONE. 2018;13:e0195413. doi: 10.1371/journal.pone.0195413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Price-Carter M., Rooker S., Collins D.M. Comparison of 45 variable number tandem repeat (VNTR) and two direct repeat (DR) assays to restriction endonuclease analysis for typing isolates of Mycobacterium bovis. Vet. Microbiol. 2011;150:107–114. doi: 10.1016/j.vetmic.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 251.Collins D.M. DNA typing of Mycobacterium bovis strains from the castlepoint area of the Wairarapa. N. Z. Vet. J. 1999;47:207–209. doi: 10.1080/00480169.1999.36145. [DOI] [PubMed] [Google Scholar]
- 252.Sun Z., Cao R., Tian M., Zhang X., Zhang X., Li Y., Xu Y., Fan W., Huang B., Li C. Evaluation of Spoligotyping and MIRU-VNTR for Mycobacterium bovis in Xinjiang, China. Res. Vet. Sci. 2012;92:236–239. doi: 10.1016/j.rvsc.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 253.Rodriguez-Campos S., Aranaz A., De Juan L., Sáez-Llorente J.L., Romero B., Bezos J., Jiménez A., Mateos A., Domínguez L. Limitations of spoligotyping and variable-number tandem-repeat typing for molecular tracing of Mycobacterium bovis in a high-diversity setting. J. Clin. Microbiol. 2011;49:3361–3364. doi: 10.1128/JCM.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Biek R., Pybus O.G., Lloyd-Smith J.O., Didelot X. Measurably evolving pathogens in the genomic era. Trends Ecol. Evol. 2015;30:306–313. doi: 10.1016/j.tree.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Casali N., Broda A., Harris S.R., Parkhill J., Brown T., Drobniewski F. Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: A retrospective observational study. PLoS Med. 2016;13:e1002137. doi: 10.1371/journal.pmed.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Bryant J.M., Schürch A.C., van Deutekom H., Harris S.R., de Beer J.L., de Jager V., Kremer K., van Hijum S.A.F.T., Siezen R.J., Borgdorff M., et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect. Dis. 2013;13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Nikolayevskyy V., Kranzer K., Niemann S., Drobniewski F. Whole genome sequencing of Mycobacterium tuberculosis for detection of recent transmission and tracing outbreaks: A systematic review. Tuberculosis. 2016;98:77–85. doi: 10.1016/j.tube.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 258.Coscolla M., Gagneux S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin. Immunol. 2014;26:431–444. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Brites D., Gagneux S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol. Rev. 2015;264:6–24. doi: 10.1111/imr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Berg S., Garcia-Pelayo M.C., Müller B., Hailu E., Asiimwe B., Kremer K., Dale J., Boniotti M.B., Rodriguez S., Hilty M., et al. African 2, a clonal complex of Mycobacterium bovis epidemiologically important in East Africa. J. Bacteriol. 2011;193:670–678. doi: 10.1128/JB.00750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Müller B., Hilty M., Berg S., Garcia-Pelayo M.C., Dale J., Boschiroli M.L., Cadmus S., Ngandolo B.N.R., Godreuil S., Diguimbaye-Djaibé C., et al. African 1, an epidemiologically important clonal complex of Mycobacterium bovis dominant in Mali, Nigeria, Cameroon, and Chad. J. Bacteriol. 2009;191:1951–1960. doi: 10.1128/JB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Smith N.H., Berg S., Dale J., Allen A., Rodriguez S., Romero B., Matos F., Ghebremichael S., Karoui C., Donati C., et al. European 1: A globally important clonal complex of Mycobacterium bovis. Infect. Genet. Evol. 2011;11:1340–1351. doi: 10.1016/j.meegid.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 263.Rodriguez-Campos S., Schürch A.C., Dale J., Lohan A.J., Cunha M.V., Botelho A., De Cruz K., Boschiroli M.L., Boniotti M.B., Pacciarini M., et al. European 2—A clonal complex of Mycobacterium bovis dominant in the Iberian Peninsula. Infect. Genet. Evol. 2012;12:866–872. doi: 10.1016/j.meegid.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 264.Rodríguez S., Bezos J., Romero B., de Juan L., Álvarez J., Castellanos E., Moya N., Lozano F., Tariq Javed M., Sáez-Llorente J.L., et al. Mycobacterium caprae infection in livestock and wildlife, Spain. Emerg. Infect. Dis. 2011;17:532–535. doi: 10.3201/eid1703.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pate M., Švara T., Gombač M., Paller T., Žolnir-Dovč M., Emeršič I., Prodinger W.M., Bartoš M., Zdovc I., Krt B., et al. Outbreak of tuberculosis caused by Mycobacterium caprae in a zoological garden. J. Vet. Med. Ser. B Infect. Dis. Vet. Public Heal. 2006;53:387–392. doi: 10.1111/j.1439-0450.2006.01000.x. [DOI] [PubMed] [Google Scholar]
- 266.Krajewska M., Zabost A., Welz M., Lipiec M., Orłowska B., Anusz K., Brewczyński P., Augustynowicz-Kopeć E., Szulowski K., Bielecki W., et al. Transmission of Mycobacterium caprae in a herd of European bison in the Bieszczady Mountains, Southern Poland. Eur. J. Wildl. Res. 2015;61:429–433. doi: 10.1007/s10344-015-0912-x. [DOI] [Google Scholar]
- 267.Sahraoui N., Müller B., Guetarni D., Boulahbal F., Yala D., Ouzrout R., Berg S., Smith N.H., Zinsstag J. Molecular characterization of Mycobacterium bovis strains isolated from cattle slaughtered at two abattoirs in Algeria. BMC Vet. Res. 2009;5:4. doi: 10.1186/1746-6148-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Yahyaoui-Azami H., Aboukhassib H., Bouslikhane M., Berrada J., Rami S., Reinhard M., Gagneux S., Feldmann J., Borrell S., Zinsstag J. Molecular characterization of bovine tuberculosis strains in two slaughterhouses in Morocco. BMC Vet. Res. 2017;13:272. doi: 10.1186/s12917-017-1165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yoshida S., Suga S., Ishikawa S., Mukai Y., Tsuyuguchi K., Inoue Y., Yamamoto T., Wada T., Study T. Mycobacterium caprae infection in a captive Borneo elephant, Japan. Emerg. Infect. Dis. 2018;24:1937–1940. doi: 10.3201/eid2410.180018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Duffy S.C., Srinivasan S., Schilling M.A., Stubre T., Danchuk S.N., Michael J.S., Venkatesan M., Bansal N., Mann S., Jindal N., et al. Zoonotic tuberculosis in India: Looking beyond Mycobacterium bovis. bioRxiv. 2019 doi: 10.1101/847715. [DOI] [Google Scholar]
- 271.van Ingen J., Rahim Z., Mulder A., Boeree M.J., Simeone R., Brosch R., van Soolingen D. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg. Infect. Dis. 2012;18:653–655. doi: 10.3201/eid1804.110888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Rahim Z., Thapa J., Fukushima Y., van der Zanden A.G.M., Gordon S.V., Suzuki Y., Nakajima C. Tuberculosis caused by Mycobacterium orygis in dairy cattle and captured monkeys in Bangladesh: A new scenario of tuberculosis in South Asia. Transbound. Emerg. Dis. 2017;64:1965–1969. doi: 10.1111/tbed.12596. [DOI] [PubMed] [Google Scholar]
- 273.O’Halloran C., Hope J.C., Dobromylskyj M., Burr P., McDonald K., Rhodes S., Roberts T., Dampney R., De la Rua-Domenech R., Robinson N., et al. An outbreak of tuberculosis due to Mycobacterium bovis infection in a pack of English Foxhounds (2016–2017) Transbound. Emerg. Dis. 2018;65:1872–1884. doi: 10.1111/tbed.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.McGill I., Saunders R., Eastwood B., Menache A., Dalzell F., Hill S., Irving B., Knight A., Jones M. Mycobacterium bovis tuberculosis in hunting hounds. Vet. Rec. 2018;183:387–388. doi: 10.1136/vr.k4053. [DOI] [PubMed] [Google Scholar]
- 275.Miller M.A., Buss P., Roos E.O., Hausler G., Dippenaar A., Mitchell E., van Schalkwyk L., Robbe-Austerman S., Waters W.R., Sikar-Gang A., et al. Fatal tuberculosis in a free-ranging African elephant and one health Implications of human pathogens in wildlife. Front. Vet. Sci. 2019;6:18. doi: 10.3389/fvets.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zachariah A., Pandiyan J., Madhavilatha G.K., Mundayoor S., Chandramohan B., Sajesh P.K., Santhosh S., Mikota S.K. Mycobacterium tuberculosis in wild Asian elephants, southern India. Emerg. Infect. Dis. 2017;23:504–506. doi: 10.3201/eid2303.161741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Cui H.H., Erkkila T., Chain P.S.G., Vuyisich M. Building international genomics collaboration for global health security. Front. Public Heal. 2015;3:264. doi: 10.3389/fpubh.2015.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.