Abstract
Innate lymphoid cells (ILCs) are critical to effective immune surveillance against pathogens, have malignant counterparts, and contribute to disease. Thus, it is important to understand ILC development. All ILCs are derived from the common lymphoid progenitor cell; however, the exact mechanisms and signals that initiate their divergence from T cells, B cells and one and other are incompletely understood. Evidence now supports a stepwise developmental process that includes distinct cellular intermediates, progressively narrowed differentiation, and some plasticity. While the current models of human and murine ILC development share many similarities, they also include some distinct differences. Together these findings have established a working dynamic model of ILC development.
Introduction
Innate lymphoid cells (ILCs) are an increasingly important component of both normal immunity and pathologic processes. Collectively, the ILC family is composed of developmentally related but functionally distinct groups of cells similar to the relationships between T cell subtypes. Mature ILCs are categorized into three groups based on their immunophenotypes such that Group 1 ILCs include non-cytotoxic ILC1s that express Tbx21 (Tbet) transcription factor (TF), as well as cytotoxic natural killer (NK) cells that also express Eomesodermin (Eomes), and both produce interferon (IFN)-γ; Group 2 ILCs consist of ILC2s that produce interleukin (IL)-5 and −13 and express GATA-3 and RAR-related orphan receptor (ROR)α TFs; and Group 3 ILCs include NK cell receptor (NCR)+ and NCR- ILC3s and lymphoid tissue inducer (LTi) cells that produce IL-22 and/or IL-17 and express RORγt and the aryl hydrocarbon receptor (AHR) TFs [1]. Physiologically, ILCs play significant roles in anti-viral, bacterial and helminth infections; they are involved in metabolic regulation of fat; and they provide immunity to cancer [2]. Their roles in disease processes continue to expand and thus our ability to understand the regulation of their development and activation pathways will provide novel avenues toward disease prevention and treatment.
Developmentally, all ILCs derive from the common lymphoid progenitor (CLP), which is capable of generating all lymphocyte subsets including ILCs as well as B and T cells. This relationship was first established when mice lacking Ikaros or the common gamma chain were found to be deficient in NK, B and T cells[3,4]. Furthermore, isolation of the multipotent CLP in mice and in humans was definitive proof that these innate and adaptive cells were derived through the same lymphoid pathway [5,6]. Although there is an abundance of similarity between mouse and human ILC development, there is evidence for some key differences in their precursor populations and pathways. Therefore, in this review we discuss mouse and human ILC development separately.
Murine ILC development
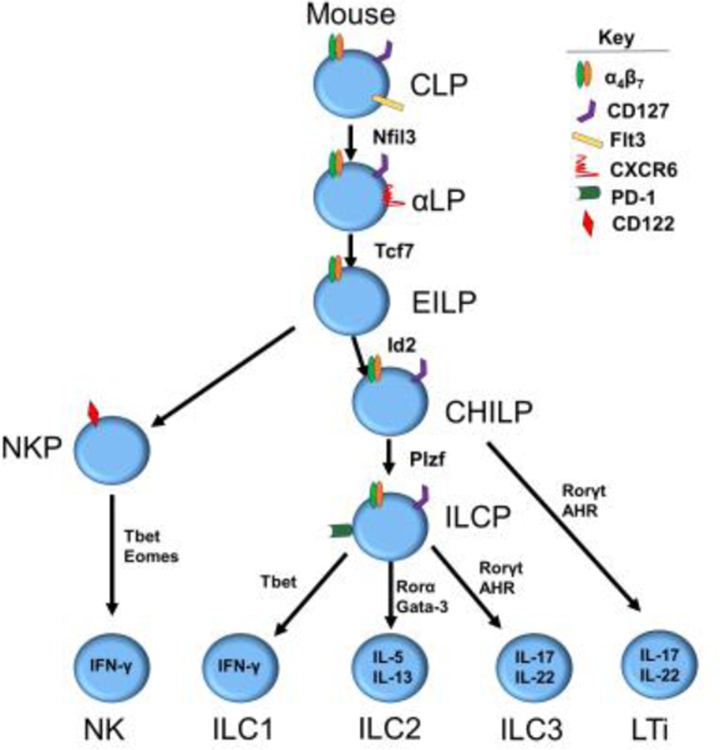
Discoveries related to murine ILC development have been augmented by robust genetic tools and in vitro clonal culture systems (Table 1) that have assisted in the identification of individual progenitors, associated TFs, and signals required for development downstream of the CLP (Figure 1). Two distinct progenitors downstream of the murine CLP have been identified, each with restricted ILC potential. These include the α-lymphoid precursor (αLP) and the early innate lymphoid progenitor (EILP) that along with the other downstream progenitors are most prevalent in murine BM [7]. The αLP was first discovered in part through the Nfil3 knockout models that demonstrated a loss of a specific CXCR6+α4β7+CD127+ cell subset (i.e., the αLP subset), in addition to the loss of the EILP and the absence of all mature ILCs, including NK cells [8]. The EILP, identified as Lin-Thy1.2-CD127-α4β7+ most notably differs from the αLP in that it lacks expression of CD127, a common marker of all most ILCs, but otherwise appears to have an identical lineage potential [9]. Identification of the EILP required labeling of the Tcf7 gene, an HMG box-containing TF, with a GFP-reporter and mice deficient in this gene lacked all ILC populations. When purified and cultured the EILP gives rise to a more restricted progenitor known as the common helper ILC progenitor (CHILP), that is functionally distinct in that it lacks NK cell developmental potential yet maintains helper ILC (ILC1, ILC2, ILC3 and LTi) potential [10]. The exact relationship and importance of the αLP and the EILP, was recently explored by Harly et al. They utilized an αLP armed with Tcf7EGFP in addition to fate mapped CD127YFP to show that GFP+ EILPs are indeed CD127 fate map positive, suggesting they originate from the CD127+ αLP that subsequently express Tcf7 [11]. These data suggest that the CD127+ αLP gives rise to the CD127- EILP, that in turn gives rise to the CD127+ CHILP (Figure 1). An additional advancement in early murine ILC development was the discovery that these multipotent ILC progenitor cells begin to acquire mature ILC fates at this early development stage. Seillet et al. recently performed extensive expression analysis on the CLP, αLP, and ILC2 progenitor (ILC2P) and found that certain subsets of the αLP express TFs associated with some mature ILC progenitors suggesting that lineage decisions, known as lineage priming, are occurring at this early stage [7,12]. Therefore, by some yet-to-be-defined mechanism, TFs that define terminal ILCs are being expressed early in development and impacting the lineage potential of that particular cell.
Table 1: Murine ILC progenitors in vitro analysis.
This table represents insight into the single cell in vitro culture conditions used in mice to discover lineage restriction and subsequent results.
| Cell | Identification (Lin-) | Progenitor Location | Transcription factors | Stromal Cell use | Length of culture (days) | Culture Cytokines | NK cell | ILC1 | ILC2 | ILC3 | Refs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| αLP | CXCR6+α4β7+CD127+ | BM | Tox, Nfil3 | OP9-DL1 | 21 | KL, IL-7 | NK1.1+Eomes+Tbet+ | NK1.1+Eomes-Tbet+ | CD127+Gata3+ Rorγt- | CD127+Gata3-Rorγt+ | 8 |
| EILP | Thy1.2-α4β7+CD127- | BM | Tox, Nfil3, Tcf7 | OP9 | 10 | KL, IL-7, IL-2 | NK1.1+Gata3-NKp46+DX5+ | NK1.1+Gata3-NKp46+DX5- | NK1.1-Gata3+ | NK1.1-Gata3-NKp46-Rorγt+ | 9 |
| CHILP | CD117+α4β7+CD127+CD25-Id2+ | BM | Tox, Tcf7, Id2 | OP9-DL1 | 14 | KL, IL-7 | NK1.1+Eomes+** | NKp46+Tbet+Rorγt- | NKp46-Gata3+Rorγt- | NKp46+/-Rorγt+ | 10 |
| ILCP | CD127+α4β7+Plzf+ | BM | Tox, Tcf7, Id2, Plzf | OP9, OP9-DL1 | 10–12 | KL, IL-7 | NK1.1+CD49a-CD49b+** | NK1.1+Bcl11b- | NK1.1-Bcl11b+ | NK1.1-Bc11b-Rorγt+ | 15, 16 |
indicates population was not tested for in single cell cultures
Figure 1.
Current pathways of murine ILC development
The CHILP, identified as Lin-Id2+CD127+CD25-α4β7+, was first discovered through its high levels of Id2 expression. Id2 is a helix-loop-helix TF that has been shown to suppress the E2A TF essential for B and T cell development [13,14]. Furthermore, the CHILP can be further separated based on expression of the Plzf TF. The Plzf+ subset loses its ability to produce LTi’s compared to the Plzf- subset [10,15]. Subsequent work has shown that the Plzf+ CHILP, also referred to as the ILC precursor (ILCp) can be identified through expression of the cell surface marker Pd-1 [16]. The identification of these latter intermediates suggests that downstream of the αLP and EILP, differentiation of helper ILCs is independent of NK development. Consistent with this, the previously described NK1.1-CD122+ NK progenitor (NKP) population [17] is absent in Tcf-1 knockout mice, but still exists in models where helper ILC lineages are disrupted such as seen in Id2 and Tox deficient mice [9,14,18]. Collectively, these published data support a widely accepted and established model of murine ILC development in which the NK cell developmental pathway diverges early from the helper ILC developmental pathways (Figure 1). Nonetheless, we note that few NK1.1+CD49a-CD49b+ NK cells were reportedly produced by the putative more committed helper ILCPs in both the Constantinides et al. and Yu et al studies [15,16]. Therefore, additional studies are likely warranted to investigate the pathways of ILC development in mice.
Human ILC Development
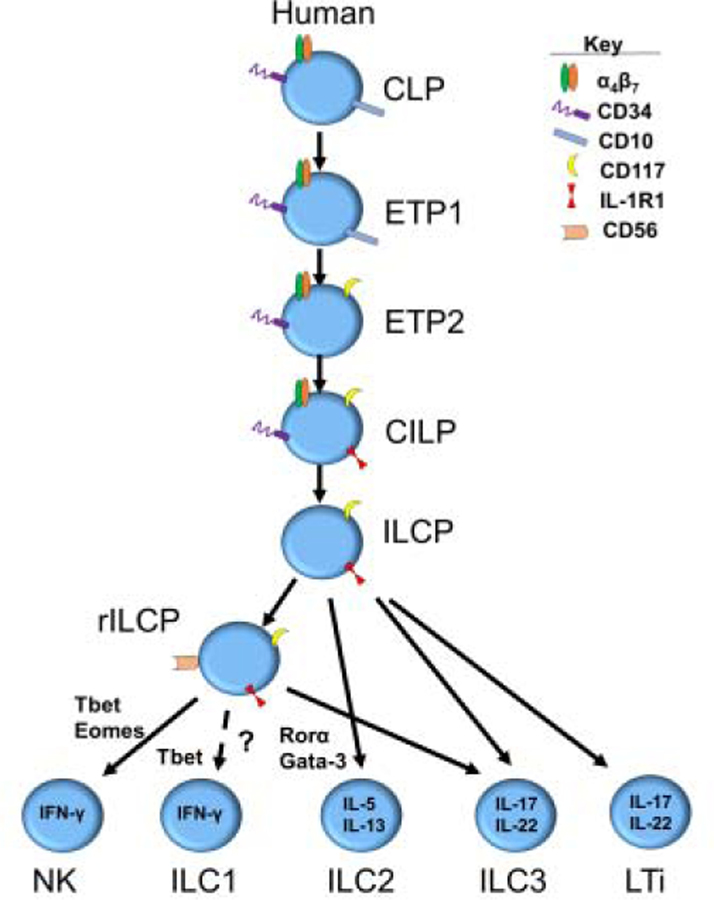
The study of human ILC development is often limited by the lack of comparable genetic and other tools that are available in animal models of lymphoid development. Nonetheless, substantial progress has been made in understanding human ILC development, sometimes inspired by and/or validated by murine studies. As was mentioned earlier murine and human development share the CLP, identified in humans as Lin-CD34+CD45RA+CD10+CD117- and as the starting point for ILC differentiation [5]. A useful framework for investigating human ILC development stems from a pre-existing model of human NK cell development based on the identification and characterization of human NK cell developmental intermediates (NKDIs) prior to the discovery of non-NK helper ILCs [22]. According to the human NK cell development model, NKDIs differentiate at least within secondary lymphoid tissues, including tonsils and lymph nodes, through functionally distinct stages based on the differential expression of multiple surface markers and TFs [23]. The most immature NKDI in this pathway, originally described as “stage 1” and “stage 2” cells by Freud et al [24], are now defined as early tonsillar progenitors (ETP) within the context of non-NK ILCs, and are identified as Lin-CD34+CD10+CD117- (ETP1) and Lin-CD34+CD10-CD117+ (ETP2), respectively. Both populations were originally found to be multipotent and to also give rise to T cells and dendritic cells (DCs) under supportive experimental conditions in vitro in bulk [24]. These data raised the possibility that each progenitor cell may be multipotent or that clonally-restricted subsets exist within each ETP population. In light of this, a recent report by Renoux et al. described a CD34+CD45RA+CD10+CD7+CD38+CD123-CD127- putatively committed NKP population [25]. Notably, however, the immunophenotype of this population overlaps with ETP1s [24], and in a later study by Chen et al. the NKPs described by Renoux et al were found to be multipotent in bulk in vitro cultures [25, 28]. Montaldo et al., described an RORγt+ population that was sorted ex vivo with the same phenotype (CD34+CD117+) as the ETP2, showing these cells preferentially gave rise to ILC3s in vitro under the conditions tested (Table 2) [26]. The authors of that study concluded that tonsillar CD34+CD117+ cells represent lineage restricted ILC3 progenitors; however, evaluation for ILC1, ILC2, T cell, or DC potential was not reported in the study, and other reports have found this population to be multipotent [24,29]. More recently, the ETP2 was found to be heterogeneous with respect to expression of the IL-1 receptor 1 (IL-1R1). The ETP2 IL-1R1- population was found to be multipotent, although more primed for ILC development compared to the ETP1, as it gave rise to proportionally more ILCs than T cells and DCs. The ETP2 IL-1R1+ population however lacks all non-ILC potential and was shown to only give rise to ILCs in vitro and in vivo in the conditions tested (Table 2). Therefore, the ETP2 IL-1R1+ cell population is developmentally the earliest known committed common innate lymphoid progenitor (CILP) that has been discovered in humans.
Table 2: Human ILC progenitors in vitro analysis.
This table represents insight into the single cell in vitro culture conditions used in humans to determine lineage restriction and subsequent results.
| Cell | Identification (Lin-) | Transcription factors | Stromal cell use | Length of culture (days) | Cytokines | NK cell | ILC1 | ILC2 | ILC3 | Refs |
|---|---|---|---|---|---|---|---|---|---|---|
| ETP1 | CD34+CD117-CD94-CD10+ | Tox | OP9-DL1 | 28 | IL-3, IL-7, KL, FL, IL-15 | CD94+IFN-γ+ | CD94-CD161+IFN-γ+ | CD94-CD161+IL-13+ | CD94-CD161+IL-22+ | 22, 29 |
| ETP2 | CD34+CD117+IL-1R1-CD94- | Tox, Id2, Nfil3 | OP9-DL1 | 28 | IL-3, IL-7,KL, FL, IL-15 | CD94+IFN-γ+ | CD94-CD161+IFN-γ+ | CD94-CD161+IL-13+ | CD94-CD161+IL-22+ | 22, 29 |
| CILP | CD34+CD117+IL-1R1+CD94- | Tox, Id2, Nfil3, AHR, Rorγt | OP9-DL1 | 28 | IL-3, IL-7, KL, FL, IL-15 | CD94+IFN-γ+ | CD94-CD161+IFN-γ+ | CD94-CD161+IL-13+ | CD94-CD161+IL-22+ | 22, 29 |
| ILCP | CD34-CD117+IL-1R1+CD94- | Tox, Tcf7, Id2, Plzf | OP9, OP9-DL4 | 14–18 | IL-2, IL-7, IL-1β, IL-23 | CD7+CD94+Eomes+* | CD7+CD94-Eomes-IFN-γ+* | CD7+CD94-IL-13+ | CD94-CD7+IL-22+and/or CD7+IL-17A+ | 27 |
| rILCP | CD34-CD117+IL-1R1+CD56+ | Tcf7, Plzf | OP9-DL1 | 28 | IL-2, IL-7, KL, FL | CD94+ | ** | CD94-CD294+ | CD94-NKp44+ | 28 |
Indicates populations were not distinguished from one another in single cell cultures
Indicates population was not detected or tested for during this study
The most recent developmental work has been within the CD34- fraction of ILC progenitors that are downstream of the CILP. One particularly relevant finding was by Lim et al. who identified a Lin-CD34-CD7+CD127+CD117+ population of ILCPs in cord blood, peripheral blood, fetal liver and other human tissues that could only generate all ILCs [27]. Building on these prior intermediates, Chen et al. recently discovered a CD56+ subset of CD34-CD117+ ILCPs with more restricted potential for Group 1 and 3 ILCs in that it could generate NK cells and ILC3s but not ILC2s; here we refer to this subset as the restricted ILCP (rILCP) [28]. In this report the authors also demonstrated that CILPs could generate ILCPs and rILCPs and furthermore that ILCPs could generate rILCPs, thus supporting a more refined model for human ILC development in secondary lymphoid tissues (Figure 2). Finally, evidence for lineage priming may also occur in early human lymphoid developmental intermediates as clonal work from ETPs, CILPs, ILCPs, and rILCPs has demonstrated bias towards specific ILC lineages [27,29].
Figure 2.
Current pathways of human ILC development
Updates on differentiation and identification of human and murine mature ILCs
When initially proposed, each ILC group was neatly classified based on the basis of TF expression, immunophenotypic markers, and functional abilities. Work since that time has continued to refine these initial models to accommodate the most recent findings. As such, our understanding of ILCs is proving to be dynamic, and plasticity among ILC populations is becoming increasingly apparent. Indeed, through the years consistent distinctions have been found within ILC subsets once thought to be homogenous populations. Examples of this include identification of natural cytotoxicity receptor (NCR) expression or the lack thereof in ILC3 phenotypes (NKp44 in humans and NKp46 in mice) [30–33]; the identification of human CD103+ intraepithelial (ie) ILC1s that express perforin and granzyme and are cytotoxic [34]; and the plasticity of ILC1 and ILC2s in generating ILC3s and vice versa [35–37]. In addition, Simoni et al. utilized mass cytometry (cytometry by time of flight or CyTOF) to investigate both normal and diseased tissues from human cord blood, tonsils, colon, omentum, and lung [38]. This comprehensive approach showed that ILC2s and ILC3s cannot alone be identified by GATA-3 and RORγt expression, respectively, as these TFs are not exclusive to those subsets, verifying previous studies [27,29,39,40]. This is not to say that TFs are not enriched within populations, but more to say that ILCs cannot therefore be identified solely based on the qualitative existence of certain TFs. Collectively, these studies capture the dynamic field of ILC development and biology.
Finally, a multitude of studies and commentaries surround the identity and existence of the ILC1 population. Challenging the existence of ILC1s in humans, Simoni et al, could not isolate ILC1-like cells similar to those described in other studies but found this population to consist of multiple lineages including T cells, CD34+ progenitors, DCs, ILC3, and NK cells [38]. This finding raises an important issue of how to define NK cells and ILC1s, especially as there has yet to be an identifying surface marker or TF that specifically distinguishes ILC1s from NK cells in humans or in mice [28,41–45]. Furthermore, and in support of ILC1 existence and importance in humans, Gao et al. have shown that ILC1s can derive from NK cells through immune-suppressive mechanisms that are operative in the tumor microenvironment [46]. Further, the NK cell phenotype can be altered following activation, such as has been documented with CD16 [47,48]. Additional work must be completed to determine how human ILC1s represent a distinct entity and if so, under what conditions.
Conclusions
A wealth of knowledge has been accumulated in recent years regarding the intricacies of both mouse and human ILC development. From identifying specific multipotent progenitor populations as well as lineage restricted precursor populations, to refining the identities of novel subsets of mature ILCs, the field is continuing to expand the knowledge surrounding ILC biology. One exciting aspect of this continued effort is the development of new methods with which to investigate these issues such as gene editing [49] and the recent development by Lopez-Lastra et al whereby human lymphoid progenitors can develop into each ILC subset in mice [50]. These efforts should further our understanding of ILC development, which in turn will provide insight for the prevention and treatment of human disease.
Highlights.
ILCs develop stepwise through intermediates with increasingly restricted potential
Murine studies have shown that NK cells develop separately from helper ILCs
Human ILCs have a more common pathway for ILCs that includes NK cells
ILCs have plasticity and are able to convert to other ILC populations
Acknowledgements
This work was supported by grants from the National Institutes of Health/National Cancer Institute (CA095426, CA210087, CA163205, CA16058, and CA068458 to M.A.C. and CA199447 and CA208353 to A.G.F.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
*special interest
**outstanding interest
- 1.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. : Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol 2013, 13:145–149. [DOI] [PubMed] [Google Scholar]
- 2.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, et al. : Innate Lymphoid Cells: 10 Years On. Cell 2018, 174:1054–1066. [DOI] [PubMed] [Google Scholar]
- 3.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A: The Ikaros gene is required for the development of all lymphoid lineages. Cell 1994, 79:143–156. [DOI] [PubMed] [Google Scholar]
- 4.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET: Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity 1995, 2:223–238. [DOI] [PubMed] [Google Scholar]
- 5.Galy A, Travis M, Cen D, Chen B: Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity 1995, 3:459–473. [DOI] [PubMed] [Google Scholar]
- 6.Kondo M, Weissman IL, Akashi K: Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997, 91:661–672. [DOI] [PubMed] [Google Scholar]
- **7.Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, Almeida FF, Shi W, Ritchie ME, Naik SH, Huntington ND, et al. : Deciphering the Innate Lymphoid Cell Transcriptional Program. Cell Rep 2016, 17:436–447.Using single cell sequencing analysis of murine ILC progenitors the authors show key relationships and transcriptional profiles each population. They also identified PD-1 as cell surface marker of the PLZF+ CHILP.
- 8.Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, Hooper LV: The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife 2014, 3. [DOI] [PMC free article] [PubMed]
- 9.Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, Wang H, Wang X, Yu S, Zhou X, et al. : TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol 2015, 16:1044–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klose CS, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. : Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 2014, 157:340–356. [DOI] [PubMed] [Google Scholar]
- **11.Harly C, Cam M, Kaye J, Bhandoola A: Development and differentiation of early innate lymphoid progenitors. J Exp Med 2018, 215:249–262.This is the first study to truly define the progenitor-progeny relationship of each known progenitor population including the αLP, EILP, and ILCPs in mice.
- 12.Zook EC, Kee BL: Development of innate lymphoid cells. Nat Immunol 2016, 17:775–782. [DOI] [PubMed] [Google Scholar]
- 13.Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P: Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 1999, 397:702–706. [DOI] [PubMed] [Google Scholar]
- 14.Boos MD, Yokota Y, Eberl G, Kee BL: Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med 2007, 204:1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A: A committed precursor to innate lymphoid cells. Nature 2014, 508:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, Brandt C, Kane L, Campos LS, Lu L, et al. : Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its developmental pathway. Nature 2016, 539:102–106.The authors demonstrate that PD-1 is a key marker of early progenitors as well as activated ILCs in mice. Furthermore, they demonstrate the clinical potential of anti-PD-1 antibodies in depleting ILCs.
- 17.Carotta S, Pang SH, Nutt SL, Belz GT: Identification of the earliest NK-cell precursor in the mouse BM. Blood 2011, 117:5449–5452. [DOI] [PubMed] [Google Scholar]
- 18.Seehus CR, Aliahmad P, de la Torre B, Iliev ID, Spurka L, Funari VA, Kaye J: The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat Immunol 2015, 16:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.Zhong C, Zhu J: Transcriptional regulators dictate innate lymphoid cell fates. Protein Cell 2017, 8:242–254.A recent detailed review of several key transcription factors associated with ILC development.
- 20.Das A, Harly C, Yang Q, Bhandoola A: Lineage specification in innate lymphocytes. Cytokine Growth Factor Rev 2018. [DOI] [PMC free article] [PubMed]
- 21.Ishizuka IE, Constantinides MG, Gudjonson H, Bendelac A: The Innate Lymphoid Cell Precursor. Annu Rev Immunol 2016, 34:299–316. [DOI] [PubMed] [Google Scholar]
- 22.Freud AG, Caligiuri MA: Human natural killer cell development. Immunol Rev 2006, 214:56–72. [DOI] [PubMed] [Google Scholar]
- 23.Scoville SD, Freud AG, Caligiuri MA: Modeling Human Natural Killer Cell Development in the Era of Innate Lymphoid Cells. Front Immunol 2017, 8:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **24.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA: Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med 2006, 203:1033–1043.This is the first study to characterize human NK cell development in secondary lymphoid tissue.
- 25.Renoux VM, Zriwil A, Peitzsch C, Michaëlsson J, Friberg D, Soneji S, Sitnicka E: Identification of a Human Natural Killer Cell Lineage-Restricted Progenitor in Fetal and Adult Tissues. Immunity 2015, 43:394–407. [DOI] [PubMed] [Google Scholar]
- 26.Montaldo E, Teixeira-Alves LG, Glatzer T, Durek P, Stervbo U, Hamann W, Babic M, Paclik D, Stölzel K, Gröne J, et al. : Human RORγt(+)CD34(+) cells are lineage-specified progenitors of group 3 RORγt(+) innate lymphoid cells. Immunity 2014, 41:988–1000. [DOI] [PubMed] [Google Scholar]
- **27.Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, et al. : Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell 2017, 168:1086–1100.e1010.This is the first study to identify a peripheral CD34- human progenitor capable of generating all ILCs
- **28.Chen L, Youssef Y, Robinson C, Ernst GF, Carson MY, Young KA, Scoville SD, Zhang X, Harris R, Sekhri P, et al. : CD56 Expression Marks Human Group 2 Innate Lymphoid Cell Divergence from a Shared NK Cell and Group 3 Innate Lymphoid Cell Developmental Pathway. Immunity 2018, 49:464–476.e464.This study describes the discovery and lineage specification of the sILCP in human tonsils and blood.
- *29.Scoville SD, Mundy-Bosse BL, Zhang MH, Chen L, Zhang X, Keller KA, Hughes T, Cheng S, Bergin SM, Mao HC, et al. : A Progenitor Cell Expressing Transcription Factor RORγt Generates All Human Innate Lymphoid Cell Subsets. Immunity 2016, 44:1140–1150.This study identified the first CD34+lineage restricted human ILC progenitor capable of generating all ILCs.
- 30.Hoorweg K, Peters CP, Cornelissen F, Aparicio-Domingo P, Papazian N, Kazemier G, Mjösberg JM, Spits H, Cupedo T: Functional Differences between Human NKp44(−) and NKp44(+) RORC(+) Innate Lymphoid Cells. Front Immunol 2012, 3:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Björklund Å, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjösberg J: The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol 2016, 17:451–460.These authors performed extensive single-cell transcriptional analysis that revealed newer subsets of human ILCs and their transcriptional signature.
- 32.Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, Di Santo JP, Eberl G: Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science 2010, 330:665–669. [DOI] [PubMed] [Google Scholar]
- 33.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Göppert N, Croxford AL, Waisman A, Tanriver Y, et al. : A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 2013, 494:261–265. [DOI] [PubMed] [Google Scholar]
- 34.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M:Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 2013, 38:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, Munneke JM, Hazenberg MD, Villaudy J, Buskens CJ, et al. : Interleukin-12 and −23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity 2015, 43:146–160. [DOI] [PubMed] [Google Scholar]
- 36.Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, Yu L, Pritchard GH, Berlin AA, Hunter CA, Bowler R, et al. : Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol 2016, 17:626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim AI, Menegatti S, Bustamante J, Le Bourhis L, Allez M, Rogge L, Casanova JL, Yssel H, Di Santo JP: IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J Exp Med 2016, 213:569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **38.Simoni Y, Fehlings M, Kløverpris HN, McGovern N, Koo SL, Loh CY, Lim S, Kurioka A, Fergusson JR, Tang CL, et al. : Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency. Immunity 2017, 46:148–161.This report is the first attempt to document all ILCs in multiple human tissues, both normal and diseased. The results from this investigation found it difficult to identify human non-NK ILC1’s that were not contaminated by other lineages
- 39.Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, Sharma S, Nakayama T, et al. : The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity 2014, 40:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J: GATA3 Regulates the Development and Functions of Innate Lymphoid Cell Subsets at Multiple Stages. Front Immunol 2017, 8:1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernink JH, Mjösberg J, Spits H: Human ILC1: To Be or Not to Be. Immunity 2017, 46:756–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ealey KN, Koyasu S: How Many Subsets of Innate Lymphoid Cells Do We Need? Immunity 2017, 46:10–13. [DOI] [PubMed] [Google Scholar]
- 43.Simoni Y, Newell EW: Toward Meaningful Definitions of Innate-Lymphoid-Cell Subsets. Immunity 2017, 46:760–761. [DOI] [PubMed] [Google Scholar]
- 44.Spits H, Bernink JH, Lanier L: NK cells and type 1 innate lymphoid cells: partners in host defense. Nat Immunol 2016, 17:758–764. [DOI] [PubMed] [Google Scholar]
- 45.Nagasawa M, Germar K, Blom B, Spits H: Human CD5+ innate lymphoid cells are functionally immature and their development from CD34+ progenitor cells is regulated by Id2. Front Immunol 2017, 8:1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **46.Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, Rautela J, Straube J, Waddell N, Blake SJ, et al. : Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol 2017, 18:1004–1015.This study describes the conversion of NK cells into a less activated ILC1 in the presence of cancer derived immunesuppression.
- 47.Romee R, Foley B, Lenvik T, Wang Y, Zhang B, Ankarlo D, Luo X, Cooley S, Verneris M, Walcheck B, et al. : NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood 2013, 121:3599–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, Freud AG, Caligiuri MA: Location and cellular stages of natural killer cell development. Trends Immunol 2013, 34:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naeimi Kararoudi M, Dolatshad H, Trikha P, Hussain SA, Elmas E, Foltz JA, Moseman JE, Thakkar A, Nakkula RJ, Lamb M, et al. : Generation of Knock-out Primary and Expanded Human NK Cells Using Cas9 Ribonucleoproteins. J Vis Exp 2018. [DOI] [PMC free article] [PubMed]
- 50.Lopez-Lastra S, Masse-Ranson G, Fiquet O, Darche S, Serafini N, Li Y, Dusséaux M, Strick-Marchand H, Di Santo JP: A functional DC cross talk promotes human ILC homeostasis in humanized mice. Blood Adv 2017, 1:601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]