Abstract
Background
Genetic research on longevity has provided important insights into the mechanism of aging and aging-related diseases. Pinpointing import genetic variants associated with aging could provide insights for aging research.
Methods
We performed a whole-genome sequencing in 19 centenarians to establish the genetic basis of human longevity.
Results
Using SKAT analysis, we found 41 significantly correlated genes in centenarians as compared to control genomes. Pathway enrichment analysis of these genes showed that immune-related pathways were enriched, suggesting that immune pathways might be critically involved in aging. HLA typing was next performed based on the whole-genome sequencing data obtained. We discovered that several HLA subtypes were significantly overrepresented.
Conclusions
Our study indicated a new mechanism of longevity, suggesting potential genetic variants for further study.
Keywords: Longevity, Centenarian, WGS
Introduction
With the development of human genomics research, a large number of studies of the genetics of longevity have been conducted. Scientists from various countries have proposed many different theories concerning the mechanisms of aging from different perspectives, involving oxidative stress, energy metabolism, signal transduction pathways, immune response, etc. [1, 2]. These mechanisms interact with each other and are influenced by heredity to some degree [2, 3]. The identification of longevity-related biological markers is critical to an in-depth understanding of the mechanisms of carrier protection against common disease and/or of the retardation of the process of aging.
Studies revealed from 300 to 750 genes related to longevity that are critically involved in a variety of life activities, such as growth and development, energy metabolism, oxidative stress, genomic stability maintenance, and neurocognition [4]. These candidate genes include mainly APOE, a gene involved in lipoprotein metabolism [5, 6]. Others are those involved in cell cycle regulation, cell growth and signal transduction, the maintenance of genome stability, and the endocrine-related pathway [7–9]. In addition, the candidates for longevity encompass genes related to drug metabolism, the ones involved in protein folding, stabilization, and degradation, as well those related to coagulation and regulation of circulation [10], etc. In most cases, these genes or their polymorphic sites were examined in multiple population replication studies, which discovered certain longevity-associated genes or pathways [4–10].
Besides, longevity is associated with immunity and inflammation [11, 12]. HLA gene, also known as the major histocompatibility complex gene, encodes the major histocompatibility complex (MHC), which is a gene family existing in most vertebrate genomes, closely related to the immune system [13]. Earlier study indicated that HLA may be the genetic basis for the specific response patterns of longevity and longevity immunity [14]. Inflammatory cytokines, such as TNF-α, IL-1β, and IL-6, may be key players [15]. IL-10 limits and terminates the inflammatory response by inhibiting the action of T cells, monocytes, and macrophages, and thus, the genetic variants of this gene may also affect the longevity phenotype [16].
However, at present, most of the investigations on longevity factors of centenarians are performed on a small number of candidate miRNAs, single tissues, or single samples, and only few studies have systematically conducted analyses of multiple tissues and copies at the whole-genome level [17–19].
Based on the results of previous cohort study, in this study, we aimed to use genome-wide sequencing technology to conduct genome-wide association studies and analysis of centenarians. Our findings would facilitate a more accurate focus on the most important genetic basis and molecular mechanisms associated with longevity. The conclusions of this study can serve as the basis for the public efforts towards the extension of the length of life. Moreover, they will provide a scientific reference for further clinical research on disease treatment and overall health care promotion.
Results
SKAT analysis revealed significantly correlated genes in aging
The sequencing platform Illumina XTen (Illumina, San Diego, CA, USA) was used for sequencing of the entire genome of 19 centenarians at an average depth of 30×. The sequencing quality metrics are provided in Supplementary Table 1. Baseline information of the centenarians is shown in Table 1. The identified variants were annotated, and non-synonymous variants affecting gene function were selected for association analysis.
Table 1.
Characteristics of the centenarians
| Age at draw | Sex | BMI | Major age-related diseases | Race |
|---|---|---|---|---|
| 104 | Male | 22.6 | None | Eastern Asiatic Mongoloid |
| 103 | Male | 21.8 | None | Eastern Asiatic Mongoloid |
| 102 | Female | 20.6 | None | Eastern Asiatic Mongoloid |
| 105 | Female | 19.8 | None | Eastern Asiatic Mongoloid |
| 116 | Female | 27.2 | None | Eastern Asiatic Mongoloid |
| 101 | Female | 22.4 | None | Eastern Asiatic Mongoloid |
| 102 | Female | 20.6 | None | Eastern Asiatic Mongoloid |
| 104 | Male | 18.6 | None | Eastern Asiatic Mongoloid |
| 103 | Male | 21.5 | None | Eastern Asiatic Mongoloid |
| 103 | Female | 20.3 | None | Eastern Asiatic Mongoloid |
| 101 | Male | 24.5 | None | Eastern Asiatic Mongoloid |
| 105 | Female | 22.5 | None | Eastern Asiatic Mongoloid |
| 103 | Male | 21.6 | None | Eastern Asiatic Mongoloid |
| 104 | Female | 20.7 | None | Eastern Asiatic Mongoloid |
| 102 | Female | 19.8 | None | Eastern Asiatic Mongoloid |
| 100 | Female | 22.3 | None | Eastern Asiatic Mongoloid |
| 103 | Male | 21.4 | None | Eastern Asiatic Mongoloid |
| 114 | Female | 20.6 | None | Eastern Asiatic Mongoloid |
| 100 | Male | 26.2 | None | Eastern Asiatic Mongoloid |
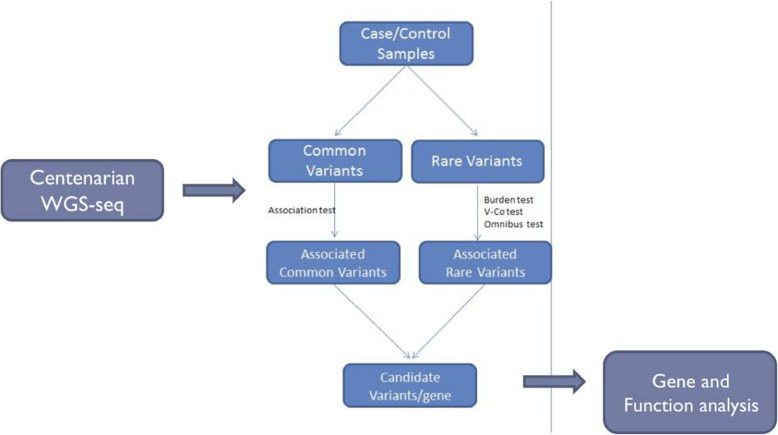
The experiment design is shown in Fig. 1. Association analysis is applied to WGS data to find important gene and pathways. All centenarian and controls were Eastern Asiatic Mongoloids ascertained to be of Chinese descent (Zhejiang Province, Southeast China). Correlative analysis involved mainly association analysis, SKAT, and Burden tests for rare variants. These methods are commonly employed for GWAS research, especially for case/control samples. They are identified by the difference in frequency of occurrence of variants between case and control samples. Variations associated with the phenotype of the disease, generally directed against common variants, are detected using this method, where the frequency of occurrence and the variation contribute to the phenotype of the disease. Annotations and literature surveys can be used for significantly related variants to further determine the effect of related genes and variants on gene function.
Fig. 1.
Overall design of the study. WGS were applied to 19 centenarians. Association test was used to select candidate variants/genes. Function analysis was then applied
Based on the whole-genome sequencing data, this analysis was performed on the variants detected with MAF > 1%. The control sample consisted of selected 1000G East Asian population data, and the total number of the control samples is 208 [20]. PCA analysis was conducted to evaluate the stratification of the case and control group (supplementary Fig 1). For rare variants with a low frequency of variation, analysis using the method Rare variants case/control association test was performed.
A total number of 41 (Supplementary Table 2) significantly correlated genes were obtained through SKAT analysis. The top 10 genes were as follows: PABPC3, BAGE2, HLA-DRB1, PDE4DIP, PADI4, CHI3L2, MUC17, WARS, HLA-DRB5, and SIRPB1 (Table 2).
Table 2.
Significantly correlated genes in SKAT analysis
| Gene | SKAT_Common.Only | SKAT_Rare.Only | SKAT_all | SKAT_Rare.pro | SKAT_Common.pro | Burden_all | Burden__Rare.pro | Burden__Common.pro |
|---|---|---|---|---|---|---|---|---|
| PABPC3 | 1.97E−27 | 4.57E−32 | 4.41E−31 | 2.72E−48 | 2.26E−51 | 2.50E−30 | 1.46E−50 | 3.13E−51 |
| BAGE2 | 1.70E−27 | 6.41E−21 | 8.76E−25 | 7.30E−36 | 1.52E−45 | 8.66E−26 | 2.20E−37 | 5.57E−30 |
| HLA-DRB1 | 0.03254211 | 1.57E−14 | 2.66E−13 | 0.029113904 | 4.24E−30 | 1.94E−13 | 1.33E−21 | 1.37E−20 |
| PDE4DIP | 4.19E−07 | 1.71E−13 | 2.01E−12 | 1.13E−09 | 3.32E−32 | 5.43E−13 | 1.40E−16 | 8.32E−16 |
| PADI4 | 0.351394973 | 4.20E−10 | 4.84E−07 | 0.564180913 | 3.26E−19 | 2.18E−08 | 3.17E−10 | 9.98E−11 |
| CHI3L2 | 0.182676324 | 2.30E−09 | 3.99E−09 | 9.37E−08 | 2.84E−24 | 3.99E−06 | 9.70E−09 | 6.54E−09 |
| MUC17 | 0.640011254 | 2.33E−09 | 4.68E−08 | 2.45E−08 | 3.01E−18 | 2.60E−06 | 2.54E−10 | 3.23E−10 |
| WARS | 0.248268983 | 2.07E−08 | 6.87E−06 | 6.88E−05 | 1.99E−15 | 8.07E−07 | 1.61E−08 | 2.34E−08 |
| HLA-DRB5 | 0.021102518 | 1.05E−06 | 5.24E−07 | 0.01776692 | 3.01E−12 | 8.72E−08 | 4.27E−11 | 5.39E−11 |
| SIRPB1 | 0.479393621 | 2.95E−06 | 3.42E−06 | 5.33E−10 | 3.26E−10 | 0.001382303 | 0.000184681 | 0.000282752 |
Immune system-related pathway was significantly enriched
The significant genes were subjected to differential pathway enrichment analysis. Then, MutsigDB was used to enrich the KEGG and Reactome pathways. As can be seen in Table 3, the associated genes were significantly enriched in the pathways related to immune and inflammatory responses, such as those of interferons, antibodies, and immunity.
Table 3.
Significantly enriched pathway
| Gene set name[#Genes(K)] | Description | #Genes in Overlap(k) | p value | FDR q value |
|---|---|---|---|---|
| REACTOME_INTERFERON_GAMMA_SIGNALING[63] | Interferon gamma signaling | 5 | 2.61E−09 | 2.81E−06 |
| KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION[89] | Antigen processing and presentation | 5 | 1.52E−08 | 8.16E−06 |
| KEGG_ALLOGRAFT_REJECTION[38] | Allograft rejection | 4 | 3.55E−08 | 1.28E−05 |
| KEGG_GRAFT_VERSUS_HOST_DISEASE[42] | Graft-versus-host disease | 4 | 5.35E−08 | 1.40E−05 |
| KEGG_TYPE_I_DIABETES_MELLITUS[44] | Type I diabetes mellitus | 4 | 6.51E−08 | 1.40E−05 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS[134] | Cell adhesion molecules (CAMs) | 5 | 1.19E−07 | 2.13E−05 |
| KEGG_AUTOIMMUNE_THYROID_DISEASE[53] | Autoimmune thyroid disease | 4 | 1.40E−07 | 2.15E−05 |
| REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE[14] | Translocation of ZAP-70 to immunological synapse | 3 | 2.21E−07 | 2.97E−05 |
| REACTOME_INTERFERON_SIGNALING[159] | Interferon signaling | 5 | 2.78E−07 | 3.32E−05 |
| REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS[16] | Phosphorylation of CD3 and TCR zeta chains | 3 | 3.39E−07 | 3.66E−05 |
HLA subtypes are correlated with aging
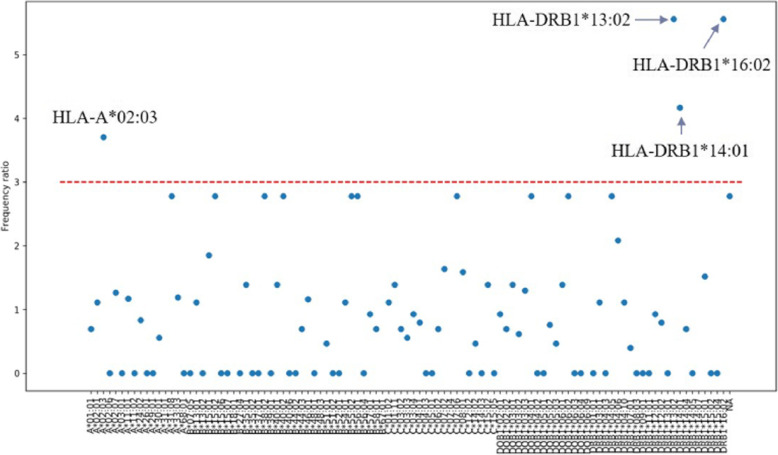
Based on the whole-genome sequencing data obtained, HLA-typing was performed. Through the analysis of HLA type distribution, as presented in Tables 3 and 4 and Fig. 2, we found that the type II HLA genes had an important relationship with longevity. Among them, the HLA DRB1 *13:02, HLA DRB1 *14:01, and HLA DRB1 *16:02 were significantly associated with longevity.
Table 4.
HLA subtype percentage in the case and control groups
| HLA type | Ratio | Control_Percentage | Case_Percentage |
|---|---|---|---|
| A*02:03 | 3.7 | 0.03 | 0.11 |
| DRB1*13:02 | 5.6 | 0.01 | 0.05 |
| DRB1*14:01 | 4.2 | 0.02 | 0.08 |
| DRB1*16:02 | 5.6 | 0.01 | 0.05 |
Fig. 2.
HLA typing’s correlation with the centinarian group. Frequency ratio of every HLA type was plotted. Four HLA types with frequency ratio larger than 3 were marked
Discussion
Researches on the genetic mechanisms of longevity have been conducted from many perspectives, including that on longevity-related genes, variants, and biological pathways [4, 10]. With the advancements in the NGS technology and analysis algorithms, increasingly more longevity-related genetic features could be found and would be useful for the understanding of mechanisms of longevity and related diseases [21, 22].
In our study, we used a small centenarian cohort to establish the association between the genetic variants and longevity. By SKAT analysis, rare variants were found that were related to the longevity phenotype. HLA-DRB1, HLA-DRB5, and PDE4 DIP have been reported to be associated with longevity [23–25]. Importantly, HLA-DRB1 variants have been specifically reported to have been significantly enriched in a French centenarian study [26]. Further, through the analysis of HLA type distribution, we found several subtypes of HLA DRB1 which have a closer relationship with longevity. Other significantly related genes that we found in this study, such as PABPC3, BAGE2, PADI4, CHI3L2, MUC17, WARS, and SIRPB1, have never been reported before, and their functions deserve further study. Pathway enrichment was performed and showed an important association of the immune-related pathway and the aging process. Previous examinations revealed that immune and inflammatory responses are closely related to ARD (aging-related diseases) [27, 28]. The significant differences in the gene enrichment of the related pathways suggest that a possible longevity mechanism may be associated with protective variants in genes that occur in the related pathways. Here, we have shown that genome-wide data can be further mined as compared with the findings of traditional SNP studies. For example, HLA-type analysis could also be associated with the phenotype, which appears to be a possibility for expanded mining of genome-wide data. Nonetheless, the relatively small sample size limited the power of our findings, and thus further validation by large cohort studies is required.
Conclusions
In conclusion, the findings of our study provide novel insights into aging mechanisms, suggesting the involvement of several genes, pathways, and specific HLA subtypes that are worth further investigations.
Materials and methods
Sample preparation
A random number table was used to randomly collect whole-blood samples from the centenarian cohort in Zhejiang Province, China. Ten percent of centenarians, that is, 19 centenarians, were chosen. All of them were free of major age-related diseases, i.e., cardiovascular or cerebrovascular disease, cancer, dementia, renal or hepatic failure, etc. They were informed about the study and signed a letter of consent, in accordance with the guidelines of the Ethics Committee of the First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, China (2015-KL-008-01). PBMC were isolated to be used for extraction of genomic DNA. Nineteen sex-matched samples with good physical condition were randomly selected from 1000G East Asian population data to serve as a control group [20]. All centenarian and controls were Eastern Asiatic Mongoloids ascertained to be of Chinese descent (Zhejiang Province, Southeast China).
Library preparation and whole-genome sequencing
Indexed Illumina NGS libraries were prepared from plasma DNA and germline genomic DNA. Next, an NGS library was prepared using the KAPA library preparation kit (Kapa Biosystems). Agencourt AMPure XP beads (Beckman-Coulter) were used to purify the extracted DNA. A 100-fold mole excess of ligation Illumina TruSeq adaptors was used for ligation at 16 °C for 16 h. Size selection of DNA fragments was performed in the 100-μL solution system, and then, the connected fragments were amplified with 500 μm Illumina backbone oligonucleotides for 4–9 rounds of PCR. After that, the DNA fragments were input.
The library concentration was assessed by Qubit and Qpcr. The fragment length was determined using a 2100 Bioanalyzer with a DNA 1000 kit (Agilent). DNA fragments were mixed with HiFi Hot Start Ready Mix (1×), and 2 × 150 bp sequencing of multiple libraries was finally performed with Illumina HiSeq X10.
Data preprocessing
Paired reads were aligned to the hg19 reference genome using the BWA (V0.7.15-r1140)-mem command [29]. Then, they were sorted and indexed using SAMtools [30]. An in-house Python script was utilized to evaluate the various statistics collected, including mapping statistics, read quality, and panels capture efficiency.
For each sample, the SAMtools pileup function was employed to generate variant candidates among the corresponding sites. We excluded the SNP sites and lower depth sites (≤ eu) among the candidates and removed the reads with low base (< Q30) and mapping qualities (< 40).
SKAT analysis
The file with the data of the centenarian variants was subjected to SKAT. Functional annotation of the genomic variation of each sample was performed, distinguishing between rare variants and variants affecting the protein function. Next, the influence of the variants in the gene was scored. We used the aforementioned three methods to test candidate genome variants to identify potential rare variants associated with the phenotype. Literature and databases were searched to find how the associated variants affected the biological processes, and speculation on disease mechanisms was carried out.
Pathway enrichment analysis
The KEGG pathway enrichment was conducted using the DAVID Functional Annotation Bioinformatics Microarray Analysis.
HLA typing
HLA typing was performed through the HLAscan algorithm [31]. HLAscan started with sequence reads in FASTQ format for mapping to IMGT/HLA data. For targeted sequencing data, sequence reads were used as direct input for HLAscan, whereas for WGS and WES data, we selected reads for HLA genes prior to running the HLAscan. In comparison with the targeted sequencing data, alignment of whole-genome/exome data directly to the IMGT/HLA database may lead to the omission of some HLA reads. Nonetheless, this algorithm was adopted because alignment of HLA reads to the IMGT/HLA database is advantageous in regard to both time and computational processing without loss of predictive accuracy. Initial alignment was performed using BWA-MEM (v0.7.10-r789) with default options. The alignment was the best fit for HLAscan in our investigation, which involved many allele sequences in IMGT/HLA and BWA-MEM. Sequence reads in the BAM file were sorted by reference coordinates using the FixMateInformation function, followed by removal of duplicate reads using MarkDuplicates in the Picard software package (version 1.68) (http://picard.sourceforge.net). Subsequently, identification of indels and re-alignment around these features were performed with the RealignerTargetCreator and IndelRealigner tools, respectively, and base-pair quality scores were recalibrated with BaseRecalibrator and PrintReads using the GATK software (version 3.3.0) [32]. Throughout these processes, sequence reads corresponding to the exonic regions of HLA genes were selected based on an initial alignment generated using GATK with a whole-genome reference (GRCh37.p13). This filtering step does not classify the sequence reads into specific HLA genes.
Supplementary information
Additional file 1: Supplementary Figure 1. PCA plot of the case and control group. PCA plot was made using the SNP genotype of centinarian group and the control group.
Additional file 2: Supplementary Table 1. Sequencing quality metrics
Additional file 3: Supplementary Table 2. Significantly correlated genes obtained through SKAT analysis.
Acknowledgements
We express our sincere gratitude to Mr. Maoyang for the valuable assistance.
Authors’ contributions
Shuhua Shen, Chao Li, Yixue Li, and Qi Huang conceived and supervised the research. Shuhua Shen and Chao Li performed all statistical analyses. Shuhua Shen, Chao Li, Luwei Xiao, Xiaoming Wang, Hang Lv, and Yuan Shi wrote the main text of the manuscript with contributions from all the authors. The authors read and approved the final manuscript.
Funding
This research was supported by the Science Research Foundation for TCM of Zhejiang Province, China (no. ZZYJ-WTRW-2018-01, no. 2017ZA045) and the National Natural Science Foundation of China (no. 81503527).
Availability of data and materials
Please contact the author for data requests.
Ethics approval and consent to participate
Nineteen centenarians were chosen. All of them were free of major age-related diseases, i.e., cardiovascular or cerebrovascular disease, cancer, dementia, renal or hepatic failure, etc. They were informed about the study and signed a letter of consent, in accordance with the guidelines of the Ethics Committee of the First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, China (2015-KL-008-01).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qi Huang and Yixue Li contributed equally to this work.
Contributor Information
Yixue Li, Email: yxli@scbit.org.
Qi Huang, Email: linda0358@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40246-020-00271-7.
References
- 1.Santos-Lozano A, Santamarina A, Pareja-Galeano H, Sanchis-Gomar F, Fiuza-Luces C, Cristi-Montero C, Bernal-Pino A, Lucia A, Garatachea N. The genetics of exceptional longevity: insights from centenarians. Matiritas. 2016;90:49–57. doi: 10.1016/j.maturitas.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Govindaraju D, Gil A, Barzilai N. Genetics, lifestyle and longevity: lessons from centenarians. Applied and Translational Genomics. 2015;4:23–32. doi: 10.1016/j.atg.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gierman HJ, Kristen F, Roach JC, Coles NS, LI H, Gustavo G, Markov GJ, Smith JD, Leroy H, Stephen CL, Kim SK. Whole-genome sequencing of the world’s oldest people. J. Plos One. 2014;9:1–10. doi: 10.1371/journal.pone.0112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budovsky A, Craig T, Wang J, Tacutu R, Csordas A, Lourenço J, Fraifeld VE, de Magalhães JP. LongevityMap: a database of human genetic variants associated with longevity. Trends Genet. 2013;29:559–560. doi: 10.1016/j.tig.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Schächter F, Faure-Delanef L, Guénot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 6.Garatachea N, Emanuele E, Calero M, Fuku N, Arai Y, Abe Y, Murakami H, Miyachi M, Yvert T, Verde Z, Zea MA, Venturini L, Santiago C, Santos-Lozano A, Rodríguez-Romo G, Ricevuti G, Hirose N, Rábano A, Lucia A. ApoE gene and exceptional longevity: insights from three independent cohorts. Exp Gerontol. 2014;53:16–23. doi: 10.1016/j.exger.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. PNAS. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mustafina OE, Nasibullin TR, Érdman VV, Tuktarova IA. Association analysis of polymorphic loci of TP53 and NFKB1 genes with human age and longevity. Adv Gerontol. 2011;24:397–404. [PubMed] [Google Scholar]
- 9.Barbieri M, Bonafè M, Franceschi C, Paolisso G. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab. 2003;285:E1064–E1071. doi: 10.1152/ajpendo.00296.2003. [DOI] [PubMed] [Google Scholar]
- 10.Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 12.Franceschi C, Capri M, Monti D, Giunta S, Olivieri F, Sevini F, Panourgia MP, Invidia L, Celani L, Scurti M, Cevenini E, Castellani GC, Salvioli S. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernández-Viña M, Geraghty DE, Holdsworth R, Hurley CK, Lau M, Lee KW, Mach B, Maiers M, Mayr WR, Müller CR, Parham P, Petersdorf EW, Sasazuki T, Strominger JL, Svejgaard A, Terasaki PI, Tiercy JM, Trowsdale J. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caruso C, Candore G, Colonna Romano G, Lio D, Bonafè M, Valensin S, Franceschi C. HLA, aging, and longevity: a critical reappraisal. Hum Immunol. 2000;61:942–949. doi: 10.1016/S0198-8859(00)00168-3. [DOI] [PubMed] [Google Scholar]
- 15.Chung HY, Cesari M, Anton S, Marzetti E, Giovannini S, Seo AY, Carter C, Yu BP, Leeuwenburgh C. Molecular inflammation: underpinnings of aging and age-related diseases. Ageing Res Rev. 2009;8:18–30. doi: 10.1016/j.arr.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lio D, Scola L, Crivello A, Colonna-Romano G, Candore G, Bonafè M, Cavallone L, Franceschi C, Caruso C. Gender-specific association between -1082 IL-10 promoter polymorphism and longevity. Genes Immun. 2002;3:30–33. doi: 10.1038/sj.gene.6363827. [DOI] [PubMed] [Google Scholar]
- 17.Noren Hooten N, Fitzpatrick M, Wood WH, 3rd, De S, Ejiogu N, Zhang Y, Mattison JA, Becker KG, Zonderman AB, Evans MK. Age-related changes in microRNA levels in serum. Aging. 2013;5:725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchis-Gomar F, Pareja-Galeano H, Santos-Lozano A, Garatachea N, Fiuza-Luces C, Venturini L, Ricevuti G, Lucia A, Emanuele E. A preliminary candidate approach identifies the combination of chemerin, fetuin-A, and fibroblast growth factors 19 and 21 as a potential biomarker panel of successful aging. Age. 2015;37:9776. doi: 10.1007/s11357-015-9776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Spoel E, Jansen SW, Akintola AA, Ballieux BE, Cobbaert CM, Slagboom PE, Blauw GJ, Westendorp RGJ, Pijl H, Roelfsema F, van Heemst D. Growth hormone secretion is diminished and tightly controlled in humans enriched for familial longevity. Aging Cell. 2016;15:1126–1131. doi: 10.1111/acel.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Akker EB, Deelen J, Slagboom PE, Beekman M. Exome and whole genome sequencing in aging and longevity. Adv Exp Med Biol. 2015;847:127–139. doi: 10.1007/978-1-4939-2404-2_6. [DOI] [PubMed] [Google Scholar]
- 23.Lio D, Pes GM, Carru C, Listì F, Ferlazzo V, Candore G, Colonna-Romano G, Ferrucci L, Deiana L, Baggio G, Franceschi C, Caruso C. Association between the HLA-DR alleles and longevity: a study in Sardinian population. Exp Gerontol. 2003;38:313-317. https://doi.org/10.1016/s0531-5565(02)00178-x. [DOI] [PubMed]
- 24.Lagaay AM, D'Amaro J, Ligthart GJ, Schreuder GM, van Rood JJ, Hijmans W. Longevity and heredity in humans. Association with the human leucocyte antigen phenotype. Ann N Y Acad Sci. 1991;621:78–89. doi: 10.1111/j.1749-6632.1991.tb16970.x. [DOI] [PubMed] [Google Scholar]
- 25.Phillips BE, Williams JP, Gustafsson T, Bouchard C, Rankinen T, Knudsen S, Smith K, Timmons JA, Atherton PJ. Molecular networks of human muscle adaptation to exercise and age. PLoS Genet. 2013;9:e1003389. doi: 10.1371/journal.pgen.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanova R, Hénon N, Lepage V, Charron D, Vicaut E, Schächter F. HLA-DR alleles display sex-dependent effects on survival and discriminate between individual and familial longevity. Hum Mol Genet. 1998;7:187–194. doi: 10.1093/hmg/7.2.187. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg EL, Dixit VD. Drivers of age-related inflammation and strategies for healthspan extension. Immunol Rev. 2015;265:63–74. doi: 10.1111/imr.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licastro F, Candore G, Lio D, Porcellini E, Colonna-Romano G, Franceschi C, Caruso C. Innate immunity and inflammation in ageing: a key for understanding age-related diseases. Immun Ageing. 2005;2:8. doi: 10.1186/1742-4933-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G. Durbin R,1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ka S, Lee S, Hong J, Cho Y, Sung J, Kim HN, Kim HL, Jung J. HLAscan: genotyping of the HLA region using next-generation sequencing data. BMC Bioinformatics. 2017;18:258. doi: 10.1186/s12859-017-1671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. PCA plot of the case and control group. PCA plot was made using the SNP genotype of centinarian group and the control group.
Additional file 2: Supplementary Table 1. Sequencing quality metrics
Additional file 3: Supplementary Table 2. Significantly correlated genes obtained through SKAT analysis.
Data Availability Statement
Please contact the author for data requests.