Summary
The habenula complex is appreciated as a critical regulator of motivated and pathological behavioral states via its output to midbrain nuclei. Despite this, transcriptional definition of cell populations that comprise both the medial (MHb) and lateral habenular (LHb) subregions in mammals remain undefined. To resolve this, we performed single-cell transcriptional profiling and highly multiplexed in situ hybridization experiments of the mouse habenula complex in naïve mice and those exposed to an acute aversive stimulus. Transcriptionally distinct neuronal cell types identified within the MHb and LHb, were spatially defined, differentially engaged by aversive stimuli and had distinct electrophysiological properties. Cell types identified in mice, also displayed a high degree of transcriptional similarity to those previously described in zebrafish, highlighting the well conserved nature of habenular cell types across the phylum. These data identify key molecular targets within habenular cell types, and provide a critical resource for future studies.
eTOc blurb
Whereas anatomy and functions of the habenula have been extensively studied, the transcriptional landscape of the mammalian habenula remains poorly defined. Multimodal analysis of single cell transcriptomes in the habenula by Hashikawa et al. provides a useful data resource to associate transcriptional diversity with functional relevance of the molecularly defined habenula cell types.
Introduction
The habenula complex serves as a neuroanatomical hub to regulate brain circuitry critical for many behavioral states related to motivation and decision-making (Hikosaka, 2010; Lecca et al., 2014; Namboodiri et al., 2016; Proulx et al., 2014; Viswanath et al., 2014). As part of the epithalamus, the habenula is well conserved across vertebrate species (Agetsuma et al., 2010; Amo et al., 2010; Bianco Isaac H and Wilson Stephen W, 2009; Lee et al., 2010; Stephenson-Jones et al., 2012), and in mammals, can be subdivided into medial (MHb) and lateral (LHb) subdivisions demarcated both anatomically and by patterned gene expression (Aizawa et al., 2012; Marburg, 1944). LHb neurons are almost entirely composed of SLC17A6 (VGlut2) expressing glutamatergic neurons, which receive synaptic inputs from regions such as the ventral pallidum (Golden et al., 2016; Knowland et al., 2017), lateral hypothalamic area (Lazaridis et al., 2019; Lecca et al., 2017; Stamatakis et al., 2016), endopeduncular nucleus/globus pallidus (Hong and Hikosaka, 2008; Li et al., 2019; Shabel et al., 2012; Wallace et al., 2017), ventral tegmental area (VTA) (Root et al., 2014; Stamatakis et al., 2013), and other regions (Hikosaka et al., 2008). Glutamatergic LHb output neurons project to the VTA where they directly and indirectly regulate the activity of dopaminergic neurons to alter appetitive and aversive behaviors (Hong et al., 2011; Jhou et al., 2009; Lammel et al., 2012; Stamatakis and Stuber, 2012). Additionally, LHb neurons also project to the dorsal raphe to regulate the serotonergic system and depression-related behavioral phenotypes (Amo et al., 2014; Andalman et al., 2019; Li et al., 2011, 2013; Neckers et al., 1979; Wang and Aghajanian, 1977; Zhou et al., 2017).
Despite its close proximity, the MHb differs dramatically from the LHb with respect to its circuit connectivity, cellular composition, and function. The MHb contains spatially defined glutamatergic and cholinergic neurons (Aizawa et al., 2012), which receive synaptic input from the septum (Viswanath et al., 2014), and largely project to the interpeduncular nucleus (Herkenham and Nauta, 1979), where acetylcholine, glutamate, and other transmitters are released (Claudio Cuello et al., 1978; Grady et al., 2009; Molas et al., 2017; Ren et al., 2011). While less is known about the function of MHb circuitry in controlling behavior, MHb neurons play a critical role in regulating nicotine reinforcement (Fowler and Kenny, 2011, 2012; Lester and Dani, 1995), novelty preference (Molas et al., 2017), and anxiety/fear related behaviors (Hsu et al., 2014, 2016; Yamaguchi et al., 2013; Zhang et al., 2016). Collectively, the MHb and LHb represent two largely independent subsystems, both of which importantly contribute towards regulating a variety of motivated behavioral states.
Despite the critical role of the habenula complex in orchestrating behavior, a comprehensive understanding of the genetically defined cell types in mammals within this region is still lacking. We used single cell RNA sequencing (scRNAseq) (Macosko et al., 2015) and multiplex in situ hybridization to generate a comprehensive dataset on the gene expression profiles of identified MHb and LHb cell types.
Results
scRNAseq identifies transcriptionally distinct clusters among neuronal and non-neuronal cells.
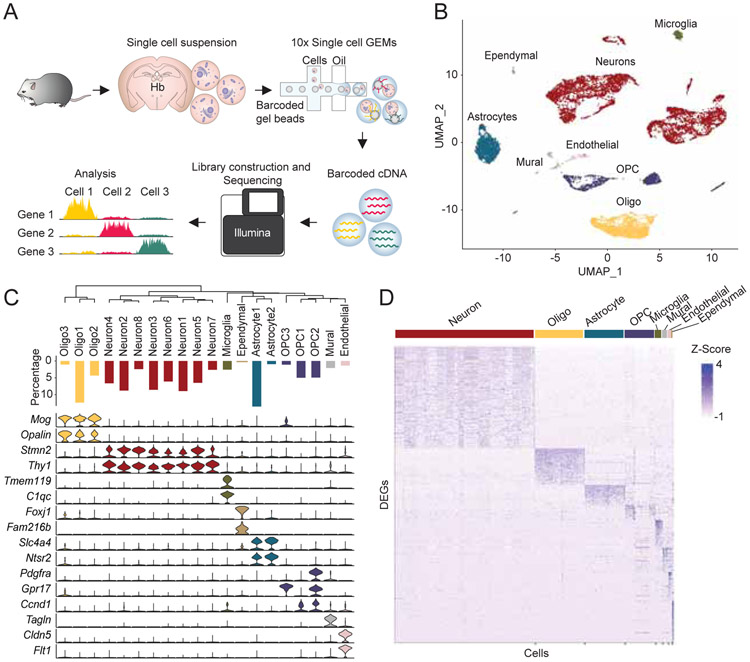
To characterize transcriptional identities of single cells in the mammalian habenula, we utilized droplet (Macosko et al., 2015; Rossi et al., 2019) based chromium technology developed by 10x Genomics (Zheng et al., 2017). We microdissected the habenula from live brain slices of adult male mice (P50-55), enzymatically digested the tissue to obtain well dissociated highly viable single cells (viability >80 %), captured single cell transcriptomes, and generated cDNA libraries for subsequent sequencing (Figure 1). After computationally removing doublets (4.0 %) (DePasquale et al., 2019), we recovered the transcriptomes from a total of 11,878 cells with 17,726 genes in total (median unique molecular identifiers (UMIs)/cell: 2,030, median genes/cell: 1,001, Figure S1). In order to minimize experimental batch effects and behaviorally induced transcripts (e.g. IEGs), a computational technique (Stuart et al., 2019) to integrate multiple datasets by implementing both canonical correlation analysis (CCA) (Butler et al., 2018) and mutual nearest neighbor analysis (MNN) (Haghverdi et al., 2018) was utilized. Dimensions of the integrated data were reduced by principle component analysis followed by graph-based clustering (Macosko et al., 2015; Stuart et al., 2019) and visualized using the UMAP algorithm (Becht et al., 2019; McInnes et al., 2018) (Figure 1B). This method identified 20 cell clusters which were then assigned to neuronal and non-neuronal cell types by their canonical gene markers (Macosko et al., 2015; Saunders et al., 2018; Wu et al., 2017; Zeisel et al., 2018) (Figure 1B and 1C). Stmn2 and Thy1 were selectively expressed in neuronal clusters, which were detected in ~ 50 % of all the cells (Figure 1C and S1), Opalin and Mog marked oligodendrocyte clusters, Slc4a4 and Ntsr2 were highly expressed in astrocyte clusters, and Pdgfra, Gpr17 or Ccnd1 were enriched in the oligodendrocyte precursor cells (OPC). This clustering method also detected less abundant cell types. Tmem119 and C1qc clearly marked microglia (2.6 % of the total cells), Tagln marked the mural cell cluster (2.1 %), Cldn5 and Flt1 marked the endothelial cells (1.4 %) and Fam216b was exclusively expressed in the ependymal cluster (0.35 %).
Figure 1. Single cell RNAseq analysis on mammalian habenula.
A. Schematic of scRNAseq experiments. B. UMAP dimensional reduction and visualization of transcriptional profiles of 11,878 habenular cells. C. Top: dendrogram showing relationships between clusters. Middle: proportions of cells in each cluster. Bottom: violin plots showing levels of expression of canonical marker genes in neuronal and non-neuronal cells. D. Heat map showing scaled expression of all marker genes in each cell class (e.g. Neuron). Related to Figure S1.
Next, we calculated differentially expressed genes (DEGs) in the major cell types (e.g. Neuron, Astrocyte). A total of 1,412 genes were differentially expressed across the major cell types (corrected with a 5% false discovery rate (FDR) and log fold change > 0.25; Neuron: 405, Oligodendrocyte: 147, Astrocyte: 89, OPC: 81, Microglia: 132, Mural: 124, Endothelial: 225, Ependymal: 209). Interestingly, some detected DEGs were known habenula neuronal markers (e.g. Tac2, Htr2c) (Aizawa et al., 2012; Namboodiri et al., 2016) and markers of non-neuronal cells (Kcnj10, enriched in habenular astrocytes; Cd68, enriched in microglia) (Cui et al., 2018; Schmitt et al., 2012; Valentinova et al., 2019) (Figure 1D). To more broadly characterize the DEG expression profiles, we assigned each DEG to specific functional domains for each cell type using gene ontology (GO) analysis (Chen et al., 2013; Kuleshov et al., 2016) (Figure S1). In Neuronal cells, the enriched GO terms were broadly suggestive of processes critical for neural function in general (e.g. syntaxin-1 binding) (Zeisel et al., 2018) as well as more specific terms indicative of habenular neurons (e.g. acetylcholine-gated cation-selective channel activity) (Ren et al., 2011). Collectively, these data outline the general cellular composition of the habenula complex and point to distinct cellular processes utilized by each cell type.
scRNAseq identifies transcriptionally distinct clusters among habenular neurons.
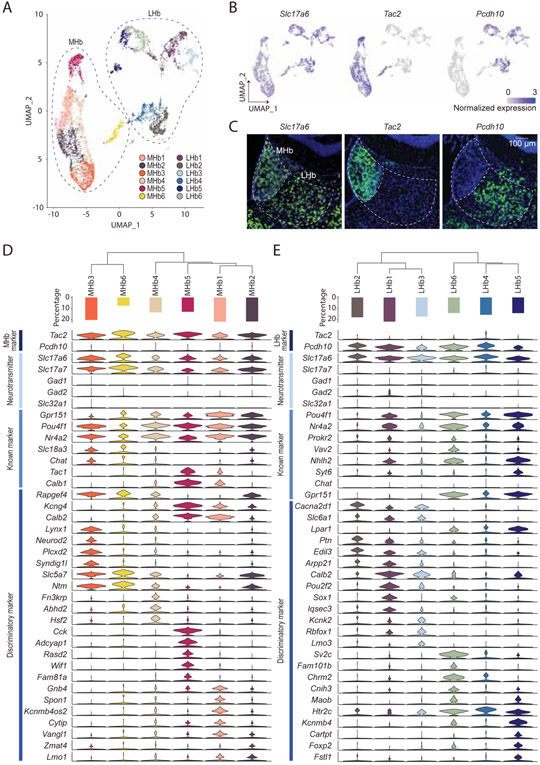
To further characterize neuronal cell types in detail, we re-clustered neuronal cells identified from the initial clustering. From this, we identified 16 discrete neuronal clusters (Figure 2A), among which 12 clusters were considered to be in the habenula due to expression of canonical gene markers (Aizawa et al., 2012; Namboodiri et al., 2016) (see methods). Visualization of canonical gene markers in UMAP space readily separated habenular neurons into 6 MHb clusters (enriched with Tac2; 2,986 cells; median UMIs/cell = 3,750, median genes/cell = 1,840) and 6 LHb clusters (enriched in Pcdh10; 2,572 cells; median UMIs/cell = 3,483, median genes/cell = 1,822) (Figure 2B, C). Consistent with previous literature (Aizawa et al., 2012; Hsu et al., 2016; Namboodiri et al., 2016), the DEG analysis between MHb and LHb clusters revealed MHb specific (170 genes; Slc17a7, Tac2) and LHb specific genes (289 genes; Pcdh10, Htr2c, Gabra1).
Figure 2. Transcriptional clusters of habenula neurons.
A. UMAP visualization of 12 MHb or LHb neuronal clusters. 4 peri-habenula clusters were removed. B. Expression plots in UMAP space illustrating normalized expression values of Slc17a6, Tac2 and Pcdh10. C. Representative FISH images for Slc17a6, Tac2 and Pcdh10 in and around the habenula. Scale bar: 100 μm. D and E. D and E correspond to the MHb and LHb respectively. Top: dendrogram showing relationships between clusters. Middle: proportions of cells in each cluster. Bottom: violin plots showing levels of expression of genes including entire MHb and/or LHb markers, major fast neurotransmitters, known markers and discriminatory markers in each cluster. Related to Figure S2 and Table S1.
To examine the transcriptional similarities between subclusters in the MHb and LHb, phylogenic trees were constructed based on a distance matrix in gene expression space (Figure 2D,E). In the MHb, subcluster #3 (MHb3) and #6 (MHb6) were closer in distance and were marked by Slc18a3 and Rapgef4 while MHb1, 2, 4 and 5 were in close proximity and were enriched with Kcng4 and Calb2. We further examined whether each MHb subcluster had unique marker genes. 16-224 genes were enriched in each MHb subcluster but few single genes were cluster specific (expressed in >30 % of cells in the enriched cluster; expressed in <10% of cells in the other subclusters). Thus, the majority of DEGs did not distinguish individual clusters alone, but their expression contributed towards a unique transcriptional profile, which when analyzed in aggregate accurately defined distinct cell types (expressed in >10% of cells in the other subclusters). Notable selective genes, Lynx1/Neurod2, Fn3krp, Cck/Adcyap1, and Kcnmb4os2 were expressed in MHb3, MHb4, MHb5 and MHb1 respectively (Figure 2D). Similarly, in the LHb, LHb1,2 and 3 were grouped with the expression of Cacna2d1 and Slc6a1 while LHb4,5 and 6 were more closely related and were marked with Gpr151, Lpar1 and Htr2c. Differential gene expression analysis revealed 77-147 enriched genes in each LHb subcluster. In contrast to the MHb, the majority of LHb subclusters could only be defined by the combination of genes except for LHb6, which was enriched with Fam101b and Sv2c (Figure 2E). Taken together, most habenula neuronal cell types can only be defined by multiple, but not single genes.
Distinct transcriptional domains between the MHb and the LHb.
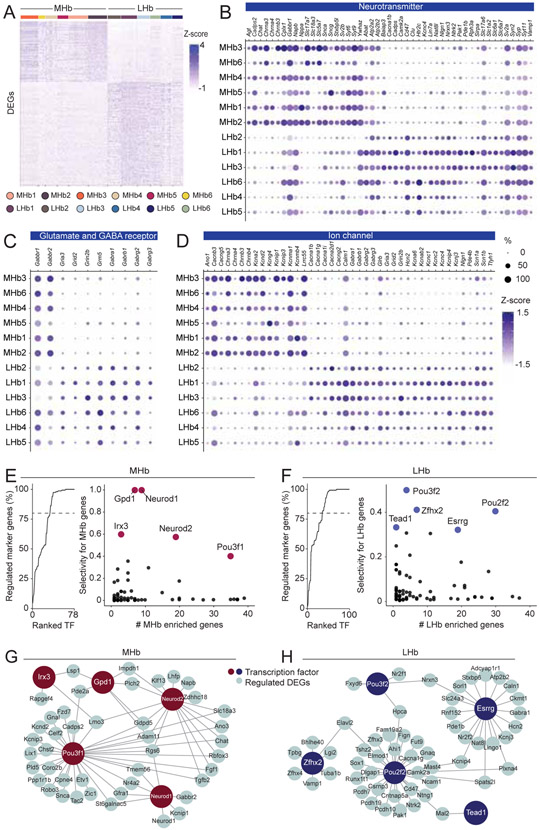
To better understand enrichment in functional domains of MHb or LHb enriched genes, we performed GO analysis on DEGs between the aggregated MHb and LHb neuronal data (the numbers of DEGs: 170 in the MHb; 289 in the LHb) (Chen et al., 2013; Kuleshov et al., 2016) (Figure 3). In the MHb, DEGs were particularly enriched in the cellular functions related to cholinergic transmission and membrane conductance (e.g. calcium ion binding, ion channels). Consistent with studies showing that the MHb is a major source of acetylcholine in the midbrain Interpeduncular nucleus (IPN) (Flumerfelt and Contestabile, 1982; Ren et al., 2011; Zhang et al., 2016), genes related to the machinery for cholinergic synthesis (Chat) and Ach gated ion channels (Chrna3, Chrna4, Chrnb3, Chrnb4) were also abundantly expressed (Figure 3 and S3). Interestingly, we found enrichments of genes related to several types of potassium channels (e.g. Kcna2, Kcnma1, Kcnip1) and calcium ion binding (e.g. Cadps2, Pcp4, Tesc, Syt15), some of which were previously well characterized in slice electrophysiology studies(Hsu et al., 2014; Quina et al., 2009) (Figure 3 and S3).
Figure 3. Comparative analysis of transcriptions and their regulations between the MHb and the LHb.
A. Heat map showing scaled expression of all marker genes for entire MHb clusters or LHb clusters. B, C and D. Dot plot illustrating scaled expression levels (color) and the proportions of expressing cells (dot size) of Neurotransmitter related genes (B), Glutamate and GABA receptor related genes (C) and Ion Channel related genes (D) in each MHb and LHb cluster. E and F. Left: Cumulative percentage of MHb (E) or LHb (F) marker genes regulated by transcription factors (TF) sorted by their selectivity to regulate marker genes. Right: Scatter plot showing the number of marker genes regulated by transcription factors (only TFs regulating <40 markers are shown) and the selectivity of gene regulation for marker genes. 5 genes with highest selectivity scores were highlighted. G and H. Schematic illustrating the regulation of marker genes by 5 TFs of high selectivity in the MHb (G) and the LHb (H). Related to Figure S3.
Ontology analysis in the LHb pointed to functions in fast neural transmission (Glutamate and GABA) and membrane conductance as well. As the LHb is innervated heavily from the entopeduncular nucleus (EPN) co-releasing Glutamate and GABA (Herkenham and Nauta, 1979; Hong and Hikosaka, 2008; Lazaridis et al., 2019; Shabel et al., 2014) and the lateral hypothalamic area (LHA; both GABAergic and Glutamatergic innervations) (Lazaridis et al., 2019; Li et al., 2019), we found enrichments in genes that are components of GABA receptors (e.g. Gabra1, Gabrb1) and Glutamate receptors (e.g. Gria3, Grid1). In addition, we found enrichments in various types of voltage gated ion channels that critically define cellular electrophysiological properties including sodium channels (e.g. Scn1a, Scn1b), calcium channels (e.g. Cacna1b, Cacna1g) (Anderson et al., 2010), potassium channels (e.g. Kcnc4, Kcna6) and hyperpolarized-activated cyclic nucleotide-gated cation channels (Hcn2) (Poller et al., 2011) (Figure 3 and S3).
Distinct transcriptional controls between the MHb and the LHb.
The divergent repertoires of gene expressions in the MHb and the LHb suggests that their expression is governed by distinct gene regulatory networks. To reconstruct gene regulatory networks in the MHb and the LHb from scRNAseq data, we performed SCENIC (single-cell regulatory network inference and clustering) analysis, which utilizes both co-expression modules between transcription factors (TFs) and candidate target genes, as well as databases of DNA binding motifs of TFs to infer significant gene regulation by transcription factors (Aibar et al., 2017; Davie et al., 2018) (Figure 3E-H). SCENIC identified 78 and 100 TFs regulating MHb enriched and LHb enriched genes, respectively (Figure 3E,F). Since some TFs ubiquitously regulated gene transcription (e.g. Elf2 and Bclaf1, which regulate over 1,000 genes), we ranked the identified TFs by their selectivity to MHb or LHb enriched genes. We found that the top 33 TFs (out of 78) in the MHb and the top 36 (out of 100) in the LHb regulated over 80% of the enriched genes (Figure 3E, F) whereas only 9 of them (25%) were common between the two subregions. This suggests that enriched genes in the MHb and LHb are controlled by distinct gene regulatory networks. In addition, the MHb and LHb enriched genes were combinatorically regulated (Figure 3G, H). The majority of enriched genes (82.4% in the MHb, 86.9% in the LHb) were regulated by multiple TFs.
Transcriptional conservation of habenular neurons between mice and fish.
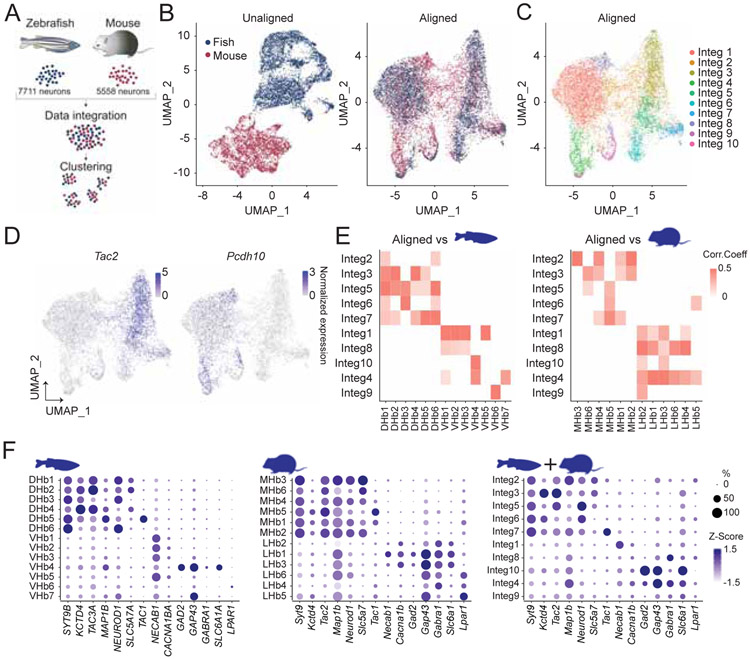
Previous studies suggest that homologous anatomical and molecular features in the habenula exist between mice and fish (Aizawa et al., 2005, 2012; Amo et al., 2010; Bianco Isaac H and Wilson Stephen W, 2009; Chou et al., 2016; Gamse et al., 2005; Herkenham and Nauta, 1979; Stephenson-Jones et al., 2012). To examine cross-species correspondence of the habenular transcriptome, we utilized Seurat V3 (Butler et al., 2018; Stuart et al., 2019) to jointly integrate the scRNAseq data sets of adult zebrafish habenula neurons (Pandey et al., 2018) (performed with chromium technology; GSE105115) and our mouse neurons (Figure 4A). Without integration, fish and mouse neuronal clusters were nearly non-overlapping (Figure 4B and S4). Seurat integration and clustering identified 10 clusters (Integ1-10), each of which consisted of both mouse and fish neurons (Figure 4B,C and S4). The expression of Tac2, a canonical marker for mouse MHb and zebrafish dorsal habenula (Aizawa et al., 2005; Gamse et al., 2005), was restricted in Integ2, 3, 5, 6 and 7 while Pcdh10, a marker for mouse LHb and zebrafish ventral habenula (Aizawa et al., 2012), marked remaining clusters (Figure 4D). To examine the extent of correspondence between integrated clusters with neural clusters in mice or zebrafish clusters (Figure S4), Pearson correlation coefficients were calculated (See methods). Consistent with canonical marker expressions, Integ2, 3, 5, 6 and 7 were moderately correlated with zebrafish dorsal habenula and mouse MHb clusters whereas Integ1, 4, 8, 9 and 10 corresponded to the ventral part or interneurons in zebrafish habenula and mouse LHb clusters (Figure 4E) with homologous gene expressions in corresponding clusters (Figure 4F), suggesting that in agreement with anatomical studies (Aizawa et al., 2005; Amo et al., 2010; Gamse et al., 2005; Herkenham and Nauta, 1979), the dorsal and ventral habenula of zebrafish are transcriptionally homologous to the MHb and the LHb of mice, respectively.
Figure 4. Integrative cross-species analysis on transcriptomes of mouse and fish habenula.
A. Schematics illustrating integrative cross species analysis of mouse and zebrafish transcriptional profiles of habenula neurons. B. UMAP dimensional reduction of transcriptional profile of jointly analyzed zebrafish and mice habenula neurons without alignment (Left) and with Seurat V3 alignment (Right). C. UMAP visualization of 10 clusters of jointly analyzed neurons. D. Expression plots showing normalized expression values for Tac2 and Pcdh10 in the UMAP space. E. Heatmap illustrating Pearson correlations between integrated clusters and zebrafish clusters (Left) and between integrated clusters and mouse clusters (Right). White, p>0.05 or correlation value<0. F. Dot plot showing scaled expression levels (color) and the proportion of expressing cells (dot size) of marker genes in zebrafish (Left), mouse (Middle) and integrated (Right) clusters. Related to Figure S4.
Visualization of transcriptionally defined habenula cell types in situ.
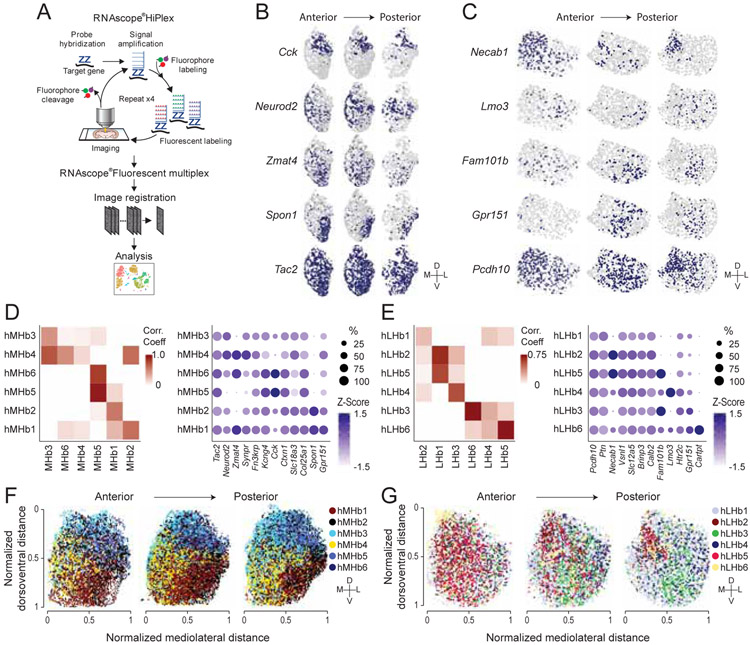
In order to cross-reference our scRNAseq data and visualize transcriptionally defined neuronal clusters in space, we performed HiPlex FISH developed by ACDbio, which permits detection of up to 15 genes within intact tissue (Figure 5A). Twelve marker genes in the MHb or LHb were detected by sequential rounds of amplification, detection, imaging and cleavage (Figure 5A-C). Because we detected 6 MHb clusters in the scRNAseq data, we next used Seurat clustering on the 22,432 cells in the HiPlex dataset to partition the data into 6 clusters (hMHb1-6) (Figure 5B, D). Correlation analysis identified corresponding clusters between the scRNAseq and HiPlex datasets with moderate to high correlation coefficient values (0.48-0.95), with the exception that MHb4 did not have a corresponding HiPlex cluster, presumably due to its lack of clear single gene markers. To examine the spatial distribution of the HiPlex clusters, anatomical coordinates of all cells were reconstructed from the raw microscopy images (Figure 5F and S5). MHb clusters were latero-medially and dorso-ventrally biased, and patterns of spatial distributions were consistent along the anterior-posterior axis. For instance, hMHb4, which was marked by Neurod2, Zmat4 and Synpr and correlated with the scRNAseq cluster MHb3, was concentrated in the medio-ventral part of the MHb while hMHb5 and 6, which were enriched with Kcng4 and Cck and corresponded with MHb5, were distributed in the dorsal MHb (Figure 5D, F). Similar approaches were applied to the LHb. In the LHb, clustering based on the expression of 12 genes in 13,151 cells identified 6 clusters (hLHb1-6) (Figure 5C, E). Correlation analysis between all the cluster pairs of the two modalities identified corresponding clusters with weak to strong correlations (0.21-0.75). Similar to the MHb clusters, reconstruction of the spatial distribution of LHb HiPlex clusters revealed clear topographical organization (Figure 5G). In addition to dorso-ventral and latero-medial bias, hLHb clusters tended to show unique distributions on antero-posterior axis as well. hLHb2, which was enriched with Necab1 and corresponded the most with LHb1 in the scRNAseq data, was most concentrated in the medial part of the anterior LHb while hLHb3, which was marked by Fam101b and corresponded with LHb6, was biased in the ventral-lateral part of the middle LHb (Figure 5E, G). Thus, HiPlex FISH analysis along with correlation analysis with scRNAseq data revealed topographical organization of transcriptionally identified clusters in the habenula.
Figure 5. Visualization of habenular transcriptional cell types in situ.
A. Schematic illustrating experimental design of HiPlex FISH. B and C. Representative ROI images showing in situ expressions of genes targeting the MHb (B) and the LHb (C). D and E. Left: Heatmap showing Pearson correlations between HiPlex clusters and scRNAseq clusters of MHb (D) and LHb (E). White, correlation value<0. Right: Dot plot illustrating the expression levels (color) and the proportions of expressing cells (dot size) of marker genes in MHb (D) and LHb (E) HiPlex clusters. F and G. Spatial distributions of MHb (F) and LHb (G) HiPlex clusters along anterior posterior axis. Related to Figure S5.
Distinct electrophysiological properties of transcriptionally defined habenula cell types.
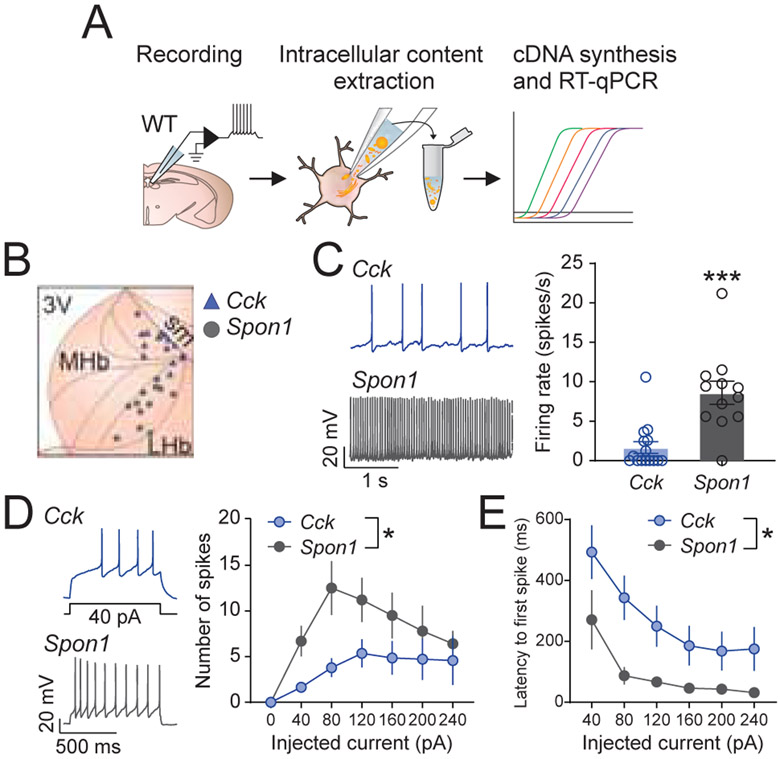
The transcriptional analysis on the habenula has revealed distinct cell types, each of which express unique patterns of genes that contribute to cellular excitability and functionality (Figure 3B-D). To test whether transcriptional identities are associated with unique functional properties, we compared the electrophysiological properties of MHb1 and MHb5 clusters because they could be readily defined by single marker genes (e.g. Spon1 and Cck, respectively) (Figure 2), and their unique spatial distributions in the MHb suggest that they form anatomically and functionally segregated units. To determine whether Cck− and Spon1−expressing neurons represent functionally distinct populations, we recorded from cells in the dorsolateral and ventrolateral MHb of wild-type mice using ex vivo patch-clamp electrophysiology. Following electrophysiological recordings, the cytosol was extracted, and cell-type identity was determined using RT-qPCR (see Methods)(Jennings et al., 2013; Stamatakis et al., 2013) (Figure 6A,B). Only cells expressing Cck, or Spon1 were included in the analysis. Each population exhibited distinct electrophysiological signatures (Figure 6 and S6). Overall, the dorsal Cck+ cells were less active at rest and less excitable than their ventral Spon1+ counterparts. Cck+ MHb neurons also exhibited lower basal firing rates (Figure 6C), spiked less in response to current injection steps (Figure 6D), and took longer to spike in response to current injection steps (Figure 6E). Collectively, these data demonstrate that transcriptionally defined MHb cell types exhibit unique functional properties.
Figure 6. Electrophysiological properties of transcriptionally defined neurons in the MHb.
A. Schematic illustrating experimental design of slice physiology experiments from molecularly defined MHb neurons. B. The spatial distributions of recorded cells. Blue: Cck (+) cells, grey: Spon1 (+) cells. C. Left: Example resting firing rate of Cck (+) (top) and Spon1 (+) (bottom) cells recorded in whole cell mode. Right: Basal firing rate was higher for Spon1 (+) cells (t-test with Welch’s correction for unequal variance: t(16.46) = 4.24, p=0.0006). D. Left: Example of Cck (+) (top) and Spon1 (+) (bottom) cells’ traces during 40 pA current injection. Right: Spon1 (+) cells spiked more in response to current injection (40 pA steps, 800 ms duration). Two-way repeated measures ANOVA [Current x Cell Type]: Main effect of Current, F(6,132)=7.99, p<0.001; Main Effect of Cell Type, F(1,22)=4.70, p=0.041; No Current x Cell Type interaction, F(6,132)=1.94, p=0.079. E. Spon1 (+) cells exhibited a shorter latency to spike during current injections. Two-way repeated measures ANOVA [Current x Cell Type]: Main effect of Current, F(5,100)=11.31, p<0.001; Main Effect of Cell Type, F(1,20)=7.35, p=0.014; No Current x Cell Type interaction, F(5,100)=0.66, p=0.65. Related to Figure S6.
Identification of MHb and LHb subclusters responding to an aversive stimulus.
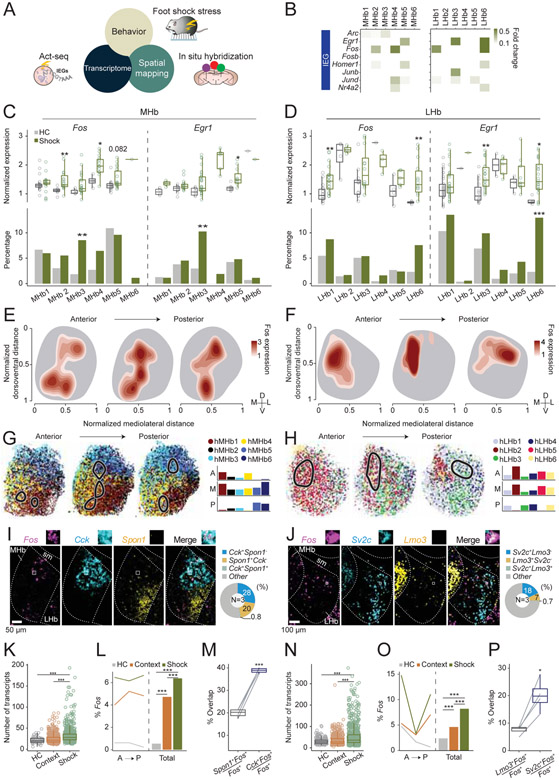
Given that both the MHb and the LHb mediate a variety of behavioral responses to aversive and appetitive stimuli including stress, pain, and addictive compounds such as nicotine and cocaine (Chou et al., 2016; Lazaridis et al., 2019; Matsumoto and Hikosaka, 2009; Stamatakis and Stuber, 2012; Velasquez et al., 2014; Yamaguchi et al., 2013; Zhang et al., 2016), we examined whether cellular activation to an aversive stimulus was transcriptionally detectable in the habenula, and if so, whether acute aversive stimulus exposure engaged transcriptionally defined cell types. Since we utilized Act-seq for our scRNAseq data acquisition (Wu et al., 2017), a technique for enhancing the signal to noise ratio of IEG detection, we could compare how acute foot shock altered IEG expression. Experimental mice received 30 foot shocks in a 60 min session while control mice remained in their home cages. We then compared IEG expression across all MHb and LHb neuronal subclusters, by examining the expression level and the proportion of Fos and Egr1 between shock and home cage groups (Figure 7A-D). This revealed that MHb2,4 and 5 showed a significant increase in Fos and/or Egr1 expression while only MHb3 showed a significant increase in the percentage of Fos and Egr1 (+) cells. For the LHb, LHb1, 3, and 6 showed increased Fos and/or Egr1 expression, but only LHb6 also showed a significant increase in the proportion of Egr1 (+) cells (Figure 7B-D). In addition to Fos and Egr1, which are widely used as surrogate neural activity markers, examinations of over 100 IEGs (Kim et al., 2019; Wu et al., 2017) revealed largely consistent activation patterns (Figure S7). To cross-validate our Act-seq results, we conducted multiplexed FISH (RNAscope) to detect stress-induced Fos and marker genes in the MHb and the LHb. Since the HiPlex FISH experiments showed spatially biased distributions of transcriptional clusters (Figure 5F-G), we generated density estimation maps of foot shock induced Fos (kernel density estimation) and projected the density estimation maps onto the anatomical gene expression maps of the HiPlex clusters (Figure 7E-H). In the MHb, consistent with scRNAseq analysis, we observed that the Fos dense area overlapped with hMHb5,6, which corresponded to MHb5 and with hMHb1,4 corresponding to MHb2,3. In the anterior and middle LHb, the Fos dense area overlapped with hLHb2,5, which corresponded to LHb1, while in the posterior LHb, Fos occupied the area enriched with hLHb3,4, corresponding to LHb3,6 (Figure 7G-H). Importantly, foot shocks induced higher Fos expression in the MHb and the LHb compared to context exposure or the home cage groups (Figure 7K, L, N, O). Since we observed spatially biased expressions of some marker genes (Figure 5B,C and S5), we examined whether there are any marker genes that better represent stress activated populations. In the MHb, Cck (+) cells representing MHb5 (Figure 2D) were colocalized with Fos more than Spon1 (+) cells, which were densely located in the ventrolateral MHb and were devoid of Fos (Figure 7I, M). Similarly in the LHb, Sv2c (+) cells, which were enriched in LHb6 (Figure 2E) overlapped with Fos (+) cells more than Lmo (+) cells (Figure 7J, P). Collectively, these results identify transcriptionally defined and genetically accessible cell types (MHb2,3,4,5 and LHb1,3,6) that are engaged following foot shock exposure, which can be prioritized for future study.
Figure 7. Activation of habenular clusters by aversive stimulus.
A. Act-seq (B-D) and multiplexed FISH (E-P) were used to identify transcriptionally defined populations responsive to aversive stimulus. B. Heatmap illustrating log fold changes in expression levels of various IEGs at each cell type comparing shock and home cage groups. White, p>0.05 or fold change<0.1. C, D. Comparison of Fos and Egr1 expression values in the expressing cells (Top) and the proportions of expressing cells (Bottom) between home cage and shock groups in the scRNAseq experiments (C: MHb and D: LHb). Wilcoxon rank sum test (Top), and Fisher’s exact test (Bottom). *p <0.05, **p <0.01, ***p <0.001. E, F. Kernel density estimation maps of foot shock induced Fos expression in the MHb (E) and the LHb (F). The outlines of the MHb or the LHb were drawn based on the HiPlex experiments. G, H. Left: Fos dense areas in the Kernel density estimation maps (level>2.5; black line) were projected onto the maps of transcriptional clusters generated by HiPlex (Figure 5F,G) in the MHb (G) and the LHb (H). Right: The percentage of cells from each cluster, which were in the Fos dense area (level>2.5). I, J. Left: Representative images of Fos and various marker genes in the MHb (I) and the LHb (J). Right. Percentage of cells expressing marker genes. K, L, N, O. Comparisons of the numbers of Fos transcripts (K: MHb, N: LHb) or the percentage of Fos (+) cells (L:MHb, O: LHb) between home cage, context and foot shock groups. Wilcoxon rank sum test (K, N), and Fisher’s exact test (L, O). ***p <0.001. M, P. The percentage of Fos cells expressing various marker genes in the MHb (M) or in the LHb (P). Paired t-test. *p <0.05, ***p <0.001. Related to Figure S7.
Discussion
Cellular diversity in the mammalian habenula.
Previous work utilizing immunohistochemistry, in situ hybridization and bulk RNA surveying techniques (e.g. microarray) (Amo et al., 2010; Hawrylycz et al., 2012; Le Foll and French, 2018; Wagner et al., 2016) have identified characteristic genes enriched in the habenula of mammals and fish. Here we conducted scRNAseq on 11,878 cells including 5,558 neuronal cells in the mammalian habenula and transcriptionally identified 12 neuronal clusters across the MHb and LHb. In the 6 MHb clusters, all of which were marked by Tac2 expression, Chat and Slc18a3, classically identified markers utilized for circuit studies targeting the MHb (Zhang et al., 2016), were only expressed in 3 clusters while Tac1 and Calb1 were expressed in the orthogonal MHb clusters as previously reported (Yamaguchi et al., 2013). Consistent with reciprocal connections between the LHb and raphe serotonergic system(Amo et al., 2014; Hikosaka et al., 2008; Vertes et al., 1999; Xie et al., 2016), Htr2c was enriched in half of the 6 LHb clusters (all were marked by Pcdh10), whereas Slc6a1 and Cacna2d1 marked the rest of clusters.
Importantly, while all 12 neuronal clusters contained enriched DEGs, few clusters had exclusively selective genes. Rather, the majority of marker genes were discriminative and expressed in multiple clusters as observed in other brain structures using similar approaches (Moffitt et al., 2018; Tasic et al., 2018). This observation motivated us to perform HiPlex FISH to expand conventional 3 color FISH into sequential detections of over 10 discriminative genes covering clusters in the MHb and LHb. Based on the marker genes for clusters identified in scRNAseq analysis, we performed HiPlex FISH to visualize 12 marker genes each for the LHb or MHb and to reconstruct clusters in situ. There was largely correspondence between scRNAseq and HiPlex clusters in both the MHb and the LHb (Figure 5). Each scRNAseq cluster was correlated with 1-2 HiPlex clusters. Moreover, HiPlex clusters showed spatially biased distributions (Figure 5). Andres et al anatomically divided the MHb and LHb into subnuclei, based on Nissl staining in rat tissue (Andres et al., 1999). 4 major subdivisions of the MHb were identified comprising superior (MHbS), central (MHbC), lateral (MHbL) and inferior (MHbI) parts while the LHb could be divided into superior (LHbS), parvocellular (LHbMPc), central (LHbMC), magnocellular (LHbMMg), lateral (LHbLO and LHbLMc) and anterior (LHbMA) parts. Our HiPlex FISH analysis suggests that some transcriptionally defined clusters have correspondence with anatomically defined subnuclei while other clusters distribute as part of a subnucleus or across subnuclei. For example, cells in hMHb2 (MHb1), which were enriched with Spon1, were in the MHbL, while hMHb4 (MHb3, Neurod2) and hMHb5/6 (MHb5, Cck) distribute in the ventral part and dorsal/dorsolateral part of the MHbC, respectively. In the LHb, hLHb5 (LHb1) is concentrated in the anterior part of LHbMc while cells in hLHb3 (LHb6, Sv2c and Fam101b) were located in the ventral part of the LHbLMc and the LHbLO.
Wallace et al recently published transcriptional analysis of the mouse habenula using a droplet based sequencing technology (Wallace et al., 2020). Consistent with our clustering results (Figure 2), Tac2 marked the whole MHb while several genes including Gap43 and Pcdh10 were enriched in the LHb. Several marker genes in the neuronal subclusters were consistently revealed including Lmo3, Spon1, Adcyap1 and Cck in the MHb and Gad2, Chrm3, Gpr151 in the LHb. In addition, some of the marker genes had topographically unique distributions in the habenula. However, although similar graph-based clustering (Seurat) was performed in both studies, our clustering revealed finer cell-types in the habenula (12 neuronal clusters in our analysis whereas 9 neuronal clusters were reported in Wallace et al paper). This might be presumably due to the technological differences (10x genomics based platform in our study detected more genes (median genes: 1,001 whereas conventional droplet based platform in Wallace et al study detected median 349 genes) (Zhang et al., 2019) and the number of cells collected (our study: MHb: 2986 cells, LHb: 2572 cells; Wallace et al study: MHb 3370 cells, LHb: 560 cells). Nevertheless, it is an important future direction to combine and further analyze both dataset to comprehensively understand habenular cellular diversity.
Activation of habenula cell types by aversive stimuli.
Previous work has demonstrated that both the MHb and LHb are engaged by a variety of aversive stimuli. To examine this further, we sought to determine whether transcriptionally distinct habenula cell types were preferentially activated (as indexed by IEG expression) following exposure to foot shock (Figure 7). Act-seq showed that a subset of neuronal clusters (MHb2,3,4,5 and LHb1,3,6) displayed elevated IEG expression following this procedure. Since scRNAseq generally detects 10 fold fewer transcripts than smFISH (Moffitt et al., 2018), we cannot rule out the activation of other habenula cell types following aversive stimulus exposure, as it is possible that other cell types are also activated to a lesser degree. To validate Act-seq results, we cross-referenced spatial maps of foot shock induced Fos detected by FISH with the spatial distributions of transcriptional clusters generated by HiPlex. This multimodal analysis revealed largely consistent Fos activation patterns between scRNAseq and FISH analysis. These data point to several cell types in the habenula that merit further attention in future studies. For instance, MHb5 could be targeted utilizing the expression of Cck, while MHb3 could be genetically targeted using an intersectional approach based on the expression of Neurod2 and Lynxl. In the LHb, LHb6 may be accessible based on the expression of Sv2c and Fam101b.
The habenula has been shown to process a variety of aversive stimuli of different modalities including painful somatosensory stimuli (e.g. foot shock) (Wang et al., 2017; Yamaguchi et al., 2013; Zhang et al., 2016), visceral malaise (e.g. LiCl) (Otsu et al., 2019; Wirtshafter et al., 1994) and predator odors (e.g. TMT) (Yang et al., 2016). Various types of aversive stimuli induced largely similar spatial distributions of Fos in the LHb (Wirtshafter et al., 1994), suggesting that a subset of neurons in the LHb encodes aversion irrespective of the modalities. It is an important future direction to examine whether aversion induced by different modalities will engage the same or different transcriptional cell types as the foot shock exposure.
Our slice electrophysiological experiments followed by single cell RT-qPCR demonstrate clear associations between transcriptional identities (MHb1 and MHb5) and electrophysiological properties. Currently, it is still unclear whether all transcriptionally defined habenula neurons display unique functional properties. Interestingly, while substantial transcriptional heterogeneity exists in both the LHb and the MHb, electrophysiological (Amo et al., 2014; Cui et al., 2018; Lecca et al., 2017; Li et al., 2011; Matsumoto and Hikosaka, 2007, 2009; Yang et al., 2018) and calcium imaging (Shabel et al., 2019) studies show that large proportions of LHb neurons (~30 to 50 %) display an increase in their firing/activity rates in response to, or following aversive stimuli exposure. Thus, it is currently unknown whether transcriptionally distinct populations in the LHb are associated with specific in vivo encoding dynamics in response to rewarding and aversive stimuli or whether transcriptionally distinct cell types display similiar activity dynamics (at least in response to simple aversive stimuli exposure). Future studies exploring how in vivo activity dynamics of habenula cell types relates to their molecular features and circuit connectivity are needed to better resolve this.
Transcriptional definition of habenular cellular properties.
The comparisons of transcriptomes between the whole units of the MHb and the LHb identified 170-289 genes related to neurotransmitters, glutamate and GABA receptors, ion channels, and morphology, which aid in defining cellular properties and functions (Figure 3 and S3). These distinct sets of genes are likely under the unique transcriptional controls in the MHb and the LHb. SCENIC analysis revealed that TFs, which regulates >80% of the DEGs, are distinct between the two nuclei. Moreover, the majority of the DEGs are regulated by the combination of multiple TFs, and some TFs regulate the expression of other TFs in the DEG (e.g. Tcf7l2 regulates Nfib in the MHb). These results suggest that cellular and functional properties of the MHb and the LHb are maintained by distinct transcriptional networks.
The unique patterns of transcription factor activities might also regulate the development of patterning and connectivity of the MHb and LHb (Quina et al., 2009). Recently, Lipiec et al found that transcription factor Tcf7l2 regulates the development of anatomical segregations and axonal patterns of the habenula through Tcf7l2 knockout assays (Lipiec et al., 2019) whereas Tcf7l2 also regulates nicotine sensitivity and nAChR function in the MHb in adult rodents (Duncan et al., 2019). Consistent with their findings, our SCENIC analysis showed that Tcf7l2 regulates a variety of DEGs including ion channels, neurotransmission and cytoskeleton related genes in the MHb and the LHb. The roles of other TFs revealed in our analysis in the habenula development remains elusive and are one of the critical future directions from our study. In addition to constitutively expressed TFs, we also observed increased levels of various TFs including Fos and Egr1 in response to aversive stimuli. Indeed, SCENIC analysis revealed 10 and 127 genes, which are respectively regulated by Fos and Egr1, suggesting that transcriptional controls by several IEGs potentially underlie experience dependent modifications of habenula circuitry (Shabel et al., 2019).
Conservation of cell types across species.
The habenula is an ancient and evolutionally conserved structure existing in all vertebrates from fish to humans (Aizawa et al., 2011). The shared features in cytoarchitecture, projection patterns and functions between species have been extensively studied. Raphe projecting neurons in zebrafish are exclusively present in the ventral habenula, which are marked by Pcdh10a expression (Amo et al., 2010) and involved in the expectation of danger (Amo et al., 2014), whereas the LHb in mice also projects to the dorsal raphe nucleus (Quina et al., 2015), is highly enriched in Pcdh10 (Amo et al., 2010) and regulates aversion and reward phenotypes. In contrast, IPN projecting habenula neurons are concentrated in the dorsal part in zebrafish (Aizawa et al., 2005; Chou et al., 2016; Gamse et al., 2005) and in the medial part in mice (Herkenham and Nauta, 1979), which are enriched in Tac2 and regulates fear and anxiety related behaviors in both species(Aizawa et al., 2012; Duboué et al., 2017; Ogawa et al., 2012; Yamaguchi et al., 2013; Zhang et al., 2016). Consistent with the previous observations, whole transcriptome analysis of mouse and zebrafish habenula showed that dorsal and ventral parts of zebrafish habenula respectively correspond to the MHb and the LHb in mice. Interestingly and consistent with the observations in cross species scRNAseq analysis (Butler et al., 2018; Hodge et al., 2019), clustering without Seurat integration generated nearly non-overlapping mice and zebrafish clusters (Figure 4B and S4), which is likely due to experimental differences between studies (e.g. cellular sequencing depth) as well as non-homologous transcriptomes between species. Structural asymmetry present in zebrafish habenula may partially underlie transcriptional diversity between species (Beretta et al., 2012; Pandey et al., 2018). Thus, the habenula is an anatomically, functionally and transcriptionally conserved structure in vertebrates.
Summary and future directions.
While single gene markers and cre driver lines have been employed to gain genetic access to habenula cells (see (Proulx et al., 2014) for discussion), in most cases, the expression of single genes does not accurately reflect habenula cell type identities. This is important to consider for future studies that will employ genetic targeting methods for manipulating habenula cell types, and presents an overall challenge for the field. The transcriptomic analysis of mammalian habenula using scRNAseq and HiPlex presented here derives a census of transcriptional diversity by unbiased clustering, provides comprehensive transcriptomic resources to the community, and infers functionalities of divergent cell types. In addition, there are largely consistent observations between distinct modalities in our dataset (e.g. scRNAseq and HiPlex; mice and zebrafish) and between our analysis and past scholarship, further suggesting the validity of our analysis. The future investigations on the electrophysiological and functional properties of transcriptionally defined cell types are expected to deepen our understanding of the habenula in the complex motivational states.
STAR★Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Garret Stuber (gstuber@uw.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The NCBI Gene Expression Omnibus accession number for the scRNAseq data reported in this paper is GSE137478. All the codes used to analyze scRNAseq, Hiplex and RNAscope data have been made publicly available at a Github repository affiliated with Stuber Laboratory group and this manuscript title (http://www.github.com/stuberlab/).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J male mice (Jackson Laboratory) were used for single-cell preparation and Fluorescent in situ hybridization (FISH) experiments (P50-55 at tissue extraction). Mice were group housed except that for the last two days before tissue isolation, they were singly housed. Mice had ad libitum access to food and water and were kept under a reverse 12 h light-dark cycle. All experiments were conducted in accordance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the University of North Carolina and the University of Washington before the start of any experiments.
METHOD DETAILS
Single-cell preparation, cDNA library construction for scRNAseq.
Shock (n=4) and Home cage (HC) (n=4) groups were handled 5 min/day for 7 days. Mice in the Shock group were habituated to the shock chamber (Med Associates Inc.) 10 min/day for 7 days prior to the behavior experiment. On the shock day, mice in the Shock group received 30 foot shocks (0.3 mA, 1 s duration, mean inter shock intervals: 116 s) in 60 min. Immediately after the shock session, mice were deeply anesthetized with intraperitoneal injection of 0.2 mL of stock solution containing sodium pentobarbital (39 mg/mL) and phenytoin sodium (5mg/mL) and transcardially perfused with ice-old NMDG-aCSF containing inhibitor cocktails which was used throughout procedures unless noted. NMDG-aCSF: 96 mM NMDG, 2.5 mM KCl, 1.35 mM NaH2PO4, 30 mM NaHCO3, 20 mM HEPES, 25 mM glucose, 2 mM thiourea, 5 mM Na+ ascorbate, 3 mM Na+ pyruvate, 0.6 mM glutathione-ethyl-ester, 2 mM N-acetyl-cysteine, 0.5 mM CaCl2, 10 mM MgSO4; pH 7.35-7.40, 300-305 mOsm, oxygenated with 95% O2 and 5% CO2. Inhibitor cocktails: 500 nM TTX, 10 μM APV, 10 μM DNQX, 5 μM actinomycin, 37.7 μM anysomycin. Mice in the control group were removed directly from the home cage. It is important to note that the aCSF used for perfusion also contained transcription and translation inhibitors and the time between i.p. injection of pentobarbital and the start of perfusion was about 1 min to minimize potential transcriptional events induced by pentobarbital injection, perfusion and brain extraction. Brains were extracted and coronal sections containing the habenula were prepared at 300 μm using a vibratome (Leica, VT1200). Slices including the habenula (3-4 slices/animal) were recovered in a chamber for 30 min on ice. Habenula samples were then punched (0.75 mm diameter, EMS) from 4 animals/group (6-8 punches/animal) pooled and enzymatically digested with 1 mg/mL pronase (Sigma-Aldrich) for 45 min at room temperature, followed by mechanical trituration with fire-polished glass capillaries (tip diameter 200-300 μm) and filtered through strainers (pore size 40 μm) to remove cellular aggregates. Dead and dying cells in the cell suspension were then removed using a dead cell removal kit (Miltenyi Biotec). After centrifugation, a small fraction of cells (~5ul) were mixed with trypan blue to manually determine the proportion of live cells and cell concentrations using a hemocytometer. Only samples of high viability (>80%) were used for subsequent library generation. Final cell concentration was adjusted to 1,000 cells/μL.
cDNA libraries were constructed following manufacture’s instruction (Chronium Single Cell 3’ Reagents Kits V2 User Guide, 10x Genomics). Briefly, ~17,000 dissociated cells were mixed with reverse transcription mix and loaded into the chip. The mRNAs of single cells were captured by barcoded beads using a Chromium controller. Reverse transcribed cDNAs were then PCR amplified, fragmented, and ligated with adaptors followed by sample index PCR. cDNA libraries were sequenced on an Illumina Nextseq 500 (v2.5) and the alignment of raw sequencing reads to the mouse genome was conducted using the 10x Genomics Cell Ranger pipeline (V2) to obtain cell by gene matrices for subsequent downstream analysis.
Multiplex/HiPlex FISH.
Mice (n=4-5) were deeply anesthetized with isoflurane, decapitated and brains were rapidly flash frozen on dry ice. Coronal sections were cut at 20 μm on a cryostat (Leica) and stored at −80 °C until use. HiPlex and subsequent Fluorescent multiplex assays (ACDBio) were performed following manufacture’s instruction (Wang et al., 2012). The HiPlex together with Fluorescent multiplex assay allows multiplex detection for up to 15 targets on a single tissue section. Briefly, sections were fixed in 4% paraformaldehyde, dehydrated with 50%, 70%, 100% ethanol, then treated with protease. In HiPlex, all gene targets are hybridized and amplified together, whereas detection is achieved iteratively in groups of 2-3 targets. Sections were incubated with pooled HiPlex probes (11 and 10 probes for MHb and LHb, respectively), and amplified with a series of amplification solutions, then detection solution was applied. MHb probes; Neurod2, Spon1, Zmat4, Kcng4, Fn3krp, Ctxn1, Tac2, Col25a1, Cck , Gpr151, Synpr. LHb probes; Necab1, Rflnb, Cartpt, Vsnl1, Lmo3, Brinp3, Slc12a5, Pcdh10, Ptn, Gpr151. Cell nuclei were counterstained with Dapi and samples were mounted. Imaging was performed as described below. After each round, samples were treated with TCEP solution to cleave fluorophores, then moved on to the next round of the fluorophore detection procedures. After completion of 4th round of HiPlex, samples were proceeded to probe hybridization step of Fluorescent multiplex (ACDBio). Samples were hybridized with probes, incubated with amplification solution, signals were visualized by applying detection solution. MHb; Slc18a3. LHb; Calb2, Hcr2c.
Images were obtained with Zeiss ApoTome2 with 20x objective using Zen (blue edition) software (Zeiss). All images from all rounds of staining were then registered to each other to generate 12 plex images using HiPlex image registration software (ACDBio). The mean fluorescent intensity was then subtracted with background and threshold which is determined for each gene. The cutoff value was defined as the mean fluorescent intensity of 3 transcripts.
Multiplex Fos FISH.
Three behavioral groups were prepared (n=2-4 mice/group). Home cage, and foot shock group received the same procedures described in the scRNAseq experiments. Context exposure group received identical procedure as the foot shock group except that they did not receive any foot shocks. 45 min after the behavioral session, the mice were deeply anesthetized with isoflurane, decapitated and brains were rapidly flash frozen on dry ice. The subsequent probe hybridization and signal detection procedures were performed following manufacture’s instruction (ACDbio). Image acquisition procedure was identical to Hiplex experiments except that only 3 genes (MHb: Fos, Cck, Spon1; LHb: Fos, Sv2c, Lmo3) were detected in each sample.
Patch-Clamp Electrophysiology.
Experimental methods were adapted from our previous study (Stamatakis et al., 2013). Male wild-type mice (n=5; 9-10 weeks old) were deeply anesthetized with pentobarbital and transcardially perfused with ice-cold sucrose cutting solution containing (in mM): 75 sucrose, 87 NaCl, 1.25 NaH2PO4, 7 MgCl2, 0.5 CaCl2, 25 NaHCO3, 304-308 mOsm. Brains were rapidly removed, and coronal sections were taken at 300 μm. Sections were incubated in aCSF at 32 degrees C containing (in mM): 126 NaCl, 2.5 KCl, 1.2 NaH2PO4, 1.2 MgCl2, 2.4 CaCl2, 26 NaHCO3, 15 glucose, 301-306 mOsm. Borosilicate pipettes were pulled at 3-3.5 MΩ and backfilled with K-gluconate solution containing (in mM): 135 potassium gluconate, 4 KCl, 10 HEPES, 4 ATP, 0.3 GTP (pH 7.35, 285 mOsm). In addition, 2 μL RNase inhibitor (ANTI-RNase, Life Technologies) were added per 1 mL of internal. Once a gigaseal was established, basal firing rate was measured in cell-attached mode for 30 s. Upon break in, cell properties were measured. Unstable recordings were immediately abandoned, and data were excluded from further analyses. Basal firing rate was measured in whole-cell mode for 30s. Rheobase was then measured by holding cells at −70 mV and injecting brief current pulses (50 ms) in 10 pA steps (1 s between sweeps). Action potential properties were measured from the rheobase spikes. 800 ms current pulses were injected at −70 mV in 40 pA steps from −80 to 240 pA (10 s between sweeps) before extracting the cytosol.
Once in whole-cell mode, cells were held for less than 3 min to minimize mRNA degradation. Following each recording, the cytoplasm was gently aspirated with negative pressure, the pipette was removed from the tissue, and the tip was broken into an RNase-free PCR tube containing lysis solution with DNase (Single cell-to-CT Kit, Invitrogen) while gentle positive pressure was applied. Between each recording, the silver chloride electrode wire was wiped with 70% ethanol to minimize cross contamination. Intracellular content aspirated immediately after slice recording was processed for real-time PCR following manufacture’s instruction (Single Cell-to CT kit, Invitrogen). Briefly, mRNA was extracted in lysis solution containing DNase I, then reverse transcribed. cDNA of target genes was preamplified using pooled TaqMan Gene Expression Assay probes (Cck, Spon1, Thermo Fisher Scientific). Rn18s, an internal reference gene, is highly abundant, therefore its probe was not included. Preamplified product was diluted and run for real-time PCR using 7300 system and SDS v1.4.1 software (Applied biosystems). Only samples that met the following criteria were included in the analysis: Amplification curve displays sigmoid shape, and absence of the curve in no template control. High consistency between technical replicates (Ct value difference <0.5). Higher Ct value than cut-off value 35. Positive for cell type marker Cck or Spon1. After quality control, a total of 15 dorsal cells expressing Cck and 12 ventral cells expressing Spon1 were included for further analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data Analysis for scRNAseq data.
Clustering, differential gene expression analysis and integrative analysis of differential conditions, cross species and cross modalities were performed using the Seurat V3 package (Stuart et al., 2019). SCENIC package was utilized to infer gene regulatory networks by transcription factors (Aibar et al., 2017).
Data preprocessing and doublet removal.
Low abundant genes (expressed in less than 3 cells) and cells of potentially low quality (total UMI<700 or total UMI>15,000, or percentage of mitochondrial genes >20%) were removed from downstream analysis. Suspected doublet cells were computationally removed by utilizing DoubletDecon package (Version 1.02.) (DePasquale et al., 2019) with the default settings.
Integrative Clustering and differential gene expression analysis.
To minimize the effects of experimental variations (batch effects and behavioral conditions) on clustering, we used Seurat V3 analysis package (Stuart et al., 2019), which utilized canonical correlation analysis (Butler et al., 2018) and mutual nearest neighbor analysis (Haghverdi et al., 2018). Briefly, gene counts were scaled by the cellular sequencing depth (total UMI) with a constant scale factor (10,000) and then natural-log transformed (log1p). 2,000 highly variable genes were selected in each sample based on a variance stabilizing transformation. Anchors between individual data were identified and correction vectors were calculated to generate an integrated expression matrix, which was used for subsequent clustering. Integrated expression matrices were scaled and centered followed by principal component analysis (PCA) for dimensional reduction. PC1 to PC30 were used to construct nearest neighbor graphs in the PCA space followed by Louvain clustering to identify clusters (resolution=0.8). For visualization of clusters, Uniform Manifold Approximation and Projection (UMAP) was generated using the same PC1 to PC30.
To identify markers (differentially expressed genes) for neuronal and non-neuronal cells, first, clusters of each cell types (neuron, astrocyte, oligodendrocyte, microglia, ependymal cells, OPC, and endothelial cells) were combined. And then, expression value of each gene in given combined clusters were compared against the rest of cells using Wilcoxon rank sum test and p-values were adjusted with the number of genes tested. Gene with log fold change >0.25 and adjusted p-value <0.05 were considered to be significantly enriched. To examine the stability and robustness of clustering results, 10-100 % of cells were randomly sub-sampled and clustered by the identical procedure described above for 10 times at each sub-sampling rate.
Based on the expression of canonical markers for neuronal cells (Stmn2 and Thy1), cells in the neuronal clusters were extracted to be re-clustered using the identical integrative clustering and differentially expressed gene analysis described above. Initial clustering resulted in 16 neuronal clusters, among which 4 clusters (436 cells) were excluded in downstream analysis due to their expressions of peri-habenula markers. Those 4 clusters were marked with one or a couple of following genes: Pdyn, Lypd6b, Lypd6, S1pr, Gbx, Ramp3, Cox6a2, Slitrk6 and Dgat2 (Figure S1). Gene ontology database at Mouse Genome Informatics was used to generate dot plots for the expression of GPCRs (GO: 0004930; G protein-coupled receptor activity; gene names containing “receptor” were used; Figure S2) and neuropeptide (GO: 0007218; neuropeptide signaling pathway; gene names containing “receptor” were excluded; Figure S2.). Similarly, Dot plots illustrating expression of neurotransmitter related genes (GO: 0042133; neurotransmitter metabolic process, GO: 0006836; neurotransmitter transport, GO: 0001504; neurotransmitter uptake, GO: 0007269; neurotransmitter secretion), Ion channel related genes (GO: 0034702; Ion channel complex), and glutamate and GABA receptor related genes (GO: 0008328; Ionotropic glutamate receptor complex, GO: 0098988; G-protein coupled glutamate receptor activity, GO: 0016917; GABA receptor activity (Gpr156 was excluded) were generated for the MHb or LHb enriched genes (Figure 3) (Ashburner et al., 2000).
Gene Ontology analysis.
Enriched ontology terms for differentially expressed genes were identified using Enrichr (Chen et al., 2013; Kuleshov et al., 2016). GO Biological Process 2018, GO Molecular Function 2018, and GO Cellular Component 2018 were referenced to identify ontology terms with the adjusted p-value<0.1 (Figure S1 and S3).
Gene regulatory network analysis.
The SCENIC package (Aibar et al., 2017) was utilized to infer transcriptional regulation on the DEGs in the MHb and LHb by transcriptional factors. SCENIC analysis consisted of two major steps: construction of co-expression network using GENIE3 and identification of direct binding by DNA-motif analysis using RcisTarget. Log-normalized expression matrix of habenular neurons generated using Seurat was used as input data. After running GENIE3, motif data set (mm9-500bp-upstream-7species.mc9nr.feather, mm9-tss-centered-10kb-7species.mc9nr.feather) was used to construct regulons for each transcription factor. To examine selectivity of transcriptional regulation on DEGs in the MHb or LHb, selectivity scores for each transcription factor was calculated by dividing the number of DEGs in the regulon by the number of all genes in the regulon. Cumulative plots illustrating the coverage of DEGs by the transcription factors were generated by using sorted transcription factors based on the selectivity score.
Cross-species analysis.
Transcriptomes of Zebrafish and mouse habenulae were joint analyzed to examine conservation of habenular transcriptome across species. The zebrafish habenula dataset was obtained from the publicly available repository (Accession number: GSE105115). Since our mouse dataset was generated from adult mice using 10x Genomics platform, we also used zebrafish dataset which was generated from adult subjects with 10x Genomics platform (GSM2818522, GSM2818523) to minimize confounding effects introduced by experimental settings. To integrate zebrafish and mouse data, first, gene names in the zebrafish were converted to mouse genes using biomaRt package (Durinck et al., 2005, 2009) and bioDBnet (biological DataBase network). Gene counts of zebrafish genes that were converted to the identical mouse genes were averaged. 8,951 genes were used for integrative analysis. Zebrafish and mouse datasets were joint clustered using Seurat V3 as described above. 100 variable genes in each sample were used to generate integrative expression matrix. PC1-PC7 were used for clustering and UMAP visualization. To establish correspondence between cross species integrated clusters and mouse or zebrafish clusters, which were generated independently by the clustering approach described in the section of “Integrative Clustering and differential gene expression analysis” (Figure S4), expression values of all cells in each cluster were averaged for each gene and then pairwise Pearson’s correlation coefficient between all pairs of clusters in each dataset was computed. In the heatmaps to visualize Pearson’s correlation of two expression matrix (Figure 4), negative correlations or correlations which were not statistically significant (p>0.05) were adjusted to 0 (White).
Analysis of electrophysiological data.
Electrophysiological data were processed using Clampfit v11.0.3.03 (Molecular Devices) and analyzed using GraphPad Prism v8.3.0. All tests were two-sided and corrected for multiple comparisons or unequal variance where appropriate. Data are presented as mean ± standard error of the mean (SEM).
Integrative analysis of HiPlex and scRNAseq data.
To cluster cells based on the detected mRNAs’ expression matrix and further establish correspondence between clusters in scRNAseq and HiPlex experiments, Seurat V3 was utilized. Cells which did not express any genes were removed for downstream analysis. Expression data were log normalized and scaled, and PCA was conducted using all the genes. PC1-PC10 were used for graph-based clustering and UMAP visualization. Since 2000 variable genes were used for clustering in scRNAseq analysis in comparison to only 12 genes in HiPlex experiments and we also found that our clustering results of scRNAseq data were stable across various parameter settings (data not shown), we assumed that clustering results in scRNAseq were more reliable than HiPlex experiments. Thus, we adjusted the granularity/resolution of clustering in HiPlex experiments to match the numbers of clusters in HiPlex experiments with those of scRNAseq analysis. In the LHb, 1 of 7 clusters was excluded from analysis as it only consisted less than 2 % of all cells. To establish correspondence between clusters in scRNAseq and HiPlex experiments, expression values of all cells in each cluster were averaged for each gene and then pairwise Pearson’s correlation coefficient between all pairs of clusters in each dataset was computed. In the heatmaps to visualize Pearson’s correlation of two expression matrix (Figure 5), negative correlations were adjusted to 0 (White). Since correlation was computed using relatively small number of genes (12), p values were not considered in the heatmap (Figure5).
IEG analysis in scRNAseq and RNAscope experiments.
Fisher’s exact test was used to test the significance of the proportion of IEG expressed cells in the shock group against the control group. Wilcoxon rank- sum tests were used as a statistical test for IEG expression levels (Figure 7C,D,K,N). P-values were Bonferroni-corrected in all data. Paired t-tests were used as a statistical test for overlap score (Figure 7M, P). Data are presented as median with 95 % confidence interval.
Statistics
Statistical details for each experiment are described in the corresponding figure legend, results, and methods. Statistical analysis for electrophysiological data was conducted using GraphPad Prism v8.3.0 after determining whether data were normally distributed. Non-parametric tests were performed to compare distributions of gene expression levels in scRNAseq and RNAscope data using R. No statistical tests were used to determine sample sizes.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Anisomycin from Streptomyces griseolus | Sigma-Aldrich | Cat#A9789 |
| Pronase | Sigma-Aldrich | Cat#10165921001 |
| Actinomycin D | Sigma-Aldrich | Cat#A1410 |
| N-Methyl-D-glucamine | Sigma-Aldrich | Cat#66930 |
| (+)-Sodium L-ascorbate | Sigma-Aldrich | Cat#A7631 |
| Sodium pyruvate | Sigma-Aldrich | Cat#P2256 |
| N-Acetyl-L-cysteine | Sigma-Aldrich | Cat#A7250 |
| SUPERase• In™ RNase Inhibitor | Invitrogen | AM2694 |
| Critical Commercial Assays | ||
| RNAscope Fluorescent Multiplex Reagent Kit | ACDBio | Cat#320850 |
| RNAscope HiPlex12 Ancillary Kit | ACDBio | Cat#324140 with RNAscope HiPlex Alternate Display Module (custom designed upon request, Cat# N/A) |
| Chromium Single Cell 3′ Library & Gel Bead Kit v2 | 10x Genomics | Cat# 120267 |
| Chromium i7 Multiplex Kit Kit | 10x Genomics | Cat#120262 |
| Chromium Single Cell A Chip | 10x Genomics | Cat#1000009 |
| Chromium Controller & Accessory Kit | 10x Genomics | Cat#120223 |
| Dead Cell Removal Kit | Miltenyi Biotec | Cat#130-090-101 |
| Illumina NextSeq 500 v2.5 | Illumina | Cat#20024907 |
| Single Cell-to-CT™ qRT-PCR Kit | Invitrogen | Cat#4458237 |
| Deposited Data | ||
| Mouse scRNA-seq data | Gene Expression Omnibus | GSE137478 |
| Adult zebrafish habenula scRNA-seq data from Pandey et al., 2018 | Gene Expression Omnibus | GSM2818522, GSM2818523 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J males | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Oligonucleotides | ||
| TaqMan® Assays-Mm-Cck | Thermo Fisher Scientific | Mm00446170_m1 |
| TaqMan® Assays-Mm-Rn18s | Thermo Fisher Scientific | Mm03928990_g1 |
| TaqMan® Assays-Mm-Spon1 | Thermo Fisher Scientific | Mm00523995_m1 |
| RNAscope® Probes, see Table S2 | ||
| Software and Algorithms | ||
| RNAscope HiPlex Image Registration Software | ACDBio | Cat#300065 |
| ImageJ (Fiji) | Schindelin et al., 2012 | RRID: SCR_002285; http://fiji.sc |
| SCENIC (v1.1.0-01) | Aibar et al., 2017 | https://aertslab.org/#scenic RRID: not found |
| R (v3.4.0 2017-04-21) | N/A | https://www.r-project.org/ RRID:SCR_001905 |
| Seurat v3 | Stuart et al., 2019 | RRID: SCR_016341; https://satijalab.org/seurat/get_started.html |
| DoubletDecon (v1.02) | DePasquale et al., 2019 |
https://github.com/EDePasquale/DoubletDecon RRID: not found |
| Enrichr | Chen et al., 2013; Kuleshov et al., 2016 |
http://amp.pharm.mssm.edu/Enrichr/ RRID:SCR_001575 |
| biomaRt | Durinck et al., 2005, 2009 |
https://bioconductor.org/packages/release/bioc/html/biomaRt.html RRID: not found |
| bioDBnet | Mudunuri et al., 2009 |
https://biodbnet-abcc.ncifcrf.gov/ RRID: not found |
| Cell Ranger | 10x Genomics | RRID: SCR_017344; https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| The Gene Ontology (GO) Project | Ashburner et al., 2000 |
http://www.informatics.jax.org/mgihome/GO/project.shtml RRID:SCR_006447 |
| GraphPad Prism v8.3.0 | GraphPad Software |
https://www.graphpad.com/ RRID:SCR_002798 |
| pClamp v11.0.3 | Molecular Devices |
https://www.moleculardevices.com/ RRID:SCR_011323 |
Highlights.
scRNAseq identifies transcriptional cell types in the mammalian habenula
Zebrafish and mouse habenula are transcriptionally conserved
Topographical and functional organization of transcriptionally defined cell types
Identification of transcriptional cell types in response to aversive stimulus
Acknowledgements
We thank Stuber lab members for critical comments on the project and the manuscript. We thank C. Trapnell for helpful discussions. We thank S. Ng-Evans for the assistance with behavioral equipment. We thank Y. Tao and Z. Hu (University of North Carolina) for assistance with 10x genomics library generation. We thank R. Ying and O. Kosyk (University of North Carolina) for mouse colony maintenance. This work was supported by the Brain and Behavior Research Foundation (NARSAD Young Investigator Awards) (K.H. and M.A.R.), the National Institutes of Health DK121883 (M.A.R.), NS007431 (M.L.B.), DA038168 and DA032750 (G.D.S.), the Foundation of Hope (G.D.S.) and P30DA048736.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agetsuma M, Aizawa H, Aoki T, Nakayama R, Takahoko M, Goto M, Sassa T, Amo R, Shiraki T, Kawakami K, et al. (2010). The habenula is crucial for experience-dependent modification of fear responses in zebrafish. Nat. Neurosci 13, 1354–1356. [DOI] [PubMed] [Google Scholar]
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Bianco IH, Hamaoka T, Miyashita T, Uemura O, Concha ML, Russell C, Wilson SW, and Okamoto H (2005). Laterotopic Representation of Left-Right Information onto the Dorso-Ventral Axis of a Zebrafish Midbrain Target Nucleus. Curr. Biol 15, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Amo R, and Okamoto H (2011). Phylogeny and Ontogeny of the Habenular Structure. Front. Neurosci 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Kobayashi M, Tanaka S, Fukai T, and Okamoto H (2012). Molecular characterization of the subnuclei in rat habenula. J. Comp. Neurol 520, 4051–4066. [DOI] [PubMed] [Google Scholar]
- Amo R, Aizawa H, Takahoko M, Kobayashi M, Takahashi R, Aoki T, and Okamoto H (2010). Identification of the Zebrafish Ventral Habenula As a Homolog of the Mammalian Lateral Habenula. J. Neurosci 30, 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amo R, Fredes F, Kinoshita M, Aoki R, Aizawa H, Agetsuma M, Aoki T, Shiraki T, Kakinuma H, Matsuda M, et al. (2014). The Habenulo-Raphe Serotonergic Circuit Encodes an Aversive Expectation Value Essential for Adaptive Active Avoidance of Danger. Neuron 84, 1034–1048. [DOI] [PubMed] [Google Scholar]
- Andalman AS, Burns VM, Lovett-Barron M, Broxton M, Poole B, Yang SJ, Grosenick L, Lerner TN, Chen R, Benster T, et al. (2019). Neuronal Dynamics Regulating Brain and Behavioral State Transitions. Cell 177, 970–985.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D, Rehak R, Hameed S, Mehaffey WH, Zamponi GW, and Turner RW (2010). Regulation of the KV4.2 complex by CaV3.1 calcium channels. Channels 4, 163–167. [DOI] [PubMed] [Google Scholar]
- Andres KH, During MV, and Veh RW (1999). Subnuclear organization of the rat habenular complexes. J. Comp. Neurol 407, 130–150. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, Mclnnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2019). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. [DOI] [PubMed] [Google Scholar]
- Beretta CA, Dross N, Gutierrez-Triana JA, Ryu S, and Carl M (2012). Habenula Circuit Development: Past, Present, and Future. Front. Neurosci 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco Isaac H, and Wilson Stephen W (2009). The habenular nuclei: a conserved asymmetric relay station in the vertebrate brain. Philos. Trans. R. Soc. B Biol. Sci 364, 1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M-Y, Amo R, Kinoshita M, Cherng B-W, Shimazaki H, Agetsuma M, Shiraki T, Aoki T, Takahoko M, Yamazaki M, et al. (2016). Social conflict resolution regulated by two dorsal habenular subregions in zebrafish. Science 352, 87–90. [DOI] [PubMed] [Google Scholar]
- Claudio Cuello A, Emson PC, Paxinos G, and Jessell T (1978). Substance P containing and cholinergic projections from the habenula. Brain Res. 149, 413–429. [DOI] [PubMed] [Google Scholar]
- Cui Y, Yang Y, Ni Z, Dong Y, Cai G, Foncelle A, Ma S, Sang K, Tang S, Li Y, et al. (2018). Astroglial Kir4.1 in the lateral habenula drives neuronal bursts in depression. Nature 554, 323–327. [DOI] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, Bravo González-Blas C, et al. (2018). A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 174, 982–998.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePasquale EAK, Schnell DJ, Van Camp P-J, Valiente-Alandí Í, Blaxall BC, Grimes HL, Singh H, and Salomonis N (2019). DoubletDecon: Deconvoluting Doublets from Single-Cell RNA-Sequencing Data. Cell Rep. 29, 1718–1727.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboué ER, Hong E, Eldred KC, and Halpern ME (2017). Left Habenular Activity Attenuates Fear Responses in Larval Zebrafish. Curr. Biol 27, 2154–2162.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A, Heyer MP, Ishikawa M, Caligiuri SPB, Liu X, Chen Z, Micioni Di Bonaventura MV, Elayouby KS, Abies JL, Howe WM, et al. (2019). Habenular TCF7L2 links nicotine addiction to diabetes. Nature 574, 372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flumerfelt BA, and Contestabile A (1982). Acetylcholinesterase histochemistry of the habenulo-interpeduncular pathway in the rat and the effects of electrolytic and kainic acid lesions. Anat. Embryol. (Berl.) 163, 435–446. [DOI] [PubMed] [Google Scholar]
- Fowler CD, and Kenny PJ (2011). Intravenous nicotine self-administration and cue-induced reinstatement in mice: Effects of nicotine dose, rate of drug infusion and prior instrumental training. Neuropharmacology 61,687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CD, and Kenny PJ (2012). Habenular Signaling in Nicotine Reinforcement. Neuropsychopharmacology 37, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamse JT, Kuan Y-S, Macurak M, Brosamle C, Thisse B, Thisse C, and Halpern ME (2005). Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development 132, 4869–4881. [DOI] [PubMed] [Google Scholar]
- Golden SA, Heshmati M, Flanigan M, Christoffel DJ, Guise K, Pfau ML, Aleyasin H, Menard C, Zhang H, Hodes GE, et al. (2016). Basal forebrain projections to the lateral habenula modulate aggression reward. Nature 534, 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady SR, Moretti M, Zoli M, Marks MJ, Zanardi A, Pucci L, Clementi F, and Gotti C (2009). Rodent Habenulo–Interpeduncular Pathway Expresses a Large Variety of Uncommon nAChR Subtypes, But Only the α3β4 and α3β3β4 Subtypes Mediate Acetylcholine Release. J. Neurosci 29, 2272–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkenham M, and Nauta WJH (1979). Efferent connections of the habenular nuclei in the rat. J. Comp. Neurol 187, 19–47. [DOI] [PubMed] [Google Scholar]
- Hikosaka O (2010). The habenula: from stress evasion to value-based decision-making. Nat. Rev. Neurosci 11, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka O, Sesack SR, Lecourtier L, and Shepard PD (2008). Habenula: Crossroad between the Basal Ganglia and the Limbic System. J. Neurosci 28, 11825–11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, and Hikosaka O (2008). The Globus Pallidus Sends Reward-Related Signals to the Lateral Habenula. Neuron 60, 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Jhou TC, Smith M, Saleem KS, and Hikosaka O (2011). Negative Reward Signals from the Lateral Habenula to Dopamine Neurons Are Mediated by Rostromedial Tegmental Nucleus in Primates. J. Neurosci 31, 11457–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-WA, Wang SD, Wang S, Morton G, Zariwala HA, Iglesia H.O. de la, and Turner EE (2014). Role of the Dorsal Medial Habenula in the Regulation of Voluntary Activity, Motor Function, Hedonic State, and Primary Reinforcement. J. Neurosci 34, 11366–11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-WA, Morton G, Guy EG, Wang SD, and Turner EE (2016). Dorsal Medial Habenula Regulation of Mood-Related Behaviors and Primary Reinforcement by Tachykinin- Expressing Habenula Neurons. ENeuro 3, ENEURO.0109-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Rizzi G, Stamatakis AM, Ung RL, and Stuber GD (2013). The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science 341, 1517–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhou TC, Geisler S, Marinelli M, Degarmo BA, and Zahm DS (2009). The mesopontine rostromedial tegmental nucleus: A structure targeted by the lateral habenula that projects to the ventral tegmental area of Tsai and substantia nigra compacta. J. Comp. Neurol 513, 566–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-W, Yao Z, Graybuck LT, Kim TK, Nguyen TN, Smith KA, Fong O, Yi L, Koulena N, Pierson N, et al. (2019). Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior. Cell 179, 713–728.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowland D, Lilascharoen V, Pacia CP, Shin S, Wang EH-J, and Lim BK (2017). Distinct Ventral Pallidal Neural Populations Mediate Separate Symptoms of Depression. Cell 170, 284–297.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM, Deisseroth K, and Malenka RC (2012). Input-specific control of reward and aversion in the ventral tegmental area. Nature 491,212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Tzortzi O, Weglage M, Märtin A, Xuan Y, Parent M, Johansson Y, Fuzik J, Fürth D, Fenno LE, et al. (2019). A hypothalamus-habenula circuit controls aversion. Mol. Psychiatry 24, 1351–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Foil B, and French L (2018). Transcriptomic Characterization of the Human Habenula Highlights Drug Metabolism and the Neuroimmune System. Front. Neurosci 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecca S, Meye FJ, and Mameli M (2014). The lateral habenula in addiction and depression: an anatomical, synaptic and behavioral overview. Eur. J. Neurosci 39, 1170–1178. [DOI] [PubMed] [Google Scholar]
- Lecca S, Meye FJ, Trusel M, Tchenio A, Harris J, Schwarz MK, Burdakov D, Georges F, and Mameli M (2017). Aversive stimuli drive hypothalamus-to-habenula excitation to promote escape behavior. ELife 6, e30697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Mathuru AS, Teh C, Kibat C, Korzh V, Penney TB, and Jesuthasan S (2010). The Habenula Prevents Helpless Behavior in Larval Zebrafish. Curr. Biol 20, 2211–2216. [DOI] [PubMed] [Google Scholar]
- Lester RA, and Dani JA (1995). Acetylcholine receptor desensitization induced by nicotine in rat medial habenula neurons. J. Neurophysiol 74, 195–206. [DOI] [PubMed] [Google Scholar]
- Li B, Piriz J, Mirrione M, Chung C, Proulx CD, Schulz D, Henn F, and Malinow R (2011). Synaptic potentiation onto habenula neurons in the learned helplessness model of depression. Nature 470, 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pullmann D, and Jhou TC (2019). Valence-encoding in the lateral habenula arises from the entopeduncular region. ELife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Zhou T, Liao L, Yang Z, Wong C, Henn F, Malinow R, Yates JR, and Hu H (2013). βCaMKII in Lateral Habenula Mediates Core Symptoms of Depression. Science 341, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipiec MA, Koziński K, Zajkowski T, Dąbrowski M, Chakraborty C, Urban-Ciećko J, Toval A, Ferran JL, Nagalski A, and Wiśniewska MB (2019). TCF7L2 is a terminal selector that regulates postmitotic differentiation programs and excitability patterns in the thalamus. BioRxiv 515–874. [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marburg O (1944). The structure and fiber connections of the human habenula. J. Comp. Neurol 80, 211–233. [Google Scholar]
- Matsumoto M, and Hikosaka O (2007). Lateral habenula as a source of negative reward signals in dopamine neurons. Nature 447, 1111–1115. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, and Hikosaka O (2009). Two types of dopamine neuron distinctly convey positive and negative motivational signals. Nature 459, 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclnnes L, Healy J, and Melville J (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv180203426 Cs Stat. [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, et al. (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molas S, Zhao-Shea R, Liu L, DeGroot SR, Gardner PD, and Tapper AR (2017). A circuit-based mechanism underlying familiarity signaling and the preference for novelty. Nat. Neurosci 20, 1260–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namboodiri VMK, Rodriguez-Romaguera J, and Stuber GD (2016). The habenula. Curr. Biol. CB 26, R873–R877. [DOI] [PubMed] [Google Scholar]
- Neckers LM, Schwartz JP, Wyatt RJ, and Speciale SG (1979). Substance P afferents from the habenula innervate the dorsal raphe nucleus. Exp. Brain Res 37, 619–623. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Ramadasan PN, Goschorska M, Anantharajah A, Ng KW, and Parhar IS (2012). Cloning and expression of tachykinins and their association with kisspeptins in the brains of zebrafish. J. Comp. Neurol 520, 2991–3012. [DOI] [PubMed] [Google Scholar]
- Otsu Y, Darcq E, Pietrajtis K, Mátyás F, Schwartz E, Bessaih T, Gerges SA, Rousseau CV, Grand T, Dieudonné S, et al. (2019). Control of aversion by glycine-gated GluN1/GluN3A NMDA receptors in the adult medial habenula. Science 366, 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Shekhar K, Regev A, and Schier AF (2018). Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-Cell RNA-Seq. Curr. Biol. CB 28, 1052–1065.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poller WC, Bernard R, Derst C, Weiss T, Madai VI, and Veh RW (2011). Lateral habenular neurons projecting to reward-processing monoaminergic nuclei express hyperpolarization-activated cyclic nucleotid-gated cation channels. Neuroscience 193, 205–216. [DOI] [PubMed] [Google Scholar]
- Proulx CD, Hikosaka O, and Malinow R (2014). Reward processing by the lateral habenula in normal and depressive behaviors. Nat. Neurosci 17, 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina LA, Wang S, Ng L, and Turner EE (2009). Brn3a and Nurrl Mediate a Gene Regulatory Pathway for Habenula Development. J. Neurosci 29, 14309–14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina LA, Tempest L, Ng L, Harris JA, Ferguson S, Jhou TC, and Turner EE (2015). Efferent Pathways of the Mouse Lateral Habenula. J. Comp. Neurol 523, 32–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Qin C, Hu F, Tan J, Qiu L, Zhao S, Feng G, and Luo M (2011). Habenula “cholinergic” neurons co-release glutamate and acetylcholine and activate postsynaptic neurons via distinct transmission modes. Neuron 69, 445–452. [DOI] [PubMed] [Google Scholar]
- Root DH, Mejias-Aponte CA, Qi J, and Morales M (2014). Role of Glutamatergic Projections from Ventral Tegmental Area to Lateral Habenula in Aversive Conditioning. J. Neurosci 34, 13906–13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MA, Basiri ML, McHenry JA, Kosyk O, Otis JM, van den Munkhof HE , Bryois J, Hübei C, Breen G, Guo W, et al. (2019). Obesity remodels activity and transcriptional state of a lateral hypothalamic brake on feeding. Science 364, 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C, Strazielle N, and Ghersi-Egea J-F (2012). Brain leukocyte infiltration initiated by peripheral inflammation or experimental autoimmune encephalomyelitis occurs through pathways connected to the CSF-filled compartments of the forebrain and midbrain. J. Neuroinflammation 9, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabel SJ, Proulx CD, Trias A, Murphy RT, and Malinow R (2012). Input to the lateral habenula from the basal ganglia is excitatory, aversive, and suppressed by serotonin. Neuron 74, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabel SJ, Proulx CD, Piriz J, and Malinow R (2014). Mood regulation. GABA/glutamate co-release controls habenula output and is modified by antidepressant treatment. Science 345, 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabel SJ, Wang C, Monk B, Aronson S, and Malinow R (2019). Stress transforms lateral habenula reward responses into punishment signals. Proc. Natl. Acad. Sci. U. S. A 116, 12488–12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AM, and Stuber GD (2012). Activation of lateral habenula inputs to the ventral midbrain promotes behavioral avoidance. Nat. Neurosci 15, 1105–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AM, Jennings JH, Ung RL, Blair GA, Weinberg RJ, Neve RL, Boyce F, Mattis J, Ramakrishnan C, Deisseroth K, et al. (2013). A Unique Population of Ventral Tegmental Area Neurons Inhibits the Lateral Habenula to Promote Reward. Neuron 80, 1039–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AM, Van Swieten M, Basiri ML, Blair GA, Kantak P, and Stuber GD (2016). Lateral Hypothalamic Area Glutamatergic Neurons and Their Projections to the Lateral Habenula Regulate Feeding and Reward. J. Neurosci. Off. J. Soc. Neurosci 36, 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson-Jones M, Floras O, Robertson B, and Grillner S (2012). Evolutionary conservation of the habenular nuclei and their circuitry controlling the dopamine and 5-hydroxytryptophan (5-HT) systems. Proc. Natl. Acad. Sci 109, E164–E173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentinova K, Tchenio A, Trusel M, Clerke JA, Lalive AL, Tzanoulinou S, Matera A, Moutkine I, Maroteaux L, Paolicelli RC, et al. (2019). Morphine withdrawal recruits lateral habenula cytokine signaling to reduce synaptic excitation and sociability. Nat. Neurosci 22, 1053–1056. [DOI] [PubMed] [Google Scholar]
- Velasquez KM, Molfese DL, and Salas R (2014). The role of the habenula in drug addiction. Front. Hum. Neurosci 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertes RP, Fortin WJ, and Crane AM (1999). Projections of the median raphe nucleus in the rat. J. Comp. Neurol 407, 555–582. [PubMed] [Google Scholar]
- Viswanath H, Carter AQ, Baldwin PR, Molfese DL, and Salas R (2014). The medial habenula: still neglected. Front. Hum. Neurosci 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner F, Bernard R, Derst C, French L, and Veh RW (2016). Microarray analysis of transcripts with elevated expressions in the rat medial or lateral habenula suggest fast GABAergic excitation in the medial habenula and habenular involvement in the regulation of feeding and energy balance. Brain Struct. Funct 221,4663–4689. [DOI] [PubMed] [Google Scholar]
- Wallace ML, Saunders A, Huang KW, Philson AC, Goldman M, Macosko EZ, McCarroll SA, and Sabatini BL (2017). Genetically Distinct Parallel Pathways in the Entopeduncular Nucleus for Limbic and Sensorimotor Output of the Basal Ganglia. Neuron 94, 138–152.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace ML, Huang KW, Hochbaum D, Hyun M, Radeljic G, and Sabatini BL (2020). Anatomical and single-cell transcriptional profiling of the murine habenular complex. ELife 9, e51271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, and Aghajanian GK (1977). Physiological evidence for habenula as major link between forebrain and midbrain raphe. Science 197, 89–91. [DOI] [PubMed] [Google Scholar]
- Wang D, Li Y, Feng Q, Guo Q, Zhou J, and Luo M (2017). Learning shapes the aversion and reward responses of lateral habenula neurons. ELife 6, e23045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang L-C, Bui S, Nielson A, Wu X, Vo H-T, Ma X-J, and Luo Y (2012). RNAscope: A Novel in Situ RNA Analysis Platform for Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagn 14, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtshafter D, Asin KE, and Pitzer MR (1994). Dopamine agonists and stress produce different patterns of Fos-like immunoreactivity in the lateral habenula. Brain Res. 633, 21–26. [DOI] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, and Hong W (2017). Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329.e6. [DOI] [PubMed] [Google Scholar]
- Xie G, Zuo W, Wu L, Li W, Wu W, Bekker A, and Ye J-H (2016). Serotonin modulates glutamatergic transmission to neurons in the lateral habenula. Sci. Rep 6, 23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Danjo T, Pastan I, Hikida T, and Nakanishi S (2013). Distinct Roles of Segregated Transmission of the Septo-Habenular Pathway in Anxiety and Fear. Neuron 78, 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang J, Xi W, Hao S, Luo B, He X, Zhu L, Lou H, Yu Y, Xu F, et al. (2016). Laterodorsal tegmentum interneuron subtypes oppositely regulate olfactory cue-induced innate fear. Nat. Neurosci 19, 283–289. [DOI] [PubMed] [Google Scholar]
- Yang Y, Cui Y, Sang K, Dong Y, Ni Z, Ma S, and Hu H (2018). Ketamine blocks bursting in the lateral habenula to rapidly relieve depression. Nature 554, 317–322. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van derZwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tan L, Ren Y, Liang J, Lin R, Feng Q, Zhou J, Hu F, Ren J, Wei C, et al. (2016). Presynaptic Excitation via GABAB Receptors in Habenula Cholinergic Neurons Regulates Fear Memory Expression. Cell 166, 716–728. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z, Huang Y, and Wang J (2019). Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems. Mol. Cell 73, 130–142.e5. [DOI] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu M-Z, Li Q, Deng J, Mu D, and Sun Y-G (2017). Organization of Functional Long-Range Circuits Controlling the Activity of Serotonergic Neurons in the Dorsal Raphe Nucleus. Cell Rep. 18, 3018–3032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NCBI Gene Expression Omnibus accession number for the scRNAseq data reported in this paper is GSE137478. All the codes used to analyze scRNAseq, Hiplex and RNAscope data have been made publicly available at a Github repository affiliated with Stuber Laboratory group and this manuscript title (http://www.github.com/stuberlab/).