Abstract
Immune checkpoint inhibitor (ICI) therapy has revolutionized the treatment of numerous types of cancer, including colorectal cancer (CRC). Patients with CRC and deficient mismatch repair or high microsatellite instability could benefit from ICI treatment, although the response rate of most patients is low. Therefore, the immune subtyping of patients with CRC is required in order to determine the subtypes suitable for ICI treatment. The present study used a cohort of patients with CRC from The Cancer Genome Atlas (TCGA) to perform molecular subtyping, with results validated in three CRC cohorts from the Gene Expression Omnibus. Non-negative matrix factorization was used to achieve consensus molecular subtyping. The tumor immune dysfunction and exclusion algorithm was used to predict potential ICI therapy responses and gene set enrichment analysis was performed to define different pathways associated with the immune response. Two distinct subtypes of CRC were finally identified in TCGA cohorts, which were characterized as significantly different prognostic subtypes (low-risk and high-risk subtypes). Higher expression of programmed death-ligand 1, higher proportion of tumor-infiltrating lymphocytes and tumor mutation burden were significantly enriched in the low-risk subtype. Further pathway analysis revealed that the low-risk subtype was associated with immune response activation and signaling pathways involved in ‘antigen processing and presentation’. Three independent CRC cohorts were used to validate the above findings. In summary, two clinical CRC subtypes were identified, which are characterized by significantly different survival outcomes and immune infiltration patterns. The findings of the present study suggest that ICI treatment may be more effective in the low-risk CRC subtype.
Keywords: CRC, immune subtyping, ICI therapy, prognosis, ICI response
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed tumor, with the second highest mortality rate globally in 2018 (1). Due to its distinct molecular characteristics and genetic heterogeneity, patients with CRC often have different natural processes and clinical outcomes. Therefore, there is an urgent requirement to explore the molecular subtypes of CRC for precise therapy. Recently, several studies have reported some distinct subtypes of CRC. However, the subclasses generated from these studies either lack in the analysis of different molecular mechanisms (2,3) or have insufficient independent samples for validation (4,5). Thus, a more effective and credible subgroup is required for the treatment of patients with CRC.
In recent years, owing to the further understanding of the interactions between the immune system and tumor cell environment, immunotherapies such as anti-programmed cell death protein 1 (PD-1), anti-programmed cell death 1 ligand 1 (PD-L1) and anti-cytotoxic T-lymphocyte protein 4 (CTLA-4) have markedly revolutionized the therapeutic approaches in several cancer types, including lung cancer (6,7), melanoma (8,9), head and neck cancer (10), bladder cancer (11), kidney cancer (12) and CRC with deficient mismatch repair (13,14). An ~50% elevated response rate was found, which is a noteworthy improvement in patients who received ICI treatment due to the reversion of an immunosuppressive microenvironment (15,16). However, only a small number of CRC patients exhibit a clinical response to ICI treatment, with the majority of patients not benefiting from the therapy (17). The therapeutic effect of ICI agents is ascribed to several important factors, such as expression of PD-L1, proportion of tumor-infiltrating lymphocytes (TILs) and tumor mutation burden (TMB). High PD-L1 expression is an essential factor in the use of pembrolizumab, which is approved by the Food and Drug Administration (FDA) for patients with non-small cell lung cancer (NSCLC) (18). Several studies have also reported the vital roles of TIL and TMB in the response to ICI agents (19–21). In summary, the exploration of the CRC subtype that is suitable to receive ICI therapy is important and urgently required.
In the present study, immune-related gene expression profiles of four independent CRC cohorts were used to identify the potential subtype which may be sensitive to immune checkpoint blockade. Findings of the present study have implications for guiding clinical immunotherapy for patients with CRC.
Materials and methods
Acquisition of genomic data
Gene expression data of 662 CRC samples with overall survival time and status from TCGA cohort (https://gdc.cancer.gov) and 619 samples of three datasets (GSE103479, GSE38832 and GSE87211) (22–24) from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) were retrospectively acquired. TCGA cohort (mean age, 66.33; age range: 31–90; sex distribution: 355 male and 307 female patients); GSE103479 dataset (mean age: 64.26; age range: 35–88; sex distribution: 83 male and 72 female patients); GSE8832 dataset (mean age: 59.63; age range: 32–89; sex distribution: 69 male and 53 female patients) and GSE87211 dataset (mean age: 63.16; age range: 38–88; sex distribution: 182 male and 160 female patients). Clinical characteristics of these four cohorts are presented in Table SI. Finally, a total of 1,281 CRC samples with gene expression data and survival information from four independent datasets were obtained (Table SII). Complete clinical data were available for all patients included in the present study. Gene expression profiles were consistently normalized to reduce variance. Expression levels of genes with multiple probes were presented as the mean expression of all probes.
Immune-related prognostic signature used for molecular subtyping
A prognostic signature was established based on a previously conducted immune-related genes study (25). The aforementioned previous study comprehensively described the immune landscape of >10,000 samples, comprising 33 different cancer types, and integrated 160 immune-related signatures containing 2,995 immune genes. Univariate Cox proportional hazards model was used to evaluate the association between the expression of these genes and the overall survival of patients with CRC. Genes with P<0.01 were included in the signature to perform molecular subtyping.
NMF clustering analysis
Molecular subtyping was performed applying non-negative matrix factorization (NMF) (26,27). A binary matrix (A) describing the expression of immune-related genes (rows) across CRC samples (columns) was established. Subsequently, the expression matrix A was factorized into two non-negative matrices (W and H; A≈WH). Matrix H was applied to cluster samples into distinct subtypes. The values of cophenetic, dispersion and silhouette coefficients were used to select the optimal number of subtypes. The NMF clustering analysis was performed with the R package ‘NMF’ v.0.21.0 (28).
Prediction of response to ICI treatment
The tumor immune dysfunction and exclusion (TIDE) algorithm (29) was applied to predict potential distinct responses to ICI therapy. TIDE is a gene expression biomarker for predicting the response to immune checkpoint blockade in patients. A low TIDE prediction score represents weak potential immune escape, and therefore these patients would potentially exhibit a greater immune therapy response. TIL proportion was analyzed using the CIBERSORT algorithm (30), which is a useful analytical tool to provide an evaluation of the abundances of 22 immune cell types in a mixed cell population, using gene expression data.
Gene set enrichment analysis (GSEA)
Patients with CRC were partitioned into two groups, according to the subtyping results clustered with the molecular expression features. ‘DEseq2’ v.1.26.0 (31) and ‘limma’ v.3.34.8 (32) packages were applied to calculate the differential t statistics of the RNA sequencing and microarray data. The t statistic was used as the input to R function in the ‘fgsea’ v.1.12.0 package (http://bioconductor.org/packages/release/bioc/html/fgsea.html) to perform GSEA. The pathway annotation signatures from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases in Molecular Signatures Database (MSigDB) (33) were used.
Normalization of gene expression data
In the present study, the molecular subtyping of CRC samples was conducted using the NMF algorithm. The essential condition for performing the NMF approach was non-negative values, therefore, the gene expression data was compressed into the range from 0–1 in all distinct platforms to achieve data normalization.
Statistical analyses
Statistical analyses were conducted with R software 3.6.1 (https://cran.r-project.org). The difference in clinical characteristics between two subtypes were compared using the χ2 test, and differences of TIDE score, PD-L1 expression and TMB were compared using the Wilcoxon rank-sum test. Survivals plot were drawn using the Kaplan-Meier method and log-rank test for comparison. The association between CRC subtypes and prognosis was analyzed with univariate and multivariate Cox proportional hazards model in the R ‘survival’ package (v.2.41–3) (https://gitub.com/therneau/survival). P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of two CRC subtypes with distinct prognoses
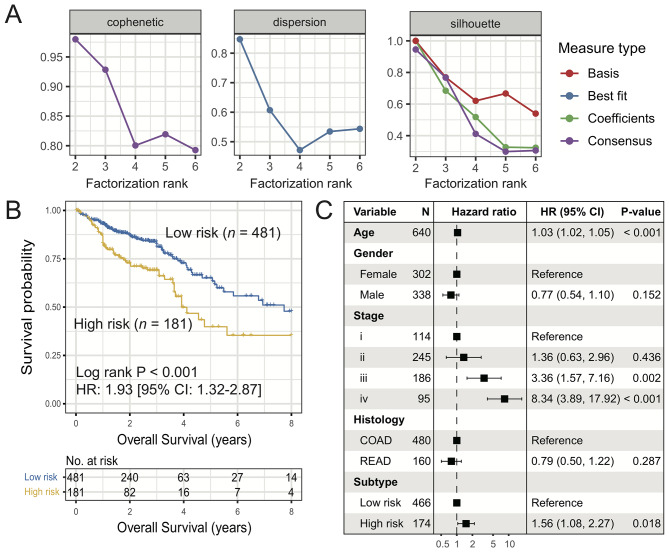
Using the univariate Cox proportional hazard model, the association between the expression of 2,995 immune-related genes and the prognosis of patients with CRC in TCGA cohort was determined. Finally, 53 genes with P<0.01 (Table SIII) were included in the prognostic signature for molecular subtyping. NMF unsupervised clustering analysis of these 53 immune genes was performed and the results showed that the cophenetic, dispersion and silhouette coefficients harbored the maximum value for the factorization rank of two subtypes (Figs. 1A and S1). The survival analysis of these two subtypes indicated that the overall survival rate of patients in the high-risk subtype was lower compared with that of patients in the low-risk subtype (HR, 1.93; 95% CI, 1.32–2.87; log-rank P<0.001; Fig. 1B). Multivariate Cox regression analysis adjusted for age, gender, stage and histology remained statistically significant (HR, 1.56; 95% CI, 1.08–2.27; P=0.018; Fig. 1C).
Figure 1.
Identification of two distinct subtypes of CRC with significant prognostic differences. (A) Association of cophenetic, dispersion and silhouette coefficients with the number of subtypes. (B) Kaplan-Meier plot of the two identified CRC subtypes (low-risk vs. high-risk). (C) Forest plot of multivariate Cox regression model of low-risk vs. high-risk subtypes with confounding clinical factors taken into consideration. CRC, colorectal cancer; COAD, colon cancer; READ, rectal cancer.
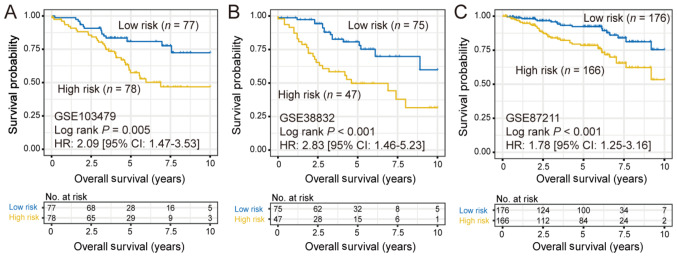
In order to verify whether the aforementioned subtyping approach would discover the same subtypes in other CRC cohorts, the same clustering approach was applied to analyze 3 independent CRC cohorts, including GSE103479 (n=155), GSE38832 (n=122) and GSE87211 (n=342). Consistently, two different subtypes were identified in all 3 additional cohorts and the high-risk subtype also exhibited a significantly poorer prognosis [GSE103479: HR, 2.09 (95% CI, 1.47–3.53; log-rank P=0.005); GSE38832: HR, 1.78 (95% CI, 1.25–3.16; log-rank P<0.001) and GSE87211: HR, 1.78 (95% CI, 1.25–3.16; log-rank P<0.001); Fig. 2].
Figure 2.
Kaplan-Meier survival plots of low-risk and high-risk colorectal cancer subtypes in (A) GSE103479, (B) GSE38832 and (C) GSE87211 datasets.
Prediction of response to ICB therapy in the two CRC subtypes
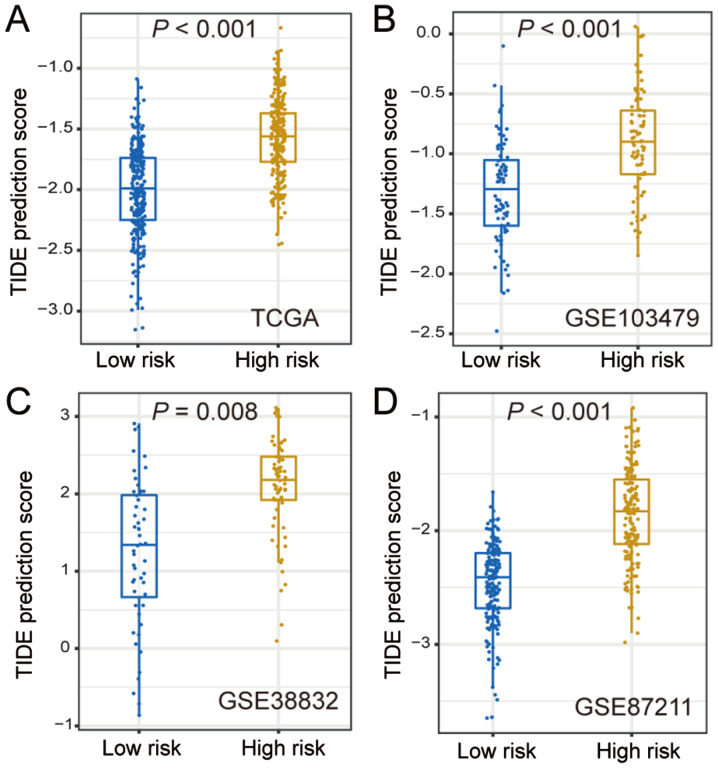
Since the two CRC subtypes were identified by the application of 53 immune-related genes, it was hypothesized that these two subtypes may have distinct responses to ICB therapy. Thus, the TIDE algorithm was used to predict the potentially different responses to ICB therapy. In TCGA cohort, the TIDE prediction score was significantly lower in the low-risk subtype compared with that in the high-risk group (P<0.001; Fig. 3A), which was further validated in the three other independent cohorts (GSE103479, GSE38832 and GSE87211; all P<0.01; Fig. 3B-D). These findings revealed that the patients of the low-risk subtype may be more sensitive to ICB treatment.
Figure 3.
Differences in TIDE prediction score between the two CRC subtypes in (A) The Cancer Genome Atlas and (B-D) three validation CRC cohorts. CRC, colorectal cancer; TIDE, tumor immune dysfunction and exclusion.
Differences in ICI response markers between the two CRC subtypes
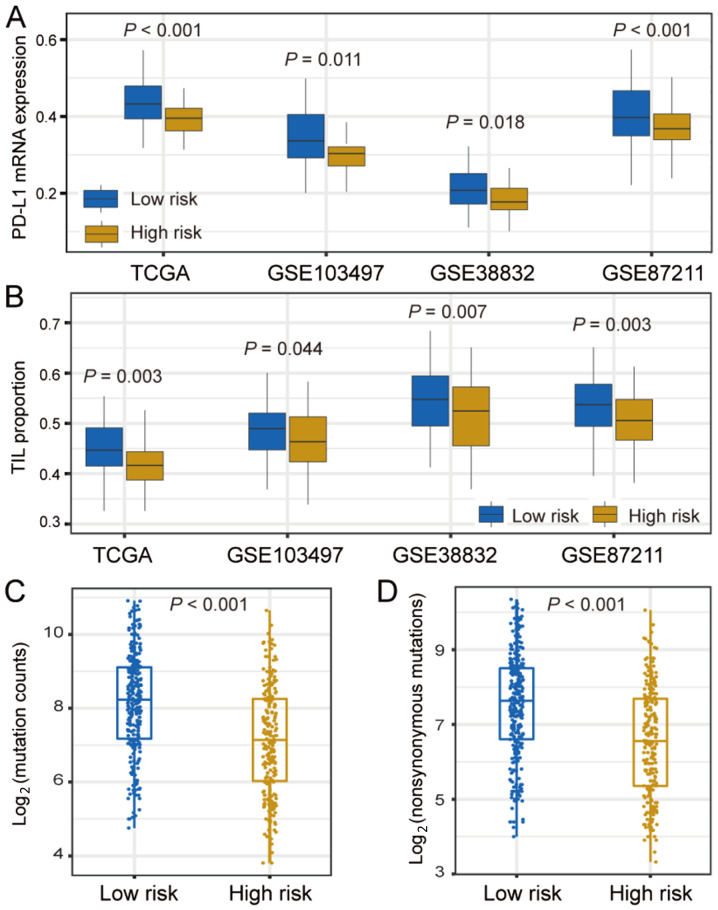
PD-L1 expression, TIL and TMB are highly important clinical factors, due to their vital roles in immunotherapy responses. Therefore, the differences in these 3 factors were compared between the two CRC subtypes. PD-L1 expression was significantly higher in the low-risk subtype compared with that in the high-risk subtype in TCGA cohort (P<0.001), which was further validated in three other independent cohorts (GSE103479, GSE38832 and GSE87211; all P<0.05; Fig. 4A).
Figure 4.
Differences in PD-L1 expression, TIL proportion and TMB in low-risk vs. high-risk CRC subtypes. Differences in (A) PD-L1 expression and (B) TIL proportion in TCGA and three independent validation cohorts, stratified by the two CRC subtypes. (C and D) Association of TMB with low-risk and high-risk CRC subtypes. CRC, colorectal cancer; TCGA, The Cancer Genome Atlas; PD-L1, programmed death-ligand 1; TIL, tumor-infiltrating lymphocyte; TMB, tumor mutation burden.
Results showed that the TIL proportion was significantly higher in the patients of the low-risk subtype compared with that in the high-risk group in TCGA cohort (P=0.003). This association was also corroborated in the three additional validation cohorts (GSE103479, GSE38832 and GSE87211; all P<0.05; Fig. 4B). The differences in the infiltration of 22 immune cells between the two identified CRC subtypes in the TCGA cohort are shown in Fig. S2. The results demonstrated that the low-risk subtype has significantly higher specific immune cells infiltration (e.g., CD4 T cells, plasma cells, natural killer cells and dendritic cells), which was consistent with the aforementioned results.
Patients with higher TMB often have a higher number of neoantigen; therefore, these patients could obtain a stronger immune response (34,35). It was found that patients in the low-risk subgroup had significantly higher TMB compared with patients in the high-risk group (P<0.001; Fig. 4C). TMB is closely associated with genomic instability. DNA damage repair (DDR)-related genes, such as BRCA1/2, TP53 and POLE, and clinical factors were therefore considered in the multivariate logistic regression model. Following adjustment of these confounding factors, the association between the low-risk subtype and higher TMB remained notably significant (OR, 2.95; 95% CI, 1.89–4.67; P<0.001; Fig. S3). Neoantigens are generated mainly owing to non-synonymous mutations. It was found that the low-risk subtype harbored a significantly higher non-synonymous mutation load compared with the high-risk group (P<0.001; Fig. 4D).
Functional annotation of the low-risk subtype
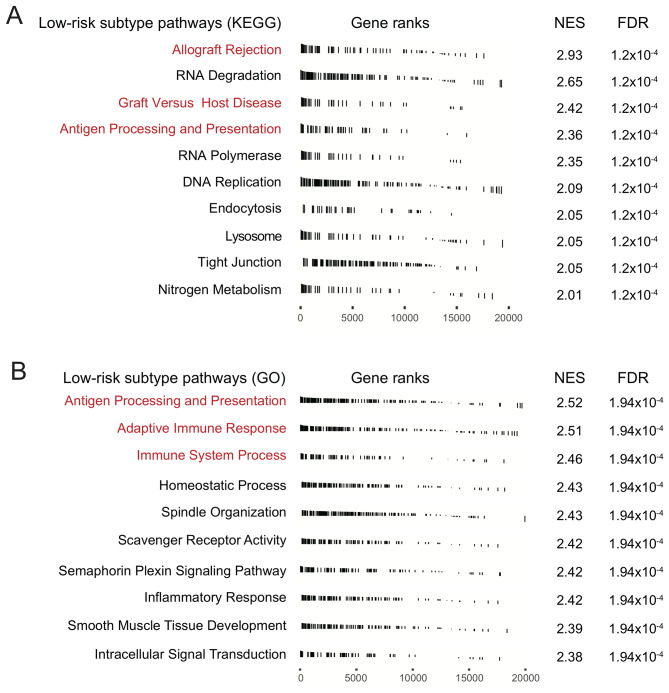
In order to verify whether the two CRC subtypes had distinct functionalities, differential analysis and GSEA pathway analysis of genome-wide gene expression data were performed on the two CRC subtypes in TCGA CRC cohort. Analysis of differentially expressed genes in the KEGG pathway demonstrated that signaling pathways associated with immune response activation, such as ‘allograft rejection’, ‘graft versus host disease’, and ‘antigen processing and presentation’ were significantly enriched in the low-risk subtype (Fig. 5A). GO analysis also revealed the enrichment of ‘antigen processing and presentation’ signaling in the low-risk subtype. Both the ‘adaptive immune response’ and ‘immune system process’ pathways were also significantly enriched in low-risk patients (Fig. 5B). These results suggested that patients in the low-risk subgroup had a better immune response.
Figure 5.
Pathway analysis of the low-risk colorectal cancer subtype, with signaling pathway annotations from (A) KEGG and (B) GO. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; NES, normalized enrichment score; FDR, false discovery rate.
Discussion
The present study identified two clinically distinct subtypes of CRC, with markedly different clinical outcomes and immune infiltration patterns, by using immune-related signatures on TCGA cohort. For responses to ICI therapy in the two CRC subtypes, the TIDE algorithm demonstrated a low score in patients of the low-risk subtype compared with that in patients of the high-risk subtype. Furthermore, higher PD-L1 expression, TIL proportion and TMB were remarkably enriched in the low-risk subtype. Finally, GSEA pathway analysis indicated immune response activation-, and ‘antigen processing and presentation’-related pathways to be significantly enriched in the low-risk subtype. Overall, the present study suggests that patients in the low-risk group have the potential to respond better to ICI treatment.
The NMF algorithm is a useful tool to perform clustering or subtyping, and extract genomic signatures; previous studies have shown its utility in distinct research directions, such as identification of image pattern, signal processing and text mining (36–38). The TIDE prediction score is a better predictor of ICI therapy compared with PD-L1 expression and TMB. The algorithm uses gene expression signatures to model two major mechanisms of tumor immune evasion: The induction of T-cell dysfunction in tumors with high infiltration of cytotoxic T lymphocytes (CTLs) and the prevention of T-cell infiltration in tumors with a low CTL level (25). In the present study, patients of the low-risk subtype harbored a significantly lower TIDE prediction score compared with patients of the high-risk subtype in TCGA and validation cohorts, suggesting that this subtype has potential for an improved response to ICI.
To date, only PD-L1 expression is confirmed by the FDA as an official criterion for ICI therapy (18). Based on the results from the KEYNOTE-001 clinical trial (18), high PD-L1 expression is now an essential condition for the use of pembrolizumab in NSCLC. The present study also demonstrated markedly higher PD-L1 expression in the low-risk CRC subtype compared with that in the high-risk subtype in TCGA and validation cohorts. TMB is emerging as a potential biomarker to predict the response to ICI therapy. Three clinical trials, including KEYNOTE-001, CHECKMATE-026 and CHECKMATE-227, demonstrated that patients with higher TMB responded more effectively to ICI (39,40). A recent study reported that high TMB was positively associated with the response to PD-1/PD-L1 inhibitors (such as pembrolizumab) in metastatic CRC, and may serve as a biomarker to predict associated immune therapy effects (41). The present study discovered that TMB in the low-risk CRC subtype was significantly higher compared with that in the high-risk subtype in TCGA cohort. Meanwhile, the non-synonymous mutation load was also markedly enriched in the low-risk CRC subgroup. Studies have reported that mutations of BRCA1/2, POLE and TP53 genes also indicate high TMB, due to the loss of DDR (42–44). Thus, multivariate analysis was conducted following the adjustment of these factors (i.e., mutations of BRCA1/2, POLE and TP53), and the association of low-risk subtype with higher TMB remained statistically significant. Several studies have revealed that TILs play crucial roles in the tumor-immune microenvironment, immune response and prognosis of CRC (44–47). In the present study, results from the CIBERSORT algorithm showed that the low-risk CRC subtype harbored a significantly higher TIL proportion compared with the high-risk group in TCGA and independent validation cohorts. Thus, TMB, TIL proportion and non-synonymous mutations load may contribute to the response to ICI in patients of the low-risk CRC subtype.
The present study has several limitations. Firstly, the gene expression data from the four validation cohorts were from different platforms, which may create bias in the analyses. Secondly, the results associated with the CRC mutation data derived from TCGA cohort were not sufficiently validated, owing to the unavailability of mutation data from the other cohorts. Thirdly, the number of CRC samples with a gene expression profile is currently insufficient to perform molecular subtyping, and thus the results could not be validated using biological experiments such as histochemical staining.
In summary, two clinically distinct CRC subtypes were identified that have significantly different survival outcomes and immune microenvironments. The low-risk CRC subtype may indicate improved response to ICI therapy, due to higher PD-L1 expression, TIL proportion and TMB. Thus, this molecular classification study and integrated multi-omic analysis of CRC may lead to a novel therapeutic approach for improving the prognosis of patients in the low-risk CRC subtype.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRC
colorectal cancer
- ICI
immune checkpoint inhibitor
- TCGA
The Cancer Genome Atlas
- NMF
non-negative matrix factorization
- TIDE
tumor immune dysfunction and exclusion
- GSEA
gene set enrichment analysis
- PD-L1
programmed death-ligand 1
- TILs
tumor-infiltrating lymphocytes
- TMB
tumor mutation burden
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and GC designed the study; GC developed the methodology and acquired the related data; GC, LW, TD, YC and CC performed the data analysis and interpretation; GC and XZ drafted and revised the manuscript; XZ supervised the study. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kamal Y, Schmit SL, Hoehn HJ, Amos CI, Frost HR. Transcriptomic differences between primary colorectal adenocarcinomas and distant metastases reveal metastatic colorectal cancer subtypes. Cancer Res. 2019;79:4227–4241. doi: 10.1158/0008-5472.CAN-18-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rashid M, Vishwakarma RK, Deeb AM, Hussein MA, Aziz MA. Molecular classification of colorectal cancer using the gene expression profile of tumor samples. Exp Biol Med (Maywood) 2019;244:1005–1016. doi: 10.1177/1535370219850788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdul Aziz NA, Mokhtar NM, Harun R, Mollah MM, Mohamed Rose I, Sagap I, Mohd Tamil A, Wan Ngah WZ, Jamal R. A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genomics. 2016;9:58. doi: 10.1186/s12920-016-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Sun X, Ge W, Qian Y, Bai R, Zheng S. A seven-gene signature predicts overall survival of patients with colorectal cancer. Oncotarget. 2016;8:95054–95065. doi: 10.18632/oncotarget.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen EEW, Soulières D, Le Tourneau C, Dinis J, Licitra L, Ahn MJ, Soria A, Machiels JP, Mach N, Mehra R, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet. 2019;393:156–167. doi: 10.1016/S0140-6736(18)31999-8. [DOI] [PubMed] [Google Scholar]
- 11.Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376:1015–1026. doi: 10.1056/NEJMoa1613683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–779. doi: 10.1200/JCO.2018.36.4_suppl.554. [DOI] [PubMed] [Google Scholar]
- 15.Motzer RJ, Rini BI, McDermott DF, Arén Frontera O, Hammers HJ, Carducci MA, Salman P, Escudier B, Beuselinck B, Amin A, et al. Nivolumab plus ipilimumab versus sunitinib in first-line treatment for advanced renal cell carcinoma: Extended follow-up of efficacy and safety results from a randomised, controlled, phase 3 trial. Lancet Oncol. 2019;20:1370–1385. doi: 10.1016/S1470-2045(19)30413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asaoka Y, Ijichi H, Koike K. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;373:1979. doi: 10.1056/NEJMc1510353. [DOI] [PubMed] [Google Scholar]
- 18.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 19.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bremnes RM, Busund LT, Kilvaer TL, Andersen S, Richardsen E, Paulsen EE, Hald S, Khanehkenari MR, Cooper WA, Kao SC, Dønnem T. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J Thorac Oncol. 2016;11:789–800. doi: 10.1016/j.jtho.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Gaedcke J, Emons G, Beissbarth T, Grade M, Jo P, Yeager M, Chanock SJ, Wolff H, Camps J, et al. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosomes Cancer. 2018;57:140–149. doi: 10.1002/gcc.22512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripathi MK, Deane NG, Zhu J, An H, Mima S, Wang X, Padmanabhan S, Shi Z, Prodduturi N, Ciombor KK, et al. Nuclear factor of activated T-cell activity is associated with metastatic capacity in colon cancer. Cancer Res. 2014;74:6947–6957. doi: 10.1158/0008-5472.CAN-14-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen WL, Dunne PD, McDade S, Scanlon E, Loughrey M, Coleman H, McCann C, McLaughlin K, Nemeth Z, Syed N, et al. Transcriptional subtyping and CD8 immunohistochemistry identifies poor prognosis stage II/III colorectal cancer patients who benefit from adjuvant chemotherapy. JCO Precis Oncol. 2018;2018 doi: 10.1200/PO.17.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Church G. Improving molecular cancer class discovery through sparse non-negative matrix factorization. Bioinformatics. 2005;21:3970–3975. doi: 10.1093/bioinformatics/bti653. [DOI] [PubMed] [Google Scholar]
- 28.Devarajan K. Nonnegative matrix factorization: An analytical and interpretive tool in computational biology. PLoS Comput Biol. 2008;4:e1000029. doi: 10.1371/journal.pcbi.1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JS, Miller VA, Lim D, Amanam I, Chao J, et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30:1096–1103. doi: 10.1093/annonc/mdz134. [DOI] [PubMed] [Google Scholar]
- 35.Birkbak NJ, Kochupurakkal B, Izarzugaza JM, Eklund AC, Li Y, Liu J, Szallasi Z, Matulonis UA, Richardson AL, Iglehart JD, Wang ZC. Tumor mutation burden forecasts outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PLoS One. 2013;8:e80023. doi: 10.1371/journal.pone.0080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng Z, Vo AH, Mao C, Clare SE, Khan SA, Luo Y. Cancer classification and pathway discovery using non-negative matrix factorization. J Biomed Inform. 2019;96:103247. doi: 10.1016/j.jbi.2019.103247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, Michailidis G. A non-negative matrix factorization method for detecting modules in heterogeneous omics multi-modal data. Bioinformatics. 2016;32:1–8. doi: 10.1093/bioinformatics/btv544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eng SWM, Aeschlimann FA, van Veenendaal M, Berard RA, Rosenberg AM, Morris Q, Yeung RSM, ReACCh-Out Research Consortium Patterns of joint involvement in juvenile idiopathic arthritis and prediction of disease course: A prospective study with multilayer non-negative matrix factorization. PLoS medicine. 2019;16:e1002750. doi: 10.1371/journal.pmed.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Billingsley CC, Cohn DE, Mutch DG, Stephens JA, Suarez AA, Goodfellow PJ. Polymerase ε (POLE) mutations in endometrial cancer: Clinical outcomes and implications for Lynch syndrome testing. Cancer. 2015;121:386–394. doi: 10.1002/cncr.29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gridelli C, Ardizzoni A, Barberis M, Cappuzzo F, Casaluce F, Danesi R, Troncone G, De Marinis F. Predictive biomarkers of immunotherapy for non-small cell lung cancer: Results from an Experts Panel Meeting of the Italian Association of Thoracic Oncology. Transl Lung Cancer Res. 2017;6:373–386. doi: 10.21037/tlcr.2017.05.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kudryavtseva AV, Lukyanova EN, Kalinin DV, Zaretsky AR, Pokrovsky AV, Golovyuk AL, Fedorova MS, Pudova EA, Kharitonov SL, Pavlov VS, et al. Mutational load in carotid body tumor. BMC Med Genomics. 2019;12(Suppl 2):S39. doi: 10.1186/s12920-019-0483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narayanan S, Kawaguchi T, Peng X, Qi Q, Liu S, Yan L, Takabe K. Tumor infiltrating lymphocytes and macrophages improve survival in microsatellite unstable colorectal cancer. Sci Rep. 2019;9:13455. doi: 10.1038/s41598-019-49878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mirjolet C, Charon-Barra C, Ladoire S, Arbez-Gindre F, Bertaut A, Ghiringhelli F, Leroux A, Peiffert D, Borg C, Bosset JF, Créhange G. Tumor lymphocyte immune response to preoperative radiotherapy in locally advanced rectal cancer: The LYMPHOREC study. Oncoimmunology. 2018;7:e1396402. doi: 10.1080/2162402X.2017.1396402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glaire MA, Domingo E, Sveen A, Bruun J, Nesbakken A, Nicholson G, Novelli M, Lawson K, Oukrif D, Kildal W, et al. Tumour-infiltrating CD8(+) lymphocytes and colorectal cancer recurrence by tumour and nodal stage. Br J Cancer. 2019;121:474–482. doi: 10.1038/s41416-019-0540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.