Abstract
In the context of animal or plant development, we tend to think of cells as small, simple, building blocks, such that complex patterns or shapes can only be constructed from large numbers of cells, with cells in different parts of the organism taking on different fates. However, cells themselves are far from simple, and often take on complex shapes with a remarkable degree of intracellular patterning. How do these patterns arise? As in embryogenesis, the development of structure inside a cell can be broken down into a number of basic processes. For each part of the cell, morphogenetic processes create internal structures such as organelles, which might correspond to organs at the level of a whole organism. Given that mechanisms exist to generate parts, patterning processes are required to ensure that the parts are distributed in the correct arrangement relative to the rest of the cell. Such patterning processes make reference to global polarity axes, requiring mechanisms for axiation which, in turn, require processes to break symmetry. These fundamental processes of symmetry breaking, axiation, patterning, and morphogenesis, have been extensively studied in developmental biology but less so at the sub-cellular level. This review will focus on developmental processes that give eukaryotic cells their complex structures, with a focus on cytoskeletal organization in free-living cells, ciliates in particular, in which these processes are most readily apparent.
Cells have anatomy
When we first learn cell biology as children, the idea is often conveyed of the cell as a watery bag of enzymes. But as one peers inside the cell, one immediately sees that the cell is chock full of highly complex internal components, organelles and other structures [1,2]. These complex structures are not just floating around at random, instead they tend to have defined positions within the cell. This is perhaps most dramatically illustrated in the ciliates, an example of which is shown in Figure 1. Here, we see the giant single-celled ciliate Stentor coeruleus [3]. This millimeter-long cell is covered with parallel rows of cilia and microtubules, with a giant array of cilia known as a membranellar band at one end, and a sticky holdfast at the other. These two structures define an anterior-posterior axis. As illustrated in Figure 1A, every structure within the cell, including the macronucleus, the contractile vacuole pore, and the oral pouch, have defined positions along this axis as well as relative to a dorsal-ventral axis defined by the pattern of cortical stripes. The complexity extends down in scale to the organization of the cortical rows of cilia (Figure 1B). Each row contains hundreds of basal bodies flanked by an orderly stack of microtubules. Below the microtubule stack is a bundle of contractile fibers that the cell uses to bend and escape predators. Going further down in scale, each of the cilia is nucleated by a basal body, also known as a centriole, which is itself a complex structure consisting of nine microtubule triplets organized around a central cartwheel. The immense complexity of just this one cell looks nothing like a watery bag of enzymes, and while Stentor may be an extreme example, it turns out that all cells have complex subcellular structure when looked at carefully. The big question is where does this structure come from. In animal or plant development, we can explain away complexity by claiming that it results from a huge number of cells each of which is capable of switching into different states based on gene expression programs. This is comforting because it lets us feel like we understand what is going on, since we know a lot about transcriptional control. At the subcellular level, we are forced to deal with the question of morphogenesis much more directly. We can break the question down into two levels. First, we ask how the individual structures of the cell are themselves assembled. Then, we can ask how the relative positions of these structures are determined. Answering both of these questions would provide a mechanistic basis for a developmental biology of single cells, but we are still far from that level of understanding.
Figure 1. Cells have complex anatomy at multiple scales.
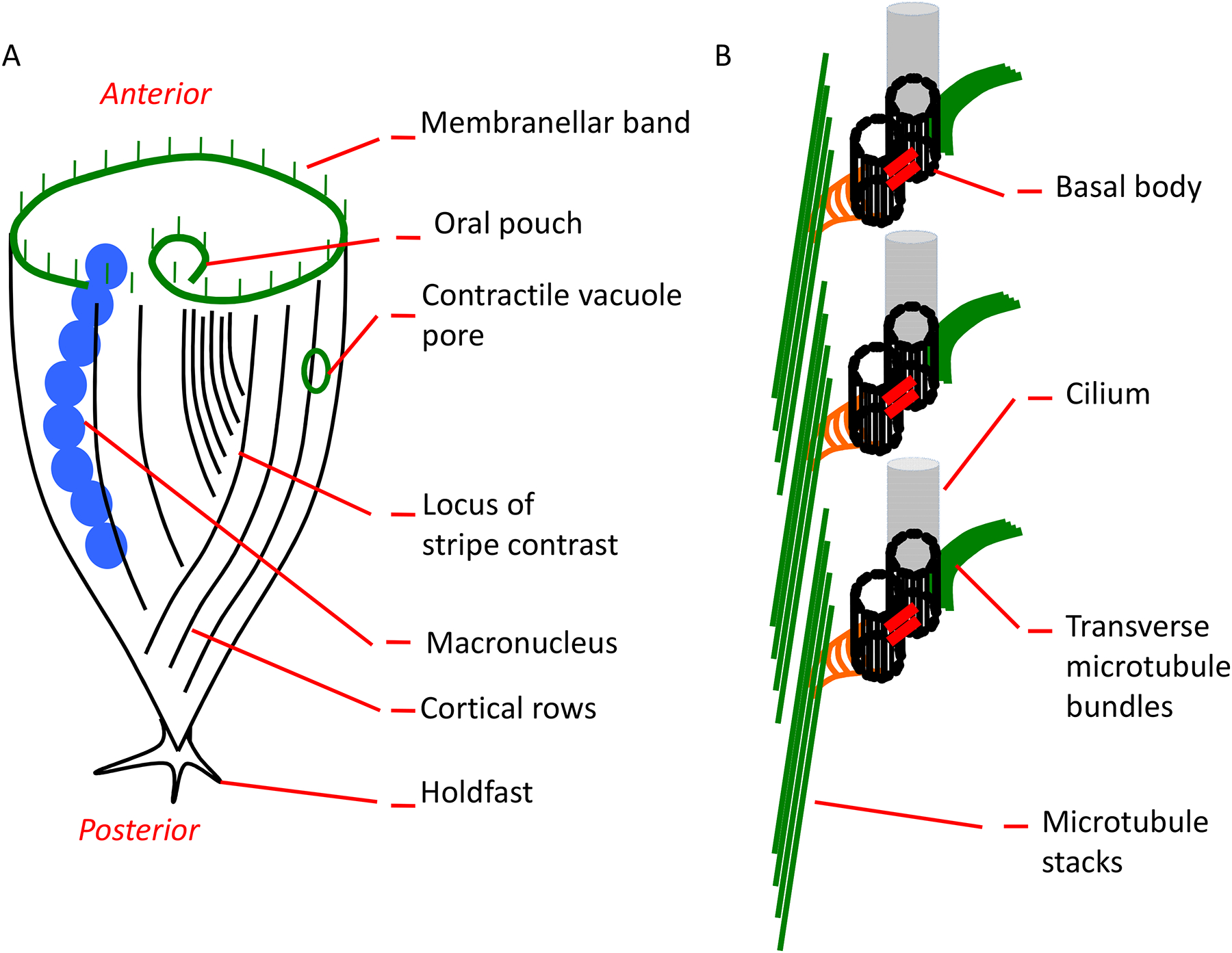
(A). The anatomy of Stentor. The anterior-posterior axis is indicated by the membranellar band at the anterior end and a holdfast at the posterior end. Longitudinal stripes of cilia run the length of the body, with variable spacing between them such that a region exists where closely spaced stripes meet widely spaced stripes. This locus of stripe contrast defines a ventral surface. Together the anterior-posterior and dorsal-ventral axes define a midline, relative to which all other cellular structures have defined locations. For example the macronucleus is always to the right (from the cell’s perspective) of the midline, while the contractile vacuole pore is always to the left.
(B). Detailed view of a cortical row showing pairs of basal bodies (centrioles), which link up to both longitudinal and transverse microtubule bundles (green).
From Physics to Shape
Although we often think about shape in terms of evolution and fitness, and say things like form implies function, the fact remains that the shape of biological structures ultimately must arise from physical forces.
A key physical principle underlying the shape of many cellular structures is energy minimization. Consider membrane bound organelles such as the lysosome. The basic form of a closed lipid bilayer comes from the fact that there is a strong energetic drive for the hydrophobic tails of the phospholipids to self-associate, shielding them from the aqueous phase. This same energetic condition means that holes in such a bilayer are not stable, so that the boundary of an organelle will always be a smooth closed surface, a manifold. However, the mere fact that the organelle is bounded by such a surface is not sufficient to predict its shape. Other factors can affect the shape of the surface. First, the surface to volume ratio determines the range of possible shapes. The minimal surface to volume ratio object would be a sphere, but if the surface to volume ratio increases, the organelle can explore a much wider range of shapes. Another factor that helps to sculpt the observed shape of an organelle is that the curvature of the membrane can be influenced by both the lipids and the proteins found embedded in it. For example, the endoplasmic reticulum contains proteins that favor binding to curved membranes and therefore drive the system to form tubules of defined radius [4]. One of the important threads in the field of differential geometry is how local curvatures of surfaces affect the global shape of the surface, and organelle shape seems to be an area ripe to apply such methods. The interplay between local forces setting curvatures, and global forces such as surface tension, can lead to some quite interesting and complex shapes, perhaps most notably the famous “parking garage” configuration of membrane connections between ER sheets [5]. Finally, we note that active forces applied by the cytoskeleton can sculpt the shape of membrane bound organelles by pulling out tethers or branches, turning a blob into a network [6].
The concept of energy minimization also underlies the recent explosion of interest in phase separation, whereby protein and protein-RNA interactions cause molecules to partition into highly concentrated droplets [7,8]. Such phase separation is a classical equilibrium behavior of polymer solutions [9].
Energy minimization is, however, an equilibrium phenomenon. Most cellular processes consume energy and are therefore non-equilibrium processes. One important difference between equilibrium and non-equilibrium systems is that equilibrium systems follow the law of detailed balance, which means that for any transition between states, the rate of the transition is equal to the rate of the reverse transition (at equilibrium). By violating detailed balance, non-equilibrium processes permit assembly to take place in a series of defined steps, one after another, whereas in an equilibrium system everything tries to stick together at once. Such sequential assembly thus allows for the assembly of much more complicated structures. Furthermore, non-equilibrium processes are capable of creating organization in a completely isotropic system, and so in some cases it may make more sense to refer to the assembly process as one of self-organization rather than self-assembly [10]. The ability of non-equilibrium systems to spontaneously break symmetry is a central theme in the complex anatomy of cells. Microtubules combined with motor proteins can spontaneously organize bipolar structures that resemble the arrangement of microtubules seen in mitotic spindles [11,12]. When actin filaments are nucleated from beads, the initially radially symmetric array of filaments spontaneously re-organizes into a linear “comet tail” that can drive motion of the bead [13–15].
Self-assembling structures inside cells
Many of the complex structures found within cells consists of multi-protein assemblies. Some of these are relatively simple, like virus capsids, in which a single type of protein self-associates through a set of defined interfaces such that the proteins can only fit together to yield a particular polyhedral shape. More complex, and less symmetric, examples include larger protein machines of the Central Dogma, such as the ribosome, the spliceosome, and the DNA replication machine. Perhaps the most classic example of such complex self-assembly is the assembly of bacteriophage like T4, which was shown to take place via a series of modular sub-assemblies which join together in a precise order [16,17]. We consider such protein machines to be self assembling in the sense that the information required to build them comes purely from intermolecular interactions, including protein-protein interactions, protein-RNA interactions, and RNA folding interactions. The fact that a structure assembles via self-assembly does not mean that the cell cannot control or regulate the process. Indeed, the size, number, and even form of self-assembling structures can be regulated by inputs from regulatory pathways that determine the rate and position of nucleation, the rate of growth, subunit availability, and eventual disassembly.
One self assembling complex that plays a particular role in establishing complex cellular patterning is the centriole. This structure, composed of a cylindrical array of nine triplet microtubules, also illustrates the complexity that can be created by self-assembly. At the core of the developing centriole is a protein-based cartwheel structure whose main structural element is the protein SAS6. Although SAS6 self-assembles into a ninefold symmetric structure [18,19], this symmetry turns out not to be required for the ninefold symmetry of the centriole itself. Elegant protein engineering studies, in which SAS6 is modified to self-assemble into structures of eight or ten fold symmetry, have found that the same constructs still yield centrioles with nine-fold symmetry when expressed in vivo, suggesting that the origin of the ninefold symmetry may lie in other parts of the centriole [20]. Recent high resolution structural studies have shown that the triplet microtubules of the centriole make extensive contacts with their neighbors via a structure called the A-C linker, and these contact may help set the angle between triplets, thus determining how many can fit into the final structure [21]). Drosophila SAS6 mutants form centrioles with fewer than nine triplets, but with gaps in the structure suggesting that adjacent triplets are trying to maintain the normal angle between them [22] On the other hand, when the cartwheel is engineered to have shorter arms, this results in a change in symmetry of the centriole from nine to eight triplets [23], suggesting that the angle between the triplets is not fixed by their interactions. It is important to note that although the centriole is a self assembling structure, this does not mean that the cell has no control over the process. Most of the time, new centrioles only assemble adjacent to pre-existing centrioles, suggesting a level of spatial control. Moreover, when a cell contains too many centrioles, the synthesis of new centrioles is inhibited [24].
Centriole structure is conserved in vastly divergent branches of the eukaryotes. For example, the centrioles of Giardia are almost indistinguishable from those of humans or green algae. Even more interesting is the high conservation of centriole associated fibers. These fibers have been perhaps most extensively characterized in Chlamydomonas. In this cell, a distal connecting fiber joins the mother and daughter centriole at their distal ends while a proximal connecting fiber joints the centrioles near their bases [25]. Four microtubule bundles known as rootlets emerge from the centriole pair and extend down the outside of the cell like lines of longitude. A so-called sinister fiber links each centriole to one of 4-member rootlets, while a dextral fiber each centriole to a 2 membered root [25]. Another fiber known as the nucleus basal body connector (NBBC) runs from the centriole to the nucleus [25].
in mammals, the basal bodies of ciliated epithelia of the airway, ependyma, and oviduct, extend two large fibrous appendages, the basal foot and striated rootlet [26]. These appendages are used to position the basal bodies and cilia relative to each other. Similar sets of fibers position and orient the basal bodies and cilia on the surface of ciliates, and indeed centriole-associated fibers appear to be a universally conserved feature of eukaryotic diversity.
The importance of these fiber systems is that they allow the centriole to act as a structural organizing center. In green algae such as Chlamydomonas, virtually every part of the cell is positioned relative to the centriole-associated rootlets. For example, the eyespot is always located adjacent the four-membered rootlet arising from the daughter basal body [27].
Other examples of apparently self-assembling or self-organizing structures within cells include the harpoon-like nematocysts of dinoflagellates [28], lens-like and reflecting eyespots of different algae that are assembled from two and three dimensional arrays of lipid containing droplets [29, 30]; and the contractile vacuole, a water-excreting organelle whose structure involves a complex network of tubes [31].
Axiation: Symmetry breaking and polarity
It isn’t enough to just build the right pieces, they also have to go in the right place. The study of animal development has largely been dominated by questions of overall arrangement of components. For example, intense research has focused on long range morphogen gradients that establish anterior-posterior ordering of organs, limbs and body segments. This focus on location of structures was brought to the forefront by the famous homeotic mutants of Drosophila, in which one body part develops in a position normally occupied by another for example legs where eyes should be. Developmental biology addresses the location of parts at four different levels: axiation, patterning, and positional information.
In animals that crawl on the ground, the anterior-posterior axis tells us which way the animal is moving, and the dorsal-ventral axis tells us which part of the animal is facing the substrate. Cells crawling on a substrate share these same axes. The “leading edge” of the cell is analogous to the anterior end of a crawling animal and the “uropod” or trailing edge is the posterior end. The equivalent of the dorsal-ventral axis is known as the basal-apical axis, with apical usually being the part of the cell away from the substrate and basal the side in contact. In some cell types, such as ciliates, the terms dorsal and ventral are used. Molecular pathways involved in symmetry breaking for these two perpendicular axes have been the subject of extensive research, and by this point many components of these pathways have been identified [32–35]. The symmetry breaking mechanisms generally seem to fall into three categories. The simplest way to break symmetry is to through some sort of singularity, meaning something that exists in the cell and that there is only one of, so wherever it is, it defined an axis. The classic example in cultured cells is the so-called nucleus-centrosome axis. If one draws an arrow from the nucleus to the centrosome, this often lies parallel to the direction of cell motion, although whether it points forward or backward depends on the cell type. In at least some cases, centrosome positioning appears to play a causal role in cell migration, polarization, or division [36–40]. But what sets the nucleus-centrosome axis? One study of migrating cells found that the orientation of this axis is set by nuclear motions driven by actin cortical flows, which position the nucleus behind the centrosome [41–42], thus arguing that the source of the directionality is not the nucleus or the centrosome, but the actin cortex. A second way to break symmetry is via rupturing of an actin-myosin cortex, such that the cortex flows towards one end of the cell, a purely physical mechanism that relies on the idea that if a sheet under tension ruptures in one spot, this rupture relieves the tension throughout the entire sheet such that further rupturing events are prevented elsewhere. A more complicated third way to break symmetry is through a reaction-diffusion system. This type of model was first proposed on purely theoretical grounds and, in its simplest form, would involve a molecule that marks a region of the cell and exerts positive feedback on itself, combined with a second molecule that tends to erase the mark created by the first molecule. Under certain conditions, such as the second molecule having more rapid diffusion than the first, such a system can spontaneously break symmetry such that the mark is present in part of the cell and absent elsewhere. The boundaries between these types of models are less clear that one might at first think, for example mechanical forces can play the role of an inhibitory signal [43]. One emerging aspect of cell axiation or polarity is the fact that some of the organelles whose position responds to the polarity are, themselves, players in the polarity establishment pathways [44].
In some cases, symmetry does not need to be broken because the anterior poster axis is already encoded in the structure of the cell cortex. This is true for ciliates as discussed above. In other cases, a polarizing signal can be provided by the external environment, for example by neighboring cells or by a basement membrane, and in such cases there does not need to be an internal spontaneous symmetry breaking mechanism. However it is important to bear in mind that the existence of external cues need not in itself mean that the system can’t spontaneously break symmetry when no cue is present.
What about left and right? Many cells look symmetrical from left to right. For example, fish keratocytes show a highly symmetrical shape [45], and most cultured cells look symmetrical. However, in some cases an inherent left-right asymmetry is present based on cell movements. The large syncytial amoeba Physarum is famous for its ability to solve mazes [46]. When individual Physarum cells are imaged moving through a T-junction, they are show a tendency to make right-hand turns [47]. It has also been reported that neutrophil-like Hl60 cells tend to polarize to the left of the nucleus-centrosome axis when exposed to uniform chemoattractant [48]. This asymmetry is likely to arise from the fact that all components of the cytoskeleton are chiral. For example, chiral movements of the actin-myosin cortex have been reported in C. elegans cells [49]. In the specific case of HL60 cells, ablation of the centriole eliminated the left-right bias [48]. Both the centriole itself, and the fibrous structures attached to it (see above) are chiral, and could thus be serving as a source of left-right asymmetry.
If cells contain an underlying left-right asymmetry, what happens when a cell divides? A tendency for sister cells to appear to be mirror images of each other was described by Albrecht-Buehler [50], who reported that in 3T3 cells, sister cells sometimes appears to be mirror images of each other. Mirror in sister cell shape was also seen in PtK2 cells [51] and in zebrafish embryonic cells [52]. At a subcellular level, mirror symmetry between sister cells has been reported for the arrangement of actin bundles [53], nucleoli [54], and chromosomes [55,56].
Patterning
In some cases, polarization or axiation is manifest primarily by the formation of a unique structure or process somewhere on the surface of the cell. Examples include bud formation in yeast cells, or the formation of a leading edge in motile cultured cells. But in other cases, the polarization extends to the interior of the cell, such that different intracellular components come to occupy defined positions relative to the cell axes. This is clearly seen in ciliates, in which many internal structures, such as the gullet (the place where food is endocytosed), the contractile vacuole, and the cytopyge (a cellular anus where digested remnants are expelled) all occupy well-defined positions along both the A-P and D-V axes [57]. In mammalian cells, the primary cilium is usually located on the apical surface, and in some cases points in a direction parallel to the A-P axis [58,59].
Many studies of pattern formation in animal development have focused on repeating structures such as the bands of pair rule gene expression. These banding patterns were initially thought to be produced by standing waves of some type, but it is now generally thought that, at least to a first approximation, they result from long range gradients combined with enhancers that are sensitive to narrow ranges of concentrations of the gradient signal. This type of pattern is also seen in in cells. One example is the positioning of denticles within single cells of Drosophila embryos. These actin based structures have a uniform spacing that scales proportionally to the size of the cell [60]. Ciliates are probably the most visibly obvious example of patterning, in which the entire surface of the cell is spanned by a parallel set of ciliary rows with defined spacing between them [61]. In a given species, the number of rows per cell is narrowly distributed, and if a cell occurs with too many or too few rows, the number is inherited by both daughters when the cell divides, but tends to correct itself back to the mean over several generations [62]. This latter effect suggests some mechanism exists to sense or control the number of rows, but how this works is not known.
Positional information and morphogen gradients
Symmetry breaking events can mark one region of the cell as being different from other regions, but if this is to lead to a globally defined arrangement of components, this information needs to be propagated to the rest of the cell, such that each region of the cell is “marked” with positional information telling which components belong in that region. A simple-sounding way to do this is via gradients of chemical signals, which could be small molecules such as lipids, macromolecules such as proteins or mRNA molecules, or else modified states such as phosphorylated proteins. The two most-discussed examples of gradients as a source of positional information in cells are those seen with RanGTP and with POM1 in fission yeast. A gradient of the small GTPase Ran forms around the mitotic spindle due to the presence of a GTP exchange factor located on the chromatin which coverts RanGDP to RanGTP [63]. As RanGTP diffuses away from the chromatin, the enzymatic activity of Ran converts GTP to GDP, such that at steady state there is a detectable gradient in the GTP/GDP ratio that is highest near the spindle and falls off with increasing distance from the spindle [64]. This gradient is though to regulate microtubule nucleation and other activities involved in sculpting the spindle [63]. The original picture of a diffusion-driven gradient of Ran is probably too simple. Ran interacts with microtubules, such that its spatial distribution is not determined solely by solution phase diffusion [65]. Because Ran interacts with microtubules, which are themselves the structure whose grows is regulated by RanGTP, both the form of the Ran gradient and its influence on the spindle can only be fully understood in terms of an interaction between a diffusing signal and a solid substrate [65].
In fission yeast, the division-regulating protein kinase POM1 forms gradient that is highest at the ends of the cell [66–69]. Pom1 regulates both the timing of division and the position of the division site, and interestingly these two functions seem to depend on different threshold levels of Pom1 [70], thus evoking a unicellular version of Wolpert’s “French Flag” model for morphogen gradient function. Establishing a reproducible gradient is challenging and can require careful balancing between rates of production and degradation of the signal.
In abnormal situations, a single cell can contain conflicting intracellular patterning signals. By studying how the cell responds to these conflicting signals, we can gain clues regarding how those signals are interpreted and generated. One example is seen in the giant ciliate Stentor which, as discussed above, has a clear anterior-posterior body axis with a feeding structure at one end and a holdfast at the other. The feeding end is considered anterior and the holdfast posterior. Stentor excels and wound healing and regeneration, which allows cutting and pasting experiments to be done [4]. Such experiments have found that if a second posterior region is grafted onto a cell, such that is has two holdfasts, these ends will gradually coalesce such that the cell can restore a single anterior-posterior body axis [3]. This experiment suggests that a cell is able to recognize the fact that it contains two conflicting sets of anterior-posterior signals, and then adjusts its morphology so as to bring the two gradients into alignment. Maintenance of a unified AP axis requires the kinase scaffold protein Mob1, such that when Mob1 is knocked down, the cell starts to develop multiple body axes, leading the normally cone-shaped cell to sprout extra tails [71]. Because Mob1 protein localizes to the posterior end of the cell, the client kinases of Mob1 would be clear candidates for components of an anterior-posterior position gradient. However, the phenotype of Mob1 knockdowns is not loss of anterior posterior information, just the loss of the ability to maintain a single unified body axis, so it is also possible that the output signal from Mob1 is not a positional cue per se but some sort of beacon indicating the presence or absence of a pre-existing posterior pole.
Another source of positional information is the cytoskeleton itself. Because microtubules are polarized structures, they confer information about direction and distance relative to the microtubule organizing center [72]. Moreover, it is possible for different microtubules within one cell to be marked with post-translation modifications that allow them to serve as tracks for different cargo [73]. An illustrative example is in the unicellular green alga Chlamydomonas (Figure 2). As discussed above, these cells contain four rootlet microtubule bundles, one of which correlates with the position of the eyespot. Another cellular structure, the contractile vacuole that the cell uses to pump out water to handle osmotic stress, also shows a consistent localization relative to the rootlet microtubules. In mutants that alter rootlet length, the distance of the eyespot from the top of the cell correlates with the length of the rootlet [74]. Another way to test a causal role of the rootlets is using mutants that alter the location of the basal bodies on the surface of the cell [75]. In the asq2 mutant, the rootlets form normally with respect to the basal bodies, but the basal bodies are mis-positioned, taking the rootlets with them. In such cells, the contractile vacuoles also show mis-positioning, which continued to be consistent with the rootlets rather than the overall cell axis, suggesting that their position was dependent on the rootlets. However, the eyespot was not mislocalized in centriole position mutants, suggesting that these cells must contain another source of positional information besides the centrioles-associated fibers and rootlets. Whey then does the position of the eyespot correlate with the length of the rootlets? One possibility is that both features are responding to some other global intracellular positional cue. In that case, the cell might be using a combination of cytoskeleton-mediated and gradient-mediated positional information. A similar type of coordinated mis-positioning experiment established that mitochondria in Tetrahymena are positioned on the cortex by the ciliary rows, such that altering the organization or orientation of the ciliary rows has a corresponding effect on the associated mitochondria [61, 76].
Figure 2. Multiple sources of positional information in one cell.
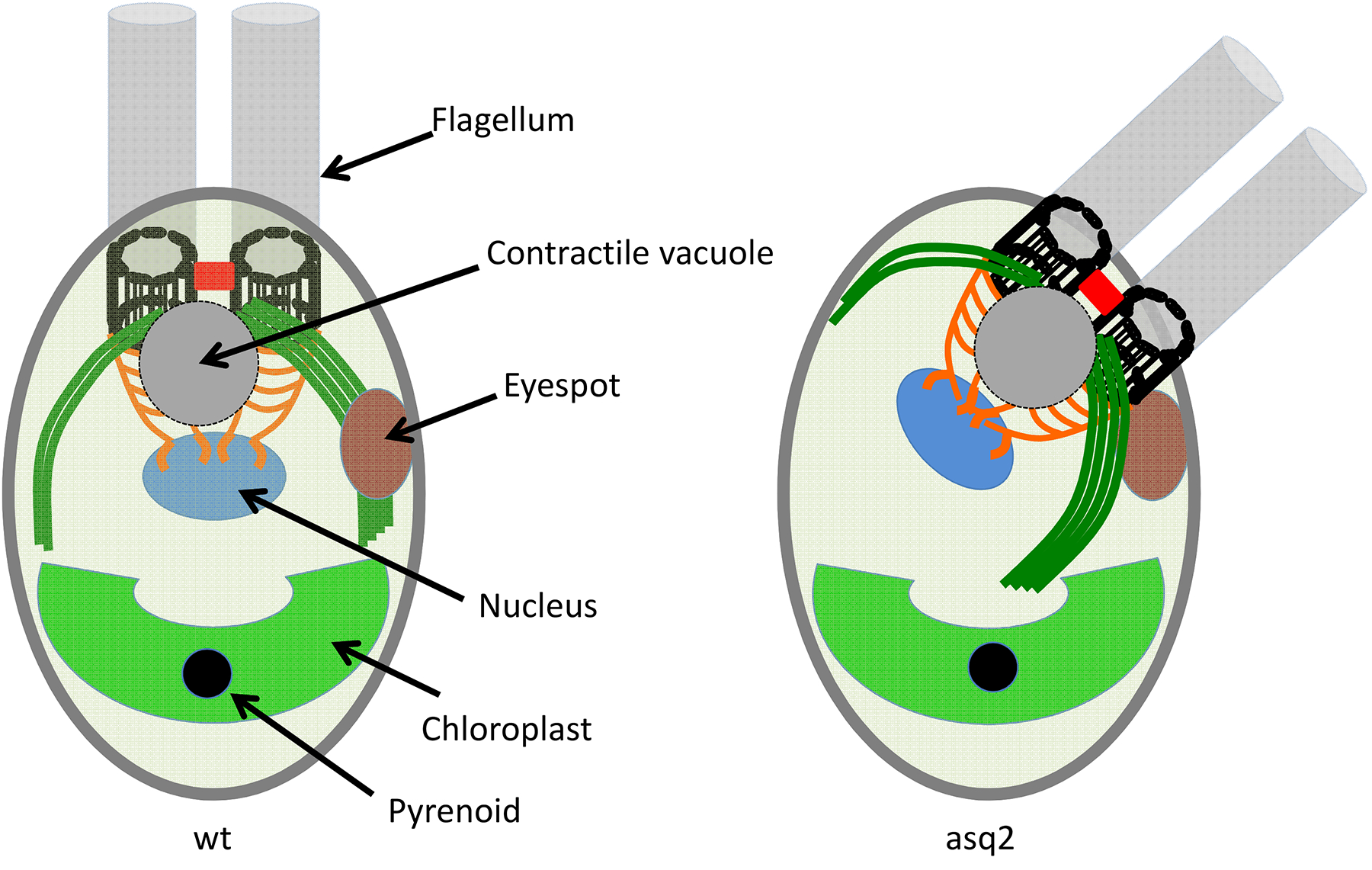
The left panel shows a wild-type Chlamydomonas cell with its complex internal anatomy. In the asq2 mutant (right panel), the basal bodies are displaced, and this results in some structures (nucleus and contractile vacuole) also being displaced, indicating that these structures gain positional information from the basal bodies or their associated fiber systems. Other structures (eyespot, chloroplast, pyrenoid) retain their normal position, indicating a positional information system that is not dependent on basal body position.
We have discussed two sources of positional information thus far: whole-cell patterning signals that extend over long ranges, and short-range local order provided by cytoskeletal structures. These do not need to be mutually exclusive. Surgical experiments in the ciliate Stylonychia shows that positional information can be provided by both the local orientation of the cortex and also by longer range directional cues, both at the same time. This cell sucks in food particles by means of a series of ciliary arrays that we will collectively refer to as feeding structures. The feeding structures are located on the anterior right half of the ventral surface, and the individual structures occupy reproducible positions with respect to anterior-poster and left-right axes. When the cell is cut in half lengthwise and then folded over, the cell builds a second set of feeding on the left half of the cell, which normally would never form such structures [77]. This presumably happens because the cortex is a stable protein lattice that retains its right side identity even after the cell is folded, bringing that patch of cortex onto the left side. Importantly, the left to right arrangement of the new feeding structures is backwards, implying that the left-right information they are using to direct their own formation is hard-wired into the cortex. However, the new feeding structures still form near the anterior end, and retain their normal anterior-posterior arrangement, which shows that the anterior-posterior information these structures use to form is coming from a global anterior-posterior body axis cue. Since this cue is retained even when the cortex is folded over, it is apparently not encoded in the cortex itself and thus may instead be a soluble gradient of some unknown signalling molecule.
The use of global and local information also takes place in the hair cells of the mammalian cochlea. These cells contain chevron-shaped arrays of stereocilia whose orientation is dictated by planar cell polarity, a long range signal that indicates direction within the plane of a developing tissue [78]. The fact that all the stereocilia are oriented relative to the planar polarity cue indicates that such cues can influence position within the cell. However, if planar cell polarity is eliminated the stereocilia still form normal-looking chevrons, although the direction is randomized with respect to the global orientation of the tissue [79]. Just as in Stylonychia, local information specifies the a sub-cellular structure (the chevron of stereocilia), while global information specifies where the structure will form and how it is oriented.
Convergent evolution of complex looking shapes
We are thus left with the picture of cellular anatomy arising from the interplay of multiple systems. Each structure has its own assembly pathway that can involve equilibrium processes of energy minimization or non-equilibrium processes of self-organization and symmetry breaking. The position of these structures responds to cell polarity and patterning processes which communication positional information over long ranges. A natural question to ask is how could such complex systems for generating cellular anatomy have evolved? The evolution of complexity is always a challenging issue, because it seems that multiple innovations must have all taken place before a complex shape is fully made, and anything short of that final structure would not be functional. This creates a sense of “irreducible complexity”. A different way to view this issue to ask just how “hard” it is to evolve a particular shape, where hard in this case is defined in terms of how many different genomic changes would be required to create a certain type of pattern. It is important, in thinking about this question, to recognize that shared motifs may appear again and again, due to deep evolutionary conservation. An excellent example has already been discussed, namely the many different structures that are formed using centrioles with associated striated fiber systems. Another example of a structural motif is formed from a ring of actin-based protrusions (stereocilia) surrounded a central microtubule-based cilium. This type of arrangement is shared between Choanoflagellates, the closest unicellular relatives of the animals, and the choanocytes of sponges [80], but is also seen in the hair cells of the mammalian cochlea, which start our as radially symmetric structures and only acquire their asymmetric structure later on in development, often accompanied by loss of the central cilium [81]. Given that these structures are typically composed of conserved protein modules, it is likely that they represent an example of something that has been inherited by common descent from something present in a common ancestor. In the case of the basal body fiber system discussed above, this common ancestor is apparently the last eukaryotic common ancestor.
An alternative way to approach the question of the evolutionary difficulty of a particular shape or form, is to ask how many times a given form has independently arisen during the course of evolution. One famous example is the “heliozoan” cell shape, which consists of a spherical cell radiating out a symmetrical array of microtubule-based projections. Such a complex, symmetrical array might seem to require substantial molecular innovation to create, but it turns out that the heliozoan shape has actually arisen in at least four completely independent lineages [82], suggesting that this type of cell bauplan may be easy to achieve with conserved molecular components.
One way that a structure may arise again and again, with potentially different molecular components, is if the shape or form arises from the physical processes. From an evolutionary point of view, the laws of physics represent a shared resource or legacy that all living things can draw on in a similar way, no matter how much their genomes may have diverged. But this is not to say that a physics-based self organization process exists outside of the genome – for example by altering the lipid composition of membranes, the curvature can be shifted.
Leveraging subcellular pattern formation to create multicellular life
We generally take for granted the idea that complex multicellular structures arise by putting lots of cells together in some defined arrangement, like LEGO building blocks. But this wasn’t always how cells were viewed in the context of organisms. A quite different view was often taken by the early cytologists, in which the organization of protoplasm into an organism was the primary activity of development, and the dividing up of that protoplasm into individual cells was a secondary process [83]. This is, of course, exactly what happens during Drosophila development, where much of the patterning is established while the embryo is still a single syncytial cell, after which the different embryonic regions are divided up into cells by ingression of plasma membrane. In fact, the developmental patterning of the Drosophila embryo is remarkably indifferent to the way in which the cytoplasm ends up being divided into cells. If cellularization is completely blocked using cytoskeletal inhibitors, pair rule genes like fushi-tarazu still show their characteristic stripes of expression, which can be seen at both the mRNA and protein levels [84]. A classic example in which patterning supersedes cellularity is provided by studies of the pronephric duct in salamanders of different ploidy. The pronephric duct is tube surrounded by a single layer of cells. Fankhauser [85] found that as ploidy was increased, such that cells because larger and larger, the diameter of the duct did not change, such that essentially the same biological structure could be composed of a far smaller number of cells than would normally be seen. When F.R. Lillie caused marine annelid eggs to develop parthenogenically, the cells underwent mitosis without cytokinesis. These organisms normally form a trochophore larvae consisting of hundreds to thousands of cells with distinct rings of cilia that they use to swim. In Lillie’s experiments, the syncytial division-blocked eggs were still able to develop ciliary rings despite being single cells [86]. This result suggests that the machinery for building a ciliary band does not require multicellular tissue organization, and indeed the appearance of a trochophore larva is remarkably similar to that of the giant ciliate Didinium, which also forms ciliary bands. Overall, the picture emerges that at least some of the development of complex animal embryos is using morphogenetic processes present within a single cell. For animals that undergo mosaic development, this fact should not be surprising. In these animals, lineages are set aside from the earliest divisions, meaning that fate determinants have already been patterned within the one-cell stage embryo. It would be surprising if the mechanisms for this patterning were completely distinct from those used in patterning of unicellular organisms. The question now is to what extent did early animal evolution rely on subcellular patterning mechanisms versus newly evolved cell-cell communication pathways. Studying the mechanisms of pattern formation and complexity in single cells may thus hold the key to understanding how plants and animals evolved.
Acknowledgments
Work on cell geometry in the author’s lab has been supported by NIH grant R35 GM130327 and by the Chan Zuckerberg Biohub.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kirschner M, Gerhart J, Mitchison T (2000). Molecular “vitalism”. Cell 100, 79–88. [DOI] [PubMed] [Google Scholar]
- 2.Rafelski SM, Marshall WF (2008). Building the cell: design principles of cellular architecture. Nat. Rev. Mol. Cell Biol 9, 593–602 [DOI] [PubMed] [Google Scholar]
- 3.Tartar V (1961). The Biology of Stentor. Pergammon Press. [Google Scholar]
- 4.Voeltz GK, Prinz WA (2007). Sheets, ribbons and tubules – how organelles get their shape. Nat. Rev. Mol. Cell Biol 8, 258–64. [DOI] [PubMed] [Google Scholar]
- 5.Terasaki M, Shemesh T, Kasthuri N, Klemm RW, Schalek R, Hayworth KJ, Hand AR, Yankova M, Huber G, Lichtman JW, Rapoport TA, Kozlov MM (2013). Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell. 154, 285–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waterman-Storer CM, Salmon ED (1998). Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol 8, 798–806 [DOI] [PubMed] [Google Scholar]
- 7.Banani SF, Lee HO, Hyman AA, Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langdon EM, Gladfelter AS (2018). A new lens for RNA localization: liquid- liquid phase separation. Annu. Rev. Microbiol 72,255–271 [DOI] [PubMed] [Google Scholar]
- 9.Keating CD (2012). Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc. Chem. Res 45, 2114–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karsenti E (2008). Self-organization in cell biology: a brief history. Nat. Rev. Mol. Cell Biol 9, 255–62 [DOI] [PubMed] [Google Scholar]
- 11.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E (1996). Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 382, 420–5 [DOI] [PubMed] [Google Scholar]
- 12.Loughlin R, Heald R, Nédélec F (2010). A computational model predicts Xenopus meiotic spindle organization. J. Cell. Biol 191, 1239–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernheim-Groswasser A, Wiesner S, Golsteyn RM, Carlier MF, Sykes C (2002). The dynamics of actin-based motility depend on surface parameters. Nature. 417, 308–11 [DOI] [PubMed] [Google Scholar]
- 14.Upadhyaya A, Chabot JR, Andreeva A, Samadani A, van Oudenaarden A (2003). Probing polymerization forces by using actin-propelled lipid vesicles. Proc. Natl. Acad. Sci. U. S. A 100, 4521–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akin O, Mullins RD (2008). Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp 2/3 complex. Cell. 133, 841–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood WB (1980). Bacteriophage T4 morphogenesis as a model for assembly of subcellular structure. Q. Rev. Biol 55, 353–67. [DOI] [PubMed] [Google Scholar]
- 17.Arisaka F, Yap ML, Kanamaru S, Rossmann MG (2016). Molecular assembly and structure of the bacteriophage T4 tail. Biophys. Rev 8, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, Steinmetz MO (2011). Structural basis of the 9-fold symmetry of centrioles. Cell. 144,364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Breugel M, Hirono MN, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich MN, Ebong IO, Robinson CV, Johnson CM, Veprintsev D, Zuber B (2011). Structures of SAS-6 suggest its organization in centrioles. Science. 331,1196–1199. [DOI] [PubMed] [Google Scholar]
- 20.Hilbert M, Noga A, Frey D, Hamel V, Guichard P, Kraatz SH, Pfreundschuh M, Hosner S, Fluckiger I, Jaussi R, Wieser RM, Thieltges KM, Deupi X, Muller DJ, Kammerer RA, Gonczy P, Hirono M, Steinmetz MO. (2016). SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nat. Cell Biol 18, 393–403. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Fernandez JJ, Marshall WF, Agard DA (2019). Electro cryo-tomography provides insight into procentriole architecture and assembly mechanism. Elife. 8 pii: e43434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C,Ferreira I, Callaini G, and Glover DM (2007). DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol 17, 1465–1472. [DOI] [PubMed] [Google Scholar]
- 23.Hiraki M, Nakazawa Y, Kamiya R, Hirono M (2007). Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol 17, 1778–83. [DOI] [PubMed] [Google Scholar]
- 24.Marshall WF (2007). Stability and robustness of an organelle number control system: modeling and measuring homeostatic regulation of centriole abundance. Biophys. J 93, 1818–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geimer S, Melkonian M (2004). The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J. Cell Sci 117, 2663–2674. [DOI] [PubMed] [Google Scholar]
- 26.Garcia G, Reiter JF (2016). A primer on the mouse basal body. Cilia. 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes JA, Dutcher SK (1989). Cellular asymmetry in Chlamydomonas reinhardtii. J. Cell Sci 94, 273–85. [DOI] [PubMed] [Google Scholar]
- 28.Gavelis GS, Wakeman KC, Tillmann U, Ripken C, Mitarai S, Herranz M, Özbek S, Holstein T, Keeling PJ, Leander BS (2017). Microbial arms race: ballistic nematocysts in dinoflagellates represent a new extreme in organelle complexity. Sci. Adv 3, e1602552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreimer G (1999). Reflective properties of different eyespot types in dinoflagellates. Protist 150, 311–23 [DOI] [PubMed] [Google Scholar]
- 30.Hayakawa S, Takaku Y, Hwang JS, Horiguchi T, Suga H, Gehring W, Ikeo K, Gojobori T (2015). Function and evolutionary origin of unicellular camera-type eye structure. PLoS One. 10, e0118415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plattner H (2015). The contractile vacuole complex of protists – new cues to function and biogenesis. Crit. Rev. Microbiol 41, 218–27. [DOI] [PubMed] [Google Scholar]
- 32.Drubin DG, Nelson WJ (1996). Origins of cell polarity. Cell. 84, 335–44 [DOI] [PubMed] [Google Scholar]
- 33.Chiou JG, Balasubramanian MK, Lew DJ (2017). Cell polarity in yeast. Annu. Rev. Cell. Dev. Biol 33, 77–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vendel KJA, Tschirpke S, Shamsi F, Dogterom M, Laan L (2019). Minimal in vitro systems shed light on cell polarity. J. Cell Sci 132, pii: jcs217554 [DOI] [PubMed] [Google Scholar]
- 35.Pickett MA, Naturale VF, Feldman JL (2019). A polarizing issue: diversity in the mechaniss underlying apico-basolateral polarization in vivo. Annu. Rev. Cell Dev. Biol 35, 285–308 [DOI] [PubMed] [Google Scholar]
- 36.de Anda FC, Meletis K, Ge X, Rei D, Tsai LH (2010). Centrosome motility is essential for initial axon formation in the neocortex. J. Neurosci 30, 10391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bienkowska D, Cowan CR (2012). Centrosomes can initiate a polarity axis from any position within one-cell C. elegans. Curr. Biol 22, 583–9. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Wang YL (2017). Centrosome defines the rear of cells during mesenchymal migration. Mol. Biol. Cell 28, 3240–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang N, Marshall WF (2012). Centrosome positioning in vertebrate development. J. Cell Sci 125, 4951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burute M, Prioux M, Blin G, Truchet S, Letort G, Tseng Q, Bessy T, Lowell S, Young J, Filhol O, Théry M (2017). Polarity reversal by centrosome repositioning primes cell scattering during epithelial-to-mesenchymal transition. Dev. Cell 40, 168–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes ER, Jani S, Gundersen GG (2005). Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121, 451–63. [DOI] [PubMed] [Google Scholar]
- 42.Luxton GW, Gomes ER, Folker ES, Worman HJ, Gundersen GG (2011). TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2, 173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD (2012). Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 148, 175–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaidžiulytė K, Coppey M, Schauer K (2019). Intracellular organization in cell polarity – placing organelles into the polarity loop. J. Cell Sci 132, pii: jcs230995. [DOI] [PubMed] [Google Scholar]
- 45.Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, and Theriot JA (2008). Mechanism of shape determination in motile cells. Nature 453, 475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagaki T, Yamada H, Toth A (2000). Maze-solving by an amoeboid organism. Nature 407, 470–470. [DOI] [PubMed] [Google Scholar]
- 47.Dimonte A, Adamatzky A, Erokhin V, Levin M (2016). On chirality of slime mould. Biosystems. 140, 23–7. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, Van Keymeulen A, Wakida NM, Carlton P, Berns MW, Bourne HR (2007). Polarity reveals intrinsic cell chirality. Proc. Natl. Acad. Sci. U. S. A 104, 9296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naganathan SR, Fürthauer S, Nishikawa M, Jülicher F, Grill SW (2014). Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. Elife. 3, e04165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albrecht-Buehler G (1977). Daughter 3T3 cells. Are they mirror images of each other? J. Cell Biol 72, 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klymkowsky MW, Miller RH, Lane EB (1983). Morphology, behavior, and interaction of cultured epithelial cells after the antibody-induced disruption of keratin filament organization. J. Cell Biol 96, 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR,Tada M, Clarke JD (2007). A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature. 446, 797–800. [DOI] [PubMed] [Google Scholar]
- 53.Delhanty P, Leung H, Locke M (1991). Paired cytoskeletal patterns in an epithelium of Siamese twin cells. Eur. J. Cell Biol 56, 443–50. [PubMed] [Google Scholar]
- 54.Locke M Leung H (1985). The pairing of nucleolar pattern in an insect epithelium as evidence for a conserved nuclear skeleton. Tissue Cell 17, 573–588. [DOI] [PubMed] [Google Scholar]
- 55.Boveri T (1909). Die blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität, Archiv für Zellforschung 3, 181–268. [Google Scholar]
- 56.Berr A, Schubert I (2007). Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics 176, 853–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aufderheide KJ, Frankel J, Williams NE (1980). Formation and positioning of surface-related structures in protozoa. Microbiol. Rev 44, 252–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albrecht-Buehler G. (1977b) Phagokinetic tracks of 3T3 cells: parallels between the orientation of track segments and of cellular structures which contain actin or tubulin. Cell. 12, 333–9. [DOI] [PubMed] [Google Scholar]
- 59.Katsumoto T, Higaki K, Ohno K, Onodera K (1994). The orientation of primary cilia during the wound response in 3Y1 cells. Biol. Cell 81, 17–21. [DOI] [PubMed] [Google Scholar]
- 60.Spencer AK, Schaumberg AJ, Zallen JA (2017). Scaling of cytoskeletal organization with cell size in Drosophila. Mol. Biol. Cell 28, 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aufderheide KJ (1980). Mitochondrial associations with specific microtubular components of the cortex of Tetrahymena thermophila. II. Response of the mitochondrial pattern to changes in the microtubular pattern. J. Cell Sci 42, 247–261. [DOI] [PubMed] [Google Scholar]
- 62.Nanney DL (1966). Corticotype transmission in Tetrahymena. Genetics 54, 955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalab P, Heald R (2008). The RanGTP gradient – a GPS for the mitotic spindle. J Cell Sci. 121, 1577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaláb P, Pralle A, Isacoff EY, Heald R, Weis K (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 440, 697–701. [DOI] [PubMed] [Google Scholar]
- 65.Oh D, Yu CH, Needleman DJ (2016). Spatial organization of the Ran pathway by microtubules in mitosis. Proc. Natl. Acad. Sci. U. S. A 113, 8729–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Padte NN, Martin SG, Howard M, Chang F (2006). The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr. Biol 16, 2480–7. [DOI] [PubMed] [Google Scholar]
- 67.Moseley JB, Mayeux A, Paoletti A, Nurse P (2009). A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 459, 857–60. [DOI] [PubMed] [Google Scholar]
- 68.Allard CAH, Opalko HE, Moseley JB (2019). Stable Pom1 clusters form a glucose-modulated concentration gradient that regulates mitotic entry. Elife. 8, pii: e46003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerganova V, Floderer C, Archetti A, Michon L, Carlini L, Reichler T, Manley S, Martin SG (2019). Multi-phosphorylation reaction and clustering tune Pom1 gradient mid-cell levels according to cell size. Elife. 8, pii: e45983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhatia P, Hachet O, Hersch M, Rincon SA, Berthelot-Grosjean M, Dalessi S, Basterra L, Bergmann S, Paoletti A, Martin SG (2014). Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle. 13, 538–52. [DOI] [PubMed] [Google Scholar]
- 71.Slabodnick MM, Ruby JG, Dunn JG, Feldman JL, DeRisi JL, Marshall WF (2014). The kinase regulator mob1 acts as a patterning protein for Stentor morphogenesis. PLoS Biol. 12, e1001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirschner M, Mitchison T (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell. 145, 329–42 [DOI] [PubMed] [Google Scholar]
- 73.Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ (2009). Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 7, e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boyd JS, Gray MM, Thompson MD, Horst CJ, Dieckmann CL (2011). The daughter four-membered microtubule rootlet determines anterior-posterior positioning of the eyespot in Chlamydomonas reinhardtii. Cytoskeleton 68, 459–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feldman JL, Geimer S, Marshall WF (2007). The mother centriole plays an instructive role in defining cell geometry. PLoS Biol. 5, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frankel J (1979). An analysis of cell-surface patterning in Tetrahymena, p. 215–246. In Subtelny S and Konigsberg IR (ed.), Determinants of spatial organization. Academic Press, Inc., New York. [Google Scholar]
- 77.Grimes GW, L’Hernault SW (1979). Cytogeometrical determination of ciliary pattern formation in the hypotrich ciliate Stylonychia mytilus. Dev. Biol 70, 372–395. [DOI] [PubMed] [Google Scholar]
- 78.Marcinkevicius E (2009). Q&A: Quantitative approaches to planar polarity and tissue organization J. Biol, 8,103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montcouquiol M, Rachel RA, Lanford PJ, Copeland NH, Jenkins NA, Kelley MW (2003). Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature 423, 173–7. [DOI] [PubMed] [Google Scholar]
- 80.Mah JL, Christensen-Dalsgaard KK, Leys SP (2014). Choanoflagellate and choanocyte collar-flagellar systems and the assumption of homology. Evol. Dev 16, 25–37 [DOI] [PubMed] [Google Scholar]
- 81.Nayak GD, Ratnayaka HS, Goodyear RJ, Richardson GP (2007). Development of the hair bundle and mechanotransduction. Int. J. Dev. Biol 5, 597–608 [DOI] [PubMed] [Google Scholar]
- 82.Gast RJ (2017). Centrohelida and other heliozoan-like protists In Handbook of the Protists, Volume 2 Archibald JM, Simpson AGB, Slamovits C, eds. Springer. [Google Scholar]
- 83.Maienschein J (2018). Changing ideas about cells as complex systems In Visions of Cell Biology, Matlin KS, Maienschein J, and Laubichler MD, eds. University of Chicago Press, Chicago: 368 pp. [Google Scholar]
- 84.Edgar B, Odell GM, Schubiger G (1987). Cytoarchitecture and the patterning of fushi tarazu expression in the Drosophila blastoderm. Genes Dev. 1, 1226–37 [DOI] [PubMed] [Google Scholar]
- 85.Fankhauser G (1945). Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment of cell number and cell shape. J. Exp. Zool 100, 445–455. [DOI] [PubMed] [Google Scholar]
- 86.Lillie FR (1902). Differentiation without cleavage in the egg of the annelid Chaetopterus pergamentaceus. Arch. Entwicklungsmechanik Org 14, 477–499. [Google Scholar]


