Abstract
Oligometastasis represents an intermediate disease stage between localized and widely metastatic cancer. Efficient identification of patients with oligometastasis remains a barrier for accrual on clinical trials of oligometastasis-directed therapy. Here we review the prospect of circulating tumor DNA (ctDNA) based monitoring to promote sensitive, specific, and cost-efficient detection of cancer recurrence during post-treatment surveillance. Thus, an impetus for the development and implementation of clinical grade ctDNA assays should be for the positive impact they will have on clinical investigations of oligometastasis-directed therapy.
Keywords: Oligometastasis, Radiotherapy, circulating tumor DNA, biomarkers
Introduction
Metastasis remains the major cause of cancer-related deaths, and rightfully a focus area of research investigation. Recent developments in cancer therapy have transformed our perspective on the survival potential of patients with metastatic cancer. Improvements in biologically-directed systemic therapies, such as Her2-targeted therapy for Her2-amplified metastatic breast cancer, continue to push the boundaries of survival benefit1,2. Additionally, the advent of immune checkpoint inhibitors have led to marked prolongation of survival for a subset of patients with previously incurable metastatic cancers3–5. Another emerging and exciting area of clinical investigation is the delivery of definitive local therapy for patients with oligometastatic disease (OMD). Technological advances in radiotherapy over the past few decades have enabled the broad application of Stereotactic Ablative Radiotherapy (SAbR) as a non-surgical alternative for oligometastasis-directed therapy (ODT)—thereby broadening the number of patients with OMD for whom ODT may be considered6–8. Recently reported randomized clinical trials of SAbR for oligometastasis have suggested the possibility of overall survival advantages9–12. These initial studies provide a glimmer of hope that the clinical impact of ODT may be substantial. However, the potential of ODT will be limited by our ability to efficiently identify OMD before it progresses into widely metastatic disease. The goal of this review is to highlight emerging roles for circulating tumor DNA biomarkers to facilitate early identification of OMD and to potentially direct the implementation of ODT.
Oligometastatic disease represents a cancer stage that falls between localized and widely disseminated disease13. Key questions remain in our understanding of how best to identify patients with OMD who will benefit from ODT. We do not know whether all patients who develop metastatic cancer pass through an OMD stage. Although a major emphasis for clinical studies is on the number of lesions that define OMD, other criteria may be equally or possibly more relevant when predicting progression risk. These include histological cancer type, initial disease stage, interval therapy, and disease-free interval—all of which may be helpful when anticipating the pace of metastatic progression14. A potentially distinct entity of an “induced” OMD-like state must also be considered, which may arise in patients with disseminated metastases who experience a near complete response to systemic therapy yet have a small number of residual or oligoprogressive lesions15. Whether ODT in such cases of induced OMD can improve clinical outcomes is not known, yet should be investigated. Recent studies have highlighted emerging genomic biomarkers that may help facilitate the identification of patients with OMD who are most likely to benefit from ODT16,17. The validation of predictive biomarkers is necessary for optimal clinical decision making, and should be integrated into ongoing and future trials of ODT.
A major obstacle to OMD-directed clinical trials, and for the broader application of ODT, is efficient patient identification. Only a minority of patients with OMD will be identified due to symptomatic presentation. Currently, the majority of OMD patients are diagnosed after biopsy confirmation of an incidental finding on initial staging or post-treatment radiological examinations. The high costs and potential for false positives from imaging tests has led to variable practice patterns in post-treatment radiographic surveillance across cancer types. Herein, we review circulating tumor DNA (ctDNA) assays and summarize the potential for ctDNA biomarkers to facilitate identification of patients with oligometastasis for ODT.
Circulating Tumor DNA Biomarkers
Circulating biomarkers of cancer have been investigated for decades, and broadly encompass three main categories: circulating tumor cells, circulating tumor associated proteins or metabolites, and circulating tumor nucleic acids. While there has been progress in all of these areas over the past decade, the field of circulating tumor nucleic acids is in the midst of a renaissance that is being driven by technological advances, including but not limited to next-generation sequencing (NGS). There are at least two distinct types of circulating tumor nucleic acid biomarkers. Circulating tumor-associated RNA can be identified in cancer patients, and their etiology entails the release of vesicle or exosome encapsulated RNA molecules from cancer cells18. In contrast, ctDNA is believed to derive from dying cancer cells—either through apoptosis, necrosis, or other cell death pathways—that release nucleosomal histone-bound DNA fragments into the circulation19. Although most commonly evaluated in plasma, ctDNA can also be present in other biofluids, such as urine, CSF, and ascites depending on the cancer type and anatomical location of disease. The past decade has witnessed an explosion in technologies for ctDNA detection, some of which are beginning to enter the clinical arena.
Clinical applications
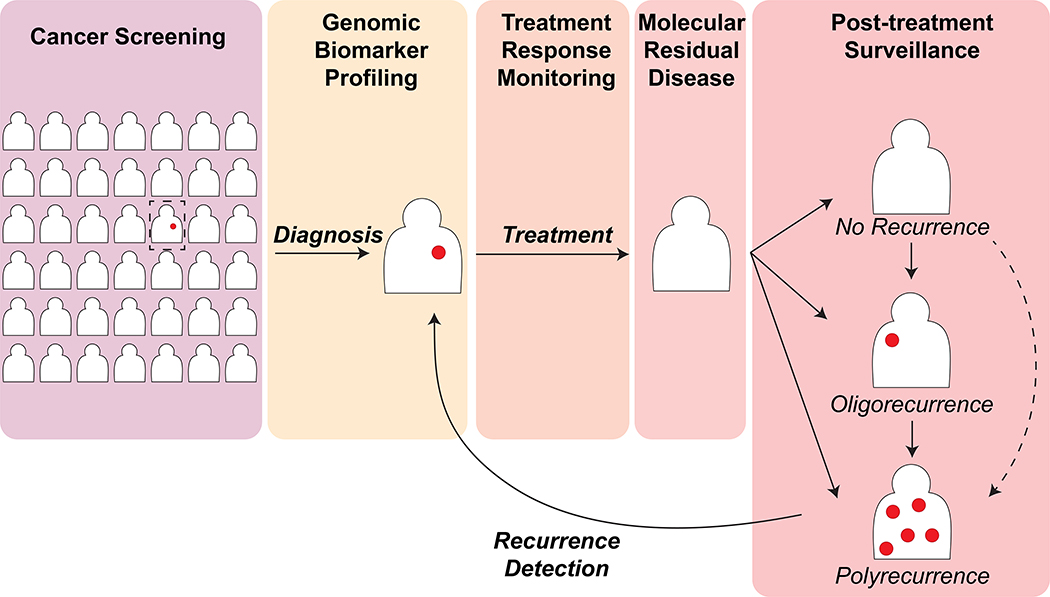
ctDNA assays may be used for cancer screening in a healthy population 20, molecular characterization of a newly diagnosed cancer when tissue diagnosis is not feasible, risk categorization, prognostication, prediction of response to therapy, non-invasive monitoring of treatment response, post-treatment surveillance and early recurrence detection, and discovery of potential resistance mechanisms after treatment failure (Figure 1).
Figure 1: Potential clinical applications of plasma ctDNA assays for detection and monitoring of cancer.
Please refer to the text for further details.
Prostate specific antigen (PSA) represents a protein-based biomarker used in precisely this fashion for more than 35 years in the management of prostate cancer and provides a framework for how ctDNA assays can be clinically impactful in the future. PSA has proven benefit as a screening test, where PSA defines indication for biopsy. Furthermore, National Comprehensive Cancer Network (NCCN) prostate cancer risk categorization is determined by PSA level in addition to other factors21. Risk categorization in turn determines intensity of recommended therapies. Monitoring of the PSA in response to androgen deprivation therapy prior to radiation has been shown to be prognostic22. PSA is used to define and detect ‘biochemical failure’ or early recurrence prior to radiologic or clinical failure, and to monitor response to metastatic directed therapies.
PSA testing is thus integral to the optimal clinical management of prostate cancer. It is difficult to imagine treating patients with prostate cancer without the availability of this test. ctDNA assays in many malignancies including but not limited to head and neck, lung, breast, gastrointestinal, gynecologic, and bladder cancers hold promise to become as integral to improve the precision of oncologic management as PSA has in prostate cancer. ctDNA assays will likely improve bladder preservation paradigms in bladder cancer, better select patients for esophagectomy after chemoradiation in esophageal cancer, and allow for intensification or omission of adjuvant systemic therapies in multiple malignancies. Furthermore, unlike PSA, ctDNA can be used to monitor specific molecular alterations over time including drug resistance.
Here we will specifically focus on attributes of ctDNA technologies that are important for post-treatment surveillance and treatment response monitoring, which are the most pertinent contexts for ctDNA assays in the identification and management of patients with OMD.
Technical considerations in ctDNA measurement
Distinguishing ctDNA from non-malignant processes:
Precise quantification of ctDNA is akin to counting needles in a proverbial haystack, where the hay is comprised of circulating cell free DNA (cfDNA) that is released from non-malignant sources, predominantly from physiologic turnover of hematopoietic cells 23. In most cases, ctDNA will comprise <1% of the plasma cfDNA biomass. Thus, all ctDNA assays must discriminate between tumor-derived and non-tumor derived cfDNA. The most common strategy for achieving specificity for ctDNA is by detecting known tumor-specific mutations within cfDNA. Thus, if genomic analysis has been performed on a patient’s tumor, identification of matching tumor-specific mutant alleles are indicative of ctDNA. De novo discovery of tumor-specific mutations through cfDNA analyses—when tissue based analysis has not been performed—remains an area of interest, yet such analyses must be interpreted with caution. There is now substantial evidence that cancer driver gene mutations can be observed in non-malignant tissues24–26, and that these can be represented in cfDNA. A recent analysis determined that a substantial percentage of candidate ctDNA mutations identified in the analysis of a large cancer patient cohort were attributed to clonal hematopoiesis rather than the index cancer of interest27. In addition to tumor-specific mutations, other properties can be used to distinguish ctDNA. Copy number changes in tumors can be detected in ctDNA, and used to estimate ctDNA proportion of total cfDNA28. Changes in DNA methylation state may also be cancer-specific, and have been incorporated into ctDNA assays29–31. Finally, the pattern of DNA fragmentation may be specific for tumor-derived DNA versus non-malignant DNA sources, and can potentially be integrated into ctDNA assays32,33. Effective ctDNA assays must use one or more of these strategies to achieve the necessary specificity and sensitivity for ctDNA measurement in a high background of non-malignant cfDNA.
ctDNA assay design: breadth versus depth
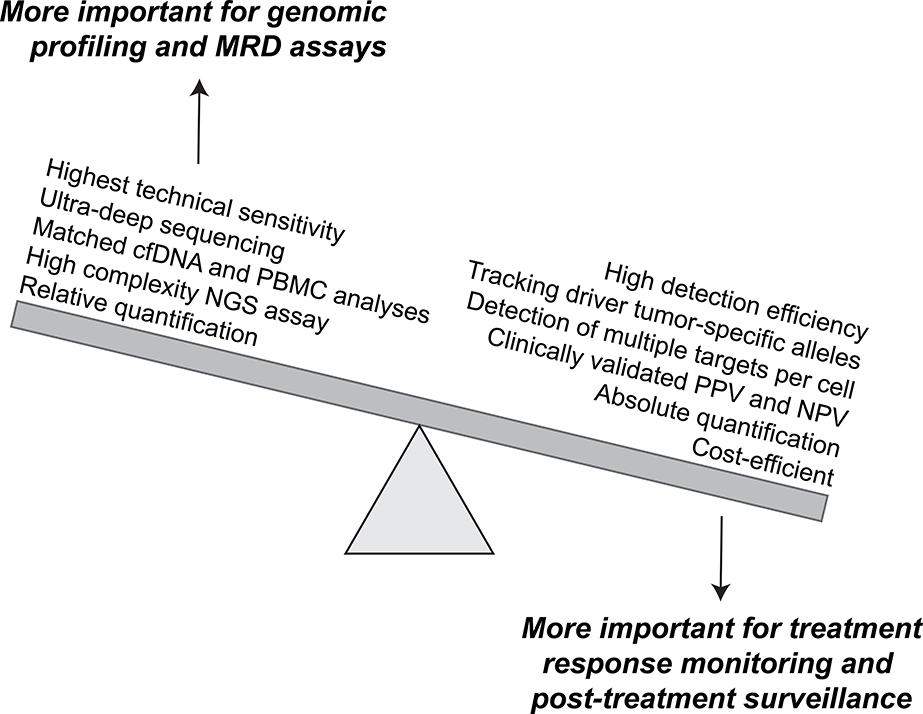
There are a wide variety of technologies and assays that have been successfully used to detect and quantify ctDNA, each of which has its own set of advantages and disadvantages. In general, there tends to be a balance between breadth and depth. On one end of the spectrum, a digital PCR (dPCR) assay that specifically detects a known driver gene mutation in a particular cancer may function well as a ctDNA monitoring assay. However, such allele-specific assays are often applicable to only limited subsets of patients, and even within a single patient may have limited utility in the setting of clonal heterogeneity. Due to these limitations, NGS-based assays are common in the ctDNA technology space. Assays vary in their complexity, breadth of genome coverage, and ability to discriminate ctDNA signals from noise. Due to a need to sequence to a much higher depth than tissue-based tumor sequencing, plasma ctDNA assays often have a more limited breadth of genomic space that is being analyzed. This often means that commonly mutated genes and hotspot mutation sites for a given cancer type are the primary target of the ctDNA assays. The vast number of ctDNA assays under development are beyond the scope of this review, and have been summarized elsewhere23. However, we will review some of the key distinguishing characteristics that are important to consider when evaluating ctDNA assays for disease surveillance (Figure 2).
Figure 2: Desired features of plasma ctDNA assays for treatment response monitoring and post-treatment surveillance.
Preferred features of ctDNA assays depend on the intended clinical application. Use of ctDNA for monitoring and surveillance applications require repeated testing. Thus, the priority of assay features is distinct from applications that require ultra-deep assessment and potential de novo discovery of rare tumor-derived mutant alleles.
Sensitivity versus efficiency of detection
NGS-based ctDNA assays have evolved in complexity in order to push the limits of sensitivity. One of the more effective strategies utilizes unique molecular tags in combination with bioinformatic “training” or error suppression34. Indeed, some of these high complexity NGS assays have demonstrated sensitivity for mutant allele fractions (MAF) as low as 0.005%35. However, such technical measurements of MAF sensitivity may not always be relevant to analyses of plasma ctDNA. The main reason for this is the limited biomass of accessible plasma cfDNA for analysis, which normally ranges between 5–100ng—corresponding to approximately 1,500 – 30,000 haploid genome equivalents (hGE’s). For this reason, plasma ctDNA assays have to be efficient in addition to being sensitive. It is worth noting that detection efficiency is typically ~100% for dPCR. In contrast, the detection efficiency of NGS-based assays can be highly variable, and is often not reported or evaluated. Standardized reporting of ctDNA assay sensitivity based on a constant assay input of 50ng would go a long way in mitigating these issues.
Detection of multiple target copies per cells
An effective strategy for enhancing the practical sensitivity of a ctDNA assay is to the increase the number of targets being detected per cancer cell. Unfortunately, due to the limited genomic target size of most ctDNA assays, the number of mutations that are tracked per tumor are typically less than 5. Some groups have resorted to whole exome sequencing of tumor-germline pairs to identify clonal tumor-specific mutations, followed by custom design of ctDNA assays for these particular alleles. The major disadvantage of this strategy is the requirement for personalized ctDNA assay development, which poses significant challenges for scaling to larger patient populations. However, this approach results in a highly focused assay that will often track 15–20 tumor-specific mutations in each patient. This density of targets can be sequenced at high depth cost-effectively, and the detection efficiency will also be high to the large number of target alleles being tracked for each cancer cell. Indeed, several papers using this strategy have recently been published, and the sensitivity and correlation with risk of disease recurrence seems very high36–39. Another context in which multiple copies of target can be monitored per cell is with tumor-associated viruses, such as EBV40 or HPV41. Since there are often multiple viral genome copies per cell—upwards of 100 in some HPV-related cancers—the sensitivity of these assays can be very high, and have proven to be effective in predicting treatment response, as well as risk of disease recurrence42–44.
Absolute quantification of ctDNA
An additional distinction between NGS and dPCR based assays is the nature of ctDNA quantification. NGS provides results in the form of relative abundance, or mutant allele frequency (MAF). Many factors can impact ctDNA MAF that are independent from tumor burden. For example, treatments that change the rate of hematopoietic cell turnover (such as chemotherapy) may artificially reduce the MAF, as well as differences in post-sampling processing that may lead to spontaneous lysis of WBCs in a sample. Although blood collection tubes with stabilizing reagents can mitigate the post-collection lysis of WBCs, it does not eliminate the problem altogether. This variability in MAF measurement has important implications for disease monitoring applications, where ctDNA levels will be used as a surrogate for tumor burden. Thus, assays that enable absolute quantification of ctDNA—measuring tumor-specific target copies per mL plasma—should be preferred when using the assay to monitor disease status in a patient. There are ongoing efforts to normalize and/or convert the allelic fraction data obtained from NGS assays to an absolute measure of copies per mL; However, these currently remain in relatively early stages of development. The ability to measure absolute target copy number represents a major advantage of digital PCR based assays over NGS assays, because the measured levels will be more resistant to variable rates of WBC lysis.
Pragmatic considerations
The utility of a ctDNA assay also depends on non-technical issues that may impact clinical implementation. Perhaps first and foremost is cost—assays that will be used for disease monitoring will often need to be performed multiple times during the treatment course of each patient. Thus, ctDNA assays that have high complexity and cost will have greater barriers to reimbursement, and thereby clinical adoption. Another relevant issue is timeliness. Assays that take several weeks before results are returned to physicians are often not acceptable for a cancer patient population. Although personalized ctDNA assays have shown promise in research settings, there remain questions regarding how best to operationalize this for broader patient populations, and in real time.
Emerging Evidence of clinical utility
Retrospective studies have established that a majority of advanced stage cancer patients have detectable plasma ctDNA, with some differences in detection rates by cancer type and location45,46. The ctDNA detection rate of early stage cancers is more variable, and likely dependent on assay sensitivity. In cancer patients with detectable ctDNA, there is also ample evidence that changes in ctDNA are reflective of treatment response. When using ctDNA for disease monitoring it is important to consider the possibility of mutational heterogeneity. In metastatic breast cancer, for example, ESR1 mutations are often acquired and do not always reflect overall disease control status due to the potential expansion of mutation negative clones after treatment47. Thus, monitoring of multiple mutant alleles that are present in most if not all of the cancerous cells is important when using ctDNA as a monitoring tool.
An exciting new application of ctDNA assays is in the post-treatment setting. Several prospectively conducted studies have shown that ctDNA detection in patients who have completed potentially curative treatments is associated with a substantially increased risk of disease recurrence. In lung cancer, ctDNA assays can help interpret and predict recurrence risk in patients who have equivocal PET scans after receiving radiotherapy48. Personalized ctDNA assays have recently been implemented to predict the likelihood of post-treatment recurrence in bladder, esophageal, and colorectal cancers37,39,49,50. Longitudinal monitoring has also demonstrated exceptional sensitivity and specificity, with molecular detection of recurrence 3–6 months earlier than routine clinical followup in breast cancer and HPV-related oropharyngeal cancer patients51,52.
Negative Predictive Value
High negative predictive value (NPV) will ensure that patients with negative ctDNA do not need further evaluation. Fortunately, most studies to date have shown nearly 95–100% NPV given the high specificity of ctDNA48. Meaning, for patients with detectable ctDNA prior to treatment, lack of detectable ctDNA after definitive management and in follow up is highly predictive of no radiologic or clinically discernable cancer. For a patient without detectable ctDNA in surveillance, no further surveillance via imaging or exam may be necessary, making follow up with blood tests alone feasible.
Again, using PSA as a model, the feasibility of such an approach becomes evident. Many clinics today follow patients after radiation therapy or surgery for prostate cancer with PSA alone, deferring routine surveillance visits, exams, and imaging for when a biochemical recurrence or change in PSA is noted. A high NPV of any test is therefore essential for clinical use to allow for confidence that a patient without detectable ctDNA indeed does not have disease present. The implications of this from a perspective of cost, telemedicine, and frequency of testing are discussed further below.
In lung cancer, Chaudhuri et al reported in the case of equivocal imaging at the time of follow up, 59% of patients had no detectable ctDNA while 41% of patients had detectable ctDNA48. Remarkably, only 6% of the patients without detectable ctDNA went on to recur, while 100% of the patients with detectable ctDNA experienced recurrence. Furthermore, ctDNA was detected in 13% of patients at the time of a scan interpreted as no evidence of disease and 100% of these patients went on to recur. Similar findings have been shown in multiple malignancies, where ctDNA outperforms cytology and cystoscopy in bladder cancer or imaging in esophageal or surveillance for colorectal cancer39,50,53.
Collectively, studies indicate post-treatment monitoring with high sensitivity ctDNA assays may facilitate early detection of disease recurrence. Once these assays are established as clinical-grade assays, how can they be used to improve clinical management, particularly with regards to identification and management of OMD but also in reducing costs and increasing the convenience of follow-up?
Reduced costs of imaging surveillance
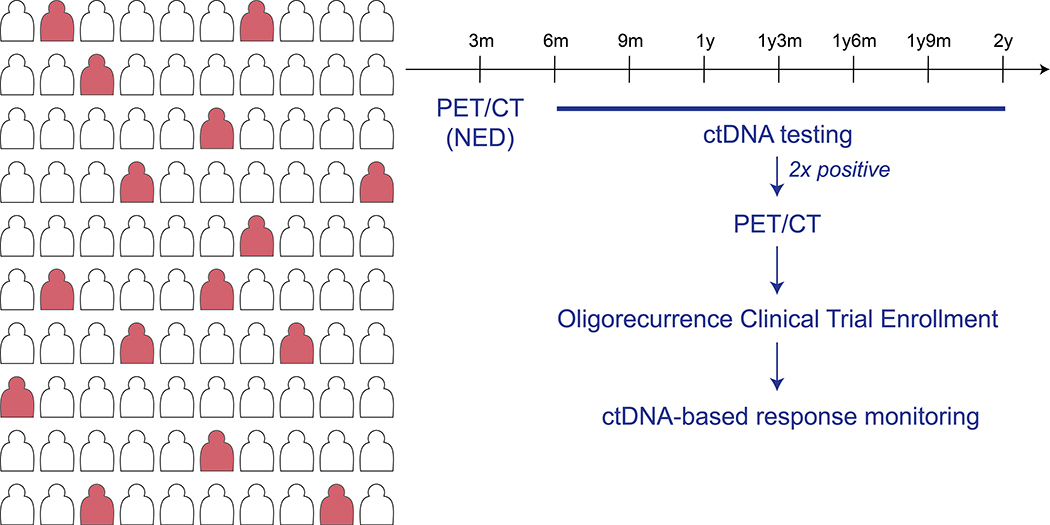
Current paradigms for imaging-based surveillance vary by disease type and recurrence risk. NCCN guidelines are often vague in the type of testing and its frequency that is most appropriate, due to a lack of supporting scientific evidence. ctDNA have the potential to dramatically improve the efficiency of radiographic surveillance. For example, in a population of patients with a 20% recurrence risk, ctDNA based screening can spare imaging costs for the majority of patients who remain ctDNA negative during follow-up (Figure 3). Overall cost savings will be based on the specific costs of the imaging and ctDNA assays, but it is possible that integration of ctDNA into post-treatment surveillance paradigms can improve early detection of recurrence without increasing overall costs.
Figure 3: Post-treatment surveillance with ctDNA assays.
For many cancer types, such as head and neck cancers, a PET/CT will be performed to establish treatment efficacy. For patients who have no evidence of disease after this re-staging scan, the majority will remain cancer-free after treatment. Considering the scenario of a ~20% disease recurrence rate, the figure illustrates how ctDNA-based surveillance enables the sparing of additional surveillance scans in the majority of patients who will not develop molecular relapse. In contrast, re-staging scans can be selectively performed in those patients with a recurrent ctDNA signal. This generalized strategy can lead to earlier detection of cancer recurrence with maximal efficiency of costly radiographic examinations. It is presumed that for most cancer types, earlier detection of cancer recurrence may increase the number of patients with oligometastatic disease.
Advanced imaging including choline PET/CT scan and PSMA have dramatically improved localization of metastases in prostate cancer. Even though these scans are highly valuable, they are costly. The sensitivity of disease detection varies greatly with PSA level, and positive radiologic findings in the setting of a low or undetectable PSA are more likely to be a false positive54. Newer radiotracers for multiple malignancies are in development. ctDNA in this context can reduce costs by allowing for biochemical detection of disease recurrence to precede and inform indication for more costly specialized imaging. A PSMA PET/CT post curative intent treatment is not indicated when PSA is undetectable, just as an E6/E7 or traditional FDG PET/CT scan in the setting of a negative HPV ctDNA assay is likely not indicated.
Facilitation of Telemedicine based Follow-up
ctDNA assays can facilitate telemedicine-based follow-up, which has been shown to have similar patient satisfaction levels with reduced the financial toxicity during survivorship care55. For many cancer types, there is limited evidence that routinely scheduled physical exams in asymptomatic patients are helpful for detecting recurrence. For patients with HPV-associated head and neck cancer, recurrences were most commonly detected upon workup of patient-reported symptoms, thereby questioning the utility of routine clinical follow-up56. In contrast, post-treatment surveillance with HPV ctDNA assays exhibits excellent sensitivity (100%) and specificity (99%) for recurrence detection43. These findings argue that transitioning from routine in-office follow-up visits to telemedicine-based follow-up with ctDNA-based surveillance has the potential to reduce costs, increase patient satisfaction, and improve early detection of cancer recurrence.
Positive Predictive Value
High PPV will reduce the risk of potential harm from false positives. However, the definition of false positive and harm require further refinement and context. ctDNA studies have now demonstrated not all patients who recur biochemically will fail clinically. Intriguingly, some patients with a detectable ctDNA in surveillance will clear this ctDNA on subsequent testing, likely due to an immune response43,48. This provides insight and incentive for patients and clinician refinement in defining the term “cure.” For example, a patient with lung cancer or HPV associated head and neck cancer with a single detectable surveillance months after definitive chemoradiation, may have undetectable ctDNA for all subsequent tests and never recur. Two patients with the same staging, same treatment and response, and both with undetectable ctDNA in early post-treatment surveillance may ultimately have discordant outcomes because of a yet to occur event or series of events, where both have molecular recurrence but one patient is able to immunologically clear the disease. However, both patients would be demonstrated to have evidence of ongoing malignancy via ctDNA. Thus, development of a single detectable ctDNA in surveillance and subsequent conversion to having no detectable ctDNA is not necessarily a false positive related to test characteristics. To address this issue and increase the PPV, any single detectable surveillance ctDNA should be repeated, at least 2–4 weeks after the initial detectable test. Studies have shown 2 serial detectable tests greatly increases the PPV in surveillance, with PPV of >90% for 2 detectable serial tests43.
Facilitation of ODT clinical trials
The precise definition of OMD varies by disease site and across clinical trials within disease sites. Furthermore, the definition of OMD often relies on imprecise methods of radiologic counting of skeletal and visceral metastases. ctDNA may allow for improved definitions of OMD by measuring the area under the curve – aiding in quantification of disease burden. At minimum ctDNA can act as a stratification for clinical trials.
The broad application of ctDNA assays in the pre and post-treatment setting will improve early detection of metastatic disease and disease recurrence, and will likely increase the number of patients identified with OMD. This will help to overcome a major barrier for ODT trials—identification of trial-eligible patients. Furthermore, ctDNA based monitoring of response may help identify patients who have durable responses to ODT.
Synchronous OMD
It is not an uncommon scenario for patients to develop metastatic disease within the first year of follow-up, where this metastatic disease was likely present but not appreciated at the time of initial diagnosis and staging. ctDNA has the potential to aid in identification of such patients with likely occult OMD in the pre-treatment setting. Generally, patients with higher concentrations of ctDNA in plasma are more likely to have distant disease present. For select patients with higher quantity of ctDNA, advanced imaging techniques will allow for identification of a significant population of trial eligible patients to investigate synchronous MDT. Advanced or whole body imaging would not be wisely applied to all patients, and ctDNA can help to identify patients with a high enough pre-test probability of distant disease to make additional imaging worthwhile.
If ctDNA indeed facilitates identification of synchronous OMD, this will allow for trials confirming the benefit of total ODT to all sites of known disease. Brook and Chang nicely summarized the rational for treatment of all sites of disease, in order to increase the likelihood of an immunogenic priming event, to release heterogeneous tumor-associated antigens potentially distinct for each disease site, and in order to promote immune cell penetration at each site overcoming local immunosuppressive contexture57. Indeed, initial clinical data demonstrate treatment of all sites of known disease reduced the time to development of new sites of metastatic disease58, suggesting active metastatic disease increases development of new metastatic disease.
Metachronous OMD
Since the early characterization of OMD as a potentially distinct disease state59, multiple reports have demonstrated a long term disease control is possible for select patients who develop OMD. Precisely defining which patients are most likely to benefit from ODT is a work in progress. However, ctDNA may be one factor which eventually aids in patient selection. While factors such as time to development of OMD may be helpful in selecting patients, ctDNA quantification and associated metrics of ctDNA doubling time could further improve patient selection.
Detectable ctDNA in surveillance has been shown to precede any radiologic or clinical evidence of disease by months. If there is indeed a window of curability for patients with more limited metastatic disease before development of diffuse metastatic disease, surveillance ctDNA makes identification of patients with OMD more likely.
There are likely many benefits to trial design and eventual treatment decision making by monitoring ctDNA after ODT. The indication for and duration of systemic therapies in OMD requires further study. For patients treated with an initial course of chemotherapy, immunotherapy, or endocrine therapy in combination with ODT, ctDNA can provide an early response assessment. Patients converting to undetectable ctDNA may be offered a holiday from systemic therapy at an earlier time-point than they otherwise would be, decreasing potential toxicity and costs.
Oligoprogressive OMD
Oligoprogressive OMD may very well become the most common clinical scenario in which patients are referred for ODT. There will be wide variation in the clinical scenarios in which patients present. For example, patients with diffuse metastatic disease may respond to targeted or immune stimulating agents with significant response at all sites except one or two, where lesions based on mutation of the cells themselves or changes in the microenvironment have escaped from the mechanism of action of these systemic therapies. ODT, especially radiation therapy, is less likely to depend on this same pathway and may be able to control the sites of oligoprogression, while the systemic agents continue to provide benefit at other sites of disease. ctDNA can help in the classification of oligoprogression and in monitoring disease response. As described above, ctDNA can also discriminate response better than imaging alone and even non-invasively identify pathways of resistance.
Another pathway to oligoprogression or OMD is in a competing paradigm to ODT, early systemic therapy for biochemical recurrence or persistence of molecular residual disease (MRD). In conjunction with trials for ODT, ctDNA detection at the MRD time point or in surveillance will allow for intervention for patients with biochemical failure alone, without discernable OMD60. ctDNA as a biomarker is a necessary component of this approach. For example, a lung cancer patient in surveillance with 2 serial detectable ctDNAs 18 months after definitive treatment may be randomized to immediate systemic therapy and delayed ODT at the time of oligoprogression versus no initial systemic therapy and ODT at the time of development of discernable OMD.
Regardless of the context and clinical scenario of ODT for oligoprogression, trial enrollment is imperative and ctDNA will be a necessary component of trial design, for patient selection and response monitoring.
Discussion and Outlook
There is an urgent and ongoing need to address the burden of mortality from metastatic cancer. The recently reported findings of clinical benefit in multiple phase 2 randomized clinical trials of ODT versus systemic therapy for patients with OMD has enthralled the cancer research community. However, confirmatory clinical trials are necessary to validate this effect, and predictive biomarkers that help to identify patients who are most likely to benefit from ODT are needed.
We propose that the remarkable advances in ctDNA technology over the past decade can directly benefit clinical research efforts in patients with OMD. A rate-limiting step for OMD clinical trials is the efficient identification of suitable patients. Integration of clinically validated ctDNA assays into post-treatment surveillance paradigms has the potential to substantially enhance our ability to identify asymptomatic OMD. We have highlighted some of the technical considerations with regards to assay sensitivity, specificity, target quantification, and cost-effectiveness that will help to determine which ctDNA assays are ideally suited for the post-treatment surveillance setting. In addition to direct benefits in earlier detection of recurrence, ctDNA-based post-treatment surveillance may help to reduce the financial toxicity of cancer survivorship, facilitate telemedicine-based follow-up, and reduce false positives associated with imaging-based surveillance. By measuring circulating tumor-specific biomarkers, ctDNA and/or ctRNA assays may facilitate discovery of prospective biomarkers that enrich for patients who are most likely to benefit from ODT, since obtaining a tissue biopsy in these patients can often be challenging.
We envision transformative changes in the management of cancer patients over the next decade. It seems plausible that ctDNA assays will allow each patient and tumor type to have its own “PSA-like” test. Such advances will facilitate our ability to select the optimal treatment type and intensity for each individual patient. After the receipt of curative-intent therapy, we envision a simplified follow-up paradigm that is facilitated by ctDNA-based surveillance and telemedicine to reduce the burden of cancer survivorship for patients, and to more efficiently use the limited resources of a complex health care system. Earlier detection of cancer recurrence will maximize the potential for salvage therapy--through ODT versus systemic therapy—based on evidence-based medicine practices. Altogether, we believe that these advances will reduce the suffering and mortality from metastatic cancer. Getting to this future from where we are now will take concerted effort by multiple research and clinical disciplines. Yet, we remain optimistic in our ability to realize this vision, particularly because the reward is simply too great to ignore.
References
- 1.Modi S, Saura C, Yamashita T, et al. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Breast Cancer. N Engl J Med 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murthy RK, Loi S, Okines A, et al. Tucatinib, Trastuzumab, and Capecitabine for HER2-Positive Metastatic Breast Cancer. N Engl J Med 2019. [DOI] [PubMed] [Google Scholar]
- 3.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodi FS, Chiarion-Sileni V, Gonzalez R, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol 2018;19:1480–92. [DOI] [PubMed] [Google Scholar]
- 6.Otake S, Goto T. Stereotactic Radiotherapy for Oligometastasis. Cancers (Basel) 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tree AC, Khoo VS, Eeles RA, et al. Stereotactic body radiotherapy for oligometastases. Lancet Oncol 2013;14:e28–37. [DOI] [PubMed] [Google Scholar]
- 8.Palma DA, Louie AV, Rodrigues GB. New Strategies in Stereotactic Radiotherapy for Oligometastases. Clin Cancer Res 2015;21:5198–204. [DOI] [PubMed] [Google Scholar]
- 9.Andrews DW, Scott CB, Sperduto PW, et al. Whole brain radiation therapy with or without stereotactic radiosurgery boost for patients with one to three brain metastases: phase III results of the RTOG 9508 randomised trial. Lancet 2004;363:1665–72. [DOI] [PubMed] [Google Scholar]
- 10.Gomez DR, Tang C, Zhang J, et al. Local Consolidative Therapy Vs. Maintenance Therapy or Observation for Patients With Oligometastatic Non-Small-Cell Lung Cancer: Long-Term Results of a Multi-Institutional, Phase II, Randomized Study. J Clin Oncol 2019;37:1558–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palma DA, Olson R, Harrow S, et al. Stereotactic ablative radiotherapy versus standard of care palliative treatment in patients with oligometastatic cancers (SABR-COMET): a randomised, phase 2, open-label trial. Lancet 2019;393:2051–8. [DOI] [PubMed] [Google Scholar]
- 12.Ruers T, Van Coevorden F, Punt CJ, et al. Local Treatment of Unresectable Colorectal Liver Metastases: Results of a Randomized Phase II Trial. J Natl Cancer Inst 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weichselbaum RR. The 46th David A. Karnofsky Memorial Award Lecture: Oligometastasis-From Conception to Treatment. J Clin Oncol 2018:JCO1800847. [DOI] [PubMed] [Google Scholar]
- 14.Foster CC, Pitroda SP, Weichselbaum RR. Staging the Metastatic Spectrum Through Integration of Clinical and Molecular Features. J Clin Oncol 2019;37:1270–6. [DOI] [PubMed] [Google Scholar]
- 15.Guckenberger M, Lievens Y, Bouma AB, et al. Characterisation and classification of oligometastatic disease: a European Society for Radiotherapy and Oncology and European Organisation for Research and Treatment of Cancer consensus recommendation. Lancet Oncol 2020;21:e18–e28. [DOI] [PubMed] [Google Scholar]
- 16.Pitroda SP, Khodarev NN, Huang L, et al. Integrated molecular subtyping defines a curable oligometastatic state in colorectal liver metastasis. Nat Commun 2018;9:1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitroda SP, Weichselbaum RR. Integrated molecular and clinical staging defines the spectrum of metastatic cancer. Nat Rev Clin Oncol 2019;16:581–8. [DOI] [PubMed] [Google Scholar]
- 18.Kalluri R The biology and function of exosomes in cancer. J Clin Invest 2016;126:1208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223–38. [DOI] [PubMed] [Google Scholar]
- 20.Chan KCA, Woo JKS, King A, et al. Analysis of Plasma Epstein-Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med 2017;377:513–22. [DOI] [PubMed] [Google Scholar]
- 21.Mohler JL, Antonarakis ES, Armstrong AJ, et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2019;17:479–505. [DOI] [PubMed] [Google Scholar]
- 22.Hallemeier CL, Zhang P, Pisansky TM, et al. Prostate-Specific Antigen After Neoadjuvant Androgen Suppression in Prostate Cancer Patients Receiving Short-Term Androgen Suppression and External Beam Radiation Therapy: Pooled Analysis of Four NRG Oncology Radiation Therapy Oncology Group Randomized Clinical Trials. Int J Radiat Oncol Biol Phys 2019;104:1057–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin RI, Chen K, Usmani A, et al. Detection of Solid Tumor Molecular Residual Disease (MRD) Using Circulating Tumor DNA (ctDNA). Mol Diagn Ther 2019;23:311–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blokzijl F, de Ligt J, Jager M, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016;538:260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunner SF, Roberts ND, Wylie LA, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature 2019;574:538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risques RA, Kennedy SR. Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet 2018;14:e1007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo H, Zhao Q, Wei W, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med 2020;12. [DOI] [PubMed] [Google Scholar]
- 30.Lam WKJ, Jiang P, Chan KCA, et al. Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. Nat Commun 2019;10:3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579–83. [DOI] [PubMed] [Google Scholar]
- 32.Sun K, Jiang P, Cheng SH, et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res 2019;29:418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016;164:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scherer F, Kurtz DM, Diehn M, Alizadeh AA. High-throughput sequencing for noninvasive disease detection in hematologic malignancies. Blood 2017;130:440–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christensen E, Birkenkamp-Demtroder K, Sethi H, et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J Clin Oncol 2019;37:1547–57. [DOI] [PubMed] [Google Scholar]
- 38.Coombes RC, Page K, Salari R, et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin Cancer Res 2019;25:4255–63. [DOI] [PubMed] [Google Scholar]
- 39.Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung SF, Zee B, Ma BB, et al. Plasma Epstein-Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol 2006;24:5414–8. [DOI] [PubMed] [Google Scholar]
- 41.Cao H, Banh A, Kwok S, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int J Radiat Oncol Biol Phys 2012;82:e351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chera BS, Kumar S, Beaty BT, et al. Rapid Clearance Profile of Plasma Circulating Tumor HPV Type 16 DNA during Chemoradiotherapy Correlates with Disease Control in HPV-Associated Oropharyngeal Cancer. Clin Cancer Res 2019;25:4682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chera BS, Kumar S, Shen C, et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-Associated Oropharyngeal Cancer. J Clin Oncol 2020:JCO1902444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Damerla RR, Lee NY, You D, et al. Detection of Early Human Papillomavirus-Associated Cancers by Liquid Biopsy. JCO Precis Oncol 2019;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Lindsay D, Chen QB, et al. Tracking plasma DNA mutation dynamics in estrogen receptor positive metastatic breast cancer with dPCR-SEQ. NPJ Breast Cancer 2018;4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Li L, Cohen JD, et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azad TD, Chaudhuri AA, Fang P, et al. Circulating Tumor DNA Analysis for Detection of Minimal Residual Disease After Chemoradiotherapy for Localized Esophageal Cancer. Gastroenterology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Murillas I, Chopra N, Comino-Mendez I, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chera BS, Kumar S, Shen C, et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-associated Oropharyngeal Cancer. J Clin Oncol 2020;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudley JC, Schroers-Martin J, Lazzareschi DV, et al. Detection and Surveillance of Bladder Cancer Using Urine Tumor DNA. Cancer Discov 2019;9:500–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans JD, Jethwa KR, Ost P, et al. Prostate cancer-specific PET radiotracers: A review on the clinical utility in recurrent disease. Pract Radiat Oncol 2018;8:28–39. [DOI] [PubMed] [Google Scholar]
- 55.Viers BR, Lightner DJ, Rivera ME, et al. Efficiency, satisfaction, and costs for remote video visits following radical prostatectomy: a randomized controlled trial. Eur Urol 2015;68:729–35. [DOI] [PubMed] [Google Scholar]
- 56.Masroor F, Corpman D, Carpenter DM, Ritterman Weintraub M, Cheung KHN, Wang KH. Association of NCCN-Recommended Posttreatment Surveillance With Outcomes in Patients With HPV-Associated Oropharyngeal Squamous Cell Carcinoma. JAMA Otolaryngol Head Neck Surg 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks ED, Chang JY. Time to abandon single-site irradiation for inducing abscopal effects. Nat Rev Clin Oncol 2019;16:123–35. [DOI] [PubMed] [Google Scholar]
- 58.Phillips R, Lim SJ, Shi WY, et al. Primary Outcomes of a Phase II Randomized Trial of Observation Versus Stereotactic Ablative RadiatIon for OLigometastatic Prostate CancEr (ORIOLE). International Journal of Radiation Oncology • Biology • Physics 2019;105:681. [Google Scholar]
- 59.Hellman S, Weichselbaum RR. Oligometastases. J Clin Oncol 1995;13:8–10. [DOI] [PubMed] [Google Scholar]
- 60.Coakley M, Garcia-Murillas I, Turner NC. Molecular Residual Disease and Adjuvant Trial Design in Solid Tumors. Clin Cancer Res 2019;25:6026–34. [DOI] [PubMed] [Google Scholar]