Abstract
Long non-coding RNA small nucleolar RNA host gene 7 (SNHG7) is involved in a variety of different types of cancer; however, the role of SNHG7 during liver cancer progression is not completely understood. The aim of the present study was to investigate the functional role and regulatory mechanism underlying SNHG7 during liver cancer. A total of 25 paired hepatocellular carcinoma (HCC) tumor tissues and adjacent normal tissues were collected. Reverse transcription-quantitative PCR and western blotting were performed to detect the expression levels of SNHG7, microRNA (miR)-34a, sirtuin 1 (SIRT1) and pyroptosis-related targets. RNA fluorescence in situ hybridization was performed to detect the expression of SNHG7 in HCC tissues. SNHG7 expression was upregulated in HCC tissues and liver cancer cells compared with normal tissues and normal liver cell lines. High expression of SNHG7 inhibited NLR family pyrin domain containing 3 (NLRP3)-dependent pyroptosis in HepG2 and SK-hep-1 cells. Bioinformatics analysis and dual-luciferase reporter assays were performed to investigate the interactions between miR-34a and SNHG7 or SIRT1. SNHG7 served as a competing endogenous RNA of miR-34a, and SIRT1 was identified as a direct target of miR-34a. Cell pyroptosis was evaluated by TUNEL and lactate dehydrogenase release assays. SNHG7 knockdown reduced SIRT1 expression, but increased the expression levels of NLRP3, caspase-1 and interleukin-1β, leading to pyroptosis. SNHG7 knockdown-induced effects were enhanced by miR-34a upregulation. In summary, the present study indicated that the SNHG7/miR-34a/SIRT1 axis contributed to NLRP3-dependent pyroptosis during liver cancer.
Keywords: small nucleolar RNA host gene 7, microRNA-34a, sirtuin 1, pyroptosis, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC), the most common type of liver cancer, is the fifth most frequently diagnosed cancer and the third leading cause of cancer-associated mortality worldwide (1). Aside from liver transplantation, hepatic resection and chemotherapy are the main effective treatment strategies used for HCC. However, despite recent advances in surgery and chemotherapy, the prognosis of HCC remains poor, with a postoperative 5-year recurrence rate of approximately 70% (2). Therefore, understanding the mechanisms underlying liver cancer and identifying novel therapeutic targets for the disease is important.
Inflammasomes are a group of intracellular multiprotein complexes that mediate host immune responses to pathogen infection and cellular damage (3). Chronic inflammation in the tumor microenvironment serves a crucial role during tumor development; however, the roles of the inflammasome during tumor progression are complex (4). The inflammasome acts as a double-edged sword in various types of cancer, serving as a tumor promoter or suppressor (5). The NLR pyrin domain containing 3 (NLRP3) inflammasome, the most highly characterized inflammasome, consists of the scaffold protein NLRP3, the adaptor apoptosis-associated speck-like protein containing a CARD and caspase-1 (6). Upon activation, NLRP3 recruits and cleaves pro-caspase-1 into its active form, which subsequently cleaves pro-interleukin (IL)-1β into mature biologically active IL-1β, ultimately resulting in inflammation (7). Activated caspase-1 also induces pyroptosis, a proinflammatory programmed cell death (6). It has been reported that the components of the NLRP3 inflammasome are significantly downregulated in HCC cells (8). Consistently, another study demonstrated that pyroptosis is inhibited in HCC tissues and cells (9), suggesting that NLRP3-dependent pyroptosis may serve crucial roles in liver cancer. Therefore, investigating the mechanism underlying NLRP3-dependent pyroptosis in liver cancer is of interest.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs that are >200 base pairs in length. lncRNAs are novel regulators of various cellular processes during tumorigenesis and tumor progression, including cell proliferation, apoptosis, pyroptosis and cell cycle progression (10,11). A genome-wide analysis of lncRNA profiles identified small nucleolar RNA host gene 7 (SNHG7) as a metastasis-associated lncRNA during HCC. It has also been reported that SNHG7 is upregulated during HCC, indicating its critical role during the development of the disease (11). Furthermore, The Cancer Genome Atlas database indicated a strong correlation between a high expression of SNHG7 and poor prognosis of patients with HCC. Patients with HCC with a higher SNHG7 expression had a poorer overall survival compared with patients with lower SNHG7 expression, indicating that SNHG7 may serve as a potential indicator of HCC clinical outcomes (12). Consistently, HCC tissues with an advanced TNM stage, high tumor grade or vascular invasion displayed higher expression levels of SNHG7 compared with corresponding controls (13). A recent study demonstrated that SNHG7 acts as an oncogenic lncRNA by competing with microRNA (miR)-34a and polypeptide N-acetylgalactosaminyltransferase 7 (GALNT7) to regulate the PI3K/Akt/mTOR signaling pathway during colorectal cancer (14). However, the biological functions and regulatory mechanisms underlying SNHG7 during liver cancer progression are not completely understood.
To the best of our knowledge, the aim of the present study was to demonstrate for the first time that SNHG7 regulated sirtuin 1 (SIRT1) expression by acting as a competing endogenous RNA (ceRNA) for miR-34a, resulting in inhibition of NLRP3-dependent pyroptosis during liver cancer.
Materials and methods
Collection of normal and HCC human tissue samples
A total of 25 paired HCC tumor and adjacent normal tissues (≥3 cm from the tumor margin) were collected post-operatively from patients with HCC (mean age, 63 years; age range, 43–72 years; 13 males and 12 females) at the People's Hospital of Deyang City between January 2016 and December 2018. HCC diagnosis was confirmed by two independent pathologists. The present study was approved by the Ethics Committee of the People's Hospital of Deyang City. Written informed consent was obtained from all patients.
Cell culture and transfection
Normal liver epithelial THLE-3 cells, human embryonic kidney 293 cells, and human hepatocellular carcinoma HepG2 and SK-hep-1 cells were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. The cell lines were authenticated by short tandem repeat DNA profiling analysis prior to purchase by The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin (all Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
The small interfering (si)RNA targeted against SNHG7, miR-34a mimic, miR-34a inhibitor and corresponding controls were obtained from Guangzhou RiboBio Co., Ltd. SNHG7 was cloned into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.), as previously described (13). Cells were seeded at a density of 2×105 cells/well in 12-well plates and were transfected using Lipofectamine® 3000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. For siRNA and miRNA transfection, 50 nM siRNA and/or 50 nM miRNA was transfected into cells. For overexpression experiments, 0.5 µg plasmid DNA was transfected into cells. pcDNA3.1 alone served as a negative control. The sequences of the siRNAs and miRNAs used in the present study are as follows: si-SNHG7-1, 5′-GUGCAGCAAUUACUCUUAAUU-3′; si-SNHG7-2, 5′-UUAGCAGAGUAAUUUGCACUU-3′; miR-34a mimic NC forward, 5′-UUCUCCGAACGUGUCAGGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′; miR-34a mimic forward, 5′-UGGCAGUGUCUUAGCUGGUUGU-3′ and reverse, 5′-AACCAGCUAAGACACUGCAAUU-3′; miR-34a inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′; and miR-34a inhibitor, 5′-ACAACCAGCUAAGACACUGCCA-3′. Cells were harvested and subjected to subsequent analysis 48 h post-transfection.
RNA isolation and reverse transcription-quantitative PCR (qRT-PCR)
Total RNA was isolated from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using the Prime-Script RT Reagent kit (Takara Biotechnology Co., Ltd.). Subsequently, qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) and the ABI PRISM 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR reaction was performed using the following conditions: Pre-denaturation at 95°C for 1 min, 40 cycles of denaturation at 95°C for 30 sec, annealing at 67°C for 30 sec and extension at 72°C for 30 sec, followed by a final extension step at 72°C for 5 min. The following primers were used for qPCR: SNHG7 forward, 5′-TTGCTGGCGTCTCGGTTAAT-3′ and reverse, 5′-GGAAGTCCATCACAGGCGAA-3′; miR-34a forward, 5′-CACGGACTCGGGGCATTTGGAGATTTT-3′ and reverse, 5′-CTGTCTAGATCGCTTATCTTCCCCTTGG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; NLRP3 forward, 5′-CACCTGTTGTGCAATCTGAAG-3′ and reverse, 5′-GCAAGATCCTGACAACATGC-3′; caspase-1 forward, 5′-CCTTAATATGCAAGACTCTCAAGGA-3′ and reverse, 5′-TAAGCTGGGTTGTCCTGCACT-3′; IL-1β forward, 5′-TACCTGTCCTGCGTGTTGAA-3′ and reverse, 5′-TCTTTGGGTAATTTTTGGGATCT-3′; SIRT1 forward, 5′-TGCTGGCCTAATAGAGTGGCA-3′ and reverse, 5′-CTCAGCGCCATGGAAAATGT-3′; and GAPDH forward, 5′-AACGTGTCAGTGGTGGACCTG-3′ and reverse, 5′-AGTGGGTGTCGCTGTTGAAGT-3′. The specificity of the fluorescent signal was verified by melting curve and 1% agarose gel electrophoresis. miRNA and mRNA expression levels were quantified using the 2−ΔΔCq method (15) and normalized to the internal reference genes U6 and GAPDH, respectively.
RNA fluorescence in situ hybridization (FISH)
RNA FISH was performed using the Fluorescent in Situ Hybridization kit (Guangzhou RiboBio Co., Ltd.), according to the manufacturer's protocol. HCC and paired adjacent normal tissues were fixed with 4% paraformaldehyde at room temperature for 8 h, embedded in paraffin and cut into 7-µm-thick sections. The Cy3-conjugated SNHG7 probe was synthesized by Guangzhou RiboBio Co., Ltd. as previously described (16). For Cy3 detection, fluorescence images (magnification, ×400) were acquired using a fluorescence confocal laser-scanning microscope with excitation at 543 nm (Olympus Corporation). Slides were mounted with Prolong Gold Antifade reagent with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.).
Western blotting
Total protein from HepG2 and SK-hep-1 cells was extracted using RIPA lysis buffer (Pierce; Thermo Fisher Scientific) and quantified using the BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of protein (40 µg/lane) were separated via 4–12% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore), which were blocked with 5% non-fat milk at room temperature for 1 h. Subsequently, the membranes were incubated at 4°C overnight with primary antibodies targeted against: SIRT1 (1:500; cat. no. sc-74504; Santa Cruz Biotechnology, Inc.), NLRP3 (1:1,000; cat. no. ab214185; Abcam), caspase-1 (1:1,000; cat. no. 4199; Cell Signaling Technology, Inc.), IL-1β (1:1,000; cat. no. sc-52012; Santa Cruz Biotechnology, Inc.) and GAPDH (1:1,000; cat. no. 5174; Cell Signaling Technology, Inc.). Following primary incubation, the membranes were incubated with HRP-conjugated goat anti-rabbit (cat. no. 31460) or goat anti-mouse (cat. no. 31430) (both 1:5,000; Invitrogen; Thermo Fisher Scientific, Inc.) secondary antibodies at room temperature for 1 h. Protein bands were visualized using SuperSignal West Pico PLUS Chemiluminescent substrate (Pierce) and exposed to X-ray film. Protein expression was quantified using ImageJ v1.52a software (National Institutes of Health) with GAPDH as the loading control.
Dual-luciferase reporter assay
The potential binding sites between SNHG7 and miR-34a, and the putative association between miR-34a and SIRT1 were predicted using bioinformatics analysis, namely TargetScan (www.targetscan.org) and miRanda (www.microrna.org/microrna/searchMirnas.do). The putative binding site between miR-34a and the 3′-untranslated region (UTR) of SNHG7 or SIRT1 was cloned into the pmirGLO vector (Promega Corporation). The mutant (MUT) 3′UTR of SNHG7 or SIRT1 was synthesized using the QuickChange Lighting Multi Site-Directed Mutagenesis kit (Agilent Technologies, Inc.) according to the manufacturer's protocol. 293 cells (2×105 cells/well) were co-transfected with 0.5 µg SNHG7-wild-type (WT), SIRT1-3′UTR-WT, SNHG7-MUT or SIRT1-3′UTR-MUT vectors and 50 nM miR-34a mimic NC or miR-34a mimic using Lipofectamine® 3000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following incubation at 37°C for 48 h, luciferase activities were measured using the Dual Luciferase Reporter assay system (Promega Corporation), according to the manufacturer's protocol. Firefly luciferase activities were normalized to Renilla luciferase activities.
Analysis of DNA fragmentation
A TUNEL assay was performed to detect DNA fragmentation of individual cells. Briefly, HepG2 cells (2×105 cells/well) were seeded onto coverslips in 12-well dishes. Following transfection, cells were fixed with 4% paraformaldehyde at room temperature for 10 min and permeabilized using 0.1% Triton X-100 at room temperature for 10 min. Subsequently, cells were incubated with TUNEL reaction mixture (Roche Diagnostics), according to the manufacturer's protocol. For one test, 45 µl TUNEL Label and 5 µl TUNEL Enzyme were prepared prior to use. Slides were incubated with TUNEL reaction mixture at 37°C for 1 h. After rinsing with PBS, slides were mounted with Prolong Gold Antifade reagent with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.). Stained cells were observed using a fluorescence confocal laser-scanning microscope (Olympus Corporation). The cells were observed at 5–10 fields of view (magnification, ×400), and the TUNEL-positive cells were counted per 100 cells in each field of view.
Cytotoxicity assay
Cytotoxicity assay was performed using HepG2 cells. Cell culture medium was collected and centrifuged at 12,000 × g at 4°C for 15 min to remove the cell debris. Pyroptotic cell death was determined by quantifying lactate dehydrogenase (LDH) content of the cell culture medium using the LDH Cytotoxicity Detection kit (Clontech Laboratories, Inc.), according to the manufacturer's protocol.
Statistical analysis
All experiments were performed at least three times. Data are expressed as the mean ± standard deviation. Differences among groups were analyzed using one-way ANOVA followed by Dunnett's post hoc test. Differences between two groups were analyzed using the paired Student's t-test. Statistical analyses were performed using GraphPad Prism software (version 6.0; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
SNHG7 is upregulated in HCC tissues and cell lines, and mediates NLRP3-dependent pyroptosis during HCC
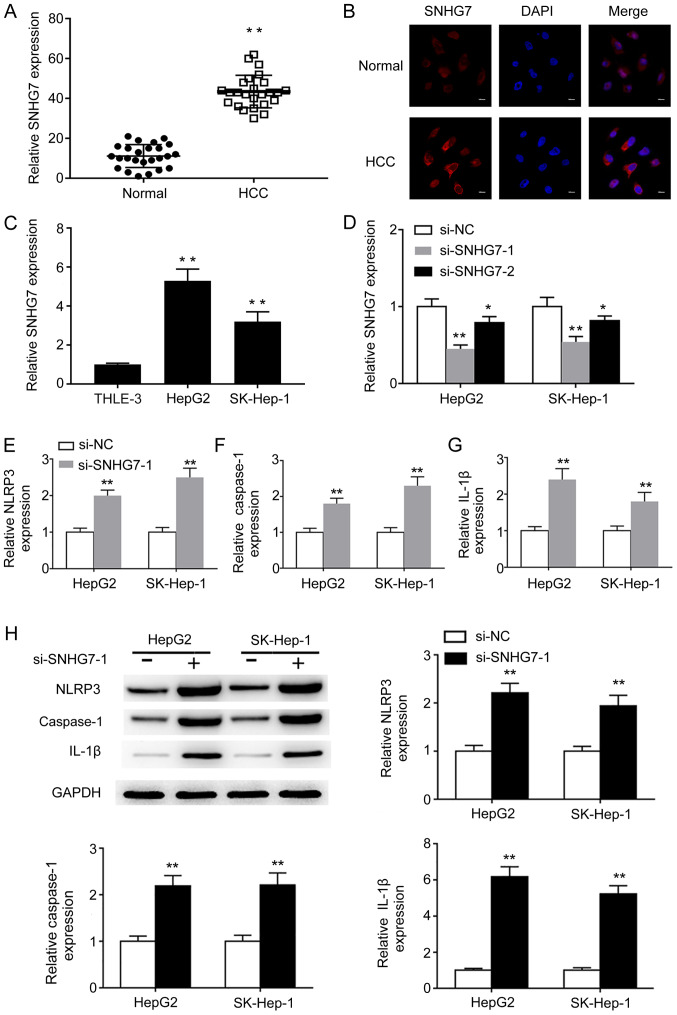
To investigate the biological function of SNHG7 during HCC, the expression of SNHG7 in HCC and adjacent normal tissues was assessed. SNHG7 expression levels were significantly higher in HCC tissues compared with adjacent normal tissues (Fig. 1A). RNA FISH confirmed that SNHG7 expression was increased in HCC tissues compared with adjacent normal tissues (Fig. 1B). Similarly, SNHG7 expression levels were significantly upregulated in liver cancer cell lines HepG2 and SK-hep-1 compared with the normal liver epithelial THLE-3 cell line (Fig. 1C), indicating that SNHG7 may serve important roles during liver cancer tumorigenesis. Furthermore, a previous study demonstrated that pyroptosis is inhibited in HCC tissues and cells (9); therefore, to investigate whether SNHG7 was involved in liver cancer cell pyroptosis, loss-of-function experiments were performed. si-SNHG7-1 and si-SNHG7-2 significantly decreased SNHG7 expression levels in HepG2 and SK-hep-1 cells compared with si-NC. However, si-SNHG7-1 silenced SNHG7 expression more effectively compared with si-SNHG-7-2, so si-SNHG7-1 was used for subsequent experiments (Fig. 1D). SNHG7 knockdown significantly increased the mRNA expression levels of pyroptosis-related NLRP3, caspase-1 and IL-1β in HepG2 and SK-hep-1 cells compared with si-NC (Fig. 1E-G). Consistently, the western blotting results indicated that the protein expression levels of NLRP3, caspase-1 and IL-1β were significantly increased in SNHG7-knockdown cells compared with si-NC-transfected cells (Fig. 1H). The results suggested that SNHG7 may serve a crucial role during NLRP3-dependent pyroptosis in liver cancer cells.
Figure 1.
SNHG7 is upregulated in HCC and mediates NLRP3-dependent pyroptosis. (A) SNHG7 mRNA expression levels in paired HCC and adjacent normal tissues were determined by RT-qPCR. (B) The expression of SNHG7 in paired HCC and adjacent normal tissues was determined by RNA FISH (scale bar, 10 µm). (C) SNHG7 mRNA expression levels in THLE-3, HepG2 and SK-Hep-1 cells were determined by RT-qPCR. mRNA expression levels of (D) SNHG7, (E) NLRP3, (F) caspase-1 and (G) IL-1β were determined by RT-qPCR. (H) Protein expression levels of NLRP3, caspase-1 and IL-1β were determined by western blotting and semi-quantified. *P<0.05 and **P<0.01 vs. the corresponding control. SNHG7, small nucleolar RNA host gene 7; HCC, hepatocellular carcinoma; NLRP3, NLR pyrin domain containing 3; RT-qPCR, reverse transcription-quantitative PCR; FISH, fluorescence in situ hybridization; IL-1β, interleukin-1β; si, small interfering RNA; NC, negative control.
SNHG7 sponges miR-34a to repress its expression in liver cancer cells
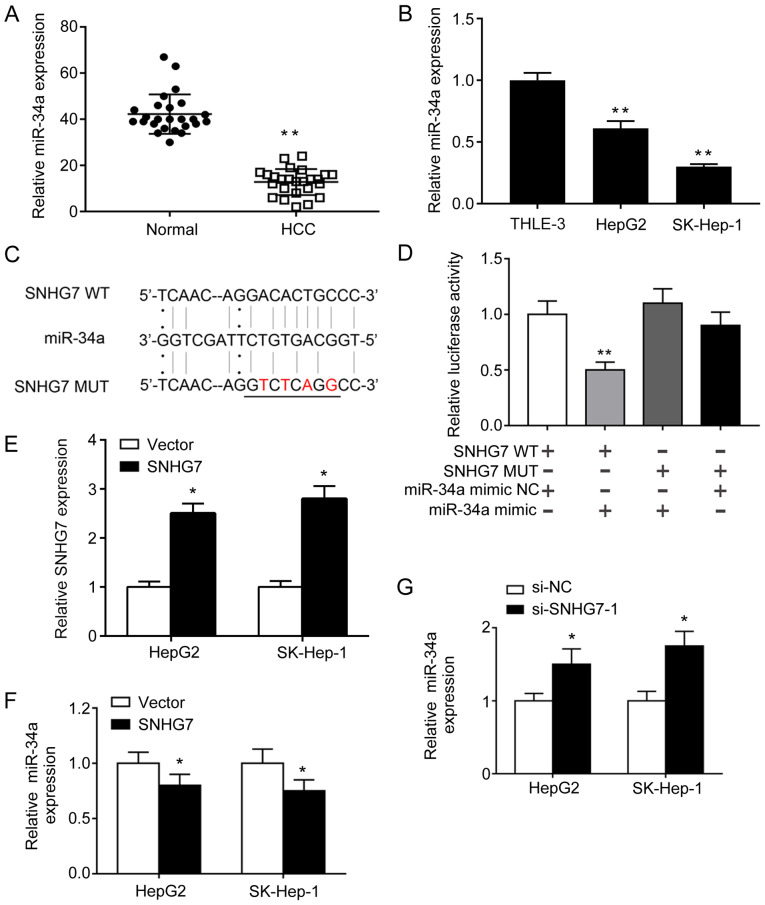
To further understand the mechanism underlying SNHG7 during NLRP3-dependent pyroptosis, bioinformatics analysis was performed to predict the putative interactions between SNHG7 and miRNAs. Among the predicted miRNAs, the expression levels of miR-34a were significantly lower in HCC tissues compared with adjacent normal tissues (Fig. 2A). Moreover, miR-34a expression levels were significantly decreased in HepG2 and SK-hep-1 cells compared with THLE-3 cells (Fig. 2B), suggesting that miR-34a was negatively regulated by SNHG7 during liver cancer; therefore, SNHG7 may function as a molecular sponge of miR-34a. A dual-luciferase reporter assay was conducted to verify the direct binding between SNHG7 and miR-34a. The potential binding sites were predicted using TargetScan and miRanda (Fig. 2C). miR-34a mimic significantly reduced the luciferase activities of the SNHG7-WT group compared with the miR-34a mimic NC. By contrast, miR-34a mimic displayed no effect on the luciferase activities of the SNHG7-MUT group compared with miR-34a mimic NC (Fig. 2D). Gain- and loss-of-function experiments were conducted. SNHG7 overexpression significantly increased the expression levels of SNHG7 in HepG2 and SK-hep-1 cells compared with the vector control (Fig. 2E). SNHG7 overexpression and knockdown experiments suggested that miR-34a was negatively regulated by SNHG7 in HepG2 and SK-hep-1 cells (Fig. 2F and G). Collectively, the results indicated that SNHG7 acted as a sponge of miR-34a to repress its expression in HepG2 and SK-hep-1 cells.
Figure 2.
SNHG7 sponges miR-34a to repress its expression in HCC cells. (A) mRNA expression levels of miR-34a in (A) paired HCC and adjacent normal tissues, as well as in (B) THLE-3, HepG2 and SK-Hep-1 cells were determined by RT-qPCR. (C) The potential binding sites between SNHG7 and miR-34a. A mutation was generated in the miR-34a binding site of the SNHG7 sequence. The mutations were indicated in red. (D) Luciferase activities were determined by a dual-luciferase reporter assay. (E) SNHG7 mRNA expression levels following SNHG7 overexpression were determined by RT-qPCR. miR-34a expression levels following SNHG7 (F) overexpression and (G) knockdown were determined by RT-qPCR. *P<0.05 and **P<0.01 vs. the corresponding control. SNHG7, small nucleolar RNA host gene 7; miR, microRNA; HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative PCR; WT, wild-type; MUT, mutant; si, small interfering; NC, negative control.
SIRT1 is a direct target of miR-34a
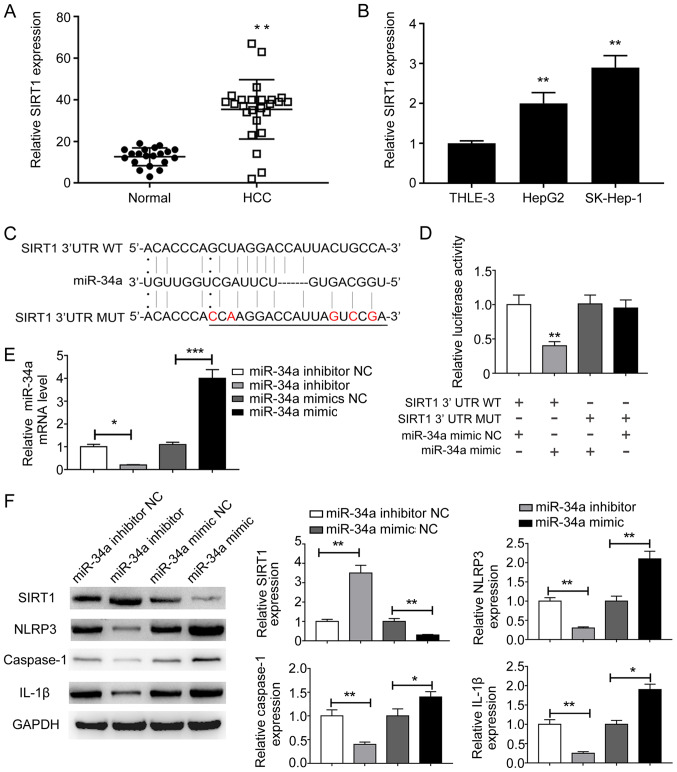
Increasing evidence has suggested that lncRNAs function as miRNA sponges and inhibit miRNA function, thus regulating miRNA target gene expression (17,18). Therefore, the present study aimed to identify the direct target of miR-34a by bioinformatics analysis. Among the putative theoretical targets, the present study focused on SIRT1 due to its reported role in NLRP3-dependent pyroptosis (19). The mRNA expression levels of SIRT1 were examined in HCC tissues and cells. SIRT1 was significantly upregulated in HCC tissues compared with adjacent normal tissues (Fig. 3A). Similarly, the mRNA expression levels of SIRT1 were significantly higher in HepG2 and SK-hep-1 cells compared with THLE-3 cells (Fig. 3B). The complementary binding site between miR-34a and the 3′UTR of SIRT1 was predicted (Fig. 3C). miR-34a mimic significantly reduced the luciferase activities of the SIRT1-3′UTR-WT group compared with miR-34a NC, indicating that SIRT1 was a direct target of miR-34a (Fig. 3D). To further investigate the effect of miR-34a on SIRT1 expression, HepG2 and SK-hep-1 cells were transfected with miR-34a inhibitor, miR-34a mimic and corresponding miRNA controls. miR-34a inhibitor and miR-34a mimic significantly decreased and increased miR-34a expression levels in HepG2 cells, respectively, compared with the corresponding controls (Fig. 3E). The protein expression levels of SIRT1 were significantly increased by miR-34a inhibitor, but significantly decreased by miR-34a mimic compared with the corresponding controls in liver cancer cells (Fig. 3F). In HepG2 cells, the expression levels of pyroptosis-related NLRP3, caspase-1 and IL-1β were significantly decreased by miR-34a inhibitor and significantly increased by miR-34a mimic compared with the corresponding controls (Fig. 3F). Similar results were observed in SK-hep-1 cells (data not shown). Collectively, the results indicated that SIRT1 was a direct target of miR-34a; therefore, miR-34a/SIRT1 may be associated with NLRP3-dependent pyroptosis in liver cancer cells.
Figure 3.
SIRT1 is a direct target of miR-34a. (A) mRNA expression levels of SIRT1 in (A) paired HCC and adjacent normal tissues, as well as in (B) THLE-3, HepG2 and SK-Hep-1 cells were determined by RT-qPCR. (C) The potential binding sites between miR-34a and the 3′UTR of SIRT1. A mutation was generated in the miR-34a binding site of the 3′UTR sequence of SIRT1. The mutations were indicated in red. (D) Luciferase activities were determined by a dual-luciferase reporter assay. (E) miR-34a expression levels following miR-34a overexpression or knockdown were determined by RT-qPCR. (F) Protein expression levels of SIRT1, NLRP3, caspase-1 and IL-1β following miR-34a overexpression or knockdown were determined by western blotting. *P<0.05 and **P<0.01 vs. the corresponding control. SIRT1, sirtuin 1; miR, microRNA; HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative PCR; 3′UTR, 3′-untranslated region; NLRP3, NLR pyrin domain containing 3; IL-1β, interleukin-1β; WT, wild-type; MUT, mutant; NC, negative control.
NLRP3-dependent pyroptosis is regulated by the SNHG7/miR-34a/SIRT1 axis during liver cancer
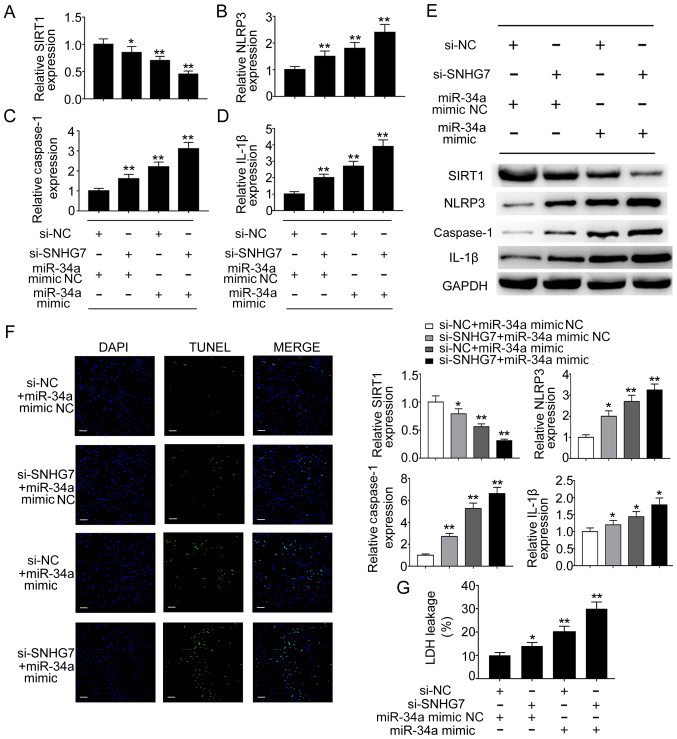
To further determine the effect of the SNHG7/miR-34a/SIRT1 axis on pyroptosis in liver cancer cells, si-SNHG7, miR-34a mimic and corresponding controls were co-transfected into liver cancer cells. Liver cancer cells transfected with si-SNHG7 or miR-34a mimic displayed significantly reduced mRNA expression levels of SIRT1 compared with the negative control group; co-transfection of si-SNHG7 and miR-34a mimic also significantly reduced SIRT1 expression levels in HepG2 cells compared with the negative control group (Fig. 4A). By contrast, the mRNA expression levels of NLRP3, caspase-1 and IL-1β were significantly increased in cells transfected with si-SNHG7 or miR-34a mimic compared with the negative control group; additionally, the co-transfection group displayed higher expression levels compared with the si-SNHG7 or miR-34a mimics group (Fig. 4B-D). The western blotting results indicated that protein expression levels displayed a similar pattern (Fig. 4E). In addition, the present study aimed to determine the effect of the SNHG7/miR-34a/SIRT1 axis on cell death. A TUNEL assay was performed to detect DNA fragmentation in liver cancer cells. Liver cancer cells transfected with si-SNHG7 or miR-34a mimic displayed an increased proportion of TUNEL-positive cells compared with the negative control group, and co-transfection of si-SNHG7 and miR-34a mimic further increased the proportion of TUNEL-positive HepG2 cells (Fig. 4F). Furthermore, pyroptosis is generally assessed by quantifying cytosolic LDH release (20). In HepG2 cells, LDH release was significantly increased in the cell culture media of cells transfected with si-SNHG7 or miR-34a mimic compared with the negative control group (Fig. 4G). LDH release was even higher in the si-SNHG7+miR-34a mimics group compared with that in the si-SNHG7 or miR-34a mimics groups (Fig. 4G). Similar results were observed in SK-hep-1 cells (data not shown). Collectively, the results indicated that the SNHG7/miR-34a/SIRT1 ceRNA network regulated NLRP3-dependent pyroptosis in liver cancer cells.
Figure 4.
NLRP3-dependent pyroptosis is regulated by the SNHG7/miR-34a/SIRT1 axis in HCC. mRNA expression levels of (A) SIRT1, (B) NLRP3, (C) caspase-1 and (D) IL-1β were determined by RT-qPCR. (E) Protein expression levels of SIRT1, NLRP3, caspase-1 and IL-1β were determined by western blotting. (F) DNA fragmentation was assessed using the TUNEL assay. Blue indicates DAPI nuclear staining and green indicates TUNEL staining (scale bar, 50 µm). (G) Pyroptotic cell death was determined by quantifying LDH release in the cell culture media. *P<0.05 and **P<0.01 vs. the corresponding control. NLRP3, NLR pyrin domain containing 3; SNHG7, small nucleolar RNA host gene 7; miR, microRNA; SIRT1, sirtuin 1; HCC, hepatocellular carcinoma; IL-1β, interleukin-1β; RT-qPCR, reverse transcription-quantitative PCR; LDH, lactate dehydrogenase; si, small interfering RNA; NC, negative control.
Discussion
The present study demonstrated that SNHG7 and SIRT1 were upregulated, and miR-34a was downregulated in liver cancer tissues and cell lines compared with adjacent normal tissues and cell lines. The results indicated that SNHG7 sponged miR-34a to suppress its expression, and SIRT1 was identified as a direct target of miR-34a in liver cancer cells. Gain- and loss-of-function experiments demonstrated that SNHG7 repressed miR-34a by acting as a ceRNA, thus enhancing the expression of SIRT1 and inhibiting NLRP3-dependent pyroptosis. Collectively, the results indicated that the SNHG7/miR-34a/SIRT1 axis may serve as a potential therapeutic target for liver cancer.
With advancements to high-throughput sequencing techniques, a large number of dysregulated lncRNAs have been identified in HCC. Functional studies have revealed that dysregulated lncRNAs serve critical roles during the tumorigenesis and progression of HCC (17,18). lncRNA SNHG7, which is located on chromosome 9q34.3, has been identified as a novel oncogenic gene and has recently received increasing attention in the field of cancer research (21). Increasing evidence has indicated that SNHG7 regulates cell proliferation, migration, invasion and apoptosis in a variety of different types of cancer, including non-small cell lung, gastric, prostate and colorectal cancer, as well as in glioblastoma (14,22–25). For example, SNHG7 promotes lung cancer cell proliferation, migration and invasion, and inhibits apoptosis by inducing Fas apoptotic inhibitory molecule 2 expression (22). Furthermore, by comparing lncRNA profiles between early recurrence HCC tissues with metastasis and late recurrence HCC tissues without metastasis, SNHG7 was identified as a metastasis-related lncRNA (12). Moreover, Cancer Cell Line Encyclopedia RNA sequencing results demonstrated that SNHG7 is highly expressed in HCC cell lines, and SNHG7 knockdown inhibits HCC cell invasion (12). However, the role of SNHG7 in liver cancer is still not completely understood. The present study indicated that SNHG7 was significantly upregulated in liver cancer tissues and cell lines compared with adjacent normal tissues and cell lines. Consistently, recent studies have also demonstrated that SNHG7 is significantly upregulated in HCC tissues and cells (13,26). Another recent study reported that lncRNA growth arrest specific 5 serves as an ovarian tumor suppressor by inducing inflammasome formation and pyroptosis (27). By contrast, the present study indicated that SNHG7 knockdown upregulated the expression of pyroptosis-related NLRP3, caspase-1 and IL-1β compared with the negative control group, which indicated that SNHG7 may display an oncogenic role by inhibiting pyroptosis in liver cancer.
Novel lncRNA/miRNA/mRNA networks based on ceRNA hypotheses have been identified in a variety of different types of cancer. For example, SNHG7 promotes glioblastoma cell proliferation, migration and invasion by inhibiting miR-5095 (25). SNHG7 also acts as a ceRNA against miR-503 to promote prostate cancer tumorigenesis (24). Recent studies have reported that SNHG7 accelerates the progression and metastasis of HCC by regulating miR-122-5p/ribosomal protein L4 or RNA binding motif protein 5 (13,26). However, the ceRNA network that regulates pyroptosis in liver cancer is not completely understood. The present study indicated that miR-34a was negatively regulated by SNHG7 in liver cancer. Previous studies have suggested that miR-34a expression is lost or downregulated in numerous types of cancer, including HCC (28,29). miR-34a inhibits HCC glycolysis by targeting lactate dehydrogenase A, leading to reduced cell proliferation and invasion (28). However, aside from glucose metabolism, the role of miR-34a during HCC has not been previously reported. The present study further indicated that SNHG7 directly bound to and negatively regulated miR-34a in HepG2 and SK-Hep-1 cells, which was consistent with a recent study that reported that SNHG7 acts as a ceRNA to regulate GALNT7 and the PI3K/Akt/mTOR signaling pathway by sponging miR-34a in colorectal cancer (14).
SIRT1, a well-studied member of the sirtuin family, serves crucial roles in various cellular events, including aging, metabolism, inflammation and tumorigenesis (30,31). SIRT1 serves as a tumor promoter or suppressor dependent on the tumor type, as well as the temporal or spatial distribution of upstream and downstream factors (32). SIRT1 is frequently upregulated in HCC tissues and cell lines; however, SIRT1 knockdown and inhibition of SIRT1 activity by the small molecule inhibitor cambinol impairs cell proliferation, increases cell differentiation and inhibits tumor growth in vivo (33). The results of the present study support the aforementioned finding, demonstrating that SIRT1 was highly expressed in HCC tissues and cell lines. Mechanistic studies suggested that miR-34a inhibited SIRT1 expression by directly binding to the 3′UTR of SIRT1 in liver cancer cells. miR-34a-mediated inhibition of SIRT1 increased the expression of NLRP3, caspase-1 and IL-1β. To the best of our knowledge, the present study was the first to indicate that miR-34a-mediated suppression of SIRT1 regulated pyroptosis in liver cancer cells. TUNEL and LDH release assays following co-transfection of si-SNHG7 and miR-34a mimic further demonstrated that SNHG7 knockdown reduced SIRT1 expression to promote inflammasome formation and pyroptosis via sponging miR-34a. The results of the present study were consistent with a previous study, which reported that lack of SIRT1 expression enhances NLRP3 inflammasome activation and caspase-1 cleavage in vascular endothelial cells (19).
The present study further demonstrated the antipyroptotic properties of SNHG7 in liver cancer. The results suggested that SNHG7 may act as an endogenous sponge of miR-34a, thereby regulating the expression of SIRT1. Moreover, the results indicated that SIRT1 induction may inhibit NLRP3 inflammasome-induced caspase-1 cleavage and IL-1β release to inhibit pyroptosis in liver cancer cells.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- lncRNA
long non-coding RNA
- SNHG7
small nucleolar RNA host gene 7
- HCC
hepatocellular carcinoma
- SIRT1
sirtuin 1
- NLRP3
NLR pyrin domain containing 3
- ceRNA
competing endogenous RNA
- LDH
lactate dehydrogenase
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZC and QZ designed the study. MH performed the western blotting experiments. JC collected the tissue samples. CL performed the RT-qPCR and cell culture experiments. All authors participated in analyzing the data and drafting the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethic Committee of People's Hospital of Deyang City. Written informed consent was obtained from all patients prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gerbes A, Zoulim F, Tilg H, Dufour JF, Bruix J, Paradis V, Salem R, Peck-Radosavljevic M, Galle PR, Greten TF, et al. Gut roundtable meeting paper: Selected recent advances in hepatocellular carcinoma. Gut. 2018;67:380–388. doi: 10.1136/gutjnl-2017-315068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhir M, Melin AA, Douaiher J, Lin C, Zhen WK, Hussain SM, Geschwind JF, Doyle MB, Abou-Alfa GK, Are C. A review and update of treatment options and controversies in the management of hepatocellular carcinoma. Ann Surg. 2016;263:1112–1125. doi: 10.1097/SLA.0000000000001556. [DOI] [PubMed] [Google Scholar]
- 3.Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greten FR, Grivennikov SI. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantono M, Guo B. Inflammasomes and cancer: The dynamic role of the inflammasome in tumor development. Front Immunol. 2017;8:1132. doi: 10.3389/fimmu.2017.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 8.Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X, Zhao W, Huai W, Guo P, Han L. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab Invest. 2014;94:52–62. doi: 10.1038/labinvest.2013.126. [DOI] [PubMed] [Google Scholar]
- 9.Chu Q, Jiang Y, Zhang W, Xu C, Du W, Tuguzbaeva G, Qin Y, Li A, Zhang L, Sun G, et al. Pyroptosis is involved in the pathogenesis of human hepatocellular carcinoma. Oncotarget. 2016;7:84658–84665. doi: 10.18632/oncotarget.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 12.Cui H, Zhang Y, Zhang Q, Chen W, Zhao H, Liang J. A comprehensive genome-wide analysis of long noncoding RNA expression profile in hepatocellular carcinoma. Cancer Med. 2017;6:2932–2941. doi: 10.1002/cam4.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Sun L, Wang L, Yao B, Mo H, Yang W. lncRNA SNHG7 accelerates the proliferation, migration and invasion of hepatocellular carcinoma cells via regulating miR-122-5p and RPL4. Biomed Pharmacother. 2019;118:109386. doi: 10.1016/j.biopha.2019.109386. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B, Jia L. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. 2018;11:89. doi: 10.1186/s13045-018-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Yu F, Dong P, Mao Y, Zhao B, Huang Z, Zheng J. Loss of lncRNA-SNHG7 promotes the suppression of hepatic stellate cell activation via miR-378a-3p and DVL2. Mol Ther Nucleic Acids. 2019;17:235–244. doi: 10.1016/j.omtn.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu S, Xu J, Xu X, Hu S, Li B, Li W. The expression of astrocyte elevated gene-1 in human non-small-cell lung cancer and its relationship with postoperative chemotherapy and radiotherapy. Histopathology. 2015;67:817–826. doi: 10.1111/his.12720. [DOI] [PubMed] [Google Scholar]
- 18.Huo X, Han S, Wu G, Latchoumanin O, Zhou G, Hebbard L, George J, Qiao L. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: Implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16:165. doi: 10.1186/s12943-017-0734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Wang P, Yang X, Wang W, Zhang J, He Y, Zhang W, Jing T, Wang B, Lin R. SIRT1 inhibits inflammatory response partly through regulation of NLRP3 inflammasome in vascular endothelial cells. Mol Immunol. 2016;77:148–156. doi: 10.1016/j.molimm.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Rayamajhi M, Zhang Y, Miao EA. Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol Biol. 2013;1040:85–90. doi: 10.1007/978-1-62703-523-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. lncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9:722. doi: 10.1038/s41419-018-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36:2673–2680. doi: 10.3892/or.2016.5105. [DOI] [PubMed] [Google Scholar]
- 23.Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S, Xia TS. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21:4613–4622. [PubMed] [Google Scholar]
- 24.Qi H, Wen B, Wu Q, Cheng W, Lou J, Wei J, Huang J, Yao X, Weng G. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:326–332. doi: 10.1016/j.biopha.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Yang Y, Xue J, Xi Z, Hu L, Pan SJ, Sun Q. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496:712–718. doi: 10.1016/j.bbrc.2018.01.109. [DOI] [PubMed] [Google Scholar]
- 26.Sun BZ, Ji DG, Feng ZX, Wang Y. Long noncoding RNA SNHG7 represses the expression of RBM5 to strengthen metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:5699–5704. doi: 10.26355/eurrev_201907_18307. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Yang C, Li Y, Chen A, Li L, You Z. lncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep. 2018;38(pii):BSR20171150. doi: 10.1042/BSR20171150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang HF, Wang YC, Han YD. MicroRNA-34a inhibits liver cancer cell growth by reprogramming glucose metabolism. Mol Med Rep. 2018;17:4483–4489. doi: 10.3892/mmr.2018.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slabáková E, Culig Z, Remšík J, Souček K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017;8:e3100. doi: 10.1038/cddis.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Kazgan N. Mammalian sirtuins and energy metabolism. Int J Biol Sci. 2011;7:575–587. doi: 10.7150/ijbs.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng T, Wang J, Jiang H, Liu L. Hippo signaling in oval cells and hepatocarcinogenesis. Cancer Lett. 2011;302:91–99. doi: 10.1016/j.canlet.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Fang Y, Nicholl MB. Sirtuin 1 in malignant transformation: Friend or foe? Cancer Lett. 2011;306:10–14. doi: 10.1016/j.canlet.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Portmann S, Fahrner R, Lechleiter A, Keogh A, Overney S, Laemmle A, Mikami K, Montani M, Tschan MP, Candinas D, Stroka D. Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Mol Cancer Ther. 2013;12:499–508. doi: 10.1158/1535-7163.MCT-12-0700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.