Abstract
The Genus Alexandrium is a widespread dinoflagellate marine phytoplankton that is the primary causative organism causing Paralytic Shellfish Poisoning (PSP) intoxications in European waters. EU food safety directives specify that EU Member States must implement a routine monitoring programme to mitigate risks associated with bio-accumulation of biotoxins by bivalve shellfish, such as those produced by Alexandrium. This strategic drive comprises of both direct testing of bivalve flesh for the presence of regulated toxins and an early warning phytoplankton monitoring programme. In the UK the flesh testing moved away from animal bio-assays to analytical chemistry techniques, whereas phytoplankton monitoring methods have seen little technological advancement since implementation. Methods currently utilize light microscopy and manual enumeration of different algal species. These methods although proven are time consuming, reliant on highly trained staff, have high limits of detection (LOD) with low specificity, unable to reliably identify Alexandrium to species level. The implications of these limitations of the techniques mean that in the case of Alexandrium the LOD is also the action limit and as such it is easy to miss positive samples affecting the efficacy of any early warning strategy. This study outlines the development, preliminary method characterisation, validation and trial implementation of an alternative early warning technique, utilizing quantitative PCR to identify water samples containing Alexandrium cells. The approach outlined in this document, showed an improved correlation with flesh toxicity, improved sensitivity, improved throughput compared to traditional light microscopy methods and there was also good correlation with higher cell abundance samples when compared to the light microscopy results. The application of this approach to routine water samples was explored and was found to demonstrate potential as a corroborative method for use during flesh intoxication episodes. This study offers potential for future improvements in the accuracy and sensitivity of phytoplankton monitoring whilst ensuring continuity of public safety, providing cost savings and offering new research opportunities.
Keywords: Harmful algal bloom, Biotoxin, Alexandrium, qPCR assay, Monitoring, TaqMan
Highlights
-
•
An inhouse qPCR assay was developed using the 18s rDNA to detect Alexandrium spp.
-
•
qPCR had reduced LOD & improved specificity when compared to light microscopy.
-
•
qPCR had a higher correlation to toxicity data when compared to light microscopy
-
•
DNA extracts were found to be stable when fixed with Lugol's for >800 days.
1. Introduction
The bioaccumulation of algal borne biotoxins in filter feeding shellfish has serious public health implications including the potential for human fatalities in the case of saxitoxins also known as Paralytic Shellfish Toxins (PST's) (Rodrigue et al., 1990, Vilariño et al., 2018, Visciano et al., 2016). European food regulations dictate that there should be both a flesh testing and an early warning programme undertaken by Member States for the safe harvesting and export of shellfish (EC Regulation No. 1664/2006). The phytoplankton monitoring program is used as an early warning tool and is typically undertaken using taxonomic identification by light microscopy (LM) in Utermohl chamber (Utermöhl, 1931). It is not possible using LM to reliably identify beyond the Genus level for many groups of phytoplankton including Alexandrium without further investigation, using alternative techniques such as electron microscopy, Fluorescent In Situ Hybridization (FISH) or molecular methods such as Polymerase Chain reaction (PCR) (Antonella and Luca, 2013, John et al., 2003) are required. These alternative techniques have notable methodological or application drawbacks such as being overly specific or high relative costs when applied to high throughput applications; which have until now prevented their widespread use for routine phytoplankton monitoring purposes (Bott et al., 2010, Simon et al., 2000). Quantitative (q)PCR has been widely utilised for highly specific identification of many bacterial, viral and algal species with human-health implications to the shellfish industry (Antonella and Luca, 2013, Gao et al., 2015, Vandersea et al., 2017). However, there is a level of complexity to algal populations which has not yet been fully clarified, as studies have identified that species-specific assays that work in one location will not necessarily work on what is thought to be the same taxonomic species elsewhere (Vandersea et al., 2017). Furthermore, the use of a species specific assay will not detect new toxic species migrating into the locality by mechanisms such as inadvertent transfer of shipping ballast water and climate change (Bolch and de Salas, 2007, Hallegraeff, 2010, Hallegraeff and Bolch, 1991, Henry, 2002). A suitable alternative strategy has been facilitated by the discovery of the genes associated with the production of Saxitoxin (sxt), which allows for molecular detection of genetic potential to produce these toxins (Stüken et al., 2011). Development and testing of such assays has shown promise; however, (unpublished) application highlighted limits of detection are above the current sensitivity limits used in the UK, specifically 20 cells/L (Murray et al., 2011), limiting their practical use for monitoring purposes. England and Wales have approximately 50 designated shellfish harvesting locations included in the routine monitoring programme with around 1000 samples per year requiring phytoplankton enumeration, running in parallel with flesh testing. Seasonal blooms of primarily A. minutum pose a risk to public safety and regularly cause the closure of shellfish harvesting sites. These blooms present an opportunity to analyse water samples by molecular techniques in parallel with these routine monitoring programmes to assess their suitability and applicability. Any such tool would have to be robust, able to detect a toxicity threat and fulfil at least the same degree of specificity currently offered by light microscopy (identification to genus level) while at the same time being cheaper, faster and allow for future developments such as the ability to perform follow up analysis for species identification by additional molecular tools such as sequencing. This study was an attempt to design, characterise and trial a method that can facilitate the progression toward a molecular technique for potential future fulfilment of EU food legislation (EC regulation No. 854/2004). It was decided, due to the need to be diverse in the target organism coverage, that a genus specific assay would be the most suitable approach to achieve this, by doing this the assay would be fit for purpose and would fulfil the level of specificity achieved by LM.
2. Materials & method
2.1. Algal cell culturing conditions
Alexandrium cultures were purchased from Marine Biological Agency (Plymouth UK) and grown in 250 mL flank (75 cm2 growth area) containing L1 medium (Guillard and Hargraves, 1993) at 17 °C and exposed to 14 h of light and 10 h of darkness per day until cell density was observed by the naked eye. Forsaken.
2.2. Laboratory reference material
For purposes of quality control and performance characterisation, a laboratory reference material (LRM) was generated. This was achieved by culturing A. fundyense, fixing with 1% Lugol's iodine fixative and dispensed into 200 × 1 mL aliquots while ensuring the culture was mixed throughout the procedure to ensure homogeneity. LRM was kept at 4 °C and long-term stability was determined up to 955 days after fixation. Analysis of DNA extracts was performed by a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, Ca, USA).
2.3. Sample preparation and DNA extraction
Seawater or culture samples were centrifuged at 4500 g for 5 min, the supernatant was carefully removed by vacuum and the resulting pellet was resuspended with the relevant DNA extraction buffer before DNA extraction. Prior to analysis samples had been fixed at time of collection in Lugol's and stored at 4 °C until DNA extraction could be undertaken. The performance of three DNA extraction kits were compared to determine the most appropriate approach to isolate nucleic acids from a range of samples. The test kits included a widely used and well-published proprietary method (e.g. Galluzzi et al., 2010, Galluzzi et al., 2004), Qiagen DNeasy Plant mini kit (Qiagen, Hilden, Germany) and two alternative kits produced at the time of testing by MoBio: Power Biofilm and Power Soil. For the DNeasy Plant kit, cell lysis was achieved using MP Bio Lysing Matrix A tubes on a Fast prep system set at maximum speed for 30 s (MPbio, Solon, OH, USA). The two alternative kits used lysis tubes as per manufacturer's protocol. Extraction efficiency was assessed for each kit by performing repeat extractions of the LRM at varying degrees of dilution to ensure that recovered DNA correlated with cell abundance. Manufacturer's instructions were followed for DNA extraction except for the DNeasy kit for which the use of liquid nitrogen was required but not available. Liquid nitrogen was replaced by flash freezing in a −80 °C freezer.
2.4. qPCR method development
After identifying a suitable multi-target assay (Catherine et al., 2009), which amplifies the 18S rDNA sequence of Alexandrium spp. It was decided that a redesign of the PCR assay was required. The new TaqMan qPCR assay includes degeneracies to help improve coverage of target species more recently included in public databases. The assay targets a 125bp region of the 18S rDNA gene and was developed by de novo alignment of 25 Alexandrium species. The primers and MGB (minor groove binding) probe for the assay are shown in Table 1. Primers and probes were synthesized by Sigma life sciences (Sigma-Aldrich St Louis, Mi, USA) and prepared in TE buffer. All qPCR analyses were performed on an MX3005P instrument (Agilent Technologies, Santa Clara, Ca, USA), with the following process: 37 °C for 10 min, 95 °C for 10 min and 50 repeat cycles of 95 °C for 15 s and 63 °C for 1 min with fluorescence measured during each 63 °C step. PCR reactions used either universal master mix or environmental master mix as specified in the results section below (Thermo Fisher, Waltham Ma, USA) with Rox as a passive reference dye.
Table 1.
TaqMan primer & probe sequences.
| Name | Sequence (5′-3′) |
|---|---|
| Alex-FWD | TGTTGCGGTTAAAAAGCTCGTAG |
| Alex-REV | TGCACTTGACTGTGTGGTGTM |
| Alex MGB Probe (FAM) | TGAGTATYTGGCACAGCC |
2.5. PCR standards
Preparation of an internal standard was undertaken by performing a conventional PCR using primers Alex-FWD and Alex-REV on an A. fundyense DNA extract. The resulting PCR product was cleaned up with CargeSwitch®-Pro PCR clean-up Kit (Invitrogen, Carlsbad. Ca, USA) according to manufacturer's instructions. The target product was subsequently observed to be the correct size by gel electrophoresis and the concentration ascertained by Q-bit fluorometer (Thermo Fisher, Waltham Ma, USA) and diluted to create a 1 × 105 copies per μL. The standard was aliquoted and stored at −20 °C for serial dilutions and amplification each time an assay was undertaken.
2.6. Determination of copy number
Using a light microscope and extruded Pasteur pipette, ten A. minutum cells were taken from a culture isolated from site one of the environmental study, this was repeated five times. The cells were then centrifuged, and the culture media washed away to remove any residual DNA that could have caused over-estimation. The five aliquots had DNA extracted and were qPCR analysed to obtain an average copy number per cell.
2.7. Exclusivity/inclusivity test
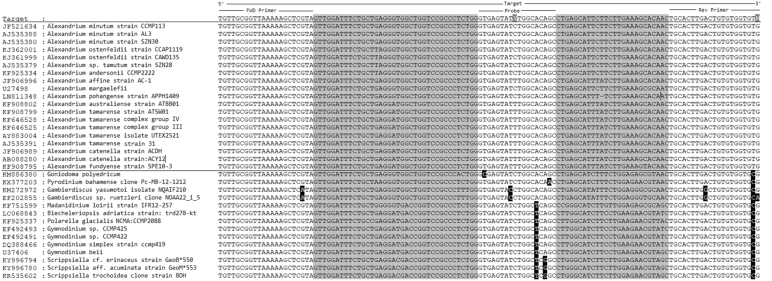
Three dinoflagellate species (Lingulodinium polyedra CCAP 1121/1, Gonyaulax spinifera CCAP118/1 and a Scrippsiella sp) which are genetically similar to Alexandrium were cultured and fixed with Lugol's. DNA was extracted, and qPCR undertaken. A BLAST search was undertaken using the target sequence to identify potential non-Alexandrium species that could also be amplified, all hits were aligned and can be seen in Table 2.
Table 2.
Blast search using target DNA sequence to indicate species that would be detectable by assay, discrepancies between primer/probe and published sequences are highlighted in black.
2.8. Comparative study
A comparison of the Taqman genus assay, cell counts, and shellfish flesh toxin levels was undertaken on samples collected from two sites in south west England in 2016 and 2017. Water samples were taken alongside routine monitoring shellfish samples and shipped directly to the laboratory for 4 °C storage until analysis. DNA was extracted from either 50 or 100 mL of water depending on sample size. The cell counts and flesh testing results were derived from the routine monitoring programme for England and Wales as previously documented (AOAC, 2005, Hatfield et al., 2016, International, 2011, Turner et al., 2009, Turner and Hatfield, 2012, Utermöhl, 1931). Briefly, semi-quantitative flesh results were generated by pre-column oxidation, Liquid Chromatography with Fluorescence detection (LC-FLD) and cell counts generated using light microscopy. Shellfish samples were collected at the same time and proximate location as water samples.
3. Results
3.1. Assessment of DNA extraction
The two MoBio DNA extraction kits (Power Biofilm and Power Soil) both had adequate extraction efficiencies and linear correlation between cell abundance and DNA yield. The Power Biofilm kit had the highest efficiency of extraction results and was therefore used throughout the rest of the study. The Qiagen DNeasy kit required adaptation by use of additional physical disruption via Lysing matrix A tubes for improved DNA extraction efficiency. However, even with the inclusion of this step, serial dilution of cell densities prior to DNA extraction returned poorer relative efficiencies for low concentrations in comparison to high, with linear plots giving slope value of m = ≥5.5.
3.2. Methodological optimisation and characterisation
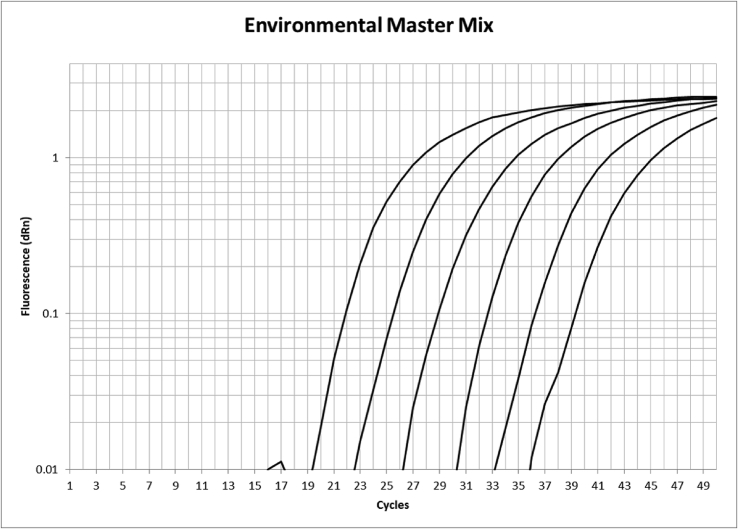
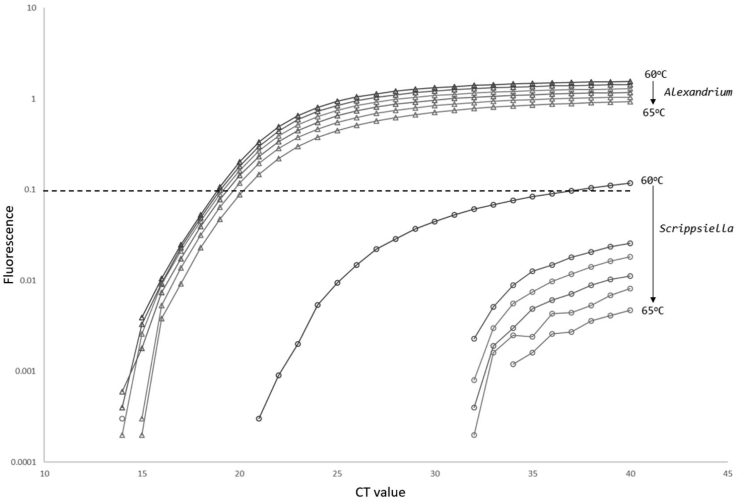
DNA extracts from all kits had an inhibitory effect on the PCR reaction and it was identified that appropriate dilution (5:1) was required to reduce this effect. However, dilution of DNA extract did not fully resolve inhibition issues, so employment of proprietary environmental master mix was tested and observed to return significantly better amplification efficiencies (Fig. 1). A low level of non-specific amplification was observed in DNA extracted from a high-density culture (approximately 106 cells forwarded to DNA extraction) of Scrippsiella. The issue was overcome by increasing the annealing temperature of the qPCR method from 59 °C to 63 °C, thus increasing the specificity of the assay to only amplify the Alexandrium cultures tested. Fig. 2 shows comparable amplification performance over a range from 60 to 65 °C of Alexandrium cells (approximately 2 × 104) and Scrippsiella cells (approximately 1 × 106). Data from the BLAST Search (see Table 2) showed that some non-Alexandrium spp shared the same genomic sequence as the forward primer site, including Scrippsiella. There were however no exact matches for the probe or reverse primer identified in any species that are not of the Genus Alexandrium. Preliminary tests indicated a high number of copies of the target gene in each Alexandrium cell, resulting in a sensitive assay. The presence of multiple copies of rDNA in dinoflagellates is well documented and is in part attributable to the very large genome present in many species (Prokopowich et al., 2003). The attempt to determine copy number from a toxic A. minutum culture isolated from site one found that within a single culture there was a high degree of variability with individual cells having estimated copy numbers per cell ranging from 1650 to 9995. An average of ±5000 target copies per cell was used as an arbitrary figure later used to determine the half LOD value used for standardisation of Fig. 3, Fig. 4.
Fig. 1.
Example serial dilution qPCR plot of standard material used to generate calibration for quantitation of 18S gene content.
Fig. 2.
qPCR amplification plot for Alexandrium and Scrippsiella at temperatures ranging between 60 and 65 °C with increments of 1 °C, threshold level is indicated by dashed line.
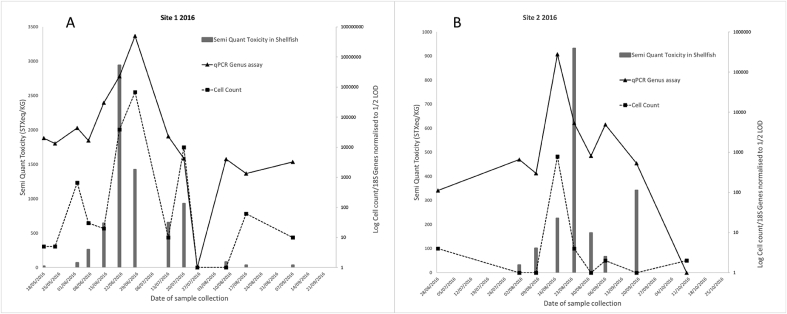
Fig. 3.
The findings of the comparative for sites A and B in 2016, with each figure showing toxicity data plotted on a linear primary scale and the cell count and qPCR result being plotted against a secondary, log scale. The secondary scale data has been normalised to half the respective LOD for that method, these being 20 cell/Lt for LM and 2500 copies/Lt, with this being an arbitrary LOD calculated from the copy number study. Semi-quantification data is used for flesh toxicity results as only samples above RL are forwarded to full quantification.
Fig. 4.
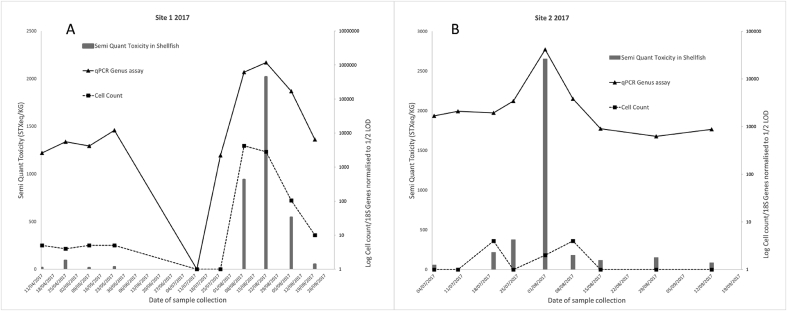
The findings of the comparative for sites A and B in 2017, with each figure showing toxicity data plotted on a linear primary scale and the cell count and qPCR result being plotted against a secondary, log scale. The secondary scale data has been normalised to half the respective LOD for that method, these being 20 cell/Lt for LM and 2500 copies/Lt, with this being an arbitrary LOD calculated from the copy number study. Semi-quantification data is used for flesh toxicity results as only samples above RL are forwarded to full quantification.
3.3. Comparative study of toxicity, cell count and genus assay
Data generated during the comparative study is shown in Fig. 3, Fig. 4. The figures provide chronological plots of cell counts and qPCR data compared to the semi-quantitative toxicity results obtained from the flesh testing programme. Results from four toxic events over two years and at two sites are shown. The semi-quantitative results tend to overestimate toxicity by a factor of 2 but are well correlated with full quantitative results and allow the tracking of low level toxicity in the shellfish on the occasions they do not undergo full quantitation (Turner et al., 2014).
As shown in Fig. 3a, data from site 1 in 2016 depicts a closure event in which both the LM and qPCR methods were able to detect the presence of Alexandrium cells prior to their being a result above the semi quantitative flesh testing toxicity reporting limit (RL) which is 400 μg STXeq/kg (half action limit). As assessment of the correlation between the flesh toxicity and the two water testing techniques was shown to be acceptable with Pearson correlation coefficients (r2 values) of 0.88 and 0.92 for qPCR and LM respectively (this is with the outlier 28/6/2016 removed). The correlation between qPCR and LM was described by an r2 of 0.91 if the outlier on the 28/6/2016 is again removed and 1.0 if it was kept. Site 2 in 2016 (Fig. 3b) represents a non-closure event, with only one flesh result breaching RL but did not exceeding the action limit when quantified. Both water testing methods identified a peak in the presence of Alexandrium cells prior to the toxin accumulation. This delay in accumulation meant that poor correlations were observed between both LM and qPCR with toxicity. If the high result is removed the correlation between toxicity and qPCR improved from an r2 value of 0.001–0.4 but correlation between qPCR and LM drops from an r2 of 1.00 to 0.18.
Site 1 results from 2017 (Fig. 4a) show a two-stage bloom, with the first incident being below RL but being detected by all methods followed by a period in which no toxicity or cells are detected and then a higher intensity second toxic event occurs which resulted in a closure. Correlation between the water sampling methods and toxicity was good, with r2 values of 0.98 and 0.62 for qPCR and LM respectively and correlation between qPCR and LM also having an acceptable r2 value of 0.70.
At site 2 in 2017 (Fig. 4b) there was a more significant toxic event when compared to 2016, during which the site was closed. There was notably very low response from the LM method and as such correlation between this data and both other metrics was poor (r2=<0.02). There was however, very good correlation between qPCR and toxicity, r2 =>0.99.
4. Discussion
This multi-disciplinary approach utilised analytical testing of shellfish flesh and light microscopy to enumerate phytoplankton cells to assess the performance of a qPCR assay applied to real world environmental biotoxin bloom events and resulting toxin bioaccumulation in shellfish. The study attempts to bridge the gap between the promise of qPCR assays and their practical applicability to routine monitoring programmes (Antonella and Luca, 2013, Bott et al., 2010, Medlin and Orozco, 2017, Zamor et al., 2012). The initial aim of this work was to develop, characterise and apply a molecular tool for the detection of all known Alexandrium spp in seawater samples. This has direct relevance for any potential future methods within the official monitoring programme. The results obtained in this study outline the potential suitability of this technique, either alongside or as an alternative tool to traditional phytoplankton testing. Overall the environmental application of the assay was promising. Firstly, qPCR data compared well with the LM phytoplankton data and there was a better correlation between qPCR results and toxicity observed in the shellfish than between LM and toxicity (Fig. 3, Fig. 4). The results generated in this study indicated that the genus-specific assay for Alexandrium spp is more sensitive, has better specificity and had a better correlation to shellfish toxicity than LM and therefore could be a suitable alternative for use in a phytoplankton monitoring programme. The costs associated LM and qPCR assay are hard to compare fairly as the methods are fundamentally different. It is fair to identify that qPCR consumable costs are significantly higher than those required of LM, however, the training required and time to undertake LM analysis are a significant factor, for these reasons no direct comparison of costs associated with the two methods are reported.
Although there have been no accounts of Gymnodinium or Pyrodinium spp causing PSP event in UK, waters it would be important to acknowledge the potential for these species to become a problem as global ocean temperatures rise (Hallegraeff, 2010, Higman et al., 2014, Townhill et al., 2018). As such if a qPCR approach was adopted as an early warning technique for routine monitoring it would be necessary to include all relevant species into any adopted strategy and would ideally be multiplexed into a single assay.
The design of the qPCR assay was based upon a published method (Catherine et al., 2009) which utilised multiple primer sets to achieve inclusivity for multiple species of Alexandrium (3 forward and 3 reverse). After testing the published assays performance and performing in silco investigation it was discovered that inclusivity and specificity could be improved by re-designing the primers and probe. The updated assay described here has single forward and reverse primers and an increased probe length. These changes proved to perform adequately, reduce costs associated with purchasing and preparing multiple primers for each master-mix preparation, and reduce complexity of the reaction.
A number of challenges were addressed during development of the method. DNA extraction was hindered by issues associated with non-linear extraction efficiencies in the Qiagen DNeasy plant kit, which was potentially due to the inability to use liquid nitrogen as a bench reagent due to health and safety limitations. Attempts to use physical lysis to overcome the issue did increase extraction efficiency but did not overcome the lower efficiency of DNA extraction in low density samples. However, alternative extraction kits subsequently tested were deemed suitable for use, providing adequate relative extraction efficiencies between differing cell densities. The Power biofilm kit (MoBio) had lower average CT values which indicates better extraction efficiency for equivalent samples and as such was selected for usage throughout the study. This information will be useful to other researchers interested in applying molecular methods such as qPCR to marine phytoplankton samples in water who do not have access to liquid nitrogen for DNA extraction.
Specificity of the assay was reassessed when low level amplification of a high cell density culture of Scrippsiella was observed. However, increasing annealing temperature was found to limit this non-specific amplification of Scrippsiella (Fig. 2). A 3 °C increase to 63 °C was adopted, as this did not impact the Alexandrium amplification. The adoption of increased annealing temperature, in conjunction with an amended cycle threshold and iterative analysis of all amplifications during the PCR assay successfully eliminated false-positive results.
Poor PCR efficiencies were observed for most samples when using universal master mix, causing potential inaccuracy in CT number and sensitivity issues for low level positive samples. This was suspected to be due to the presence of PCR-inhibitors as a dilution of template DNA often reduced the effect. The use of proprietary Environmental Master mix overcame these issues and no further PCR efficiency issues were observed throughout the study. This finding may be of general significance for researchers using qPCR on environmental marine phytoplankton samples.
Due to the large variances observed in cell densities in bloom conditions, logarithmic scales to display data were adopted and as such both LM and qPCR data was displayed accordingly. This however posed a problem in allowing comparison with such differing datasets, so data was normalised to the LOD with negative samples plotted as half LOD.
For qPCR data an arbitrary LOD was chosen using the average copy number from the toxic culture isolated from Site 1. Ongoing use of the assay will require a more robust calculation of the assay's LOD and would require year-round analysis to ensure that false positives results are not generated.
Stability of Lugol's fixed samples was tested using the LRM and in contrast to a recent publication (Eckford-Soper and Daugbjerg, 2015) no decrease in amplification was observed throughout the study period, one day and 855 day samples had ct values of 17.18 and 17.05 respectively, indicating long-term stability. Furthermore, analysis of DNA extracts indicated DNA molecular weight was high and therefore suitable for long read sequencing techniques such as Nano-pore technology utilised by Minion (Feng et al., 2015). Many Alexandrium spp are non-toxin producing and can co-occur in the same waters as toxic strains which complicates both traditional LM and molecular analysis (Cho et al., 2008, Toebe et al., 2013). However, by sequencing the PCR target region of toxic and non-toxic strains, it should be possible to develop a secondary multiplex probe that could for identification of toxic and non-toxic Alexandrium spp in a single qPCR assay (Nagai, 2011). Alternatively, coupling this approach to toxin-specific (e.g. STX genes) (Murray et al., 2011, Stüken et al., 2011) or species-specific molecular targets (Metfies et al., 2006, Otten and Genetics, 2017, Toebe et al., 2013) could offer the sensitivity, robustness and specificity required for reliable monitoring of toxin events in near real-time. This therefore represents a valid alternative to other molecular techniques such as whole cell ELISA, with the benefit of having improved sensitivity and the benefit of being able to multiplex additional species or genera (Carrera et al., 2010, Gas et al., 2009). Although the data presented here is promising, the application of these approaches using a larger collection of bloom events is required to determine the robustness of these methods on real environmental samples before the approach could be implemented in any official phytoplankton monitoring programmes.
5. Conclusion
This study aimed to investigate if a molecular technique could fulfil the requirements of the UK routine monitoring programme of seawater for Alexandrium spp. The qPCR assay performed well in comparison to LM in the environmental study with improvements in specificity, sensitivity, linearity with shellfish toxicity and a reduction in analysis time observed. There are however noted limitations to the assay, no resolution of taxonomy beyond Genus level and variability of copy number being the most significant issues that must be overcome before the assay could be implemented into an official monitoring programme. Additionally, more experiments using a larger collection of bloom events are needed to confirm and determine the robustness of this method.
Application of the assay could potentially be used strategically as a screening procedure coupled with additional analysis of positive samples. In addition, with high-throughput Sequencing costs steadily reducing, and DNA barcode databases becoming more comprehensive, DNA extract analysis could include additional tools added as they become available (Muir et al., 2016), facilitating accurate speciation and quantification of bloom events beyond that which is currently possible.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Cefas Seedcorn for primary funding this body of work under project code DP374, AlertoxNet for additional funding and Food Standards Agency for allowing us to use the samples, without which this work would not have been possible. Furthermore, thanks are also given to Dr Lisa Cross for technical assistance.
Footnotes
Transparency document related to this article can be found online at https://doi.org/10.1016/j.toxcx.2019.100011
Transparency document
References
- Anon Regulation (EC) No 854/2004 of the European Parliament and of the Council of 29 April 2004 laying down specific rules for the organisation of official controls on products of animal origin intended for human consumption. Off. J. Eur. Union L. 2004;139:206–320. [Google Scholar]
- Antonella P., Luca G. The quantitative real-time PCR applications in the monitoring of marine harmful algal bloom (HAB) species. Environ. Sci. Pollut. Res. 2013;20:6851–6862. doi: 10.1007/s11356-012-1377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AOAC AOAC official method 2005.06 paralytic shellfish poisoning toxins in shellfish prechromatographic oxidation and liquid chromatography with fluorescence detection. J. AOAC Int. 2005 [PubMed] [Google Scholar]
- Bolch C.J.S., de Salas M.F. A review of the molecular evidence for ballast water introduction of the toxic dinoflagellates Gymnodinium catenatum and the Alexandrium “tamarensis complex” to Australasia. Harmful Algae. 2007;6:465–485. doi: 10.1016/j.hal.2006.12.008. [DOI] [Google Scholar]
- Bott N.J., Ophel-Keller K.M., Sierp M.T., Herdina, Rowling K.P., Mckay A.C., Loo M.G.K., Tanner J.E., Deveney M.R. Toward routine, DNA-based detection methods for marine pests. Biotechnol. Adv. 2010;28:706–714. doi: 10.1016/j.biotechadv.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Carrera M., Garet E., Barreiro A., Garcés E., Pérez D., Guisande C., González-Fernández Á. Generation of monoclonal antibodies for the specific immunodetection of the toxic dinoflagellate Alexandrium minutum Halim from Spanish waters. Harmful Algae. 2010;9:272–280. doi: 10.1016/j.hal.2009.11.004. [DOI] [Google Scholar]
- Catherine C., Jennifer G., Lyndsay B., Eileen B. 2009. Evaluation of Quantitative Real-Time TaqMan PCRs for Detection and Quantification of Alexandrium Species in Scottish Waters; pp. 1–13. [Google Scholar]
- Cho Y., Hiramatsu K., Ogawa M., Omura T., Ishimaru T., Oshima Y. Non-toxic and toxic subclones obtained from a toxic clonal culture of Alexandrium tamarense (Dinophyceae): toxicity and molecular biological feature. Harmful Algae. 2008;7:740–751. doi: 10.1016/j.hal.2008.02.008. [DOI] [Google Scholar]
- EC Commission Regulation (EC) No 1664/2006 of 6 November 2006 amending Regulation (EC) No 2074/2005 as regards implementing measures for certain products of animal origin intended for human consumption and repealing certain implementing measures. Off. J. Eur. Union L. 2006;320:13–45. [Google Scholar]
- Eckford-Soper L.K., Daugbjerg N. Examination of six commonly used laboratory fixatives in HAB monitoring programs for their use in quantitative PCR based on Taqman probe technology. Harmful Algae. 2015;42:52–59. doi: 10.1016/j.hal.2014.12.007. [DOI] [Google Scholar]
- Feng Y., Zhang Y., Ying C., Wang D., Du C. Nanopore-based fourth-generation DNA sequencing technology. Genom. Proteom. Bioinform. 2015;13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Bertozzini E., Penna A., Perini F., Garcés E., Magnani M. Analysis of rRNA gene content in the Mediterranean dinoflagellate Alexandrium catenella and Alexandrium taylori: implications for the quantitative real-time PCR-based monitoring methods. J. Appl. Phycol. 2010;22:1–9. doi: 10.1007/s10811-009-9411-3. [DOI] [Google Scholar]
- Galluzzi L., Penna A., Bertozzini E., Garcés E., Magnani M., Vila M., Garce E. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate) Appl. Environ. Microbiol. 2004;70:1199–1206. doi: 10.1128/AEM.70.2.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yu R.C., Murray S.A., Chen J.H., Kang Z.J., Zhang Q.C., Kong F.Z., Zhou M.J. High specificity of a quantitative PCR assay targeting a saxitoxin gene for monitoring toxic algae associated with paralytic shellfish toxins in the Yellow Sea. Appl. Environ. Microbiol. 2015;81:6973–6981. doi: 10.1128/AEM.00417-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gas F., Pinto L., Baus B., Gaufres L., Crassous M. Monoclonal antibody against the surface of Alexandrium minutum used in a whole-cell. ELISA. 2009;8:538–545. [Google Scholar]
- Guillard R.R.L., Hargraves P.E. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–236. doi: 10.2216/i0031-8884-32-3-234.1. [DOI] [Google Scholar]
- Hallegraeff G.M. Ocean climate change, phytoplankton community responses, and harmful algal blooms: a formidable predictive challenge. J. Phycol. 2010;46:220–235. doi: 10.1111/j.1529-8817.2010.00815.x. [DOI] [Google Scholar]
- Hallegraeff G.M., Bolch C.J. Transport of toxic dinoflagellate cysts via ships' ballast water. Mar. Pollut. Bull. 1991;22:27–30. doi: 10.1016/0025-326X(91)90441-T. [DOI] [Google Scholar]
- Hatfield R.G., Punn R., Algoet M., Turner A.D. A rapid method for the analysis of paralytic shellfish toxins utilizing standard pressure HPLC: refinement of AOAC 2005.06. J. AOAC Int. 2016;99:475–480. doi: 10.5740/jaoacint.15-0080. [DOI] [PubMed] [Google Scholar]
- Henry C. 2002. Impacts of Climate Change on Human Health 2013; pp. 257–262. [Google Scholar]
- Higman W.A., Turner A., Veszelovszki M.A., Davidson K. 2014. Research to Support the Development of a Monitoring Programme for New or. [Google Scholar]
- International A. AOAC Int. 2011. 2011. AOAC official method 2011.02 paralytic shellfish toxins in mussels, clams, oysters, and scallops. Post-column oxidation (PCOX) method first action 2011. [PubMed] [Google Scholar]
- John U., Cembella A., Hummert C., Elbrächter M., Groben R., Medlin L. Discrimination of the toxigenic dinoflagellates Alexandrium tamarense and A. ostenfeldii in co-occurring natural populations from Scottish coastal waters. Eur. J. Phycol. 2003;38:25–40. doi: 10.1080/0967026031000096227. [DOI] [Google Scholar]
- Medlin L.K., Orozco J. Molecular techniques for the detection of organisms in aquatic environments, with emphasis on harmful algal bloom species. Sensors (Switzerland) 2017;17 doi: 10.3390/s17051184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metfies K., Töbe K., Scholin C., Medlin L.K. 24 laboratory and field applications of ribosomal RNA probes to aid the detection and monitoring 24 . 2 ribosomal RNA sequences as markers for phylogenetic. Harmful Algae. 2006;189:1–2. [Google Scholar]
- Muir P., Li S., Lou S., Wang D., Spakowicz D.J., Salichos L., Zhang J., Weinstock G.M., Isaacs F., Rozowsky J., Gerstein M. The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol. 2016;17:1–9. doi: 10.1186/s13059-016-0917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S.A., Wiese M., Stüken A., Brett S., Kellmann R., Hallegraeff G., Neilan B.A. SxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Appl. Environ. Microbiol. 2011;77:7050–7057. doi: 10.1128/AEM.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S. Development of a multiplex PCR assay for simultaneous detection of six Alexandrium species (Dinophyceae) J. Phycol. 2011;47:703–708. doi: 10.1111/j.1529-8817.2011.00976.x. [DOI] [PubMed] [Google Scholar]
- Otten T., Genetics B. 2017. Application of DNA ‐ Based Tools for Algal Bloom Monitoring. [Google Scholar]
- Prokopowich C.D., Gregory T.R., Crease T.J. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50. doi: 10.1139/g02-103. [DOI] [PubMed] [Google Scholar]
- Rodrigue D.C., Etzel R.A., Hall S., De Porras E., Velasquez O.H., Tauxe R.V., Kilbourne E.M., Blake P.A. Lethal paralytic shellfish poisoning in Guatemala. Am. J. Trop. Med. Hyg. 1990;42:267–271. doi: 10.4269/ajtmh.1990.42.267. [DOI] [PubMed] [Google Scholar]
- Simon N., Campbell L., Örnolfsdottir E., Groben R., Guillou L., Lange M., Medlin L.K. Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J. Eukaryot. Microbiol. 2000;47:76–84. doi: 10.1111/j.1550-7408.2000.tb00014.x. [DOI] [PubMed] [Google Scholar]
- Stüken A., Orr R.J.S., Kellmann R., Murray S.A., Neilan B.A., Jakobsen K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toebe K., Alpermann T.J., Tillmann U., Krock B., Cembella A., John U. Molecular discrimination of toxic and non-toxicAlexandriumspecies (Dinophyta) in natural phytoplankton assemblages from the Scottish coast of the North Sea. Eur. J. Phycol. 2013;48:12–26. doi: 10.1080/09670262.2012.752870. [DOI] [Google Scholar]
- Townhill B.L., Tinker J., Jones M., Pitois S., Creach V., Simpson S.D., Dye S., Bear E., Pinnegar J.K. Original Article Harmful algal blooms and climate change: exploring future distribution changes. 2018;75:1882–1893. doi: 10.1093/icesjms/fsy113. [DOI] [Google Scholar]
- Turner A.D., Dhanji-Rapkova M., Baker C., Algoet M. Assessment of a semiquantitative liquid chromatography-fluorescence detection method for the determination of paralytic shellfish poisoning toxin levels in bivalve molluscs from Great Britain. J. AOAC Int. 2014;97:492–497. doi: 10.5740/jaoacint.13-381. [DOI] [PubMed] [Google Scholar]
- Turner A.D., Hatfield R.G. Refinement of AOAC official MethodSM 2005.06 liquid chromatography-fluorescence detection method to improve performance characteristics for the determination of paralytic shellfish toxins in king and queen scallops. J. AOAC Int. 2012;95:129–142. doi: 10.5740/jaoacint.11-184. [DOI] [PubMed] [Google Scholar]
- Turner A.D., Norton D.M., Hatfield R.G., Morris S., Reese A.R., Algoet M., Lees D.N. Refinement and extension of AOAC method 2005.06 to include additional toxins in mussels: single-laboratory validation. J. AOAC Int. 2009;92:190–207. [PubMed] [Google Scholar]
- Utermöhl H. Neue Wege in der quantitativen Erfassung des Plankton. (Mit besonderer Berücksichtigung des Ultraplanktons.) SIL Proc. 1931;1922–2010(5):567–596. doi: 10.1080/03680770.1931.11898492. [DOI] [Google Scholar]
- Vandersea M.W., Kibler S.R., Van Sant S.B., Tester P.A., Sullivan K., Eckert G., Cammarata C., Reece K., Scott G., Place A., Holderied K., Hondolero D., Litaker R.W. qPCR assays for Alexandrium fundyense and A. ostenfeldii (Dinophyceae) identified from Alaskan waters and a review of species-specific Alexandrium molecular assays. Phycologia. 2017;56:303–320. doi: 10.2216/16-41.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño N., Louzao M., Abal P., Cagide E., Carrera C., Vieytes M., Botana L. Human poisoning from marine toxins: unknowns for optimal consumer protection. Toxins (Basel) 2018;10:324. doi: 10.3390/toxins10080324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visciano P., Schirone M., Berti M., Milandri A., Tofalo R., Suzzi G. Marine biotoxins: occurrence, toxicity, regulatory limits and reference methods. Front. Microbiol. 2016;7:1–10. doi: 10.3389/fmicb.2016.01051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamor R.M., Glenn K.L., Hambright K.D. Incorporating molecular tools into routine HAB monitoring programs: using qPCR to track invasive Prymnesium. Harmful Algae. 2012;15:1–7. doi: 10.1016/j.hal.2011.10.028. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.