Abstract
Dysfunctional dopamine (DA) signaling has been associated with a broad spectrum of neuropsychiatric disorders, prompting investigations into how midbrain DA neuron heterogeneity may underpin this variety of behavioral symptoms. Emerging literature indeed points to functional heterogeneity even within anatomically defined DA clusters. Recognizing the need for a systematic classification scheme, several groups have used single-cell profiling to catalog DA neurons based on their gene expression profiles. Here we aim to synthesize points of congruence, but also highlight key differences, between the molecular classification schemes derived from these studies. In doing so, we hope to provide a common framework that will facilitate investigations into the functions of dopaminergic neuron subtypes in the healthy and diseased brain.
Keywords: dopaminergic system, molecular diversity, single-cell RNAseq, intersectional, neuroanatomy, cell type
Heterogeneity within the dopaminergic midbrain system
Since their discovery, neurotransmitters and their associated molecular machinery have been used to differentiate one type of neuron from another. This trend has been reinforced by thousands of neuropharmacological studies which have associated diverse behaviors, mental states, or diseases to specific neurotransmitter systems. Too often however, neuronal populations possessing a common neurotransmitter were assumed to be homogeneous, even though they differed significantly based on morphological and physiological properties. Historically, this has been the case for neurons releasing the neurotransmitter dopamine (DA), a small population of less than 500,000 cells in the human brain [1].
The cellular and molecular properties of DA neurons have attracted great interest, especially since the depletion of DA in the caudate putamen (CP) was shown to underpin the locomotor symptoms of Parkinson’s disease (PD) [1]. Since then, dysfunctional DA signaling has been implicated in numerous neuropsychiatric disorders, including depression, chronic pain, drug addiction, schizophrenia, and attention deficit and hyperactivity disorder [2]. Until recently, it remained puzzling how a single neurotransmitter system could be responsible for such disparate symptoms and apparently unrelated diseases. This quandary could in part be resolved by demonstrating the existence of distinct populations of DA neurons, or “subtypes”, each possessing unique molecular, physiological, and functional properties. Although this is a somewhat Panglossian point of view since DA-associated neuropsychiatric disorders involve neural impairments that extend beyond DA circuits, identifying subtypes of DA neurons could provide a greater understanding of both the breadth of DA-related symptoms, as well as potentially reveal novel avenues for more precisely targeted pharmaceutical interventions.
The majority of DA neurons are located in three anatomical clusters in the midbrain, namely the substantia nigra pars compacta (SNc), the ventral tegmental area (VTA), and retrorubral area (RR) [2,3]. For decades, neuroscientists have searched for, and identified, markers that were expressed in subpopulations of midbrain DA neurons within these clusters (reviewed in [1,4,5]). Although these studies propelled the field forward in establishing DA diversity, the limited ability to simultaneously look at a multitude of markers made it difficult, if not impossible, to define DA neuron subtypes with certainty since the expression of a single gene is rarely sufficient to identify a molecular subtype. Bulk profiling experiments comparing SNc and VTA neurons did indeed yield some interesting gene expression differences [6–9], but suffered from the caveat that they could not distinguish heterogeneous populations within each anatomical cluster. Indeed, the identification of molecularly distinct neuronal populations requires the measurement of multiple markers in individual cells (reviewed in [10–12]).
Fortunately, in the last decade, advances in the field of single-cell transcriptomics and related bioinformatics made it possible to detect coordinated gene expression profiles within individual cells (Box 1). Such approaches have been used to classify molecularly distinct cell types in an unbiased manner in several brain structures, including midbrain DA neuron clusters. Molecular heterogeneity within the murine DAergic system has been investigated by at least six independent groups, all attempting to classify DA neurons based on gene expression [13–18]. Although these studies differed significantly in their experimental parameters and analysis methods, they identified partially overlapping subtypes, some of which were located in a manner agnostic to traditional anatomical boundaries (SN, VTA, & RR). Unfortunately, the use of different defining markers and naming schemes across studies has made it difficult to investigate and discuss the properties of specific DA subtypes. In this review, we highlight congruencies and incongruencies in the putative DA neuron subtypes identified in these studies. In the two studies that did not validate all the single cell clustering data [17,18], we present our best interpretation of their classification scheme. We then undertake the challenging task of synthesizing a classification scheme for mouse midbrain DA neurons based on all available studies, and when possible, correlate this scheme with known functional and anatomical properties of DA neurons. Finally, we briefly discuss how this cellular diversity is generated during development.
Box 1: Strengths and limitations of single-cell gene profiling technologies.
The transcriptional signature of individual cells is currently obtained through single-cell RNA sequencing (scRNA-Seq). As reviewed recently [100,101], single-cell RNAseq experiments initially require the isolation of individual cell and the capture of these cells RNA to generate a library. Depending on the protocol being used, these libraries can be biased toward the 3’ or 5’ end of transcripts, but recent approaches have successfully captured full length RNAs. Analyzing scRNA-Seq data involves reducing the dimensionality by projecting the data into a smaller-dimensionality mathematical space, using methods such as principal component analysis (PCA, linear) and T-distributed stochastic neighbor embedding (t-SNE, non-linear). This facilitates visualization of cell populations by grouping them on the basis of genes expression. This technology is now being used to classify neurons in various brain regions [10–12]. In the future, we envision that a combination of anatomical and sequencing technologies, such as spatial transcriptomics, will allow even greater refinements of cellular identity by simultaneously describing molecular subtypes and their location [102,103].
Single-cell RNA-Seq suffers from limitations, as it has for instance a low capture efficiency compared to “bulk” RNA-Seq, leading to dropout events, that is – a failure to detect the expression of a gene [10,101]. scRNA-Seq data is noisier and more variable than “bulk” RNA-Seq, since cDNA synthesis will only be possible for a fraction of the total transcripts due to Poisson sampling [104], biased towards the most highly expressed genes in a cell. In the context of neural cell-type profiling, this might explain the overabundance of highly expressed genes like neuropeptides, and underwhelming presence of genes with lower expression like transcription factors, in the list of genes defining cell types. This may compromise the ability to segregate closely related subtypes, and could underpin some of the incongruencies in DA neuron classification studies. Another limitation of classifying neurons using single-cell RNAseq results from the imperfect correlation between mRNA and protein levels, such that the transcriptome of a cell provides only a limited insight into protein levels. Strengths and weaknesses of scRNA-Seq have been reviewed in-depth elsewhere [100,101].
Defining dopamine neuron subtypes based on their molecular profiles
It is generally agreed upon that groups of neurons that possess a core number of distinctly expressed genes can be classified as the same cell type [10–12]. However, classifying neurons into subtypes is no easy task, particularly when the types are closely related. In the case of midbrain DA neurons, all display a typical neuron molecular signature, in addition to expressing genes necessary for the synthesis, packaging and release of DA (Box 2). Superimposed on this generic midbrain DA neuron signature, which also include mesodiencephalic floor plate transcription factors, is the expression of other genes confined to subpopulations of DA neurons, that can be used to categorize them into distinctive subtypes. Multiple groups have profiled the gene expression of individual midbrain DA neurons in the mouse (Figure 1; [13–18]). These groups observed several differences, both in terms of the number of DA neuron subtypes they identified, as well as the genes used to define them (Table 1). These differences can partly be explained by variations in experimental conditions such as using different cell sources, isolation procedures, developmental stages, technical approaches, and classification criteria (Table 2). Here, we attempt to synthesize the current literature and extract a classification of DA neurons derived from all published single-cell classification studies and other supporting data. We have focused almost exclusively on neurons expressing the dopamine transporter (Dat; Slc6a3). This is because the majority of DA neurons, as defined by immunolabeling for the rate limiting enzyme involved in DA synthesis tyrosine hydroxylase (TH), are recombined in the adult Dat-IRES-Cre mouse [19]. While Th mRNA+ neurons that do not express Dat are also observed in the midbrain, it is not clear if these produce functional amounts of DA, and thus they have been omitted from this review [15,19,20] (see Box 2). After assessing the current literature, here we propose seven putative DA neuron subtypes, defined by a unique molecular signature, whose existence is supported by multiple studies. Four of these are more robustly identified by diagnostic markers, whereas the three others are more ambiguous. We next describe each of these subtypes, the data supporting their existence, and when available, their distinctive properties (Figure 1). Here, the list of genes refers to mRNA detected within a subtype and is used for definition purposes (i.e. the functions of the gene product are not necessarily relevant to this discussion).
Box 2: Molecular signature of a midbrain dopamine neuron.
A DAergic neuron is typically defined as one that releases the neurotransmitter DA. This functional definition is straightforward, and thus well accepted [2,105–108]. However, defining the molecular signature of a DA neuron is more difficult, as many canonical markers of DA neurons are not specific to these neurons. To begin to molecularly define DA neurons, one can postulate that DA neurons are cells that express: 1) a set of genes common to all neurons, and 2) a set of genes that is necessary for the synthesis of DA (or found exclusively in cells that synthesize DA). Both these sets of genes are vital to DA neuron identity, since DA synthesis pathway are present in unrelated cell types of the gut and immune system (e.g. [109,110]). Neuron-specific transcriptional programs have been well characterized and include cohorts of expressed genes that underlie axonal and presynaptic functions. The transcriptional program involved in the synthesis and neurotransmission of DA includes genes encoding tyrosine hydroxylase (Th) and Dopa decarboxylase (Ddc) that are necessary for the stepwise production of DA from its precursor L-Tyrosine, and vesicular transporter Vmat2 (Slc18a2) that is necessary for the packaging of DA into vesicles [2]. Since these genes are also expressed in other monoaminergic cell types such as noradrenergic neurons, the absence of dopamine beta hydroxylase (Dbh) expression is a requirement to define a DA neuron. Complicating the matter further, a gene such as Th, often used to describe DA neurons, shows discrepancies between mRNA and protein in that the mRNA is more broadly detected than the protein in the midbrain as well as other brain regions [19,60]. This raises the question whether all neurons harboring Th mRNA have sufficient levels of the protein to synthesize DA, leading some to propose that in the midbrain, Dat (Slc6a3) expression, rather than Th, represents a more accurate descriptor of bona-fide midbrain DA neurons [19] ( see also Box 3).
Besides neuronal identity genes and DA synthesis genes, a third axis is required to define midbrain DA neurons: genes that are characteristic of a developmental origin in the mesodiencephalic floor plate. These would include developmental genes like Foxa1/2, Lmx1a/b, Nurr1 that are expressed in all midbrain DA neurons, although possibly at varying levels [72,79,111–116]. These genes are not coordinately expressed in hypothalamic or olfactory DA neurons (A11–16), which are not floor plate derived [67,111], and appear to be quite distinct from midbrain DA neurons [6,16,117]. Conversely, these genes are also expressed in non-DA neurons, like those located in the rostral linear nucleus, as well as in neighboring floor plate derived populations like the subthalamic nucleus (STN) and ventral premammillary nucleus (PMv), and thus by themselves are not sufficient to describe midbrain DA neurons [75,76]. Expression of Engrailed 1 (En1) and its downstream target Pitx3 [75–77,118,119], when added to the analysis, can be used to exclude diencephalic floor plate derived neurons (e.g. STN and PMv neurons), although not neurons of the rostral linear nucleus [75]. In summary, all midbrain DA neurons, located in three clusters RR, SNc, and VTA (A8–10) have a characteristic signature defined by an overlap of neuron-specific genes, DA pathway genes, and midbrain floor plate derived transcriptional cascades.
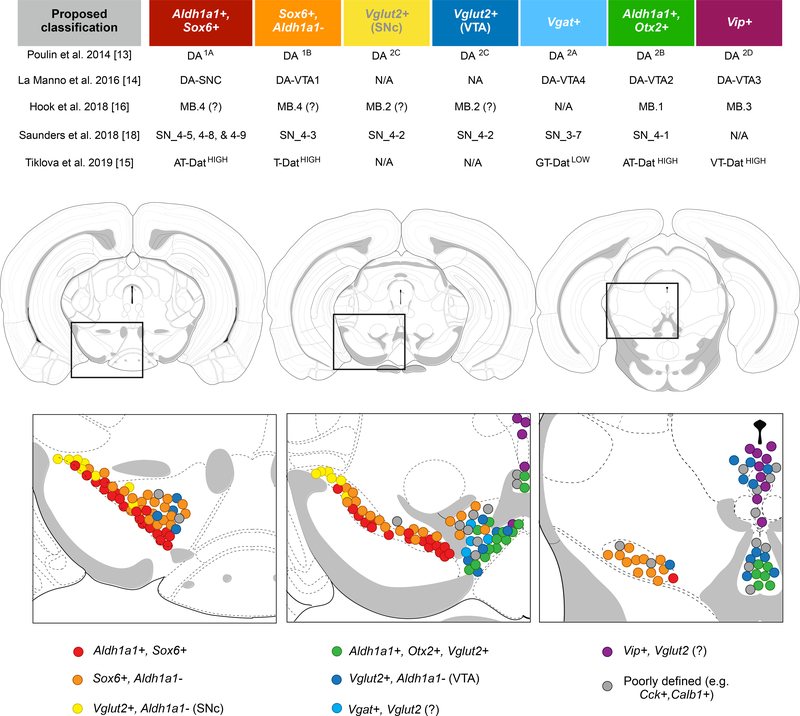
Figure 1.
Molecularly defined DA neuron subtypes.
Our interpretation of putative DA neuron subtypes across several studies with their potential neuroanatomical locations in the adult mouse midbrain. Of note, the subclusters SN_4–4 (Th, C1ql3), SN_4–6 (Th, Cadm2), and SN_4–7 (Th, Nefl) from [18] are not mentioned, as we were unable to unambiguously locate these groups. In relation to [15], we have omitted neurons that have low Th and Dat expression (N-DatLOW, G-DatLOW). NT-DatLOW, defined by Npxh4, likely represents neurons in the rostral linear region, as depicted in that study. For discussion of results from a sixth related study, [17], please see main text; findings from this study have not been listed in this figure due to the limited information on clusters.
Table 1.
List of dopamine neuron subtypes and markers with highest variance identified by single-cell gene profiling studies.
| Studies | Developmental stages | DA Subcluster (Markers) |
|---|---|---|
| La Manno et al., 2016 [14] | E11.5–E18.5 | mDA0 (Th+, Cck+, Prkca+) |
| mDA1 (Th+, Dat+, Calb1+) | ||
| mDA2 (Sox6+, Th+, Dat+, Calb1+, Aldh1a1+, Lmo3+, Bnc2+) | ||
| mNbM (Sox6+, Neurod1+, Neurog2+, Ebf2+, Rnd3+, Cnfn2+, Nhlh1+, Igfbpl1+, Pitx3-) | ||
| mNbMDA (Pbx1+, Ebf2+, Cdk14+) | ||
| P28–P56 | mDA-SNC (Sox6+, Calb1−, Aldh1a1+, Aldh1a7+, Anxa1+, Ndnf+) | |
| mDA-VTA1 (Sox6+, Calb1+, Ndnf+) | ||
| mDA-VTA2 (Sox6−, Calb1+, Aldh1a1+, Aldh1a7+, Neurod6+, Grp+, Gpr83+, Otx2+, Lpl+, Anxa1+, Cbln4+) | ||
| mDA-VTA3 (Sox6−, Calb1+, Vip+, Cck+, Cbln1+, Cbln4+) | ||
| mDA-VTA4 (Sox6−, Calb1+, Vgat+, Cbln1+) | ||
| Hook et al., 2018 [16] | P7 | P7.MB.1 (Gpr83+, Otx2+, Cck+, Neurod6+, Lpl+, Tacr3+) |
| P7.MB.2 (Lhx9+, Ldb2+, Dat−, Vglut2+, Wnt7b+) | ||
| P7.MB.3 (Vip+, Calb1+) | ||
| P7.MB.4 (Sox6+, Ndnf+, Lmo3+, Aldh1a1+, Aldh1a7+, Anxa1+, Sncg+) | ||
| E15 | E15.MB.1 (Lhx9+, Ldb2+, Vglut2+) | |
| E15.MB.2 (High Th+, high Dat+, Vmat2+, Pitx3+, Ddc+, En1+) | ||
| Poulin et al., 2014 [13] | P4 | DA1A (Aldh1a1+, Sox6+, Ndnf+) |
| DA1B (Aldh1a1−, Sox6+, Ndnf+, Tacr3+) | ||
| DA2A (Vgat+, Calb1+, Cck+, Vglut2+) | ||
| DA2B (Lpl+, Adcyap1+, Otx2+, Aldh1a1+, Vglut2+, Calb1, Cck+, Grp+, Tacr3+) | ||
| DA2C (Aldh1a1−, Calb1+, Cck+, Vglut2+, Tacr3+) | ||
| DA2D (Vip+, Calb1+, Cck+, Vglut2+, Sox6−, Aldh1a1−) | ||
| Tiklova et al., 2019 [15] | P1-P90 | N-Datlow (Nxph4+, Th−, C1ql1+, Fam19a2+, Car10+) |
| NT-Datlow (Nxph4+, Th+ high, C1ql1+, Fam19a2+, Car10+) | ||
| G- Datlow (Gad2+, Th−, Vgat+, Wnt7b+, Crhbp+) | ||
| GT-Datlow (Gad2+, Th+ high, Crhbp+, Vgat+, Wnt7b+) | ||
| T-Dathigh (Th+ high, Aldh1a1−, Sncg+) | ||
| AT-Dathigh (Aldh1a1+, Th+ high, Otx2+, Grp+, Cck+, Lpl+) | ||
| VT-Dathigh (Vip+, Th+, Dlk1+, Cck+, Gipr+, Pou2f2+) | ||
| Kramer DJ et al., 2018 [17] | P26-P34 | Cluster 1 SNC (Aldh1a7+, Igf1+, Bsn+, Ntsr1+, Kcns3+) |
| Cluster 4 SNC (Aldh1a7+, Igf1+, Bsn+, Ntsr1+, Kcns3+, Anxa1+) | ||
| Cluster 2 VTA (Ubqln2+, Ids+, Slc12a5+) | ||
| Cluster 5 VTA (Necab1+, Cthrc1+, Cbln1+) | ||
| Cluster 6 VTA (Necab1+, Cthrc1+, Cbln1+, Gucy1a3+, Ryr2+, Uri1+) | ||
| Cluster 8 VTA (Neurod6+, Grp+, Gpr83+, Cbln4+, Igfbp4+) | ||
| Saunders et al., 2018 [18] | P60-P70 | SN_3–7 (Vglut2+, Gad2+, Crhbp+, Vgat+, Wnt7b+) |
| SN_4–1 (Lpl+, Vglut2+, Cbln4+, Aldh1a1+, Neurod6+, Otx2+, Calb1+, Tacr3+) | ||
| SN_4–2 (Cbln1+, Vglut2+, Calb1+, Wnt7b+) | ||
| SN_4–3 (Cyp26b1+, Sox6+, Tacr3+) | ||
| SN_4–4 (C1ql3+, Calb1+)* | ||
| SN_4–5 (Vcan+, Aldh1a1+, Sox6+) | ||
| SN_4–6 (Cadm2+)* | ||
| SN_4–7 (Nefl+, Nefm+)* | ||
| SN_4–8 (Aldh1a1+, Anxa1+, Sox6+, Igfbp2+) | ||
| SN_4–9 (Grin2c+, Aldh1a1+, Sox6+) |
Notes:
1) Genes in bold represents those explicitly discussed in the papers, whereas those not in bold represent our own mining of underlying data
2) Asterisk (*) indicates populations of neurons in [18] which we were unable to definitively determine the correspondence to subtypes in other papers, or the localization in the mouse brain.
Table 2.
Key methodological aspects of single-cell dopamine neuron classification studies.
| Studies | Mouseline | Age | Isolation | Technique | # of cells |
|---|---|---|---|---|---|
| Poulin et al., 2014 [13] | Dat-IRES-Cre, Ai9 | P4 | FACS for tdTomato | sc-qPCR | 159 |
| La Manno et al., 2016 [14] | Dat-IRES-Cre, Ai9 | P28-P56 | FACS for tdTomato | scRNA-Seq | 245 |
| Hook et al., 2018 [16] | Th-eGFP | P7 | FACS for eGFP | scRNA-Seq | 80 |
| Saunders et al., 2018 [18] | wildtype | P60-P70 | No sorting | Drop-Seq | 919 |
| Kramer et al., 2018 [17] | Dat-IRES-Cre, Ai9 | P26–34 | FACS for tdTomato | scRNA-Seq | 232 |
| Tiklova et al., 2019 [15] | Pitx3-eGFP | P1-P90 | FACS for eGFP | scRNA-Seq | 1106 |
Aldh1a1+, Sox6+ DA neurons are located in the ventral SNc
Aldh1a1 and Sox6 mRNAs are the defining markers for this DA neuron subtype, a population overtly represented in two studies: DA1A in [13] and DA-SNC in [14]. These cells also display Aldh1a7 [14,17] and Ndnf [13,14] mRNA expression. In [15], we believe that this population has been grouped together with Aldh1a1+, Sox6- cells, possibly because Sox6 mRNA was not robustly detected in this study. In contrast, this population was split into two distinct groups in [17] based on the expression of Anxa1 mRNA. In [18], the Sox6+, Aldh1a1+ population seems to have been split into at least three subpopulations named SN_4–5, SN_4–8, and SN_4–9. The defining genes for these populations are Vcan, Anxa1 (also characterized by the highest expression levels for Aldh1a1), and Grin2c respectively, although these markers have yet to be validated by secondary methods.
Prominent projections of ALDH1A1+ DA neurons of the SNc have been observed in the dorsal caudate putamen (CP; [20–22]), with fibers being more dense in rostral sections and less so in caudal regions. In the dorsal CP, these neurons preferentially innervate some μ-opioid receptor+ (MOR) patches (commonly known as striosomes), although this is not exclusive [20–22]. Interestingly, ALDH1A1 belongs to the aldehyde dehydrogenases family and its presence in DA neurons may alter their function in at least four ways. First, aldehyde dehydrogenases catalyze the conversion of retinaldehyde to retinoic acid (RA), which may be implicated in maintenance of MOR expression in patches, potentially via transsynaptic RA signaling [23]. Second, ALDH1A1 may also mediate the oxidation of the cytotoxic side product of dopamine synthesis, DOPAL, into a less reactive species. Third, ALDH1A1 might enable the synthesis of GABA through a non-conventional enzymatic pathway [24]. Finally, this enzyme could also play a role in modulating DA release, as there is a reduction in DA release by the ALDH1A1+ fibers targeting the striosomes compared to the matrix, an effect not observed in Aldh1a1 null mice [21]. A recent report supports a function of Aldh1a1+ DA neurons in the acquisition of skilled movements, as ablation of these neurons results in significant rotarod learning deficits [22]. In addition, Aldh1a1+ DA neurons of the SNc are selectively decreased in a mouse model of PD [13], as well as in human brains affected by the disease [25]. Taken together, these studies strongly suggest that these neurons of the ventral SNc have distinctive properties, some of which are conferred by Aldh1a1 function.
Sox6+, Aldh1a1- DA neurons are located in the SNc, parabrachial VTA, and RR
While this subtype is somewhat more ambiguous, the presence of Sox6 mRNA and the absence of Aldh1a1 transcript was considered to be its defining signature. Such neurons have been identified in three studies (DA-VTA1 in [14], DA1B in [13], and SN_4–3 in [18]). These cells also co-express high levels of many of the mRNAs aforementioned in the previous population (Ndnf, Igf1 and Sncg), but in contrast, some of these neurons also express Calb1 [13,14], Lypd1 [14], Tacr3 [13,18], and Cyp26b1 [18] mRNAs. In addition to lacking Aldh1a1 mRNA, they also do not express Anxa1 [14,17,18] and Aldh1a7 [14,17,18]. It is possible that [16] may have grouped this subtype together with Aldh1a1+ cells based on high mRNA Sox6 expression and overall similarity in gene signature. In [15], these cells are likely represented within the population of Th+, Aldh1a1- neurons displaying high expression of Dat (referred to as T-Dat-high). In immunolabeling studies, there is a substantial SOX6+ population that is ALDH1A1- [13,14,26]; these cells are located more dorsal to the ALDH1A1+ neurons in the SNc, the parabrachial pigmented (PBP) region of the VTA and the RR. Because Aldh1a1 mRNA expression is one of the few diagnostic features separating the two Sox6+ cell types, further work will be required to define additional markers in order to facilitate their investigation in vivo.
The functional roles of the Sox6+, Aldh1a1+ and Sox6+, Aldh1a1- cohorts might at least be influenced by the expression of ALDH1A1, and its cognate enzymatic functions like retinoic acid synthesis or DA degradation [21,22]. However, it remains to be seen if these subtypes can be differentiated by other features, such as their physiological properties. With regard to projections, the overall SOX6+ cohort projects densely to the rostral CP and lateral shell and core divisions of the nucleus Accumbens (ACB), whereas the projections to the caudal CP appear less dense [20]. Sox6+ projections, which encompasses Aldh1a1+ projections, show significant innervation of the medial and ventral CP. Comparing projections of SNc DA neurons labeled using Sox6-Cre and Aldh1a1-CreERT2 drivers, it could be inferred that Sox6+, Aldh1a1- neurons which are located mainly in the PBP region, spanning the junction between the SNc and VTA [20], project to the medial and ventral CP and lateral shell of the ACB. Additional identification of unique molecular markers, as well as the generation of a Cre drivers, will validate the existence and facilitate the investigation of this putative subtype.
Vgat+ DA subtype is located mostly in the VTA
A DA subtype distinguished by the expression of Vgat mRNA (Slc32a1), was identified in four studies: DA2A in [13], DA-VTA4 in [14], GT-Dat-Low in [15], and based on our interpretation, SN_3–7 in [18]. This group of cells is enriched for Vgat [13–15,18], Calb1 [13,14], Crhbp [15,18], Gad2 [15,18] and Wnt7b [15,18] mRNAs. In [18], this subtype was grouped with GABAergic neurons of the SNc rather than with the DA clusters, possibly because this subtype has molecular features of both DA and GABA neurons. Many of these neurons also express Vglut2 (a.k.a. Slc17a6), a glutamate vesicular transporter [13,18], and also have lower Dat and Th expression [15]. This latter observation may explain why these neurons were not observed in [16], which utilized a Th-eGFP transgenic line for isolating DA neurons (Table 2). Anatomically, Vgat+ DA neurons have been mainly observed in the VTA [13–15]. While these cells were numerically abundant in single-cell isolation/profiling, the proportion of this population observed by in situ hybridization and immunofluorescence is significantly less, with one study suggesting that these comprise less than 5% of all adult midbrain DA neurons [15].
The projections and function of this subtype has not been directly determined, although these might correspond to a population of DA neurons GABA co-releasing and projecting to the medial shell of the ACB [27]. Alternatively, they might correspond to the mesohabenular Gad2+, Th+ neurons described in Stamatakis et al. [28], although these neurons might not have the capacity to release DA. Unlike the non-canonical GABA synthesis and vesicular loading that has been proposed for GABA co-release from SNc neurons [24,29,30], these transcriptomic studies collectively open the possibility that this VTA enriched subtype may synthesize and co-transmit GABA through canonical mechanisms involving GAD and VGAT.
Otx2+, Aldh1a1+ DA neurons in the ventromedial VTA
Cells co-expressing Aldh1a1 and Otx2 mRNAs were observed in six studies and named DA2B [13], DA-VTA2 [14], SN_4–1 [18], and MB.1 [16]. Cluster #8 of [17] also appears to match this subtype, which likely correspond to previously VTA neurons observed in immunolabeling-based studies [31,32], as well as those profiled in ACB projection-based transcriptomic analyses [33]. Based on co-expression of other genes in this subtype, we derived the following mRNA expression signature: Aldh1a1 [13,14,17,18], Lpl [13,14,18], Otx2 [13,14,16,18], Neurod6 [13,14,16–18], Gpr83 [14,17,18], Grp [13,17,18], and Cbln4 [17,18]. In [15], it is possible that all Aldh1a1 expressing DA neurons, from both SNc and VTA, were lumped together (named AT-Dat high). Otx2+, Aldh1a1+ neurons are located in the ventromedial VTA [13,14,17,31,32,34], and they roughly comprise between 12–36% of VTA DA neurons, depending on the method of quantification. Multiplex in situ hybridization suggested that both NeuroD6 and Grp transcripts might not be found in all neurons of this subtype, with Neurod6 transcript being more restricted [17]. Additionally, one study showed that only 12% of Neurod6+ cells co-express Vglut2 [35], although two other studies suggested that a substantial number of these cells were Vglut2+ [13,18], the difference possibly explained by sensitivity of techniques used.
This population has been accessed using Aldh1a1 and NeuroD6 based drivers, and it appears to direct major projections to the medial shell of the ACB, and minor projections to the lateral septum [17,20,22,36]. In Neurod6 KO mice, DA projections to the lateral septum are reduced, suggesting a dependence on NEUROD6 function [36]. Another study, using Aldh1a1-CreERT2 strain to access this population, postulated some projections to the dorsomedial striatum [22]. Physiologically, DA neurons labeled with Neurod6-Cre have a smaller Ih, more depolarized membrane potential, higher membrane resistance, less pronounced afterhyperpolarization, and shorter action potential height when compared to SNc neurons [17]. These characteristics are consistent with those observed for medial shell projecting DA neurons [37]. When optogenetically stimulated, Neurod6+ neurons induce place preference behavior [35]. Furthermore, these neurons appear to co-release glutamate [35], consistent with a larger body of work showing glutamate co-release in the medial shell [38–45]. In studies using NeuroD6 as a driver, however, it has been dutifully reported that it is expressed in non-DA neurons of the VTA [17,35,36]; thus experiments based on these drivers must be interpreted with caution.
Vip+ DA neurons are the most caudally located population
Four studies identified a subpopulation of DA neurons expressing high levels of the neuropeptide VIP: DA-VTA3 in [14], DA2D in [13], VT-Dat-high in [15], MB.3 in [16]. On a neuroanatomical basis, this subtype is located in the periaqueductal gray (PAG) and dorsal raphe (DR) regions, accounting for about 49% of DA neurons [13] in these regions, but more sparsely in the VTA [13–15,46](Figure 1). Based on the concordance of other genes co-expressed in Vip+ neurons [13–16], we developed the following mRNA signature for Vip neurons: Vip+ [13–16], Gipr+ [15], Calb1+ [13,14], Sox6- [13,14], Aldh1a1- [13,14]. These neurons are also Vglut2+ at neonatal timepoints [13], consistent with functional data showing glutamate co-transmission from PAG/DR DA neurons in the adult [47,48]. However, since the PAG/DR contains both Vip+ and Vip- DA neurons, definitive proof showing adult Vip+ neurons co-releasing glutamate requires further experimentation.
The projections of these neurons are peculiar in that they are particularly focal – they have been mapped to the bed nucleus of the stria terminals (BST) and central nucleus of the amygdala [13,20], and appear to innervate a much smaller target volume than other DA neurons (e.g. [49] and [50]). The function of the Vip+ DA subtype has never been examined in isolation. However, some studies have investigated the functions of DA neurons located in the PAG/DR and have found that they were involved in wakefulness and social behaviors [47,48,51]. DA neurons in this region also might be embedded in the circuits that transduce the antinociceptive effects of morphine [52], as well as in fear-induced learning [53]. Since Vip+ DA neurons constitute a fraction of TH+ neurons of the PAG/DR, direct testing will be necessary to know if Vip+ and Vip- DA neurons of this region have distinct roles in behavior.
Vglut2+,Calb1+ DA neurons are ambiguous and likely encompass two or more distinct populations
A Vglut2+, Calb1+ population is represented in at least three of the studies, although this remains somewhat ambiguous owing to the lack of more specific markers [13,16,18]. In [13], this population, named DA2C, was somewhat indistinct, characterized by Vglut2 and Calb1 mRNA expression, but low or sporadic expression of transcription factors like Otx2 and Sox6, and little to no expression of Aldh1a1 and Vip. In [16], this was described as a neuroblast-like population, although we interpret it as an adult neuronal population, since a population expressing these diagnostic markers is observed in the adult brain [18,20]. In [18], this population is likely to be represented within cluster SN_4–2, and is also defined by the marker Cbln1. It is unclear whether this represents a single population or multiple DA subtypes. One possibility is that the Vglut2+, Calb1+ population encompasses at least two subtypes, one located in the SNc and the other in the VTA. The first of these populations may be found in the dorsolateral part of the SNc, including a region known as the pars lateralis (SNpl) [20]. Indeed, in recent studies, a population of cells in the dorsal SNc and SNpl could be labeled by viral injections into the SNc of Vglut2-Cre, Th-Flpo mice [20,54]; only 8% of these were reported to be Aldh1a1+[20]. These cells show a predominant projection to the tail of the striatum, but also to more discrete regions in rostral striatum [20,54]. Functionally, these cells are likely to match a recently described class of tail of striatum-projecting SNc neurons, that have distinctive inputs and show responses to aversive stimuli [55–57], but direct and careful testing is required to confirm this prediction. In toxin models of PD, Vglut2+ DA neurons in the SNc are particularly resistant to degeneration [58,59], and VGLUT2 itself may play a neuroprotective role by inducing a BDNF/TRKB dependent signaling cascade [59].
A second, closely related population of mRNA Vglut2+, Calb1+ DA neurons is found in the VTA. This is distinct from the Otx2+, Aldh1a1+ subtype mentioned previously, judged by the absence of many markers including Aldh1a1 expression. Comparing the projections of Vglut2+, Aldh1a1- and Vglut2+, Aldh1a1+ VTA cohorts, we observed significant overlap in the ACB, olfactory tubercle (OT), and lateral septum (LS). In comparison, we observed projections in the BLA, entorhinal and prefrontal cortex, exclusively from Aldh1a1-, Vglut2+ projections [20]. However, additional analysis will be required to define these neurons on a molecular basis, and thereby unambiguously distinguish this subtype from Aldh1a1+ DA neurons of the VTA, and from Vglut2+ cells in the SNc. Overall, these data are consistent with the abundant literature on TH+ VTA neurons that can release glutamate in the ACB [38,43,45,60–63], but also in the basolateral amygdala, entorhinal and prefrontal cortex [43,54,64,65]. Interestingly, there is evidence of Vglut2 and Vmat2 segregation in Vglut2+ DA axons in the ACB, suggesting distinct release sites and mechanisms for these two neurotransmitters [41,42,66].
Generating midbrain DA neurons diversity during embryonic development
The embryonic primordia of midbrain DA neurons is well defined. These neurons originate from the mesodiencephalic floorplate (FP), a region at the ventral midline defined by the mRNA expression of Shh+ and Foxa2+ [67–73]. The boundaries of this primordium have been well defined by transcription factor expression. The caudal extent is defined by the Otx2/Gbx2 boundary coincident with the morphological mid-hindbrain boundary [74]. The rostral extent is defined by the En1/Dbx1 gene expression boundary which is located in the diencephalic region [75–78]. The dorsal extent is defined by the Lmx1a/Nkx6.1 gene expression boundary [71,79–81]. The primordium of the majority of midbrain DA neurons therefore can be described as the ventral mesodiencephalic region progenitors that express Foxa1/2, Lmx1a/b, Otx2 and En1. Current evidence suggests that most, if not all, midbrain DA neurons observed in the adult mouse brain arise from this primordium. While the DA primordium has been precisely demarcated, how and when DA neuron heterogeneity is generated remains to be clearly elucidated.
A starting point for answering this question is to determine whether the DA primordium can be subdivided further into molecularly discrete regions that foreshadow subtype identity. Consistent with this possibility, several genes are known to be expressed in a non-homogeneous manner in the DA primordium [82]. Some genes like Shh are dynamically expressed [67,70]; other genes like Wnt1 and Aldh1a1 are expressed specifically in lateral regions of the primordium [83,84], whereas Corin is expressed in more medial regions [85]. Some genes, for instance Wnt1 and En1, are expressed in graded fashion along the rostro-caudal axis, being more robustly expressed near the mid-hindbrain boundary. How these non-homogeneous expression patterns in the progenitor zone correlate with diversity remains to be determined. Some recombinase-based lineage studies have been performed to address this question, but differences in Cre drivers, TAM regimens, and mosaicism have provided only a blurry picture of DA subtype specification [67,70,83,86,87]. More recently, Sox6 has been shown to be expressed in the medial aspect of the progenitor pool, and based on immunolabeling experiments, this progenitor pool has been proposed to be the source of the majority of the SNc but not the VTA [26]. Lineage tracing approaches will be required to test this model. In addition to the aforementioned lineage studies, knockouts of various transcription factors and other developmental molecules have also begun to illuminate the developmental basis of DA neuron diversity, and this has been reviewed recently [82,88].
Single-cell transcriptomics has recently been applied to developing DA neurons and has begun to provide a picture of the emerging diversity within maturing DA neurons. For instance, [14] identified three types of embryonic DA neurons (E11.5-E18.5) in the mouse: 1) an immature type expressing Th and other key markers of DA neurons, 2) a more mature neuron type expressing both Th and Dat, and 3) neurons with a similar signature but also expressing Aldh1a1 and Lmo3. By contrast, [16] identified only two embryonic types (E15.MB.1 and E15.MB.2) representing a neuroblast population and a more mature DA neuron. Finally, [15] suggests that elements of DA neuron diversification might be present as early as E13, but further diversification continues throughout development. In the authors’ elegant pseudotime plot of developmental trajectories, it is apparent that DA neuron subtypes from older mice (P90) are further apart compared to similar cells obtained from younger animals (E18.5 to P7). This indicates that, as DA neurons are maturing and integrating into defined neural circuits, they continue to refine their molecular signature. Supporting the possibility of extrinsically driven refinement of gene expression, a recent study suggested that contact with striatal neurons could modulate the expression of Vglut2 in DA neurons [42]. Taken together, it appears that DA neuron diversification begins to emerge early and probably occurs in a stepwise fashion. However, because of the limitations of single cell transcriptomics (Box 1; reviewed in [10]), the complete developmental dendrogram of midbrain DA neurons remains to be further refined. Additionally, how much of this diversity is encoded in progenitors, or determined by later intrinsic or extrinsic factors, remains to be resolved.
Concluding remarks and perspectives
Single-cell gene expression profiling has enabled progress in defining DA neuron subtypes, thus revising traditional anatomically based classification schemes. Parsing midbrain DA subtypes is a challenging endeavor since these neurons all share a common developmental origin in the floor plate and share the expression of many terminal selector genes and their downstream cascades. Nonetheless, and although single-cell studies have discrepancies in resulting subtypes, these studies have begun to provide a more granular picture of molecular heterogeneity in the midbrain DAergic system, a significant advance from anatomically based definitions. Additional studies, using larger numbers of cells and ones that are sequenced at greater depth, will further refine or clarify these subpopulations. Spatial transcriptomics provides a methodology to simultaneously locate these subpopulations in situ and will be valuable tool towards generating a complete list of midbrain DA neurons. Ultimately, anatomical and functional analyses will be required to corroborate molecularly based classification schemes, to determine if each subtype aligns with a distinct set of physiological and functional features [61,89–94]. Towards this end, selective intersectional genetic approaches have been developed to target DA neuron subtypes that are defined by a combination of genes [20] (Box 3), and initial studies have suggested that indeed DA neuron subtypes expressing given markers have distinctive projection patterns even within a given target region. Such paradigms can be used to target subtypes with intersectional expression of sensors/effectors like opsins [54,95], DREADDs [96,97], or GCaMP [98], in order to study their function in isolation. We envision that a more complete midbrain DA taxonomy can be derived by concerted single-cell profiling studies and functional studies that reinforce, rebut or refine each other.
Box 3: Genetic targeting of midbrain dopamine neurons.
Investigating DA neuron function is best accomplished by genetic targeting using site-specific recombinases. For instance, Cre recombinase can be driven by transcriptional regulatory elements active in midbrain DA neurons and can be used to activate a genetically encoded effector within a viral vector or a mouse reporter. A thorough characterization of the driver used is critical for a proper interpretation of experimental results. For instance, two distinct Th-Cre drivers have been generated and used to drive expression of opsins. In one of these lines [120], Cre recombinase has been knocked into the 3’UTR of the tyrosine hydroxylase (Th) locus, and by virtue of the internal ribosome entry site (IRES) sequence, has the advantage of preserving two functional alleles of the gene. For most genes, mRNA and protein levels are highly correlated, however in the case of Th, many midbrain cells show mRNA expression in the absence of protein (e.g. [20]). Accordingly, Th-IRES-Cre mice have displayed recombination in cells where TH protein could not be detected [19]. For this reason, Dat-IRES-Cre is now the driver of choice as it does not appear to drive recombination in TH immunonegative neurons in the midbrain. However, some TH- cells are labeled in the nearby premammillary ventral nucleus [75,121] and Dat-IRES-Cre might not completely recombine neurons that express low levels of Dat, depending on the reporter used. Further, DAT levels are decreased in the available strains [122,123], possibly interfering with the interpretation of behavioral experiments.
Genetic targeting of DA neuron subpopulations can be achieved by two methods: 1) projection specific retrograde viruses or 2) intersectional genetic approaches. Projection specific approaches typically involve injection of viruses that show retrograde capabilities, into a target region of choice, allowing the efficient segregation of DA neuron subtypes based on their projections; indeed, these approaches have been used to segregate functionally distinct subpopulations within the VTA and SNc with extraordinary success [61,91,92,94]. Projection-specific approaches typically are performed in wild type mice, or in mice harboring only a single allele (e.g. Dat-IRES-Cre) and therefore require minimal husbandry. However, a caveat is that these approaches require accurate and reproducible placement of viruses, often at multiple sites. Additionally, these approaches are predicated on the idea that each target region receives innervation from a single DA subtype, which does not always appear to be the case [20]. Intersectional genetic approaches require more than one recombinase to achieve the required specificity. One way to accomplish this is by using a combination of Cre and Flp recombinases, thus activating a genetically encoded effector that is defined by two drivers [124]. Genetic targeting using Boolean logic is highly flexible and exploits the numerous Cre and Flp drivers available in mouse repositories. To facilitate prospective targeting of DA neuron subtypes, we previously generated a Th-2A-Flpo driver [20]. Combining this Flp driver with a subtype specific Cre line and an intersectional reporter [125] or virus [95] allows genetic targeting of subsets of DA neurons. Another intersectional strategy to target DA neuron subtypes makes use of the tetracycline expression system, in conjunction with Cre recombinase. To this effect, a Dat-tTA mouse was generated in which the tetracycline-controlled transactivator protein (tTA) is driven by the Dat gene and can be turned off by doxycycline injection [126]. Combining this driver with a subtype specific Cre line and a TRE reporter that is Cre-dependent allows genetic targeting of subsets of DA neurons [20]. While intersectional approaches require mice with multiple targeted alleles, their main strength is that they are predicated on molecularly defined DA subpopulations, rather than solely by projection targets. In the future, we envision even greater selectivity of targeting by combining projection specific approaches with intersectional approaches using the plethora of new recombinases available.
Establishing a consensus of DA neuron subtypes, is a first step toward a thorough understanding of the DAergic system and its role in diseases. Having an accurate, molecularly defined list of DA subtypes will be crucial for 1) understanding molecular cascades underpinning selective vulnerability in PD, 2) pharmacologically manipulating the DA neurons in a subtype specific fashion towards treatment of DA related disorders like depression and chronic pain, 3) rational and accurate differentiation of iPS cells towards DA subtypes, and 4) evolutionary comparisons across species, including humans. All these goals would be facilitated by a common nomenclature, which will allow for easier comparison of findings across laboratories, thus providing a shared framework to understand DA neuron diversity. Once a taxonomy of DA subtypes is established, the field will need to attribute specific functional roles to these molecularly defined subtypes. Although it is often assumed that behaviors would neatly segregate between molecularly defined neuronal populations, a recent study of hypothalamic cell types suggests that it is not always that simple [99]. Indeed, in the ventromedial hypothalamus, few transcriptionally defined cell types exhibited a clear correspondence with behavior-specific activation or connectivity. In the DAergic system however, we have observed distinct but partly overlapping projection patterns for molecularly defined types, although these projections often included multiple targets [20]. In the future, mapping behavior-specific activation of genetically defined DA circuits will be an important goal, that might also help understand how disruptions of these circuits can lead to specific symptoms of disease. Although circuit alterations in most DA-associated disorders are by no means restricted to the DAergic system, identifying the specific DA subtypes that are affected may offer insight into molecular or circuit-level mechanisms underlying these diseases.
Acknowledgments
RA is supported by NIH grants R01NS096240, R01MH110556, and P50 DA044121. JFP is supported by HBHL/CFREF.
References
- 1.Brichta L and Greengard P (2014) Molecular determinants of selective dopaminergic vulnerability in Parkinson’s disease: an update. Front Neuroanat 8, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björklund A and Dunnett SB (2007) Dopamine neuron systems in the brain: an update. Trends Neurosci. 30, 194–202 [DOI] [PubMed] [Google Scholar]
- 3.Dahlstroem A and Fuxe K (1964) Evidence for the existence of monoamine-containing neurons in the central nervous system. i. demonstration of monoamines in the cell bodies of brain stem neurons. Acta Physiol Scand Suppl [PubMed] [Google Scholar]
- 4.Anderegg AM et al. (2015) Molecular heterogeneity of midbrain dopaminergic neurons--Moving toward single cell resolution. FEBS Lett. 589, 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brignani S and Pasterkamp RJ (2017) Neuronal Subset-Specific Migration and Axonal Wiring Mechanisms in the Developing Midbrain Dopamine System. Front Neuroanat 11, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm J et al. (2004) Molecular basis for catecholaminergic neuron diversity. Proc. Natl. Acad. Sci. U.S.A. 101, 13891–13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung CY et al. (2005) Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum. Mol. Genet. 14, 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene JG et al. (2005) Gene expression profiling of rat midbrain dopamine neurons: implications for selective vulnerability in parkinsonism. 18, 19–31 [DOI] [PubMed] [Google Scholar]
- 9.Brichta L et al. (2015) Identification of neurodegenerative factors using translatome-regulatory network analysis. Nat. Neurosci. 18, 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulin J-F et al. (2016) Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 11.Zeng H and Sanes JR (2017) Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546 [DOI] [PubMed] [Google Scholar]
- 12.Tasic B (2018) Single cell transcriptomics in neuroscience: cell classification and beyond. Curr. Opin. Neurobiol. 50, 242–249 [DOI] [PubMed] [Google Scholar]
- 13.Poulin J-F et al. (2014) Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep 9, 930–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Manno G et al. (2016) Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiklova K et al. (2019) Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat Commun 10, 581–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hook PW et al. (2018) Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease. Am. J. Hum. Genet. 102, 427–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer DJ et al. (2018) Combinatorial Expression of Grp and Neurod6 Defines Dopamine Neuron Populations with Distinct Projection Patterns and Disease Vulnerability. eNeuro 5, ENEURO.0152–18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders AB et al. (2018) Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lammel S et al. (2015) Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron 85, 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poulin J-F et al. (2018) Mapping projections of molecularly defined dopamine neuron subtypes using intersectional genetic approaches. Nat. Neurosci. 21, 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sgobio C et al. (2017) Aldehyde dehydrogenase 1-positive nigrostriatal dopaminergic fibers exhibit distinct projection pattern and dopamine release dynamics at mouse dorsal striatum. Sci Rep 7, 5283–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J et al. (2019) Distinct Connectivity and Functionality of Aldehyde Dehydrogenase 1a1-Positive Nigrostriatal Dopaminergic Neurons in Motor Learning. Cell Rep 28, 1167–1181.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan J et al. (2019) ALDH1A1 regulates postsynaptic μ-opioid receptor expression in dorsal striatal projection neurons and mitigates dyskinesia through transsynaptic retinoic acid signaling. Sci Rep 9, 3602–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J-I et al. (2015) Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons. Science 350, 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu G et al. (2014) Aldehyde dehydrogenase 1 defines and protects a nigrostriatal dopaminergic neuron subpopulation. J. Clin. Invest. 124, 3032–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panman L et al. (2014) Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell Rep 8, 1018–1025 [DOI] [PubMed] [Google Scholar]
- 27.Berrios J et al. (2016) Loss of UBE3A from TH-expressing neurons suppresses GABA co-release and enhances VTA-NAc optical self-stimulation. Nat Commun 7, 10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis AM et al. (2013) A unique population of ventral tegmental area neurons inhibits the lateral habenula to promote reward. Neuron 80, 1039–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tritsch NX et al. (2012) Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature 490, 262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tritsch NX et al. (2014) Midbrain dopamine neurons sustain inhibitory transmission using plasma membrane uptake of GABA, not synthesis. Elife 3, e01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Salvio M et al. (2010) Otx2 expression is restricted to dopaminergic neurons of the ventral tegmental area in the adult brain. Int. J. Dev. Biol. 54, 939–945 [DOI] [PubMed] [Google Scholar]
- 32.Di Salvio M et al. (2010) Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat. Neurosci. 13, 1481–1488 [DOI] [PubMed] [Google Scholar]
- 33.Ekstrand MI et al. (2014) Molecular profiling of neurons based on connectivity. Cell 157, 1230–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viereckel T et al. (2016) Midbrain Gene Screening Identifies a New Mesoaccumbal Glutamatergic Pathway and a Marker for Dopamine Cells Neuroprotected in Parkinson’s Disease. Sci Rep 6, 35203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bimpisidis Z et al. (2019) The NeuroD6 Subtype of VTA Neurons Contributes to Psychostimulant Sensitization and Behavioral Reinforcement. eNeuro 6, ENEURO.0066–19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan S et al. (2017) Survival of a Novel Subset of Midbrain Dopaminergic Neurons Projecting to the Lateral Septum Is Dependent on NeuroD Proteins. J. Neurosci. 37, 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lammel S et al. (2008) Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron 57, 760–773 [DOI] [PubMed] [Google Scholar]
- 38.Stuber GD et al. (2010) Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J. Neurosci. 30, 8229–8233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi T et al. (2011) Mesocorticolimbic glutamatergic pathway. J. Neurosci. 31, 8476–8490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendez JA et al. (2008) Developmental and target-dependent regulation of vesicular glutamate transporter expression by dopamine neurons. J. Neurosci. 28, 6309–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S et al. (2015) Dopaminergic and glutamatergic microdomains in a subset of rodent mesoaccumbens axons. Nat. Neurosci. 18, 386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortin GM et al. (2019) Segregation of dopamine and glutamate release sites in dopamine neuron axons: regulation by striatal target cells. FASEB J. 33, 400–417 [DOI] [PubMed] [Google Scholar]
- 43.Mingote S et al. (2015) Functional Connectome Analysis of Dopamine Neuron Glutamatergic Connections in Forebrain Regions. J. Neurosci. 35, 16259–16271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hnasko TS et al. (2010) Vesicular glutamate transport promotes dopamine storage and glutamate corelease in vivo. Neuron 65, 643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tecuapetla F et al. (2010) Glutamatergic signaling by mesolimbic dopamine neurons in the nucleus accumbens. J. Neurosci. 30, 7105–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dougalis AG et al. (2012) Functional properties of dopamine neurons and co-expression of vasoactive intestinal polypeptide in the dorsal raphe nucleus and ventrolateral periaqueductal grey. Eur. J. Neurosci. 36, 3322–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthews GA et al. (2016) Dorsal Raphe Dopamine Neurons Represent the Experience of Social Isolation. Cell 164, 617–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho JR et al. (2017) Dorsal Raphe Dopamine Neurons Modulate Arousal and Promote Wakefulness by Salient Stimuli. Neuron 94, 1205–1219.e8 [DOI] [PubMed] [Google Scholar]
- 49.Matsuda W et al. (2009) Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J. Neurosci. 29, 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prensa L and Parent A (2001) The nigrostriatal pathway in the rat: A single-axon study of the relationship between dorsal and ventral tier nigral neurons and the striosome/matrix striatal compartments. J. Neurosci. 21, 7247–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu J et al. (2006) Identification of wake-active dopaminergic neurons in the ventral periaqueductal gray matter. J. Neurosci. 26, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C et al. (2016) Mu Opioid Receptor Modulation of Dopamine Neurons in the Periaqueductal Gray/Dorsal Raphe: A Role in Regulation of Pain. Neuropsychopharmacology 41, 2122–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Groessl F et al. (2018) Dorsal tegmental dopamine neurons gate associative learning of fear. Nat. Neurosci. 21, 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuhma N et al. (2018) Dopamine neuron glutamate cotransmission evokes a delayed excitation in lateral dorsal striatal cholinergic interneurons. Elife 7, 9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menegas W et al. (2015) Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. Elife 4, e10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menegas W et al. (2018) Dopamine neurons projecting to the posterior striatum reinforce avoidance of threatening stimuli. Nat. Neurosci. 21, 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto M and Hikosaka O (2009) Two types of dopamine neuron distinctly convey positive and negative motivational signals. Nature 459, 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steinkellner T et al. (2018) Role for VGLUT2 in selective vulnerability of midbrain dopamine neurons. J. Clin. Invest. 128, 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen H et al. (2018) Genetic deletion of vesicular glutamate transporter in dopamine neurons increases vulnerability to MPTP-induced neurotoxicity in mice. Proc. Natl. Acad. Sci. U.S.A. 115, E11532–E11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi T et al. (2015) Glutamatergic and dopaminergic neurons in the mouse ventral tegmental area. Eur. J. Neurosci. 41, 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang H et al. (2018) Nucleus Accumbens Subnuclei Regulate Motivated Behavior via Direct Inhibition and Disinhibition of VTA Dopamine Subpopulations. Neuron 97, 434–449.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mingote S et al. (2019) Dopamine-glutamate neuron projections to the nucleus accumbens medial shell and behavioral switching. Neurochem. Int. 129, 104482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papathanou M et al. (2018) Targeting VGLUT2 in Mature Dopamine Neurons Decreases Mesoaccumbal Glutamatergic Transmission and Identifies a Role for Glutamate Co-release in Synaptic Plasticity by Increasing Baseline AMPA/NMDA Ratio. Front Neural Circuits 12, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabanova A et al. (2015) Function and developmental origin of a mesocortical inhibitory circuit. Nat. Neurosci. 18, 872–882 [DOI] [PubMed] [Google Scholar]
- 65.Lutas A et al. (2019) State-specific gating of salient cues by midbrain dopaminergic input to basal amygdala. Nat. Neurosci. 22, 1820–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silm K et al. (2019) Synaptic Vesicle Recycling Pathway Determines Neurotransmitter Content and Release Properties. Neuron 102, 786–800.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joksimovic M et al. (2009) Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc. Natl. Acad. Sci. U.S.A. 106, 19185–19190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joksimovic M et al. (2009) Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat. Neurosci. 12, 125–131 [DOI] [PubMed] [Google Scholar]
- 69.Kittappa R et al. (2007) The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 5, e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blaess S et al. (2011) Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andersson E et al. (2006) Identification of intrinsic determinants of midbrain dopamine neurons. Cell 124, 393–405 [DOI] [PubMed] [Google Scholar]
- 72.Metzakopian E et al. (2012) Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors. Development 139, 2625–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bonilla S et al. (2008) Identification of midbrain floor plate radial glia-like cells as dopaminergic progenitors. Glia 56, 809–820 [DOI] [PubMed] [Google Scholar]
- 74.Brodski C et al. (2003) Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain-hindbrain organizer. J. Neurosci. 23, 4199–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nouri N and Awatramani RB (2017) A novel floor plate boundary defined by adjacent En1 and Dbx1 microdomains distinguishes midbrain dopamine and hypothalamic neurons. Development 144, 916–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kee N et al. (2017) Single-Cell Analysis Reveals a Close Relationship between Differentiating Dopamine and Subthalamic Nucleus Neuronal Lineages. Cell Stem Cell 20, 29–40 [DOI] [PubMed] [Google Scholar]
- 77.Lahti L et al. (2012) Cell-autonomous FGF signaling regulates anteroposterior patterning and neuronal differentiation in the mesodiencephalic dopaminergic progenitor domain. Development 139, 894–905 [DOI] [PubMed] [Google Scholar]
- 78.Smits SM et al. (2013) Molecular marker differences relate to developmental position and subsets of mesodiencephalic dopaminergic neurons. PLoS ONE 8, e76037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan CH et al. (2011) Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J. Neurosci. 31, 12413–12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakatani T et al. (2010) Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev. Biol. 339, 101–113 [DOI] [PubMed] [Google Scholar]
- 81.Anderegg AM et al. (2013) An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 9, e1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bodea GO and Blaess S (2015) Establishing diversity in the dopaminergic system. FEBS Lett. 589, 3773–3785 [DOI] [PubMed] [Google Scholar]
- 83.Brown A et al. (2011) Molecular organization and timing of Wnt1 expression define cohorts of midbrain dopamine neuron progenitors in vivo. J. Comp. Neurol. 519, 2978–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wallén A et al. (1999) Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp. Cell Res. 253, 737–746 [DOI] [PubMed] [Google Scholar]
- 85.Ono Y et al. (2007) Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 134, 3213–3225 [DOI] [PubMed] [Google Scholar]
- 86.Bodea GO et al. (2014) Reelin and CXCL12 regulate distinct migratory behaviors during the development of the dopaminergic system. Development 141, 661–673 [DOI] [PubMed] [Google Scholar]
- 87.Hayes L et al. (2011) Timing of Sonic hedgehog and Gli1 expression segregates midbrain dopamine neurons. J. Comp. Neurol. 519, 3001–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blaess S and Ang S-L (2015) Genetic control of midbrain dopaminergic neuron development. Wiley Interdiscip Rev Dev Biol 4, 113–134 [DOI] [PubMed] [Google Scholar]
- 89.Evans RC et al. (2017) Dopamine Inhibition Differentially Controls Excitability of Substantia Nigra Dopamine Neuron Subpopulations through T-Type Calcium Channels. J. Neurosci. 37, 3704–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farassat N et al. (2019) In vivo functional diversity of midbrain dopamine neurons within identified axonal projections. Elife 8, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lerner TN et al. (2015) Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell 162, 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beier KT et al. (2015) Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell 162, 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Howe MW and Dombeck DA (2016) Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature 535, 505–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lammel S et al. (2012) Input-specific control of reward and aversion in the ventral tegmental area. Nature 491, 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fenno LE et al. (2014) Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11, 763–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ray RS et al. (2011) Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science 333, 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Plummer NW et al. (2015) Expanding the power of recombinase-based labeling to uncover cellular diversity. Development 142, 4385–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Madisen L et al. (2015) Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim D-W et al. (2019) Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior. Cell 179, 713–728.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen G et al. (2019) Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front Genet 10, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haque A et al. (2017) A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med 9, 75–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Codeluppi S et al. (2018) Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods 15, 932–935 [DOI] [PubMed] [Google Scholar]
- 103.Wang X et al. (2018) Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Islam S et al. (2014) Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 [DOI] [PubMed] [Google Scholar]
- 105.Morales M and Margolis EB (2017) Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nat. Rev. Neurosci. 18, 73–85 [DOI] [PubMed] [Google Scholar]
- 106.Roeper J (2013) Dissecting the diversity of midbrain dopamine neurons. Trends Neurosci. 36, 336–342 [DOI] [PubMed] [Google Scholar]
- 107.Lammel S et al. (2014) Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology 76, 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trudeau L-E et al. (2014) The multilingual nature of dopamine neurons. Prog. Brain Res. 211, 141–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pinoli M et al. (2017) Dopaminergic Regulation of Innate Immunity: a Review. J Neuroimmune Pharmacol 12, 602–623 [DOI] [PubMed] [Google Scholar]
- 110.Buttarelli FR et al. (2011) The dopaminergic system in peripheral blood lymphocytes: from physiology to pharmacology and potential applications to neuropsychiatric disorders. Curr Neuropharmacol 9, 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pristerà A et al. (2015) Transcription factors FOXA1 and FOXA2 maintain dopaminergic neuronal properties and control feeding behavior in adult mice. Proc. Natl. Acad. Sci. U.S.A. 112, E4929–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Doucet-Beaupré H et al. (2016) Lmx1a and Lmx1b regulate mitochondrial functions and survival of adult midbrain dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 113, E4387–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wever I et al. (2019) Lmx1b Influences Correct Post-mitotic Coding of Mesodiencephalic Dopaminergic Neurons. Front Mol Neurosci 12, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Laguna A et al. (2015) Dopaminergic control of autophagic-lysosomal function implicates Lmx1b in Parkinson’s disease. Nat. Neurosci. 18, 826–835 [DOI] [PubMed] [Google Scholar]
- 115.Andersson EKI et al. (2007) Ngn2 and Nurr1 act in synergy to induce midbrain dopaminergic neurons from expanded neural stem and progenitor cells. Exp. Cell Res. 313, 1172–1180 [DOI] [PubMed] [Google Scholar]
- 116.Stott SRW et al. (2013) Foxa1 and foxa2 are required for the maintenance of dopaminergic properties in ventral midbrain neurons at late embryonic stages. J. Neurosci. 33, 8022–8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Romanov RA et al. (2017) Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smits SM et al. (2005) Molecular and cellular alterations in the Pitx3-deficient midbrain dopaminergic system. Mol. Cell. Neurosci. 30, 352–363 [DOI] [PubMed] [Google Scholar]
- 119.Veenvliet JV et al. (2013) Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 140, 3373–3384 [DOI] [PubMed] [Google Scholar]
- 120.Lindeberg J et al. (2004) Transgenic expression of Cre recombinase from the tyrosine hydroxylase locus. Genesis 40, 67–73 [DOI] [PubMed] [Google Scholar]
- 121.Soden ME et al. (2016) Genetic Isolation of Hypothalamic Neurons that Regulate Context-Specific Male Social Behavior. Cell Rep 16, 304–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bäckman CM et al. (2006) Characterization of a mouse strain expressing Cre recombinase from the 3’ untranslated region of the dopamine transporter locus. Genesis 44, 383–390 [DOI] [PubMed] [Google Scholar]
- 123.Zhuang X et al. (2005) Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J. Neurosci. Methods 143, 27–32 [DOI] [PubMed] [Google Scholar]
- 124.Awatramani RB et al. (2003) Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat. Genet. 35, 70–75 [DOI] [PubMed] [Google Scholar]
- 125.Jensen P and Dymecki SM (2014) Essentials of recombinase-based genetic fate mapping in mice. Methods Mol. Biol. 1092, 437–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen L et al. (2015) A53T human α-synuclein overexpression in transgenic mice induces pervasive mitochondria macroautophagy defects preceding dopamine neuron degeneration. J. Neurosci. 35, 890–905 [DOI] [PMC free article] [PubMed] [Google Scholar]