Abstract
Snake species within the Bothrops complex (sensu lato) are of medical relevance in Latin America, but knowledge on their venom characteristics is limited, or even unavailable, for some taxa. Perú harbors 17 species of pit vipers, within the genera Bothrops, Bothriechis, Bothrocophias, Porthidium, Crotalus, and Lachesis. This study compared the venoms of twelve species, through chromatographic and electrophoretic profiles, as well as proteolytic and phospholipase A2 (PLA2) activities. Also, proteomic profiles were analyzed for nine of the venoms using a shotgun approach. Results unveiled conspicuous differences in the expression of venom PLA2s among species, six of them presenting scarce levels as judged by RP-HPLC profiles. Since most species within the bothropoid lineage possess venoms with high to intermediate abundances of this protein family, our findings suggest the existence of a phenotypic duality in the expression of venom PLA2s within the Bothrops (sensu lato) complex. Bothrops barnetti and Bothrocophias andianus venoms, very scarce in PLA2s, were shown to lack significant myotoxic activity, highlighting that the observed variability in PLA2 expression bears toxicological correlations with effects attributed to these proteins. Finally, an attempt to identify phylogenetic relationships of bothropoid species from Perú presenting low- or high-PLA2 venom phenotypes showed an interspersed pattern, thus precluding a simple phylogenetic interpretation of this venom compositional dichotomy.
Keywords: Snake venom, Viperidae, Phospholipase A2, Bothropoid, Phenotype, Dichotomy
Graphical abstract
Highlights
-
•
Venoms from 12 viperids of Perú were compared.
-
•
Conspicuous differences in the expression of PLA2 were found.
-
•
Venoms presenting scarce levels of PLA2 lack myotoxicity.
-
•
A new phenotypic dichotomy in venom PLA2 expression is described within Bothrops (sensu lato).
1. Introduction
In the Americas, an average of 6.2 snakebites per 100,000 population (about 57,500 accidents) is estimated to occur annually, leading to a fatality rate of 0.04 per 100,000 population (Chippaux, 2017). The genus Bothrops (lanceheads), distributed from Mexico to southern Argentina, is responsible for the majority of envenomings in Latin America (Chippaux, 2017). Classified within the Viperidae family, this genus includes 45 species listed in the Reptile Database (http://www.reptile-database.org) and has been the subject of dynamic taxonomic reclassifications within the bothropoid lineages in the last two decades (Wüster et al., 2002, Fenwick et al., 2009, Carrasco et al., 2012, Carrasco et al., 2016, Alencar et al., 2016). Despite the medical relevance of species within the Bothrops complex (sensu lato), knowledge on the characteristics of their venoms is still limited, or unavailable, for some species.
The growing body of information on the composition of viperid venoms, as provided by “omics” approaches, has revealed the existence of a limited number of protein families that are generally shared among conspecific species, with both qualitative and quantitative variations (Calvete, 2013, Calvete, 2017, Lomonte et al., 2014, Göçmen et al., 2015, Tan et al., 2015, Viala et al., 2015, Serrano et al., 2018). Knowledge of this variability is essential for the production of broad-coverage therapeutic antivenoms to confront snakebite envenomings. Also, the study of inter- and intraspecific venom variability may shed light on understanding the evolutionary pathways that have shaped the present-day landscape of the snake venom toxin arsenals.
Perú harbors a rich herpetofauna, which includes 34 species or subspecies of venomous snakes distributed in two families, Elapidae and Viperidae. Sixteen coral snakes represent the former, all included in the genus Micrurus of subfamily Elapinae, and a single species of sea snake, Hydrophis platurus within subfamily Hydrophiinae. Viperidae includes 17 pitviper species (Carrillo de Espinoza and Icochea, 1995) within the genera Bothrops, Bothriechis, Bothrocophias, Porthidium, Crotalus, and Lachesis (taxonomical nomenclature in the present work follows the proposal of Carrasco et al., [2016] for the Bothrops complex, sensu lato). Viperidae species are responsible for the vast majority of the estimated 1500–2500 snakebites/year recorded in Perú (Ministerio de Salud, 2017), resulting in a mortality rate of not less than 0.043/100,000 population (Chippaux, 2017).
Previous studies have examined various biochemical aspects and toxic activities of the venoms of Viperidae snakes inhabiting Perú (Sanz et al., 2008, Núñez et al., 2009, Jimenez et al., 2010, Kohlhoff et al., 2012, Guerra-Duarte et al., 2015, Rodrigues et al., 2018), and their neutralization (Laing et al., 2004, Rojas et al., 2005, García et al., 2008, Estevao-Costa et al., 2016). However, information on most venoms of these species is still partial, and more efforts are needed to gain a broader knowledge of their general properties. In the present study, we have comparatively explored the chromatographic and electrophoretic profiles of the venoms of twelve species of pitvipers, as well as their proteolytic and phospholipase A2 (PLA2) activities. Besides, proteomic profiles were analyzed for nine of the venoms using a shotgun approach. These comparative analyses revealed important differences in the expression of PLA2s among the species, which relate to variations in toxicological activities attributed to these enzymes and their related PLA2-like homologs.
2. Materials and methods
2.1. Venoms
Venom pools were obtained from adult specimens collected in Perú, from the species and locations indicated in Table 1. In some comparative experiments, venoms of species from other regions of Latin America were included: Bothrops alternatus (Coronel Pringles, Argentina), B. neuwiedii (Tacuarembó, Uruguay), B. asper (Pacific versant, Costa Rica), and Bothriechis schlegelii (Costa Rica).
Table 1.
Venoms from Viperidae species of Perú analyzed in the present study, and their localities.
| Speciesa | Common name | nb | District | Province | Department |
|---|---|---|---|---|---|
| Bothrops atrox | jergón de la selva | 60 | Llata, Chaglia, Puerto Inca, Codo del Pozuzo | Humalíes, Pachitea, Puerto Inca | Huánuco |
| Bothrops barnetti | macanche | 20 | Quebrada Honda, Olmos | Talara, Lambayeque | Piura, Lambayeque |
| Bothrops bilineatus | loro machaco | 10 | Pichanaki, Vitoc, Chanchamayo | Chanchamayo | Junin |
| Bothrops taeniata (castelnaudi) | jergón de árbol | nd | Satipo | Jauja | Junin |
| Bothrops chloromelas | – | 8 | Huancabamba | Huancabamba | Piura |
| Bothrops oligolepis (peruvianus) | jergón negro | 8 | Satipo | Jauja | Junin |
| Bothrops pictus | jergón de la costa | 5 | Pachacamac | Lima | Lima |
| Bothrocophias andianus | jergona | 4 | Macchu Picchu | Urubamba | Cusco |
| Bothrocophias hyoprora | jergón shushupe | 10 | Puerto Pakuy, Imaza | Bagua | Amazonas |
| Bothrocophias microphtalmus | jergón pudridora | nd | Pichanaki | Chanchamayo | Junin |
| Bothriechis schlegelii | loro | 4 | Reserva de Tumbes | Zarumilla | Tumbes |
| Lachesis muta | shushupe | 10 | Alto Marañón, Nazareth, Pijuayal | Bagua, Condorcanqui | Amazonas |
Taxonomical nomenclature used in this work follows the proposal of Carrasco et al. (2016), which synonimizes Rhinocerophis, Bothropoides, and Bothriopsis with Bothrops, while maintaining Bothrocophias and assigning Bothrops andianus to that genus.
n: number of individuals in the venom pool; nd: not determined.
2.2. SDS-PAGE
Venom proteins were separated by SDS-PAGE using pre-cast gradient gels (4–20%; Bio-Rad), either unreduced or after reduction with 2-mercaptoethanol (5 min at 95 °C). Samples (30 μg) were separated at 150 V, along with molecular weight standards (Bio-Rad). Proteins were visualized by Coomassie blue R-250 staining and electrophoretic patterns were recorded with the ImageLab™ software (Bio-Rad).
2.3. Reverse phase-HPLC profiling
Chromatographic profiles of venoms were obtained by RP-HPLC using a C18 column (250 × 4.6 mm, 5 μm particle diameter; Phenomenex) in a model 1220 instrument (Agilent) monitored at 215 nm. Venoms (1.5–2.0 mg) were dissolved in purified water (18 MΩ/cm) containing 0.1% trifluoroacetic acid (TFA; solution A), injected, and eluted with a gradient toward acetonitrile with 0.1% TFA (solution B), at a flow rate of 1 mL/min, for a total time of 94 min: 0% B for 5 min, 0–15% B in 10 min, 15–45% B in 60 min, 45–70% B in 10 min, and 70% B for 9 min (Lomonte and Calvete, 2017).
2.4. Phospholipase A2 activity
Phospholipase A2 (PLA2) activity of the venoms was assayed using the synthetic substrate 4-nitro-3-octanoyloxy-benzoic acid (NOBA), as described (Mora-Obando et al., 2014a). Various amounts of venoms (5, 10, or 20 μg), in 25 μL, were added to 25 μL of NOBA (1 mg/mL acetonitrile) and 200 μL of 10 mM Tris, 10 mM CaCl2, 0.1 M NaCl (pH 8.0), in 96-well microplates. Blanks in which venom was replaced with buffer were included. The mixtures were incubated at 37 °C for 60 min, and final absorbances at 450 nm were measured in a microplate reader (Multiskan FC, Thermo), using three replicates for each venom concentration.
2.5. Proteolytic activity
Proteolytic activity of the venoms was determined using azocasein (10 mg/mL in 50 mM Tris-HCl, 0.15 M NaCl, 5 mM CaCl2, pH 8.0) as substrate, as previously described (Jiménez-Charris et al., 2015). Various amounts of venoms (5, 10, or 20 μg) were incubated with azocasein at 37 °C for 90 min, in a total volume of 100 μL, and the reaction was stopped by adding 200 μL of 5% trichloroacetic acid (TCA). Blanks in which venom was omitted were included. After centrifugation of the microplates, 150 μL of the supernatant was transferred to clean wells, and 150 μL of 0.5 M NaOH was added for color development. Absorbances were measured at 450 nm in a microplate reader (Multiskan FC, Thermo), using three replicates for each venom concentration.
2.6. Proteomic profiling
Venoms from nine species were analyzed by a bottom-up shotgun proteomic approach. Samples of 15 μg were reduced with 10 mM dithiothreitol for 30 min at 56 °C, alkylated with 50 mM iodoacetamide for 20 min in the dark, and digested with sequencing grade trypsin at 37 °C overnight, in a total volume of 40 μL. After the addition of 0.5 μL of formic acid, the resulting tryptic peptide mixtures were centrifuged and separated by RP-HPLC on a nano-Easy 1200 chromatograph (Thermo) on-line with a Q-Exactive Plus® mass spectrometer (Thermo). Twelve μL of peptide mixture, containing 0.7 μg, were loaded on a C18 trap column (75 μm × 2 cm, 3 μm particle; PepMap, Thermo), washed with 0.1% formic acid (solution A), and separated at 200 nL/min on a C18 Easyspray® column (75 μm × 15 cm, 3 μm particle; PepMap, Thermo). A gradient toward solution B (80% acetonitrile, 0.1% formic acid) was developed in a total of 120 min (1–5% B in 1 min, 5–26% B in 84 min, 26–80% B in 30min, 80–99% B in 1 min, and 99% B in 4 min). MS spectra were acquired in positive mode at 2.0 kV, with a capillary temperature of 200 °C, using 1 μscan in the range 400–1600 m/z, maximum injection time of 50 msec, AGC target of 1 × 106, and resolution of 70,000. The top 10 ions with 2–5 positive charges were fragmented with AGC target of 3 × 106, minimum AGC 2 × 103, maximum injection time 110 msec, dynamic exclusion time 5 s, and resolution 17,500. MS/MS spectra were processed against protein sequences contained in the UniProt/SwissProt database (Serpentes, October 2019) using Peaks X® (Bioinformatics Solutions) and matches were assigned to known protein families by similarity. Cysteine carbamidomethylation was set as a fixed modification, while deamidation of asparagine or glutamine and methionine oxidation were set as variable modifications, allowing up to 3 missed cleavages by trypsin. Parameters for match acceptance were set to FDR<0.1%, detection of at least one unique peptide, and −10lgP protein score ≥100.
2.7. Myotoxicity assay
The myotoxic effect of the venoms of Bothrops barnetti and Bothrocophias andianus (selected as examples of venoms nearly devoid of PLA2s) was tested in mice. The venom of B. asper from the Pacific versant of Costa Rica, known to be abundant in PLA2s (Alape-Girón et al., 2008), was included in this assay as a positive control. Groups of five CD-1 mice (18–20 g body weight) received an intramuscular venom injection (50 μg/50 μL) in the gastrocnemius. After 3 h, blood was obtained from the tip of the tail into heparinized capillary tubes, centrifuged, and plasma (4 μL) was assayed for creatine kinase (CK) activity using a kinetic commercial kit (CK-Nac, Biocon Diagnostik). A control group of mice received a similar injection of phosphate-buffered saline (PBS; pH 7.2) instead of venom. This experiment followed the ethical guidelines of the Institutional Committee for the Use and Care of Animals (CICUA) of the University of Costa Rica (No. 021–17). Statistical significance of the differences between groups was determined by ANOVA, followed by Tukey-Kramer post-hoc test, using the InStat v.3 (GraphPad) software.
3. Results
3.1. Electrophoresis
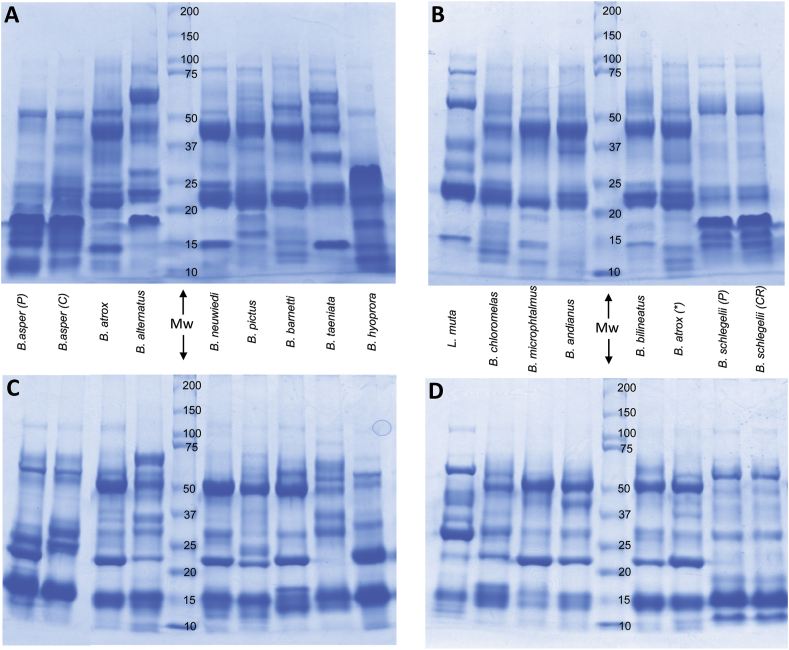
Venom SDS-PAGE patterns, under non-reducing and reducing conditions, are shown in Fig. 1. Although none of the inter-species patterns are entirely identical, significant resemblances are evident between several venoms, for example, Bothrops atrox, B. barnetti, B. pictus, and B. neuwiedii, especially after disulfide bond reduction. More differences appear in the unreduced electrophoretic patterns than in reduced ones, suggesting inter-species variations in the oligomeric association of subunits of similar molecular mass. Notably, the electrophoretic pattern of Bothriechis schlegelii venom from Perú appears indistinguishable from that of the same species originating from Costa Rica, under both conditions (Fig. 1). All major venom proteins distributed below ~100 kDa. The most prominent reduced protein bands migrated at ~50, ~23, and ~15 kDa.
Fig. 1.
SDS-PAGE profiles of venoms from twelve viperid species of Perú, unreduced (A, B) or reduced (C, D). Venoms of some species from other locations are included: Bothrops asper from the Pacific (P) or Caribbean (C) regions of Costa Rica, B. alternatus from Argentina, B. neuwiedi from Uruguay, and B. schlegelii from Costa Rica (CR). An additional sample of B. atrox venom from Perú is indicated with an asterisk. Molecular weight (Mw) markers are indicated in kDa. Proteins were stained with Coomassie brilliant blue G-250. The venom of B. oligolepis could not be analyzed in this technique due to insufficient amount. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
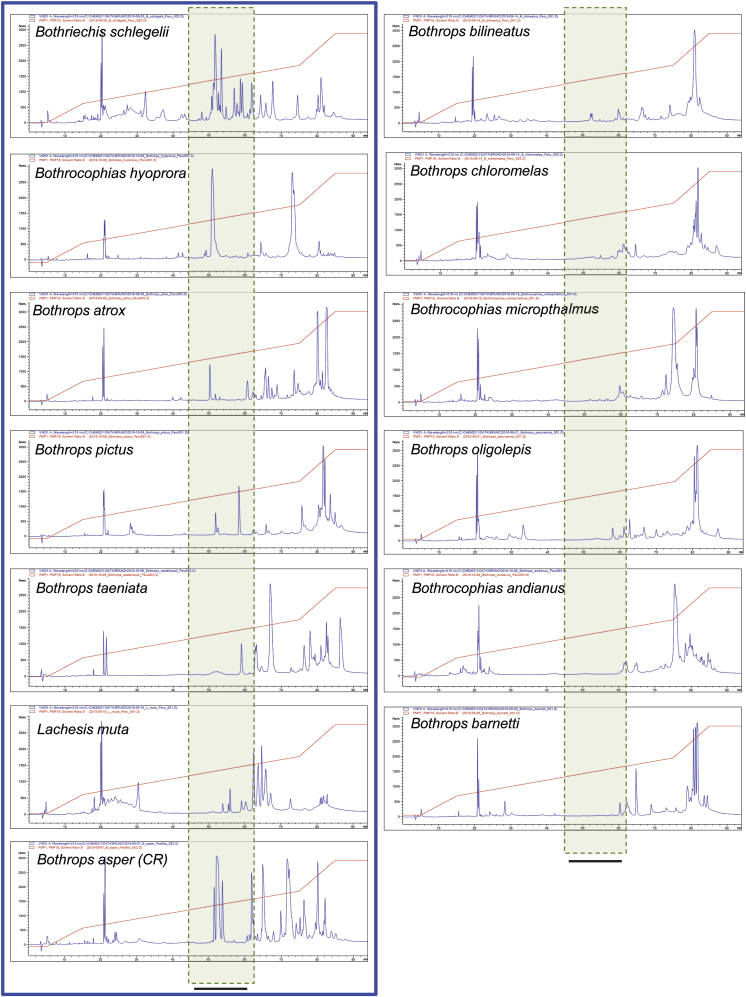
3.2. RP-HPLC profiles
Venoms were analyzed using a previously standardized C18 RP-HPLC separation protocol (Lomonte and Calvete, 2017). Its application in several proteomic studies on snake venoms has shown that monomeric PLA2s and PLA2-like homologs elute within the range indicated with a shadowed dotted box in Fig. 2. Venoms varied largely regarding the presence of chromatographic peaks within this region, and can be separated into two types, namely those presenting conspicuous peaks (blue frame in Fig. 2) and those having barely detectable peaks. The venoms of Bothriechis schlegelii and Bothrocophias hyoprora showed the most intense peaks in this region, comparable to those of Bothrops asper venom from Costa Rica, used as a reference for the elution of PLA2s (Fig. 2).
Fig. 2.
C18 reverse-phase HPLC profiles of venoms from twelve viperid species of Perú. The venom of Bothrops asper from Costa Rica was included as a reference. The dotted light green shaded area indicates the known elution region for PLA2/PLA2-like proteins in viperid venoms in this standardized chromatographic gradient. Venoms were divided into two groups, those with evident peaks in such region (enclosed within the blue frame) and those with very minor peaks. For reference to colors, the reader is referred to the web version of this article. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
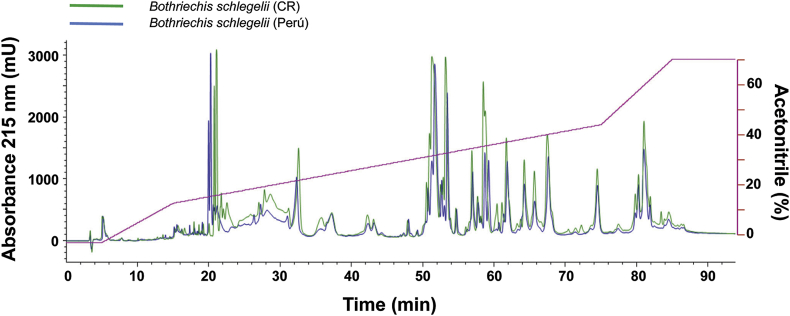
Since electrophoretic analysis showed a very high similarity of Bothriechis schlegelii venoms collected in Perú and Costa Rica (Fig. 1), their RP-HPLC profiles were determined and compared in Fig. 3. As shown, the two profiles are virtually superimposable, confirming their remarkable high similarity.
Fig. 3.
Superposition of the C18 reverse-phase HPLC profiles of the venoms of Bothriechis schlegelii from Costa Rica (CR; green trace) and Perú (blue trace). For reference to colors, the reader is referred to the web version of this article. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
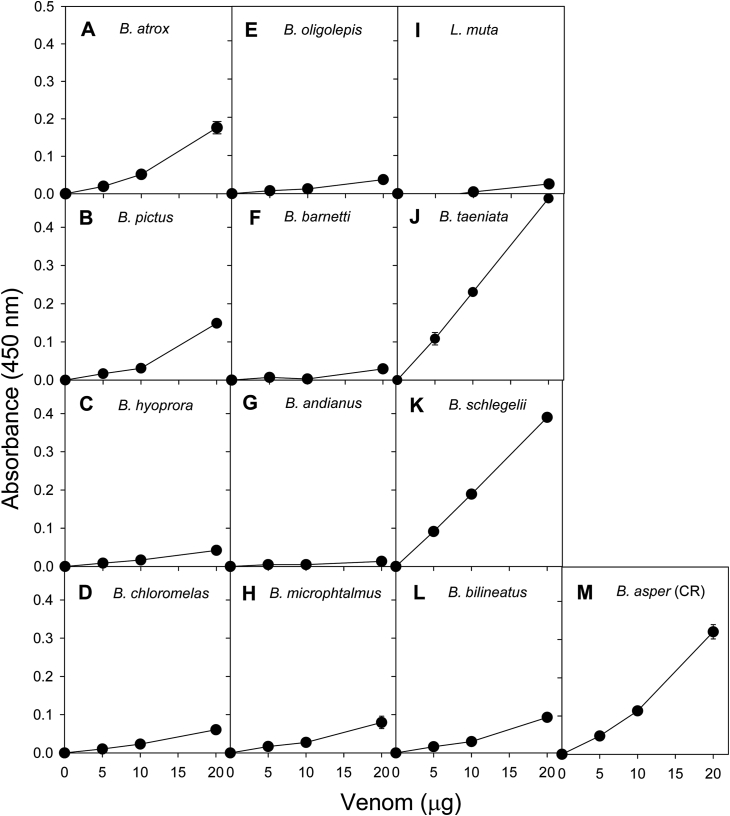
3.3. Phospholipase A2 and proteolytic activities
PLA2 activity of the venoms on the NOBA substrate was highly variable, as presented in Fig. 4. This activity was strikingly low in the case of Bothrops oligolepis, B. barnetti, Bothrocophias andianus, Bothrocophias hyoprora, and Lachesis muta venoms. In contrast, Bothrops atrox, B. pictus, B. taeniata, and Bothriechis schlegelii venoms were highly active in this assay, while low to intermediate activities were recorded for Bothrops chloromelas, B. bilineatus, and Bothrocophias microphtalmus venoms.
Fig. 4.
Phospholipase A2 activity of venoms from twelve viperid species of Perú on 4-nitro-3-octanoyloxy-benzoic acid. The venom of Bothrops asper from Costa Rica was included as a reference. Each point represents the mean ± SD of three replicates.
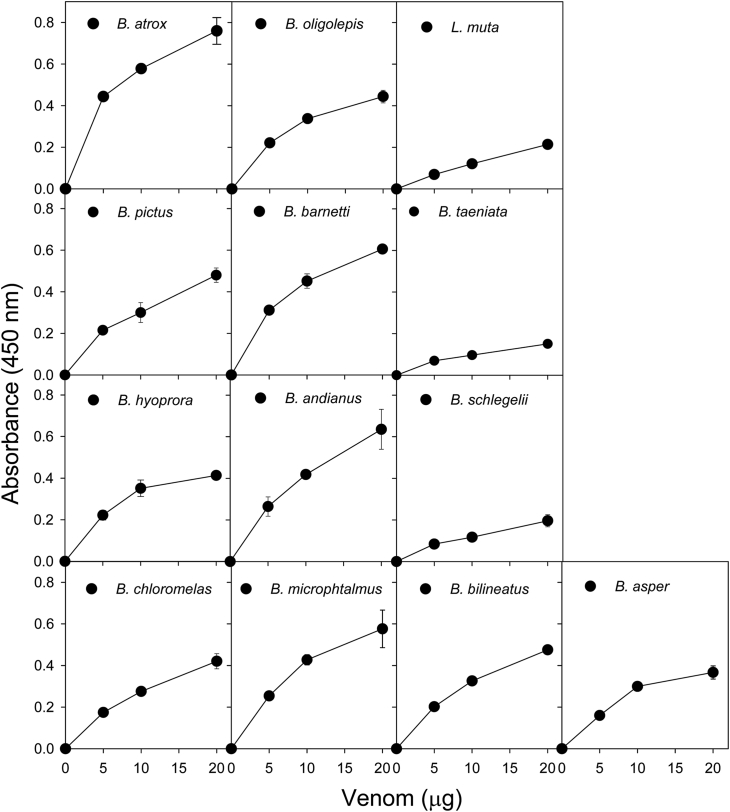
On the other hand, proteolytic activity on the azocasein substrate was more similar among the different venoms (Fig. 5), but Bothriechis schlegelii, Bothrops taeniata, and Lachesis muta venoms showed a slightly lower proteolytic activity than the rest.
Fig. 5.
Proteolytic activity of venoms from twelve viperid species of Perú on azocasein. The venom of Bothrops asper from Costa Rica was included as a reference. Each point represents the mean ± SD of three replicates.
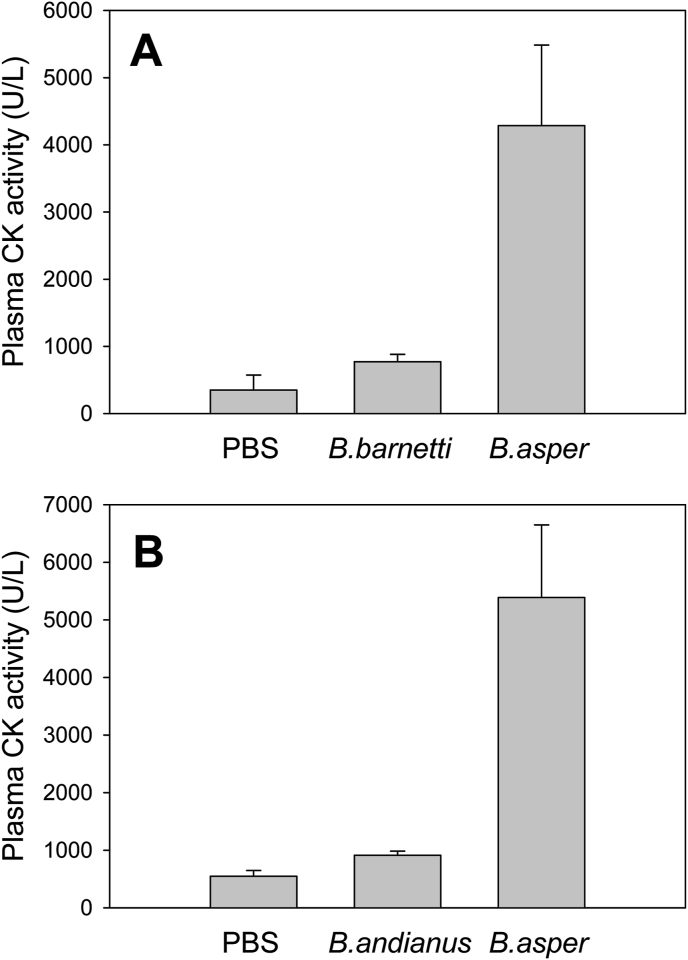
3.4. Myotoxicity
The venoms of Bothrops barnetti and Bothrocophias andianus were selected as examples of venoms virtually devoid of PLA2s in the RP-HPLC profiles, to test whether this phenotypic trait would correlate with toxic activities that are known to depend mainly on these proteins, such as in vivo myotoxicity. As shown in Fig. 6, intramuscular injection of these two venoms in mice did not induce a significant increment of plasma CK activity, in contrast to a venom (Bothrops asper, used as a reference) known to contain abundant PLA2 (Asp49) and PLA2-like (Lys49) myotoxins.
Fig. 6.
Comparative evaluation of the myotoxic activity of the venoms of (A)Bothrops barnetti and (B)Bothrocophias andianus (Perú), compared to Bothrops asper (Costa Rica) in mice. Venoms were injected by the intramuscular route into groups of five mice. A control group received an injection of vehicle (PBS) alone. After 3 h the plasma creatine kinase (CK) activity was determined. Each bar represents the mean ± SD (n = 5). No statistical difference (p > 0.05) was observed between CK levels of PBS and B. barnetti venom, or PBS and B. andianus venom. B. asper venom was significantly different from both groups, in both independent experiments.
3.5. Shotgun proteomic profiling
Nine of the twelve venoms here studied were subjected to a bottom-up shotgun proteomic profiling. The complete dataset of matches with known proteins in the Uniprot “Serpentes” database is presented in Supplemental Table S1, and the different protein families identified in each venom are summarized in Table 2. Venom protein families detected by this qualitative approach ranged from 9 (Bothrops barnetti) to 13 (B. atrox, B. oligolepis, B. chloromelas). The most variable protein family detected corresponded to hyaluronidase (HYA), found in three out nine venoms, and vascular endothelial growth factor (VEGF), found in five out of nine (Table 2). Nerve growth factor (NGF) was present in seven venoms, while phospholipase B (PLB) phosphodiesterase (PDE) and cysteine-rich secretory proteins (CRISP) were found in eight. Other major protein families commonly found in viperid venoms were detected in all nine species.
Table 2.
Protein families identified in nine venoms from Viperidae species of Perú by bottom-up shotgun proteomics.
| Snake species | MP* | SP | PLA2 | LAAO | CTL | NUC | GCY | CRiSP | PDE | PLB | NGF | VEGF | HYA | Total N° |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bothrops atrox | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 13 |
| Bothrops oligolepis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 13 |
| Bothrops chloromelas | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 13 |
| Bothrocophias andianus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | (−) | 12 |
| Bothrops pictus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | (−) | ✓ | ✓ | ✓ | ✓ | (−) | 11 |
| Bothrocophias microphtalmus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | (−) | (−) | 11 |
| Bothrocophias hyoprora | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | (−) | ✓ | (−) | (−) | 10 |
| Bothrops bilineatus | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | (−) | (−) | (−) | 10 |
| Bothrops barnetti | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | (−) | ✓ | (−) | (−) | (−) | 9 |
| Family presence | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 89% | 89% | 89% | 77% | 56% | 33% |
* Abbreviations: MP: metalloproteinase; SP: serine proteinase; PLA2: phospholipase A2; LAAO: L-amino acid oxidase; VEGF: vascular endothelial growth factor; NGF: nerve growth factor; CTL: C-type lectin/lectin-like; NUC: nucleotidase; PDE: phosphodiesterase; GCY: Glutaminyl cyclotransferase; PLB: phospholipase B; CRiSP: cysteine-rich secretory protein; HYA: hyaluronidase. For a detailed summary of protein matches refer to Supplemental Table S1.
√: detected; (−) not detected.
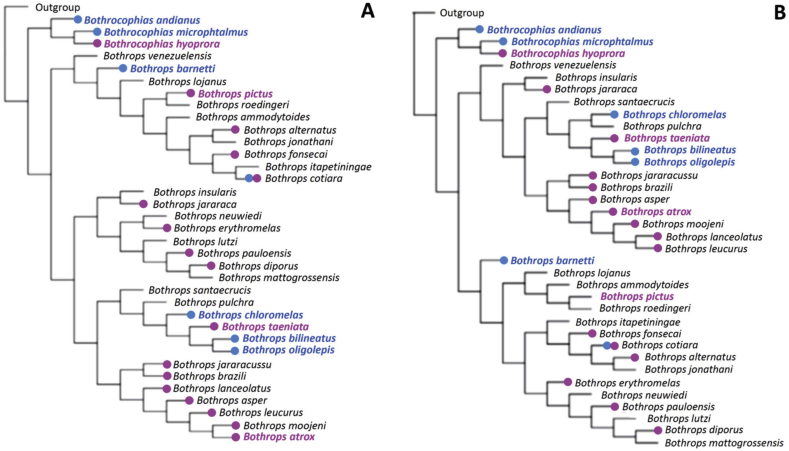
3.6. Relationship of the phospholipase A2 low/high phenotypes to phylogeny
Differences in the expression of PLA2s in viperids of the bothropoid lineage were mapped onto the partial phylogeny described by Carrasco et al. (2016) in Fig. 7, using results of the present study together with data from the literature (evaluated either by proteomic analyses or by reported isolation of PLA2s). The obtained cladogram shows that the majority of bothropoid venoms studied to date typically present PLA2s (indicated with pink dots). One notable exception is Bothrops cotiara from Brazil, reported lacking these enzymes (Tashima et al., 2008). However, B. cotiara has also been reported to have PLA2s as the most abundant venom components in the case of specimens collected in Argentina (de Roodt et al., 2018), and was therefore indicated with both pink and blue symbols in Fig. 7.
Fig. 7.
Partial phylogenies of bothropoid viperids showing the relationships of species reported to produce venoms with high (pink dots) and low (blue dots) phospholipase A2 contents. The species from Perú analyzed in the present study are indicated by names in corresponding pink or blue colors. The phylogenetic trees were generated by Carrasco et al. (2016) using a total evidence approach from morphological, ecological, and mtDNA sequences (12 S and 16 S rRNA). Phylogenetic reconstruction included (A) or excluded (B) sequences from ND4 and cytb genes. For reference to colors, the reader is referred to the web version of this article. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
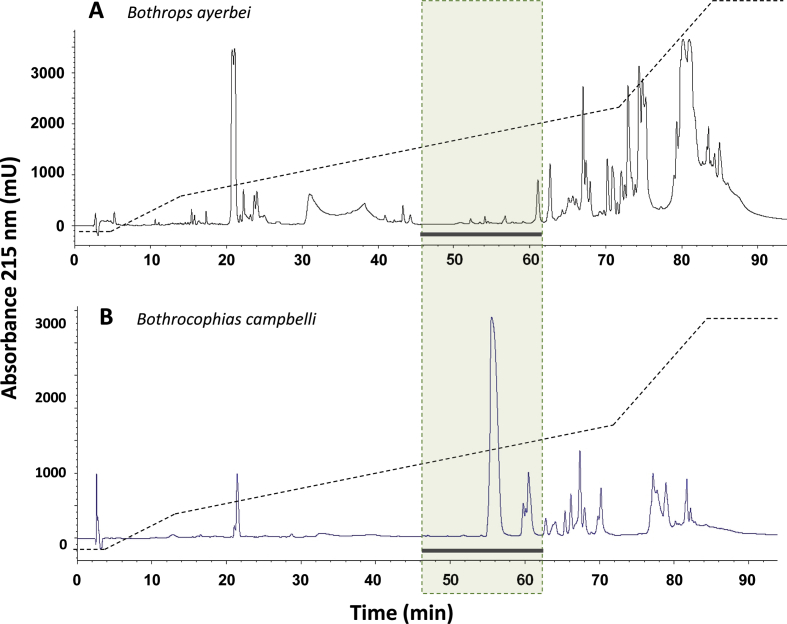
In addition to B. cotiara from Brazil, another bothropoid species with barely detectable amounts of venom PLA2s (0.7%) is Bothrops ayerbei (Supplemental Fig. S1; Mora-Obando et al., 2014b), a lineage of the B. asper species complex (Silva-de-França et al., 2019).
In contrast to the widespread presence of PLA2s in characterized venoms of most species included in the genus Bothrops (sensu lato) (Fig. 7), the chromatographic profiles of six out of the twelve venoms of species from Perú here studied revealed a phenotype in which PLA2s are notably scarce: Bothrops barnetti, at the base of the “alternatus” clade (Carrasco et al., 2016), Bothrocophias andianus and B. microphtalmus, clustered in the “microphtalmus” clade, and Bothrops chloromelas, B. bilineatus, and B. oligolepis, in the “taeniata” clade (Fig. 7). The venom of Bothrocophias hyoprora differed sharply in this regard from those of other Bothrocophias species (B. andianus and B. microphtalmus), showing a major protein peak in the PLA2-eluting region, and rather resembling the venom of Bothrocophias campbelli (Supplemental Fig. S1; Salazar-Valenzuela et al., 2014), a species not represented in the phylogenetic trees displayed in Fig. 7.
4. Discussion
Compositional, biochemical, and toxicological characteristics of the venoms of several species of the Bothrops complex (sensu lato) remain unexplored. In this study, the venoms of twelve viperid species that inhabit Perú were analyzed using electrophoretic, chromatographic, and enzymatic profiling for PLA2 and proteolytic activities in vitro. Also, their proteomic qualitative profiles were determined using a shotgun approach. Results provide additional information to previous studies on biochemical and toxicological aspects of snake venoms from this South American country (Sanz et al., 2008, Núñez et al., 2009, Jimenez et al., 2010, Kohlhoff et al., 2012, Guerra-Duarte et al., 2015).
In general, venom SDS-PAGE profiles did not deviate from the typical patterns observed in viperids by this technique but revealed some resemblances and moderate differences among the species. Remarkably, the venom of B. schlegelii from Perú showed identical electrophoretic and chromatographic profiles to that of the same species found in Costa Rica, separated by a distance of more than 2000 km. This was unexpected, considering that a previous comparison of B. schlegelii venoms from Costa Rica and Colombia reported significant intra-specific differences in electrophoretic and toxicological parameters (Prezotto-Neto et al., 2016). Also, there are marked differences in external characters, mainly scutellation and color patterns, between the Central American populations and those at the southern extreme of the distribution of this species in Ecuador and Perú (Campbell and Lamar, 2014). Moreover, venoms of Bothriechis are known for their extremely high inter-species variability (Fernández et al., 2010, Lomonte et al., 2008, Lomonte et al., 2012, Lomonte et al., 2014, Calvete, 2017, Pla et al., 2017).
Comparison of venom RP-HPLC profiles revealed an unexpected feature since six out of the twelve species studied presented only minor protein peaks in the elution region known to correspond to PLA2s as observed in several proteomic studies based on this chromatographic protocol (reviewed by Lomonte et al., 2014, Lomonte and Calvete, 2017). This particular characteristic had been observed in venoms from only a few of the bothropoid species studied until now, such as B. ayerbei from Colombia (Mora-Obando et al., 2014b), and B. cotiara from Brazil (Tashima et al., 2008). Most commonly, venoms of species within the Bothrops complex display prominent to medium-intensity peaks in the PLA2/PLA2-like eluting region. In contrast, venoms from six species of Perú here studied presented a low-PLA2 phenotypic feature. These observations point to an apparent dichotomy among bothropoid venoms, regarding the occurrence of high to scarce levels of PLA2 expression, and may bear practical relevance in selecting species whose venoms are used for antivenom production.
Venom compositional dichotomies are not infrequent in other taxonomic groups of venomous snakes. For example, rattlesnakes (Crotalus species) display either type I or type II venoms, defined as containing inverse proportions of metalloproteinases and PLA2s, respectively (Mackessy, 2008, Mackessy, 2010). Another example relates to venoms of New World coral snakes (Micrurus species), which express either ‘PLA2-rich’ or ‘3FTx (three-finger toxin)-rich’ phenotypes (Fernández et al., 2015, Lomonte et al., 2016, Sanz et al., 2019). In contrast to the streamlined 3FTx neurotoxic venoms of non-spitting, particularly African, cobras (Naja haje, N. nivea, N. annulifera) (Malih et al., 2014; Silva de Roodt et al., 2018, Tan et al., 2019a; unpublished results), variable expression of PLA2s has also been observed in cytotoxic venom proteomes among species of African (N. nubiae, N. pallida, N. nigricollis, N. mossambica) (Petras et al., 2011) and Asian (N. sumatrana, N. siamensis, N. philippinensis) (Chong et al., 2019, Tan et al., 2019a, Tan et al., 2019b) spitting cobras. The emergence of venom spitting in cobras is tightly linked with convergent upregulation of PLA2 toxins, which potentiate the nociceptive action of their venoms compared to those of their non-spitting counterparts (Kazandjian et al., 2019). Enhanced pain caused by spitting cobras is explicitly associated with the defensive use of venom, rather than for prey subjugation (Kazandjian et al., 2019).
The variable expression of PLA2/PLA2-like proteins in the Peruvian Bothrops venoms here studied showed only partial correlation with a functional assay for enzymatic hydrolysis of the NOBA substrate. This result is expected, since protein peaks in the known PLA2-eluting RP-HPLC region may correspond to either Asp49 PLA2 enzymes (catalytically active) or enzymatically inactive (but highly myotoxic) Lys49 PLA2-like homologs (Lomonte and Rangel, 2012). Moreover, catalytic efficiency (specific activity) may vary among active PLA2s. On the other hand, and in contrast to the highly variable expression of PLA2s, proteolytic activity did not vary widely among the studied venoms.
Regardless of their relative abundances, both PLA2s and metalloproteinases were detected by shotgun proteomics in all the studied venoms, together with serine proteinases, C-type lectins/lectin-like, and L-amino acid oxidases, as typical of viperids. Other protein families that appeared common to all venoms were nucleotidases and glutaminyl cyclotransferases. In contrast, some differences were observed among them regarding the detection of cysteine-rich secretory proteins, phosphodiesterases, nerve growth factors, phospholipases B, vascular endothelial growth factors, and hyaluronidases.
An inference of present results revealing the low expression of PLA2s in several bothropoid venoms from Perú is that they should display low toxicity for effects that depend mainly on this protein family. One of these known effects is myotoxicity (Gutiérrez and Lomonte, 2013). To test this prediction, the venoms of Bothrops barnetti and Bothrocophias andianus were selected as representatives of the ‘low-PLA2’ phenotype and assayed in vivo for induction of muscle necrosis. Results demonstrated their negligible myotoxic effect. Whether this lack of myotoxicity confers any restriction on the trophic function of snake venoms that express it is still unknown, since information on diet and basic ecology is still very fragmented. Also, it is reasonable to expect that these differences in myotoxicity among bothropoid venoms, shown experimentally in mice, should translate to clinical envenomings. Envenomings by Bothrops (sensu lato) species are sometimes described in the Latin American medical literature as a single entity having shared general features. However, present results predict that important differences exist depending on the species, e.g., myonecrosis would not be expected in envenomings by species that express minimal amounts of venom PLA2s.
An indirect myotoxic action has been reported for some hemorrhagic metalloproteinases (Queiroz et al., 1985, Gutiérrez et al., 1995), but their relative contribution to myonecrosis, in comparison to directly myotoxic PLA2s, has not been clearly established. Present results obtained in mice with B. barnetti and B. andianus venoms suggest that acute muscle damage would be largely driven by PLA2s, and are in agreement with studies that showed almost complete inhibition of myonecrosis when the venom of B. asper was neutralized by PLA2-specific polyclonal (Lomonte et al., 1990) or monoclonal (Lomonte et al., 1992) antibodies.
In addition to its interest from a medical perspective, from a biological standpoint, the study of inter-species variations might contribute to understanding the evolutionary events that shaped the venom proteomes of extant species. In this work, we attempted to visualize the positions and relationships of bothropoid species from Perú presenting low- or high-PLA2 venom phenotypes, based on the partial phylogeny generated by Carrasco et al. (2016). Although the interspersed pattern of the two phenotypes precludes reaching a simple phylogenetic deduction, it is clear that bothropoid venoms rich in PLA2s are more common than those presenting scarce PLA2s, and that the latter tend to cluster partially in the “microphtalmus” clade (Bothrocophias species), and the “taeniata” clade. The basal phylogenetic position of Bothrops barnetti (having a “low-PLA2″ venom) for the “alternatus” clade is interesting and leads to note that all clades encompassing a combination of the two phenotypes present a “low-PLA2″ basal species in these phylogenetic trees. If this observation holds after examining a more significant number of species, it would be tempting to suggest that the increased expression of PLA2s could be an evolutionarily more derived phenotypic feature. However, since the available phylogenetic information is still incomplete and limited in species coverage, further studies will be required to understand the complex relationships between venom phenotypes and phylogeny. Currently enigmatic, understanding the selective forces that originated the highly variable expression of PLA2s in venoms of the Bothrops complex anticipates a challenging research task.
5. Concluding remarks
Venom is an intrinsically ecological trophic trait crucial for the foraging success of the organisms that produce it. The evolution of venom phenotypes does not appear to be phylogenetically constrained, but more likely shaped by ecological filtering that permits a small number of optimal solutions involving combinations of only a few major toxin families (Barua and Mikheyev, 2019). Of particular note is the occurrence of congeneric, even conspecific, divergent venom phenotypes, such as the 3FTx-rich/PLA2-predominant venoms described in venoms of the New World genus Micrurus (Elapinae) and sea snake (Hydrophiinae) lineages (Sanz et al., 2016, Lomonte et al., 2016); the PIII-SVMP (type-I)/β-neurotoxic heterodimeric PLA2 (type-II) venom compositional dichotomy found within Crotalus (Mackessy, 2008, Massey et al., 2012, Calvete, 2017, Strickland et al., 2018) and Bothriechis (Pla et al., 2017); and the divergent 3FTx/Kunitz-type dendrotoxins venom phenotypes among Dendroaspis (Ainsworth et al., 2018). The high/low PLA2 venom compositional divergence described here among venoms of Peruvian Bothrops species represents a new dichotomy. The finding of similar patterns of venom variability among evolutionarily distant snake lineages suggests that dichotomic venom phenotypes may constitute a more general trend than previously thought.
Venom dichotomy has different evolutionary origins in Nearctic, Central American, and South American rattlesnakes. Differential expression of type-I or type-II venom phenotypes in North American rattlesnakes appears to be due to recent lineage-independent losses of the genes coding for the subunits of the heterodimeric β-neurotoxin PLA2 (Dowell et al., 2016, Dowell et al., 2018). In contrast, in Central American C. simus it is ontogenetic and post-transcriptionally modulated by miRNAs (Durban et al., 2013). It represents a pedomorphic trait driven by the increased gain of neurotoxicity and lethal venom activity to mammals (rodents) in South American Crotalus durissus (Calvete et al., 2010). The 3FTx/PLA2 dichotomy among micrurine venoms may have been shaped by balancing selection (Sanz et al., 2019), an evolutionary scenario where local adaptation depends on the frequencies of the two toxin classes, with the hetero- (i.e. 3FTx + PLA2) phenotype having an advantage over the homo-phenotypes (Llaurens et al., 2017, Wang and Mitchell-Olds, 2017). In contrast, the streamlined 3FTx venom of the aquatic coral snake M. surinamensis may have evolved under strong pressure to quickly immobilize aquatic prey (Sanz et al., 2019, Silva et al., 2018). The different evolutionary paths responsible for generating variation in venom composition underscore the importance of considering venom diversification in the proper historical scenario.
Diversification of venoms within Bothrops should be understood in the context of the evolution of this clade. The early colonization by the ancestor of all Bothrops had its roots in South America during the middle Miocene, 14.07 Mya (CI95% 16.37–11.75 Mya), and was followed much later by Bothriechis schlegelii (7–6.6 Mya), Porthidium (3.5 Mya), and Crotalus durissus (1 Mya) (Wüster et al., 2002, Parkinson et al., 2002; Wüster et al., 2008, Alencar et al., 2016). Bothrops represents the most successful South American pitviper radiation, with extant lineages exhibiting extremely diverse morphological and ecological traits (Martins et al., 2002). Except for southwestern South America, the extreme highlands of the Andes, and southernmost Patagonia, extant bothropoid pitvipers are widely distributed in tropical Latin America, from northeastern Mexico to Argentina, and the southern parts of the lower Caribbean islands, where they inhabit a broad spectrum of ecoregions, such as lowland evergreen forests, montane semideciduous forests, savannas, and open montane formations, from sea level to 3000 m of elevation (Campbell and Lamar, 1989; 20104; Carrasco et al., 2012). Diversification of Bothrops should be understood in the context of the unique historical contingencies of the evolution of this clade, the absence of other pitvipers and placental carnivores as potential competitors or predators when their lineage radiated throughout South America (Wüster et al., 2002) and the orogenic processes that created vicariant events (Hamdan et al., 2020, Salazar-Valenzuela et al., 2019). For example, in South America the uplift of the Central Andes mountain range (which began in the late Oligocene to early Miocene (23 Mya) with the formation of the highest peaks during the late Middle Miocene 11–12 Mya and continued until around 2–3 Mya, contemporaneous with deformation in the Santa Bárbara system to the east) (Folguera et al., 2011; Hamdan et al., 2020), the uplift of the Guanacaste, Central, and Talamanca mountain ranges in Mesoamerica since the late Miocene or early Pliocene (8-5 Mya) to the Pliocene, and the closure of the Isthmus of Panama 3.2 Mya (Gregory-Wodzicki, 2000, O'Dea et al., 2016, Bergoeing, 2017), are some of the main vicariance events involved in the cladogenesis of the lancehead vipers as a consequence of the fragmentation of populations and allopatric speciation (Kattan et al., 2004, Saldarriaga-Córdoba et al., 2017, Hamdan et al., 2020). Recent phylogeographic analyses have shown that demographic processes and ecological differentiation in montane habitats would also be reliable drivers implicated in the divergence of different taxa along altitudinal gradients (Salazar-Valenzuela et al., 2019).
An increasing number of studies support the idea that snake venom evolution is driven by diet-related selection pressures leading to local adaptations (Daltry et al., 1996, Barlow et al., 2009, Smiley-Walters et al., 2019, Davies and Arbuckle, 2019, Jackson et al., 2019). Adaptive venom variability represents a common phenomenon at all taxonomic levels among individuals of the same or different geographic locations, dietary habits, genders and across the life history stages of an individual (Calvete, 2013, Calvete, 2017). Although the mechanisms of venom variation remain elusive, ecological release seemingly promotes increased phenotypic variance (Van Valen, 1965, Bolnick et al., 2007). From a functional perspective, venom from a generalist viperid may have the capability to subdue different prey items, although with varying degrees of effectiveness. However, other dimensions, such as non-consumptive habitat use, or dietary decisions, define the trophic niche of the organism and have significant implications on the individual energy budget and the outcomes of interactions between sympatric taxa (Jackson et al., 2004, Vincent and Brown, 2005, Beermann et al., 2018).
Besides providing insights into the selective pressures that resulted in local adaptation and species-level divergence in venoms, understanding the evolutionary processes and the ecological constraints that moulded snake venoms to their present-day variation may also provide compelling insights into venom clinical toxicology. Thus, toxins that have the highest prey incapacitation activity to a mammalian model are often also the most medically important molecules in the context of a human envenoming. Identifying the molecular basis of venomous snake adaptation to their natural ecosystems may assist in the identification of those toxins that must be neutralized to reverse the effects of venom, thereby guiding the rational development of next-generation snakebite therapeutics.
Credit author statement
Conceptualization: BL, AZ, MSalas, MSasa. Data curation and Formal analysis: BL, CD, FCH, JF, JJC. Funding acquisition: BL, AZ. Investigation and Methodology: CD, FCH, JF, MR. Resources: AZ, MSalas. Writing and editing: BL, CD, JJC, MS.
Declaration of competing interest
The authors declare that they have no competing interests related to this work.
Acknowledgements
We thank Instituto Nacional de Salud (Perú) for providing venoms through an agreement signed with Universidad Peruana Cayetano Heredia. Support to the Proteomics Laboratory of Instituto Clodomiro Picado by Vicerrectoría de Investigación, Universidad de Costa Rica, is also gratefully acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.toxcx.2020.100044.
Ethical statement
The authors declare that they have no competing interests related to this work.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Fig. S1.
RP-HPLC profiles of the venoms of Bothrops ayerbei from Colombia (Mora-Obando et al., 2014b) and Bothrocophias campbelli from Ecuador (Salazar-Valenzuela et al., 2014), showing the large variability in the expression of phospholipase A2 proteins. These venoms are not included in the phylogenetic trees shown in Fig.7 and therefore their positions and relationships to other bothropoid species cannot be indicated. As indicated in the legend of Fig.2, the dotted light green shaded area indicates the known elution region for PLA2/PLA2-like proteins in viperid venoms in this standardized chromatographic gradient. These two species exemplify the evident dichotomy between venoms with “high-" and “low-PLA2″ phenotypes among bothropoids. For reference to colors, the reader is referred to the web version of this article.
References
- Ainsworth S., Petras D., Engmark M., Süssmuth R.D., Whiteley G., Albulescu L.O., Kazandjian T.D., Wagstaff S.C., Rowley P., Wüster W., Dorrestein P.C., Arias A.S., Gutiérrez J.M., Harrison R.A., Casewell N.R., Calvete J.J. The medical threat of mamba envenoming in sub-Saharan Africa revealed by genus-wide analysis of venom composition, toxicity and antivenomics profiling of available antivenoms. J. Proteomics. 2018;172:173–189. doi: 10.1016/j.jprot.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Alape-Girón A., Sanz L., Escolano J., Flores-Díaz M., Madrigal M., Sasa M., Calvete J.J. Snake venomics of the lancehead pitviper Bothrops asper: geographic, individual, and ontogenetic variations. J. Proteome Res. 2008;7:3556–3571. doi: 10.1021/pr800332p. [DOI] [PubMed] [Google Scholar]
- Alencar L.R.V., Quental T.B., Grazziotin F.G., Alfaro M.L., Martins M., Venzon M., Zaher H. Diversification in vipers: phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 2016;105:50–62. doi: 10.1016/j.ympev.2016.07.029. [DOI] [PubMed] [Google Scholar]
- Barlow A., Pook C.E., Harrison R.A., Wüster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. Biol. Sci. 2009;276:2443–2449. doi: 10.1098/rspb.2009.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua A., Mikheyev A.S. Many options, few solutions: over 60 My snakes converged on a few optimal venom formulations. Mol. Biol. Evol. 2019;36:1964–1974. doi: 10.1093/molbev/msz125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann J., Boos K., Gutow L., Boersma M., Peralta A.C. Combined effects of predator cues and competition define habitat choice and food consumption of amphipod mesograzers. Oecologia. 2018;186:645–654. doi: 10.1007/s00442-017-4056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergoeing J.P. Elsevier; 2017. Geomorphology and Volcanology of Costa Rica. [Google Scholar]
- Bolnick D.I., Svanback R., Araujo M.S., Persson L. Comparative support for the niche variation hypothesis that more generalized populations also are more heterogeneous. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10075–10079. doi: 10.1073/pnas.0703743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete J.J., Sanz L., Cid P., de la Torre P., Flores-Díaz M., Dos Santos M.C., Borges A., Bremo A., Angulo Y., Lomonte B., Alape-Girón A., Gutiérrez J.M. Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 2010;9:528–544. doi: 10.1021/pr9008749. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. Snake venomics: from the inventory of toxins to biology. Toxicon. 2013;75:44–62. doi: 10.1016/j.toxicon.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. Venomics: integrative venom proteomics and beyond. Biochem. J. 2017;474:611–634. doi: 10.1042/BCJ20160577. [DOI] [PubMed] [Google Scholar]
- Campbell J.A., Lamar W.W. Cornell University Press; Ithaca, New York: 2014. The Venomous Reptiles of the Western Hemisphere. [Google Scholar]
- Carrasco P.A., Mattoni C.I., Leynaud G.C., Scrocchi G.J. Morphology, phylogeny and taxonomy of South American bothropoid pitvipers. Zool. Scripta. 2012;41:109–124. [Google Scholar]
- Carrasco P.A., Venegas P.J., Chaparro J.C., Scrocchi G.J. Nomenclatural instability in the venomous snakes of the Bothrops complex: implications in toxinology and public health. Toxicon. 2016;119:122–128. doi: 10.1016/j.toxicon.2016.05.014. [DOI] [PubMed] [Google Scholar]
- Carrillo de Espinoza N., Icochea J. Lista taxonómica preliminar de los reptiles vivientes del Perú. Publ Museo Historia Nat UNMSM. 1995;49:1–27. [Google Scholar]
- Chippaux J.P. Incidence and mortality due to snakebite in the Americas. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H.P., Tan K.Y., Tan N.H., Tan C.H. Exploring the diversity and novelty of toxin genes in Naja sumatrana, the equatorial spitting cobra from Malaysia through de novo venom-gland transcriptomics. Toxins. 2019;11:E104. doi: 10.3390/toxins11020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daltry J.C., Wüster W., Thorpe R.S. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- Davies E.L., Arbuckle K. Coevolution of snake venom toxic activities and diet: evidence that ecological generalism favours toxicological diversity. Toxins. 2019;11:E711. doi: 10.3390/toxins11120711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roodt A., Fernández J., Solano D., Lomonte B. A myotoxic Lys49 phospholipase A2-homologue is the major component of the venom of Bothrops cotiara from Misiones, Argentina. Toxicon. 2018;148:143–148. doi: 10.1016/j.toxicon.2018.04.026. [DOI] [PubMed] [Google Scholar]
- Dowell N.L., Giorgianni M.W., Kassner V.A., Selegue J.E., Sanchez E.E., Carroll S.B. The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 2016;26:2434–2445. doi: 10.1016/j.cub.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell N.L., Giorgianni M.W., Griffin S., Kassner V.A., Selegue J.E., Sanchez E.E., Carroll S.B. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr. Biol. 2018;28:1016–1026. doi: 10.1016/j.cub.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J., Pérez A., Sanz L., Gómez A., Bonilla F., Rodríguez S., Chacón D., Sasa M., Angulo Y., Gutiérrez J.M., Calvete J.J. Integrated ‘omics’ profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genom. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevao-Costa M.I., Gontijo S.S., Correia B.L., Yarleque A., Vivas-Ruiz D., Rodrigues E., Chávez-Olortegui C., Oliveira L.S., Sanchez E.F. Neutralization of toxicological activities of medically-relevant Bothrops snake venoms and relevant toxins by two polyvalent bothropic antivenoms produced in Peru and Brazil. Toxicon. 2016;122:67–77. doi: 10.1016/j.toxicon.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Fenwick A.M., Gutberlet R.L., Evans J.A., Parkinson C.L. Morphological and molecular evidence for phylogeny and classification of South American pitvipers, genera Bothrops, Bothriopsis, and Bothrocophias (Serpentes: Viperidae) Zool. J. Linn. Soc. 2009;156:617–640. [Google Scholar]
- Fernández J., Lomonte B., Sanz L., Angulo Y., Gutiérrez J.M., Calvete J.J. Snake venomics of Bothriechis nigroviridis reveals extreme variability among palm viper venoms: different evolutionary solutions for the same trophic purpose. J. Proteome Res. 2010;9:4234–4241. doi: 10.1021/pr100545d. [DOI] [PubMed] [Google Scholar]
- Fernández J., Vargas N., Pla D., Sasa M., Rey-Suárez P., Sanz L., Gutiérrez J.M., Calvete J.J., Lomonte B. Snake venomics of Micrurus alleni and Micrurus mosquitensis from the Caribbean region of Costa Rica reveals two divergent compositional patterns in New World elapids. Toxicon. 2015;107:217–233. doi: 10.1016/j.toxicon.2015.08.016. [DOI] [PubMed] [Google Scholar]
- García P.J., Yarlequé A., Bonilla-Ferreira C., Pessah S., Vivas D., Sandoval G.A., Lazo F. Características bioquímicas y evaluación preclínica de un antiveneno botrópico liofilizado contra el veneno de la serpiente Bohtrops atrox. Rev. Peru. Med. Exp. Salud Pública. 2008;25:386–390. [Google Scholar]
- Göçmen B., Heiss P., Petras D., Nalbantsoy A., Süssmuth R.D. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon. 2015;107:163–174. doi: 10.1016/j.toxicon.2015.09.013. [DOI] [PubMed] [Google Scholar]
- Gregory-Wodzicki K.M. Uplift history of the central and northern Andes: a review. Geol. Soc. Am. Bull. 2000;112:1091–1105. [Google Scholar]
- Guerra-Duarte C., Lopes-Peixoto J., Fonseca-de-Souza B.R., Stransky S., Oliveira D., Schneider F.S., Lopes-de-Souza L., Bonilla C., Silva W., Tintaya B., Yarleque A., Chávez-Olórtegui C. Partial in vitro analysis of toxic and antigenic activities of eleven Peruvian pitviper snake venoms. Toxicon. 2015;108:84–96. doi: 10.1016/j.toxicon.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Lomonte B. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon. 2013;62:27–39. doi: 10.1016/j.toxicon.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Romero M., Núñez J., Chaves F., Borkow G., Ovadia M. Skeletal muscle necrosis and regeneration after injection of BaH1, a hemorrhagic metalloproteinase isolated from the venom of the snake Bothrops asper (Terciopelo) Exp. Mol. Pathol. 1995;62:28–41. doi: 10.1006/exmp.1995.1004. [DOI] [PubMed] [Google Scholar]
- Hamdan B., Guedes T.B., Carrasco P.A., Melville J. A complex biogeographic history of diversification in Neotropical lancehead pitvipers (Serpentes, Viperidae), Zool. Scr. 2020;49:145–158. [Google Scholar]
- Jackson A.C., Rundle S.D., Attrill M.J., Cotton P.A. Ontogenetic changes in metabolism may determine diet shifts for a sit-and-wait predator. J. Anim. Ecol. 2004;73:536–545. [Google Scholar]
- Jackson T.N.W., Jouanne H., Vidal N. Snake venom in context: neglected clades and concepts. Front. Ecol. Evol. 2019;7:332. doi: 10.3389/fevo.2019.00332. [DOI] [Google Scholar]
- Jimenez K.L., Zavaleta A.I., Izaguirre V., Yarleque A., Inga R.R. Clonaje y caracterización molecular in silico de un transcrito de fosfolipasa A2 aislado del veneno de la serpiente peruana Lachesis muta. Rev. Peru. Med. Exp. Salud Pública. 2010;27:532–539. doi: 10.1590/s1726-46342010000400007. [DOI] [PubMed] [Google Scholar]
- Jiménez-Charris E., Montealegre-Sánchez L., Solano-Redondo L., Mora-Obando D., Camacho E., Castro-Herrera F., Fierro-Pérez L., Lomonte B. Proteomic and functional analyses of the venom of Porthidium lansbergii lansbergii (Lansberg's hognose viper) from the Atlantic Department of Colombia. J Proteomics. 2015;115:287–299. doi: 10.1016/j.jprot.2014.11.016. [DOI] [PubMed] [Google Scholar]
- Kattan G.H., Franco P., Rojas V., Morales G. Biological diversification in a complex region: a spatial analysis of faunistic diversity and biogeography of the Andes of Colombia. J. Biogeogr. 2004;31:1829–1839. [Google Scholar]
- Kazandjian T., Petras D., Robinson S., Undheim E., Arbuckle K., Whiteley G., Albulescu L.-O., Ainsworth S., Wagstaff S., Wüster W., Harrison R., Vetter I., Calvete J., Casewell N. 2019. Convergent Evolution of Defensive Venom Components in Spitting Cobras. 20th World Congress of the International Society on Toxinology. Buenos Aires, Argentina, Wednesday September 11, Session 5B: Student Invited Presentations. [Google Scholar]
- Kohlhoff M., Borges M.H., Yarleque A., Cabezas C., Richardson M., Sanchez E.F. Exploring the proteomes of the venoms of the Peruvian pit vipers Bothrops atrox, B. barnetti and B. pictus. J Proteomics. 2012;75:2181–2195. doi: 10.1016/j.jprot.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Laing G.D., Yarlequé A., Marcelo A., Rodriguez E., Warrell D.A., Theakston R.D. Preclinical testing of three South American antivenoms against the venoms of five medically-important Peruvian snake venoms. Toxicon. 2004;44:103–106. doi: 10.1016/j.toxicon.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Llaurens V., Whibley A., Joron M. Genetic architecture and balancing selection: the life and death of differentiated variants. Mol. Ecol. 2017;26:2430–2448. doi: 10.1111/mec.14051. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Rangel J. Snake venom Lys49 myotoxins: from phospholipases A2 to non-enzymatic membrane disruptors. Toxicon. 2012;60:520–530. doi: 10.1016/j.toxicon.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Calvete J.J. Strategies in 'snake venomics' aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017;23:26. doi: 10.1186/s40409-017-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte B., Gutiérrez J.M., Carmona E., Rovira M.E. Equine antibodies to Bothrops asper myotoxin II: isolation from polyvalent antivenom and neutralizing ability. Toxicon. 1990;28:379–384. doi: 10.1016/0041-0101(90)90075-i. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Gutiérrez J.M., Ramírez M., Díaz C. Neutralization of myotoxic phospholipases A2 from the venom of the snake Bothrops asper by monoclonal antibodies. Toxicon. 1992;30:239–245. doi: 10.1016/0041-0101(92)90866-4. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Escolano J., Fernández J., Sanz L., Angulo Y., Gutiérrez J.M., Calvete J.J. Snake venomics and antivenomics of the arboreal neotropical pitvipers Bothriechis lateralis and Bothriechis schlegelii. J. Proteome Res. 2008;7:2445–2457. doi: 10.1021/pr8000139. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Tsai W.C., Bonilla F., Solórzano A., Solano G., Angulo Y., Gutiérrez J.M., Calvete J.J. Snake venomics and toxicological profiling of the arboreal pitviper Bothriechis supraciliaris from Costa Rica. Toxicon. 2012;59:592–599. doi: 10.1016/j.toxicon.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Fernández J., Sanz L., Angulo Y., Sasa M., Gutiérrez J.M., Calvete J.J. Venomous snakes of Costa Rica: biological and medical implications of their venom proteomic profiles analyzed through the strategy of "snake venomics. J Proteomics. 2014;105:323–339. doi: 10.1016/j.jprot.2014.02.020. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Rey-Suárez P., Fernández J., Sasa M., Pla D., Vargas N., Bénard-Valle M., Sanz L., Corrêa-Netto C., Núñez V., Alape-Girón A., Alagón A., Gutiérrez J.M., Calvete J.J. Venoms of Micrurus coral snakes: evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon. 2016;122:7–25. doi: 10.1016/j.toxicon.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Mackessy S.P. Venom composition in rattlesnakes: trends and biological significance. In: Hayes W.K., Beaman K.R., Cardwell M.D., Bush S.P., editors. The Biology of Rattlesnakes. Loma Linda University Press; 2008. pp. 495–510. [Google Scholar]
- Mackessy S.P. Evolutionary trends in venom composition in the western rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon. 2010;55:1463–1474. doi: 10.1016/j.toxicon.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Malih I., Rusmili M.R., Tee T.Y., Saile R., Ghalim N., Othman I. Proteomic analysis of Moroccan cobra Naja haje legionis venom using tandem mass spectrometry. J. Proteomics. 2014;96:240–252. doi: 10.1016/j.jprot.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Martins M., Marques O.A.V., Sazima I. Ecological and phylogenetic correlates of feeding habits in Neotropical pitvipers of the genus Bothrops. In: Schuett G.W., Höggren M., Douglas M.E., Greene H.W., editors. Biology of the Vipers. Eagle Mountain Publishing; Eagle Mountain, Utah: 2002. pp. 307–328. 13: 978-0972015400. [Google Scholar]
- Massey D.J., Calvete J.J., Sánchez E.E., Sanz L., Richards K., Curtis R., Boesen K. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteomics. 2012;75:2576–2587. doi: 10.1016/j.jprot.2012.02.035. [DOI] [PubMed] [Google Scholar]
- Ministerio de Salud . MINSA; Perú: 2017. Mapa de Ofidismo por distritos, Perú. Centro Nacional de Epidemiología, Prevención y Control de Enfermedades.www.dge.gob.pe/portal/docs/vigilancia/sala/2017/SE27/ofidismo.pdf Consulted at: [Google Scholar]
- Mora-Obando D., Díaz C., Angulo Y., Gutiérrez J.M., Lomonte B. Role of enzymatic activity in muscle damage and cytotoxicity induced by Bothrops asper Asp49 phospholipase A2 myotoxins: are there additional effector mechanisms involved? Peer J. 2014;2:e569. doi: 10.7717/peerj.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Obando D., Guerrero-Vargas J.A., Prieto-Sánchez R., Beltrán J., Rucavado A., Sasa M., Gutiérrez J.M., Ayerbe S., Lomonte B. Proteomic and functional profiling of the venom of Bothrops ayerbei from Cauca, Colombia, reveals striking interspecific variation with Bothrops asper venom. J Proteomics. 2014;96:159–172. doi: 10.1016/j.jprot.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Núñez V., Cid P., Sanz L., De La Torre P., Angulo Y., Lomonte B., Gutiérrez J.M., Calvete J.J. Snake venomics and antivenomics of Bothrops atrox venoms from Colombia and the Amazon regions of Brazil, Perú and Ecuador suggest the occurrence of geographic variation of venom phenotype by a trend towards paedomorphism. J. Proteomics. 2009;73:57–78. doi: 10.1016/j.jprot.2009.07.013. [DOI] [PubMed] [Google Scholar]
- O'Dea A., Lessios H.A., Coates A.G., Eytan R.I., Restrepo-Moreno S.A., Cione A.L., Collins L.S., De Queiroz A., Farris D.W., Norris R.D. formation of the Isthmus of Panama. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson C., Campbell J., Chippindale P. Multigene phylogenetic analyses of pitvipers; with comments on the biogeographical history of the group. In: Schuett M.H.G.W., Greene H.W., Douglas M.E., editors. Biol. Vipers. Eagle Mountain Publishing; Eagle Mountain, Utah: 2002. pp. 93–110. [Google Scholar]
- Petras D., Sanz L., Segura A., Herrera M., Villalta M., Solano D., Vargas M., León G., Warrell D.A., Theakston R.D., Harrison R.A., Durfa N., Nasidi A., Gutiérrez J.M., Calvete J.J. Snake venomics of African spitting cobras: toxin composition and assessment of congeneric cross-reactivity of the pan-African EchiTAb-Plus-ICP antivenom by antivenomics and neutralization approaches. J. Proteome Res. 2011;10:1266–1280. doi: 10.1021/pr101040f. [DOI] [PubMed] [Google Scholar]
- Pla D., Sanz L., Sasa M., Acevedo M.E., Dwyer Q., Pérez A., Rodríguez Y., Lomonte B., Calvete J.J. Proteomic analysis of venom variability and ontogeny across the arboreal palm-pitvipers (genus Bothriechis) J Proteomics. 2017;152:1–12. doi: 10.1016/j.jprot.2016.10.006. [DOI] [PubMed] [Google Scholar]
- Prezotto-Neto J.P., Kimura L.F., Alves A.F., Gutiérrez J.M., Otero R., Suárez A.M., Santoro M.L., Barbaro K.C. Biochemical and biological characterization of Bothriechis schlegelii snake venoms from Colombia and Costa Rica. Exp. Biol. Med. 2016;241:2075–2085. doi: 10.1177/1535370216660214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz L.S., Santo-Neto H., Assakura M.T., Reichl A.P., Mandelbaum F.R. Pathological changes in muscle caused by hemorrhagic and proteolytic factors from Bothrops jararaca snake venom. Toxicon. 1985;23:341–345. doi: 10.1016/0041-0101(85)90158-8. [DOI] [PubMed] [Google Scholar]
- Rodrigues C.R., Teixeira-Ferreira A., Vargas F.F.R., Guerra-Duarte C., Costal-Oliveira F., Stransky S., Lopes-de-Souza L., Dutra A.A.A., Yarlequé A., Bonilla C., Sanchez E.F., Perales J., Chávez-Olórtegui C. Proteomic profile, biological activities and antigenic analysis of the venom from Bothriopsis bilineata smaragdina ("loro machaco"), a pitviper snake from Peru. J. Proteomics. 2018;187:171–181. doi: 10.1016/j.jprot.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Rojas E., Quesada L., Arce V., Lomonte B., Rojas G., Gutiérrez J.M. Neutralization of four Peruvian Bothrops sp. snake venoms by polyvalent antivenoms produced in Perú and Costa Rica: preclinical assessment. Acta Trop. 2005;93:85–95. doi: 10.1016/j.actatropica.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Salazar-Valenzuela D., Mora-Obando D., Fernández M.L., Loaiza-Lange A., Gibbs L.H., Lomonte B. Proteomic and toxicological profiling of the venom of Bothrocophias campbelli, a pitviper species from Ecuador and Colombia. Toxicon. 2014;90:15–25. doi: 10.1016/j.toxicon.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Salazar-Valenzuela D., Kuch U., Torres‐Carvajal O., Valencia J.H., Gibbs H.L. Divergence of tropical pitvipers promoted by independent colonization events of dry montane Andean habitats. J. Biogeogr. 2019;46:1826–1840. [Google Scholar]
- Saldarriaga-Córdoba M., Parkinson C.L., Daza J.M., Wüster W., Sasa M. Phylogeography of the central American lancehead Bothrops asper (serpentes: Viperidae) PloS One. 2017;12 doi: 10.1371/journal.pone.0187969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Escolano J., Ferretti M., Biscoglio M.J., Rivera E., Crescenti E.J., Angulo Y., Lomonte B., Gutiérrez J.M., Calvete J.J. Snake venomics of the South and Central American Bushmasters. Comparison of the toxin composition of Lachesis muta gathered from proteomic versus transcriptomic analysis. J. Proteomics. 2008;71:46–60. doi: 10.1016/j.jprot.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Sanz L., Pla D., Pérez A., Rodríguez Y., Zavaleta A., Salas M., Lomonte B., Calvete J.J. Venomic analysis of the poorly studied desert coral snake, Micrurus tschudii tschudii, supports the 3FTx/PLA₂ dichotomy across Micrurus venoms. Toxins. 2016;8:E178. doi: 10.3390/toxins8060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Quesada-Bernat S., Ramos T., Casais-e-Silva L.L., Corrêa-Netto C., Silva-Haad J.J., Sasa M., Lomonte B., Calvete J.J. New insights into the phylogeographic distribution of the 3FTx/PLA2 venom dichotomy across genus Micrurus in South America. J Proteomics. 2019;200:90–101. doi: 10.1016/j.jprot.2019.03.014. [DOI] [PubMed] [Google Scholar]
- Serrano S.M.T., Zelanis A., Kitano E.S., Tashima A.K. Analysis of the snake venom peptidome. Methods Mol. Biol. 2018;1719:349–358. doi: 10.1007/978-1-4939-7537-2_23. [DOI] [PubMed] [Google Scholar]
- Silva F.M., Prudente ALdaC., Machado F.A., Santos M.M., Zaher H., Hingst-Zaher E. Aquatic adaptations in a Neotropical coral snake: a study of morphological convergence. J. Zool. Syst. Evol. Res. 2018;56:382–394. [Google Scholar]
- Silva-de-França F., Villas-Boas I.M., Serrano S.M.T., Cogliati B., Chudzinski S.A.A., Lopes P.H., Kitano E.S., Okamoto C.K., Tambourgi D.V. Naja annulifera snake: new insights into the venom components and pathogenesis of envenomation. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley-Walters S.A., Farrell T.M., Gibbs H.L. High levels of functional divergence in toxicity towards prey among the venoms of individual pigmy rattlesnakes. Biol. Lett. 2019;15:20180876. doi: 10.1098/rsbl.2018.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland J.L., Smith C.F., Mason A.J., Schield D.R., Borja M., Castañeda-Gaytán G., Spencer C.L., Smith L.L., Trápaga A., Bouzid N.M., Campillo-García G., Flores-Villela O.A., Antonio-Rangel D., Mackessy S.P., Castoe T.A., Rokyta D.R., Parkinson C.L. Evidence for divergent patterns of local selection driving venom variation in Mojave Rattlesnakes (Crotalus scutulatus) Sci. Rep. 2018;8:17622. doi: 10.1038/s41598-018-35810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan N.H., Fung S.Y., Tan K.Y., Yap M.K., Gnanathasan C.A., Tan C.H. Functional venomics of the Sri Lankan Russell's viper (Daboia russelii) and its toxinological correlations. J Proteomics. 2015;128:403–423. doi: 10.1016/j.jprot.2015.08.017. [DOI] [PubMed] [Google Scholar]
- Tan C.H., Wong K.Y., Tan N.H., Ng T.S., Tan K.Y. Distinctive distribution of secretory phospholipases A₂ in the venoms of afro-asian cobras (subgenus: Naja, afronaja, boulengerina and uraeus) Toxins. 2019;11:E116. doi: 10.3390/toxins11020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.H., Wong K.Y., Chong H.P., Tan N.H., Tan K.Y. Proteomic insights into short neurotoxin-driven, highly neurotoxic venom of Philippine cobra (Naja philippinensis) and toxicity correlation of cobra envenomation in Asia. J. Proteomics. 2019;206:103418. doi: 10.1016/j.jprot.2019.103418. [DOI] [PubMed] [Google Scholar]
- Tashima A.K., Sanz L., Camargo A.C., Serrano S.M., Calvete J.J. Snake venomics of the Brazilian pitvipers Bothrops cotiara and Bothrops fonsecai. Identification of taxonomy markers. J Proteomics. 2008;71:473–485. doi: 10.1016/j.jprot.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Van Valen L. Morphological variation and width of ecological niche. Am. Nat. 1965;99:377–390. [Google Scholar]
- Viala V.L., Hildebrand D., Fucase T.M., Sciani J.M., Prezotto-Neto J.P., Riedner M., Sanches L., Nishimura P.J., Oguiura N., Pimenta D.C., Schlüter H., Betzel C., Arni R.K., Spencer P.J. Proteomic analysis of the rare Uracoan rattlesnake Crotalus vegrandis venom: evidence of a broad arsenal of toxins. Toxicon. 2015;107:234–251. doi: 10.1016/j.toxicon.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Vincent T.L., Brown J.S. Cambridge University Press; 2005. Evolutionary Game Theory, Natural Selection, and Darwinian Dynamics. [Google Scholar]
- Wang B., Mitchell-Olds T. Balancing selection and trans-specific polymorphisms. Genome Biol. 2017;18:231. doi: 10.1186/s13059-017-1365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüster W., Salomão M.G., Quijada-Mascareñas J.A., Thorpe R.S. Origin and evolution of the South American pitviper fauna: evidence from mitochondrial DNA sequence data. In: Schuett G.W., Höggren M., Douglas M.E., Greene H.W., editors. Biology of the Vipers. Eagle Mountain Publishing; USA: 2002. pp. 111–128. [Google Scholar]
- Wüster W., Peppin L., Pook C.E., Walker D.E. A nesting of vipers: phylogeny and historical biogeography of the Viperidae (Squamata: serpentes) Mol. Phylogenet. Evol. 2008;49:445–459. doi: 10.1016/j.ympev.2008.08.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.