Abstract
Background
Artemisia annua exerts powerful effects in non-small cell lung carcinoma (NSCLC). Some studies have shown that Artemisia annua possesses the characteristics of new therapeutic drugs for NSCLC patients. However, the underlying molecular mechanism of Artemisia annua anti-NSCLC is not yet fully elucidated because Artemisia annua contains hundreds of ingredients. This study aimed to conduct network pharmacological analysis on the mechanism of action of Artemisia annua against NSCLC.
Material/Methods
The active ingredients and corresponding potential targets of Artemisia annua were searched and screened in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP). Then through The Cancer Genome Atlas (TCGA) and the National Center for Biotechnology Information (NCBI) databases to establish NSCLC related targets. Based on the matching results of Artemisia annua potential targets and NSCLC targets, a protein–protein interaction (PPI) network was constructed to analyze the interactions between these targets and topologically screen the central targets. Furthermore, Gene Ontology (GO) biological functions analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathways enrichment were carried out.
Results
There were 19 main active ingredients of Artemisia annua screened for target prediction; 40 NSCLC-related common targets were identified via multiple NSCLC databases. The node area and corresponding degree value of AKT1, MYC, CCND1, VEGFA, JUN, MAPK1, EGFR, and ESR1 were large and could be easily found in the PPI network. The aforementioned results were further verified by the analysis of GO biological function and KEGG enrichment analysis.
Conclusions
The network pharmacology analysis reveals the molecular biological mechanism of Artemisia annua anti-NSCLC via multiple active components, multi-channels, and multi-targets. This suggests that Artemisia annua might be developed as a promising anti-NSCLC drug.
MeSH Keywords: Antineoplastic Agents; Carcinoma, Non-Small-Cell Lung; Medicine, Chinese Traditional; Protein Interaction Maps
Background
Lung cancer is one of the most lethal and most common malignant tumors in the world. The incidence of non-small cell lung cancer (NSCLC) in lung cancer overall incidence is about 80%, and it is the main cause of cancer-related deaths in China [1]. Despite advances in treatment options, including radiation, surgery, targeted therapies, and chemotherapy, prognosis remains poor due to the presence of locally advanced or extensively metastatic tumors at diagnosis in most patients [2]. Therefore, scientists are persistently looking for novel drugs to improve the effectiveness of NSCLC treatment.
Traditional Chinese medicines (TCMs), unique biomedical and pharmaceutical resources have been widely used in the prevention and treatment of NSCLC [3–5]. Artemisia annua is an ancient antipyretic traditional Chinese medicine, which is used as tea or press juice to treat malaria. Since 2015 when the Nobel Prize in Physiology or Medicine was conferred to a Chinese scientist Youyou Tu, Artemisia annua has drawn worldwide attention [6]. Artemisia annua and its derivatives have been shown to have the anti-cancer activity in vitro and in vivo [4–6]. Mechanisms for anti-cancer activity of Artemisia annua rely mainly on the generation of reactive oxygen species (ROS) resulting from the interaction of the endoperoxide bridge with either free ferrous iron or heme iron [7,8]. Increased requirements for iron in cancer cells and higher intake of iron through transferrin receptors, typically upregulates in cancer cells, making them more susceptible to damage by ROS from an iron-catalyzed reaction with Artemisia annua [9–11]. Some fundamental research studies have shown that Artemisia annua possesses qualities of a novel therapeutic for patients with NSCLC [12,13]. In addition, Artemisia annua and its derivatives also exert synergistic anti-tumor effects on NSCLC with a wide array of clinically established drugs [14–16]. However, Artemisia annua contains hundreds of ingredients; its active ingredients, targets, and mechanism of anti-NSCLC effects have not been apparent.
Due to the multi-chemical components, multi-pharmacological effects, and multi-action targets of TCM in disease treatment, it is difficult for traditional research methods to explain its mechanism of action adequately [3,6]. However, in recent years, network pharmacology generated by the combination of bioinformatics and pharmacology can clarify the principle of action of these herbs [17,18]. Therefore, we used the network pharmacology method to study the mechanism and material basis of Artemisia annua treatment of NSCLC to provide a new theoretical basis and direction for the systematic development of Artemisia annua.
Material and Methods
Database and software
The databases involved in this study were the following: Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://tcmspw.com/tcmsp.php), PubChem (http://pubchem.ncbi.nlm.nih.gov), Unipot (https://www.uniprot.org), STRING (https://string-db.org/), National Center for Biotechnology Information (NBCI) (https://www.ncbi.nlm.nih.gov/), and The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). The data analysis software includes Cytoscape 3.7.2 (plug-ins ClueGo 2.5.2 and CluePedia 1.5.5), discovery studio, and Venn diagrams (OmicShare tools).
Screening the active ingredients of Artemisia annua and predicting the component-targets
All the active ingredients of Artemisia annua were collected from the TCMSP platform, and the screening conditions were based on drug-likeness (DL) ≥0.18 and oral bioavailability (OB) ≥30%. Enter the active ingredients into PubChem to search for the corresponding molecular structure and record the corresponding PubChem CID. Then, based on TCMSP database, the potential targets of the active components were matched one by one, and then the target protein and gene information were corrected through UniProt database to select the object of “Homo sapiens,” which is the prediction target of the main active ingredients of Artemisia annua.
Seeking out NSCLC-related targets
With “Non-small-cell lung carcinoma” as the keywords, NBCI (https://www.ncbi.nlm.nih.gov/) and TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) were used to search and screen the known disease-targets for the subsequent study, and the repeated targets in the search results were discarded. UniProtKB (http://www.uniprot.org/) was used to get the standard targets’ names with the organism selected as “homo sapiens.” Then, the predicted targets of Artemisia annua primary active ingredients and NSCLC related targets were intersected, and the Wayne map was drawn to extract common target genes.
Construction of a protein–protein interaction (PPI) network
The common target genes were uploaded to the STRING platform to obtain protein interaction data. Then use Cytoscape software to establish and analyze the protein-protein interaction (PPI) network, and classify according to the MCODE algorithm, which is a method to automatically find molecular complexes in large protein interaction networks [19]. The size and color depth of the nodes were set according to the degree, that is, the larger the node was, the higher the degree of the target gene corresponding to the darker the color was. The thickness of the edge expressed the score of the relationship value between the 2 target genes, that is, the higher the combination score, the thicker the line of the edge.
Gene Ontology (GO) biological function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
Using the ClueGo and CluePedia plug-ins on the Cytoscape software, and using P<0.05 as the screening condition for statistical differences, Gene Ontology (GO) biological function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the potential targets of Artemisia annua against NSCLC were performed. Furthermore, the main signal pathway and the biological process involved in the pharmacological action of Artemisia annua against NSCLC were obtained, and the topological network diagram of the active component target genes of Artemisia annua was constructed.
Results
Active ingredients and target selection of Artemisia annua
After screening, a total of 19 main active ingredients were included in the study, target prediction was carried out for these 19 ingredients, and 510 targets were extracted after de-duplication, as shown in Table 1.
Table 1.
Main active components of artemisia annua.
| TCMSP ID | PubChem CID | compound | Target number |
|---|---|---|---|
| MOL000006 | 5280445 | Luteolin | 57 |
| MOL000098 | 5280343 | Quercetin | 154 |
| MOL000354 | 5281654 | Isorhamnetin | 37 |
| MOL000359 | 12303645 | Sitosterol | 3 |
| MOL000422 | 5280863 | Kaempferol | 63 |
| MOL000449 | 5280794 | Stigmasterol | 31 |
| MOL002235 | 5317287 | EUPATIN | 16 |
| MOL004083 | 5281699 | Tamarixetin | 15 |
| MOL004112 | 5281678 | Patuletin | 11 |
| MOL004609 | 158311 | Areapillin | 17 |
| MOL005229 | 5320351 | Artemetin | 23 |
| MOL007274 | 188323 | Skrofulein | 11 |
| MOL007401 | 160237 | Cirsiliol | 10 |
| MOL007404 | N/A | Vitexin_qt | 15 |
| MOL007412 | 5281603 | DMQT | 10 |
| MOL007415 | 10026486 | [(2S)-2-[[(2S)-2-(benzoylamino)-3-phenylpropanoyl]amino]-3-phenylpropyl] acetate | 5 |
| MOL007423 | N/A | 6,8-di-c-glucosylapigenin_qt | 16 |
| MOL007424 | N/A | Artemisinin | 15 |
| MOL007426 | N/A | Deoxyartemisinin | 1 |
Screening of NSCLC targets
The database of NCBI and TCGA were used to retrieve 1520 and 1042 NSCLC related targets respectively, and then the human gene name transformation was carried out through the UniProt database. The NCBI as aforementioned related target genes and 185 predicted target genes, were input into Venn Diagrams (OmicShare tools) software for intersection and construction of Venn diagrams as shown in Figure 1. There were 40 common target genes, namely Artemisia annua potential anti-NSCLC targets obtained as shown in Table 2.
Figure 1.
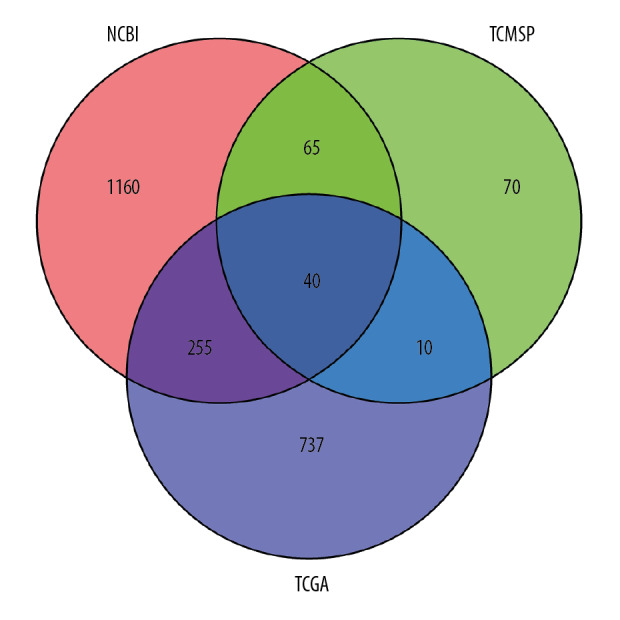
Screening of potential anti-NSCLC targets of Artemisia annua. NSCLC – non-small cell lung cancer.
Table 2.
The potential anti-non-small cell lung cancer target of artemisia annua.
| UniprotID | Target protein | Name | Degree |
|---|---|---|---|
| P31749 | RAC-alpha serine/threonine-protein kinase | AKT1 | 36 |
| P01106 | Myc proto-oncogene protein | MYC | 35 |
| P15692 | Vascular endothelial growth factor A | VEGFA | 34 |
| P24385 | G1/S-specific cyclin-D1 | CCND1 | 34 |
| P05412 | Transcription factor AP-1 | JUN | 33 |
| P28482 | Mitogen-activated protein kinase 1 | MAPK1 | 32 |
| P00533 | Epidermal growth factor receptor | EGFR | 32 |
| P03372 | Estrogen receptor | ESR1 | 31 |
| Q07817 | Bcl-2-like protein 1 | BCL2L1 | 28 |
| P04626 | Receptor tyrosine-protein kinase erbB-2 | ERBB2 | 28 |
| Q16665 | Hypoxia-inducible factor 1-alpha | HIF1A | 27 |
| P38936 | Cyclin-dependent kinase inhibitor 1 | CDKN1A | 26 |
| Q07820 | Induced myeloid leukemia cell differentiation protein Mcl-1 | MCL1 | 26 |
| Q00987 | E3 ubiquitin-protein ligase Mdm2 | MDM2 | 26 |
| O15519 | Caspase-8 | CASP8 | 25 |
| P37231 | Peroxisome proliferator activated receptor gamma | PPARG | 22 |
| P42224 | Signal transducer and activator of transcription 1-alpha/beta | STAT1 | 22 |
| P09874 | Poly [ADP-ribose] polymerase 1 | PARP1 | 21 |
| P22301 | Interleukin-10 | IL10 | 20 |
| P06400 | Retinoblastoma-associated protein | RB1 | 19 |
| P35968 | Vascular endothelial growth factor receptor 2 | KDR | 18 |
| P49841 | Glycogen synthase kinase-3 beta | GSK3B | 17 |
| P04049 | RAF proto-oncogene serine/threonine-protein kinase | RAF1 | 17 |
| O14757 | Serine/threonine-protein kinase Chk1 | CHEK1 | 17 |
| P08581 | Hepatocyte growth factor receptor | MET | 17 |
| P25963 | NF-kappa-B inhibitor alpha | NFKBIA | 15 |
| O96017 | Serine/threonine-protein kinase Chk2 | CHEK2 | 15 |
| P01344 | Insulin-like growth factor II | IGF2 | 15 |
| P10415 | Apoptosis regulator Bcl-2 | BCL2 | 14 |
| Q13950 | Runt-related transcription factor 2 | RUNX2 | 14 |
| O14920 | Inhibitor of nuclear factor kappa-B kinase subunit beta | IKBKB | 13 |
| Q07812 | Apoptosis regulator | BAX | 13 |
| P21860 | Receptor tyrosine-protein kinase erbB-3 | ERBB3 | 13 |
| P11387 | DNA topoisomerase 1 | TOP1 | 13 |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | NFE2L2 | 12 |
| Q9H3D4 | Cellular tumor antigen p53 | TP63 | 11 |
| P02452 | Collagen alpha-1 (I) chain | COL1A1 | 11 |
| P19793 | Retinoic acid receptor RXR-alpha | RXRA | 9 |
| Q15596 | Nuclear receptor coactivator 2 | NCOA2 | 7 |
| P06213 | Insulin receptor | INSR | 6 |
Construction of PPI network
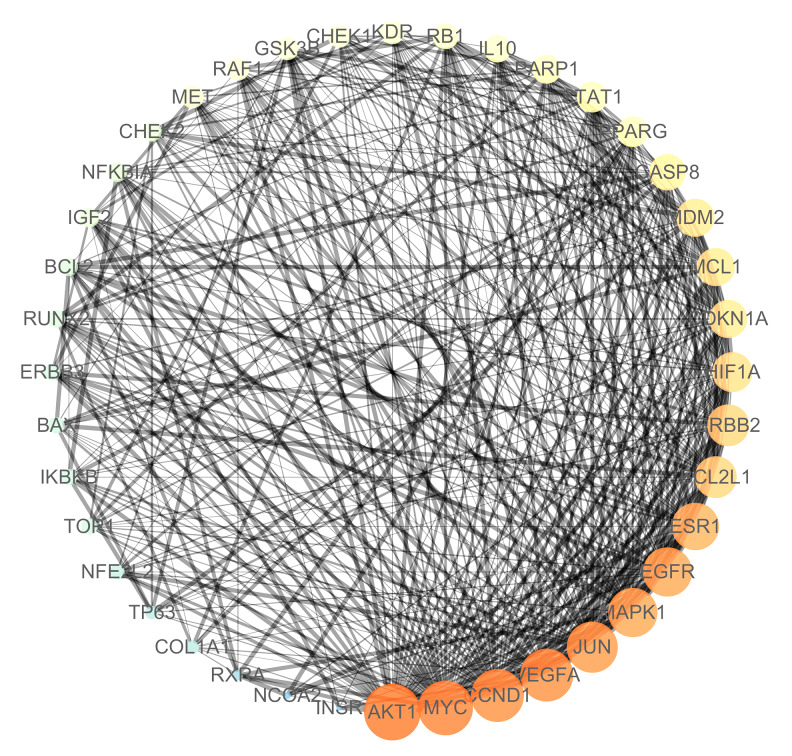
As shown in Figure 2, the aforementioned 40 target genes were uploaded to the STRING platform, and the species was set as “Homo sapiens” and the rest parameters were defaulted to obtain the protein interaction data information; then, they were imported into the software of Cytoscape to draw the PPI network. In the PPI network, 25 nodes (target genes) interact through 412 edges. The area of a node was positively related to the degree of the node that indicates the number of relationships between the node and other nodes in the network. AKT1, MYC, CCND1, VEGFA, JUN, MAPK1, EGFR, and ESR1 have a large node area and can be easily found in the PPI network (Figure 1), and their corresponding degrees were 36, 35, 34, 33, 32, 32, and 31 respectively (Table 2).
Figure 2.
PPI network of Artemisia annua potential target for NSCLC treatment. PPI – protein–protein interactions; NSCLC – non-small cell lung cancer.
Construction of anti-NSCLC target network of Artemisia annua active ingredients
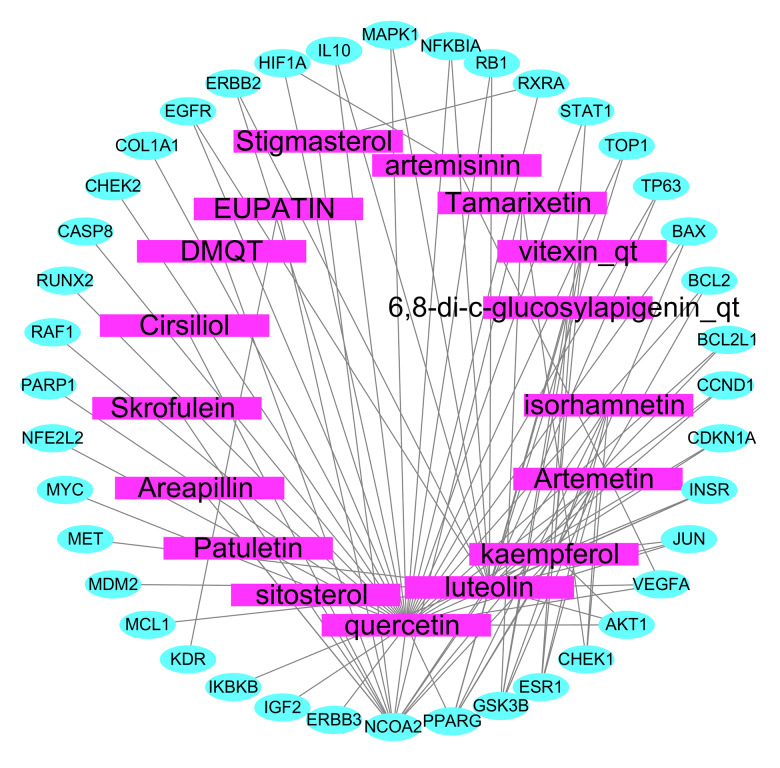
We took the 40 common targets as the anti-NSCLC targets of Artemisia annua (Table 2), and we import them into Cytoscape 3.7.2 to construct the network diagram of active components and targets of Artemisia annua for NSCLC treatment (Figure 3). The rectangle represents the active ingredient of Artemisia annua for NSCLC treatment, and the ellipse represents the target. The same related biological processes and molecular functions were obtained.
Figure 3.
Network map of active ingredients and targets of Artemisia annua in the treatment of NSCLC. NSCLC – non-small cell lung cancer.
GO biological process analysis
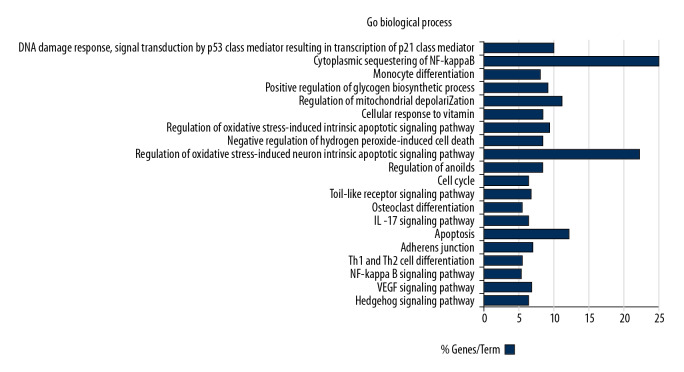
Based on the plug-ins “ClueGo” of Cytoscape software platform, GO biological process enrichment analysis was carried out on the anti-NSCLC target points of Artemisia annua obtained in “2.4”, 10 biological processes with P<0.05 were screened, as shown in Table 3, and the visualization results are shown in Figure 4. The biological process of Artemisia annua against NSCLC mainly involved the regulation of oxidative stress-induced intrinsic apoptotic signaling pathway, regulation of anoikis, negative regulation of hydrogen peroxide-induced cell death, and DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator. This suggested that the active ingredients of Artemisia annua may act on NSCLC by intervening in the above biological processes.
Table 3.
The biological process of artemisia annua on non-small cell lung cancer target.
| P-value | Description | Group genes |
|---|---|---|
| 6.51E-06 | Regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | AKT1, MCL1, NFE2L2, PARP1 |
| 8.93E-05 | Regulation of mitochondrial depolarization | BCL2, KDR, PARP1 |
| 1.85E-04 | Cellular response to vitamin | COL1A1, MDM2, PPARG |
| 1.85E-04 | Negative regulation of hydrogen peroxide-induced cell death | IL10, MET, NFE2L2 |
| 1.85E-04 | Regulation of anoikis | BCL2, CHEK2, MCL1 |
| 3.91E-04 | Cytoplasmic sequestering of NF-kappaB | IL10, NFKBIA |
| 4.02E-04 | Regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway | MCL1, PARP1 |
| 8.23E-04 | Monocyte differentiation | JUN, PPARG |
| 1.27E-03 | Ositive regulation of glycogen biosynthetic process | AKT1, INSR |
| 1.57E-03 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | CDKN1A, CHEK2 |
Figure 4.
GO biological process analysis. Go – Gene Ontology.
Enrichment analysis of KEGG signaling pathway
Based on the plug-ins “ClueGo” of Cytoscape software platform, the enrichment analysis of KEGG signal path of Artemisia annua anti-NSCLC target obtained in “2.4” was carried out, and signal paths with P<0.05 were screened, as shown in Table 4, and the visualization results are shown in Figure 5. The signal pathways involved in Artemisia annua anti-NSCLC included PI3K/Akt signal pathway, apoptosis, JAK/STAT signal pathway, VEGF signal pathway, etc.
Table 4.
Enrichment analysis of KEGG signal pathway of artemisia annua anti-small cell lung cancer targets (top 20).
| KEGG ID | Pathway | P-value | Group genes |
|---|---|---|---|
| KEGG: 04151 | PI3K-Akt signaling pathway | 3.28E-17 | AKT1, BCL2, BCL2L1, CCND1, CDKN1A, COL1A1, EGFR, ERBB2, ERBB3, GSK3B, IGF2, IKBKB, INSR, KDR, MAPK1, MCL1, MDM2, MET, MYC, RAF1, RXRA |
| KEGG: 04210 | Apoptosis | 4.69E-11 | AKT1, BAX, BCL2, BCL2L1, CASP8, IKBKB, JUN, MAPK1, MCL1, NFKBIA, PARP1, RAF1 |
| KEGG: 04012 | ErbB signaling pathway | 2.45E-10 | AKT1, CDKN1A, EGFR, ERBB2, ERBB3, GSK3B, JUN, MAPK1, MYC, RAF1 |
| KEGG: 04115 | p53 signaling pathway | 1.76E-09 | BAX, BCL2, BCL2L1, CASP8, CCND1, CDKN1A, CHEK1, CHEK2, MDM2 |
| KEGG: 04625 | C-type lectin receptor signaling pathway | 1.79E-09 | AKT1, CASP8, IKBKB, IL10, JUN, MAPK1, MDM2, NFKBIA, RAF1, STAT1 |
| KEGG: 04510 | Focal adhesion | 3.56E-09 | AKT1, BCL2, CCND1, COL1A1, EGFR, ERBB2, GSK3B, JUN, KDR, MAPK1, MET, RAF1 |
| KEGG: 04630 | JAK-STAT signaling pathway | 6.86E-09 | AKT1, BCL2, BCL2L1, CCND1, CDKN1A, EGFR, IL10, MCL1, MYC, RAF1, STAT1 |
| KEGG: 04068 | FoxO signaling pathway | 1.72E-08 | AKT1, CCND1, CDKN1A, EGFR, IKBKB, IL10, INSR, MAPK1, MDM2, RAF1 |
| KEGG: 04218 | Cellular senescence | 1.05E-07 | AKT1, CCND1, CDKN1A, CHEK1, CHEK2, MAPK1, MDM2, MYC, RAF1, RB1 |
| KEGG: 04066 | HIF-1 signaling pathway | 6.47E-07 | AKT1, BCL2, CDKN1A, EGFR, ERBB2, HIF1A, INSR, MAPK1 |
| KEGG: 04110 | Cell cycle | 2.95E-06 | CCND1, CDKN1A, CHEK1, CHEK2, GSK3B, MDM2, MYC, RB1 |
| KEGG: 04620 | Toll-like receptor signaling pathway | 1.28E-05 | AKT1, CASP8, IKBKB, JUN, MAPK1, NFKBIA, STAT1 |
| KEGG: 04380 | Osteoclast differentiation | 4.29E-05 | AKT1, IKBKB, JUN, MAPK1, NFKBIA, PPARG, STAT1 |
| KEGG: 04657 | IL-17 signaling pathway | 8.36E-05 | CASP8, GSK3B, IKBKB, JUN, MAPK1, NFKBIA |
| KEGG: 04215 | Apoptosis | 2.21E-04 | BAX, BCL2, BCL2L1, CASP8 |
| KEGG: 04520 | Adherens junction | 2.65E-04 | EGFR, ERBB2, INSR, MAPK1, MET |
| KEGG: 04658 | Th1 and Th2 cell differentiation | 7.11E-04 | IKBKB, JUN, MAPK1, NFKBIA, STAT1 |
| KEGG: 04064 | NF-kappa B signaling pathway | 7.37E-04 | BCL2, BCL2L1, IKBKB, NFKBIA, PARP1 |
| KEGG: 04370 | VEGF signaling pathway | 1.31E-03 | AKT1, KDR, MAPK1, RAF1 |
| KEGG: 04340 | Hedgehog signaling pathway | 6.15E-03 | BCL2, CCND1, GSK3B |
Figure 5.
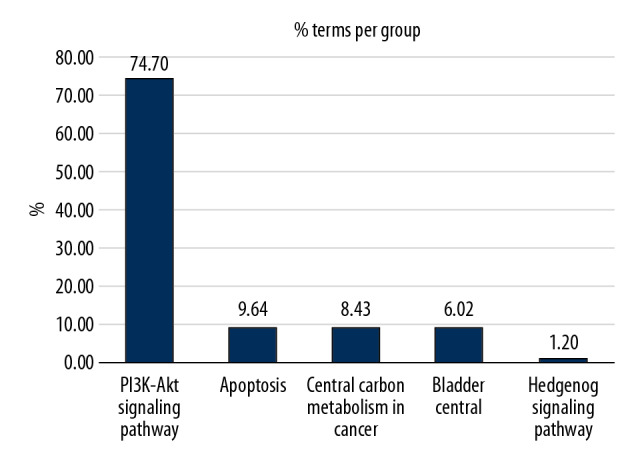
Enrichment analysis of KEGG signaling pathway. KEGG – Kyoto Encyclopedia of Genes and Genomes.
Discussion
The effectiveness of traditional Chinese medicine (TCM) and its derivatives in treating cancer has been more and more affirmed, and it has been widely used in the clinic [3,20]. A large number of clinical studies have reported the beneficial effects of TCM on the survival, immune modulation, and quality of life of cancer patients, which indicates that TCM has a significant therapeutic value and represents an excellent resource for drug discovery [21,22]. As we all know, TCM contains complex ingredients to enhance their therapeutic effects or reduce their toxicity. Therefore, it is crucial to find out and make rational use of the characteristics of TCM with multi-components, multi-targets, and multi-channels.
Network pharmacology looks at biological networks, analyzing the links among drugs, targets, and diseases in these networks, which provides a comprehensive and systematic study with the characteristics of a holistic view; the holistic view is the main feature of traditional Chinese medicine. Therefore, network pharmacology can be used to explore the synergistic effects of various targets and pathways of traditional Chinese medicine [17,18]. Artemisia annua and its derivatives have been found to have anti-cancer activity in vitro and in vivo [4,22–24]. Artemisia annua and its derivatives are not only widely used for the treatment of various tumors, but also the prevention of tumors [6,23–25]. Artemisia annua and its derivatives also exert synergistic anti-tumor effects with a wide array of clinically established drugs [26–28]. However, the target and mechanism of Artemisia annua against NSCLC have not been apparent.
In this study, 19 active ingredients of Artemisia annua were screened out via TCMSP database as potential targets. These potential ingredients were as follows: Luteolin, Quercetin, Isorhamnetin, Sitosterol, Kaempferol, Stigmasterol, EUPATIN, Tamarixetin, Patuletin, Areapillin, Artemetin, Skrofulein, Cirsiliol, vitexin_qt, DMQT, [(2S)-2-[[(2S)-2-(benzoyl amino)-3-phenylpropanoyl]amino]-3-phenylpropyl] acetate, 6,8-di-c-glucosylapigenin_qt, Artemisinin, and deoxyartemisinin. Then, we screened out 40 common target genes that might have potential anti-NSCLC function by the integration of a multi-source database.
In networks, the basic unit is a node, which can be a disease, target, or a drug; the connection between any 2 nodes is termed an edge. A particular group of related nodes is called a module and the connection between any 2 modules is termed a bridge. The degree and hubs are promising targets or parameters for drug discovery, and the complex relationships among diseases, targets, and drugs in network pharmacology can thus be visualized. In this study, we established a visual PPI network to analyze the interaction between these targets, and we based on the matching results between the potential targets of Artemisia annua and NSCLC tumor targets, screen the central targets by topology. We found that AKT1, MYC, CCND1, VEGFA, JUN, MAPK1, EGFR, ESR1 nodule area, and corresponding degree values were large, and could be easily found in the PPI network. We also found that AKT1, MYC, CCND1, VEGFA, JUN, MAPK1, EGFR, ESR1, and other genes are involved in 2 or more pathways as crosstalk genes, suggesting that these genes or pathways may be the main targets of Artemisia annua against NSCLC. As we all know, these pathways and genes are mainly involved in tumor cell proliferation, apoptosis, cell cycle regulation, epithelial-stromal transformation, angiogenesis, tumor invasion and metastasis, tumor signal transduction, immune regulation, drug resistance and other critical biological processes [27–30]. More importantly, these pathways and genes might cooperate in promoting metastasis of NSCLC through crosstalk between cancer and immune microenvironment [29–31].
Furthermore, GO enrichment analysis was used to analyze the biological process of Artemisia annua’s potential anti-NSCLC targets. We found that Artemisia annua’s anti-NSCLC target involved 10 biological processes, including the regulation of oxidative stress-induced intrinsic apoptotic signaling pathway, regulation of mitochondrial depolarization, negative regulation of hydrogen peroxide-induced cell death, regulation of anoikis, cytoplasmic sequestering of NF-kappaB, regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway, and monocyte differentiation. These biological processes are related to the occurrence of NSCLC, reflecting the characteristics of Artemisia annua as a multi-component, multi-target, and multi-channel treatment of NSCLC.
According to the analysis of KEGG signal pathway enrichment, there were 68 signal pathways related to the effect of Artemisia annua on NSCLC, including PI3K, Akt, JAK stat, erbB, FOXO, p53, HIF-1, toll-like receptor, TNF, and VEGF, which are strictly related to the pathogenesis of cancer. According to reports, inhibition of NSCLC mediated by IFN-g is regulated by PI3K-Akt signal transduction, and the inhibition of PI3K can downregulate the expression of PD-L1 and enhance the anti-tumor effect of IFN-g, which indicates that the inhibition of PI3K can maximize the anti-tumor impact mediated by IFN-g [32]. Xu et al. found that the expression of JAK2 gene was upregulated in cancer tissues, and was related to lymph node metastasis, and upregulated expression of JAK2 could enhance the ability of tumor cell proliferation, metastasis, and invasion. In contrast, downregulated expression of JAK2 had the opposite effect [33]. Kachroo et al. found that interleukin-27 (IL-27) could induce epithelial cell transformation and inhibit the generation of angiogenic factors through STAT1 in NSCLC cell lines [34]. Another study found that STAT3 could induce MDSCs and macrophages to kill CD4+ and CD8+T cells, induce the growth and inhibition of dendritic cells (DCS), and make macrophages differentiate from M1 to M2 in lung tumorigenesis, thus promoting the development of lung cancer [35]. Quercetin is the active ingredient in Artemisia annua. It exerts anticancer effects on cancer cells and tumors by regulating PI3K/Akt/mTOR, Wnt/b-catenin, and MAPK/ERK1/2 pathways. Quercetin promotes the loss of cell viability, apoptosis, and autophagy in cancer by reducing the stability of b-catenin and HIF-1a, inducing caspase-3 activation and inhibiting Akt, mTOR, and ERK phosphorylation [36]. Kaempferol is the active ingredient that also inhibits apoptosis, autophagy, and proliferation in NSCLC cells, which might function through the upregulation of PTEN and inactivation of the PI3K/AKT pathway [37]. Therefore, it is speculated that Artemisia annua might exert an anti-NSCLC effect by acting on the related signaling pathways or targets.
Although the present research revealed the mechanism of Artemisia annua in the treatment of NSCLC from the perspective of network pharmacology, it still has several limitations. The main limitation of this study is the lack of experimental verification, which will be further expanded in future research.
Conclusions
In summary, bioinformatics and network pharmacology analysis reveals the molecular biological mechanism of Artemisia annua anti-NSCLC via multiple active components, multi-channels, and multi-targets. The targets identified included AKT1, MYC, CCND1, VEGFA, JUN, MAPK1, EGFR, ESR1, et al. in the network of AN and NSCLC, indicating potential as therapeutic biomolecules for treating NSCLS. It is suggested that Artemisia annua might be developed as a promising anti-NSCLC drug.
Footnotes
Conflicts of interest
None.
Source of support: The present study was supported in part by research grants (No. 81873248 and No. 81673903) from the National Natural Science Foundation of China
References
- 1.Zhang XC, Wang J, Shao GG, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun. 2019;10:1772. doi: 10.1038/s41467-019-09762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altorki NK, Markowitz GJ, Gao DC, et al. The lung microenvironment: An important regulator of tumor growth and metastasis. Nat Rev Cancer. 2019;19(1):9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasymjanova G, Tran AT, Cohen V, et al. The use of a standardized Chinese herbal formula in patients with advanced lung cancer: A feasibility study. J Integr Med. 2018;16(6):390–95. doi: 10.1016/j.joim.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Efferth T. From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin Cancer Biol. 2017;46:65–83. doi: 10.1016/j.semcancer.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Abba ML, Patil N, Leupold JH, et al. Prevention of carcinogenesis and metastasis by Artemisinin-type drugs. Cancer Lett. 2018;429:11–18. doi: 10.1016/j.canlet.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Liang Y, He C. Anticancer activities and mechanisms of heat-clearing and detoxicating traditional Chinese herbal medicine. Chin Med. 2017;12:20. doi: 10.1186/s13020-017-0140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Neill PM, Posner GH. A medicinal chemistry perspective on artemisinin and related endoperoxides. J Med Chem. 2004;47(12):2945–64. doi: 10.1021/jm030571c. [DOI] [PubMed] [Google Scholar]
- 8.Wang JG, Zhang CJ, Chia WN, et al. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat Commun. 2015;6:10111. doi: 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shterman N, Kupfer B, Moroz C. Comparison of transferrin receptors, iron content and isoferritin profile in normal and malignant human breast cell lines. Pathobiology. 1991;59(1):19–25. doi: 10.1159/000163611. [DOI] [PubMed] [Google Scholar]
- 10.Kwok J, Richardson DR. The iron metabolism of neoplastic cells: Alterations that facilitate proliferation? Crit Rev Oncol Hematol. 2002;42(1):65–78. doi: 10.1016/s1040-8428(01)00213-x. [DOI] [PubMed] [Google Scholar]
- 11.Lai H, Singh NP. Selective cancer cell cytotoxicity from exposure to dihydroartemisinin and holotransferrin. Cancer Lett. 1995;91(1):41–46. doi: 10.1016/0304-3835(94)03716-v. [DOI] [PubMed] [Google Scholar]
- 12.Zhai DD, Supaibulwatana K, Zhong JJ. Inhibition of tumor cell proliferation and induction of apoptosis in human lung carcinoma 95-D cells by a new sesquiterpene from hairy root cultures of Artemisia annua. Phytomedicine. 2010;17(11):856–61. doi: 10.1016/j.phymed.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Rassias DJ, Weathers PJ. Dried leaf Artemisia annua efficacy against non-small cell lung cancer. Phytomedicine. 2019;52:856–61. doi: 10.1016/j.phymed.2018.09.167. [DOI] [PubMed] [Google Scholar]
- 14.Abba ML, Patil N, Leupold JH, et al. Prevention of carcinogenesis and metastasis by Artemisinin-type drugs. Cancer Lett. 2018;429:11–18. doi: 10.1016/j.canlet.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Zhang ZY, Yu SQ. [Artesunate combined with vinorelbine plus cisplatin in treatment of advanced non-small cell lung cancer: A randomized controlled trial]. Zhong Xi Yi Jie He Xue Bao. 2008;6(2):134–38. doi: 10.3736/jcim20080206. [DOI] [PubMed] [Google Scholar]
- 16.Li SG, Chen HY, Ou-Yang CS, et al. The efficacy of Chinese herbal medicine as an adjunctive therapy for advanced non-small cell lung cancer: A systematic review and meta-analysis. PLoS One. 2013;8(2):e57604. doi: 10.1371/journal.pone.0057604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Li XY, Xu XL, Yang NX. Mechanisms of Paeonia lactiflora in treatment of ulcerative colitis: A network pharmacological study. Med Sci Monit. 2019;25:7574–80. doi: 10.12659/MSM.917695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo B, Que ZJ, Zhou ZY, et al. Feiji recipe inhibits the growth of lung cancer by modulating T-cell immunity through indoleamine-2,3-dioxygenase pathway in an orthotopic implantation model. J Inegr Med. 2018;16(4):283–89. doi: 10.1016/j.joim.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Min S, Chao G, Liu MZ, et al. Therapeutic targets of vitamin C on liver injury and associated biological mechanisms: A study of network pharmacology. Int Immunopharmacol. 2019;66:383–87. doi: 10.1016/j.intimp.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 20.Luo H, Vong CT, Chen HB, et al. Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine. Chin Med. 2019;14:48. doi: 10.1186/s13020-019-0270-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen SJ, Zhang YH, Gu XX, et al. Yangfei Kongliu formula, a compound Chinese herbal medicine, combined with cisplatin, inhibits growth of lung cancer cells through transforming growth factor-b1 signaling pathway. J Inegr Med. 2017;15(3):242–51. doi: 10.1016/S2095-4964(17)60330-3. [DOI] [PubMed] [Google Scholar]
- 22.Rasheed SA, Efferth T, Asangani IA, Allgayer H. First evidence that the antimalarial drug artesunate inhibits invasion and in vivo metastasis in lung cancer by targeting essential extracellular proteases. Int J Cancer. 2010;127:1475–85. doi: 10.1002/ijc.25315. [DOI] [PubMed] [Google Scholar]
- 23.Odaka Y, Xu BS, Luo Y, et al. Dihydroartemisinin inhibits the mammalian target of rapamycin-mediated signaling pathways in tumor cells. Carcinogenesis. 2014;35(1):192–200. doi: 10.1093/carcin/bgt277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong Y, Liu Y, Zheng H, et al. Artemisinin and its derivatives can significantly inhibit lung tumorigenesis and tumor metastasis through Wnt/beta-catenin signaling. Oncotarget. 2016;7:31413–28. doi: 10.18632/oncotarget.8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi YJ, Geng GJ, Zou ZZ, et al. Dihydroartemisinin inhibits glucose uptake and cooperates with glycolysis inhibitor to induce apoptosis in non-small cell lung carcinoma cells. PLoS One. 2015;10:e0120426. doi: 10.1371/journal.pone.0120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeed MEM, Breuer E, Hegazy MEF, Efferth T. Retrospective study of small pet tumors treated with Artemisia annua and iron. Int J Oncol. 2020;56(1):123–13. doi: 10.3892/ijo.2019.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efferth T. Cancer combination therapies with artemisinin-type drugs. Biochem Pharmacol. 2017;139:56–70. doi: 10.1016/j.bcp.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Petrek H, Yu AM. MicroRNAs in non-small cell lung cancer: Gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacol Res Perspect. 2019;7(6):e00528. doi: 10.1002/prp2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dragoj M, Bankovic J, Podolski-Renic A, et al. Association of overexpressed MYC gene with altered PHACTR3 and E2F4 genes contributes to non-small cell lung carcinoma pathogenesis. J Med Biochem. 2019;38(2):188–95. doi: 10.2478/jomb-2018-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao MY, Xu P, Liu Z, et al. Dual roles of miR-374a by modulated c-Jun respectively targets CCND1-inducing PI3K/AKT signal and PTEN-suppressing Wnt/b-catenin signaling in non-small-cell lung cancer. Cell Death Dis. 2018;9(2):78. doi: 10.1038/s41419-017-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang XC, Wang J, Shao GG, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun. 2019;10:1772. doi: 10.1038/s41467-019-09762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi G, Yang JJ, Cai YX, et al. IFN-g-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143(4):931–43. doi: 10.1002/ijc.31357. [DOI] [PubMed] [Google Scholar]
- 33.Xu XJ, Jin J, Xu JW, et al. JAK2 variations and functions in lung adenocarcinoma. Tumour Biol. 2017;39(6) doi: 10.1177/1010428317711140. 1010428317711140. [DOI] [PubMed] [Google Scholar]
- 34.Kachroo P, Lee MH, Zhang L, et al. IL-27 inhibits epithelial-mesenchymal transition and angiogenic factor production in a STAT1-dominant pathway in human non-small cell lung cancer. J Exp Clin Cancer Res. 2013;32:97. doi: 10.1186/1756-9966-32-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou JJ, Qu ZX, Sun F, et al. Myeloid STAT3 promotes lung tumorigenesis by transforming tumor immunosurveillance into tumor-promoting inflammation. Cancer Immunol Res. 2017;5(3):257–68. doi: 10.1158/2326-6066.CIR-16-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marjorie RF, Catalina CP. The anti-cancer effect of quercetin: molecular implications in cancer metabolism. Int J Mol Sci. 2019;20(13):3177. doi: 10.3390/ijms20133177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X, Liu CF, Gao N, et al. Kaempferol suppresses proliferation but increases apoptosis and autophagy by up-regulating microRNA-340 in human lung cancer cells. Biomed Pharmacother. 2018;108:809–16. doi: 10.1016/j.biopha.2018.09.087. [DOI] [PubMed] [Google Scholar]