Abstract
Diffuse large B cell lymphoma (DLBCL) is the most common hematological malignancy and is one of the most frequent non-Hodgkin lymphomas. Large-scale genomic studies have defined genetic drivers of DLBCL and their association with functional and clinical outcomes. However, the lymphomagenesis of DLBCL is yet to be fully understood. In the present study, four computational tools OncodriveFM, OncodriveCLUST, integrated Cancer Genome Score and Driver Genes and Pathways were used to detect driver genes and driver pathways involved in DLBCL. The aforementioned tools were also used to perform an integrative investigation of driver genes, including co-expression network, protein-protein interaction, copy number variation and survival analyses. The present study identified 208 driver genes and 31 driver pathways in DLBCL. IGLL5, MLL2, BTG2, B2M, PIM1, CARD11 were the top five frequently mutated genes in DLBCL. NOTCH3, LAMC1, COL4A1, PDGFRB and KDR were the 5 hub genes in the blue module that were associated with patient age. TP53, MYC, EGFR, PTEN, IL6, STAT3, MAPK8, TNF and CDH1 were at the core of the protein-protein interaction network. PRDM1, CDKN2A, CDKN2B, TNFAIP3, RSPO3 were the top five frequently deleted driver genes in DLBCL, while ACTB, BTG2, PLET1, CARD11, DIXDC1 were the top five frequently amplified driver genes in DLBCL. High EIF3B, MLH1, PPP1CA and RECQL4 expression was associated with decreased overall survival rate of patients with DLBCL. High XPO1 and LYN expression were associated with increased overall survival rate of patients with DLBCL. The present study improves the understanding of the biological processes and pathways involved in lymphomagenesis. The driver genes, EIF3B, MLH1, PPP1CA, RECQL4, XPO1 and LYN, pave the way for developing prognostic biomarkers and new therapeutic strategies for DLBCL.
Keywords: driver gene, driver pathway, WGCNA, PPI network, CNV, overall survival rate, diffuse large B cell lymphoma
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive non-Hodgkin lymphoma (1). The incidence rate is 6.3%, with an estimated 25,380 new cases in the United States in 2016 (2). DLBCL can be classified into three molecular subtypes; the germinal center B cell-like subtype, the activated B cell-like subtype and primary mediastinal B cell lymphoma (1). Cyclophosphamide, doxorubicin, vincristine and prednisolone (CHOP) are the standard treatment for non-Hodgkin lymphoma (3). The 3-year overall survival rate is ~60% following CHOP treatment in patients with DLBCL (4).
Large genomics studies have been conducted to characterize the genetic alterations in DLBCL genomes, which provide an unbiased view of the landscape of mutations and the pathogenesis of the disease (5–7). Lohr et al (5) performed exome sequencing on 55 paired tumor and normal samples of primary DLBCL and identified 58 significantly mutated genes, such as CD79b molecule (CD79B), Tumor Protein P53 (TP53), caspase recruitment domain family member 11 (CARD11), MYD88 innate immune signal transduction adaptor (MYD88) and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2). Reddy et al (6) identified 150 genetic drivers by performing an integrative analysis of whole exome sequencing in a cohort of 1,001 patients with DLBCL. In the previously mentioned study, CRISPR-based knockout of 35 driver genes resulted in the decreased viability of DLBCL cells, suggesting these driver genes serve an oncogenic function. MYC proto-oncogene (MYC), CD79B and zinc finger and AT-hook domain containing (ZFAT) mutations were significantly associated with poor overall survival rate of patients with DLBCL. While, mutations in neurofibromin 1 and serum/glucocorticoid regulated kinase 1 were associated with favorable overall survival rate of patients with DLBCL (5). Chapuy et al (7) integrated genetic drivers using consensus clustering and identified 5 distinct DLBCL subgroups associated with distinct pathogenic mechanisms and clinical outcomes. For example, tumors in the activated B cell (ABC) and germinal center B cell (GCB)-independent group are characterized by biallelic inactivation of TP53, cyclin dependent kinase inhibitor 2A loss, and associated with genomic instability (7).
Driver genes that are recurrently mutated in a large cohort of cancer samples have consistently been a focus of the previously published studies of DLBCL; however, the mutation frequency of numerous driver genes may remain relatively low (e.g. <1%) in tumors (8). Few studies have been conducted on the driver genes with low mutation frequency in DLBCL (5,6). In the present study, four computational tools were used to identify driver genes and conduct integrative analyses on these genes in 48 DLBCL samples. The study aimed to detect novel driver genes, driver pathways and their association with clinical characteristics of patients with DLBCL; enhancing the understanding of this disease and providing potential therapeutic targets in DLBCL.
Materials and methods
Data for analysis of somatic mutations in DLBCL
In total, 16,918 somatic mutations of 8,672 genes were identified in 48 patients with DLBCL (22 men and 26 women; age range, 23–82 years; mean age, 56.27 years) and obtained from The Cancer Genome Atlas (TCGA) database (9). The functional impact of somatic mutations was evaluated using Ensembl Variant Effect Predictor (10) and the mutations were classified into 9 categories according to their functional impact, including frame shift insertions and deletions (indels), in frame indels, missense mutation, nonsense mutation, RNA, silent and splice site. RNA denotes somatic mutations that are located in the 5′-untranslated region (UTR) or 3′-UTR and may be functional, but probably act by impacting RNA levels.
Prediction of driver genes and pathways
Driver genes were predicted using 4 distinct computational tools, including OncodriveCLUST v.0.4.1 (11), OncodriveFM v.0.0.1 (12), The integrated Cancer Genome Score (iCAGES,) (13) and Drivers Genes and Pathways (DrGaP v.0.1.0) (14). The parameters were set to default values. Driver genes were determined based on the following criteria: Genes with q-values <0.05 were considered as driver genes using OncodriveCLUST and OncodriveFM; genes with iCAGES gene scores >0.5 were determined as drivers using iCAGES and genes or pathways with P-values <0.05 were regarded as driver genes or pathways using DrGaP. To further annotate the driver genes, the list of driver genes were compared with curated ONGene (15) and TSGene (16) databases.
Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses
In order to characterize the functional enrichment of all driver genes, GO (17) biological process terms and KEGG pathway enrichment analyses (18) were performed with The Database for Annotation, Visualization and Integrated Discovery (DAVID) (19). Driver genes were considered to be significantly enriched in GO terms or KEGG pathways using a cut-off of Benjamini adjusted P-value of <0.05.
Co-expression network analysis in patients with DLBCL
Normalized read counts of driver genes of 48 patients with DLBCL were downloaded from TCGA database (http://firebrowse.org/?cohort=DLBC&download_dialog=true). Co-expression networks were constructed with the R package of weighted gene co-expression network analysis (WGCNA version 1.67) using normalized read counts of driver genes (20). The softpower and minimum number of genes were set as 7 and 10 respectively, all other parameters were set to the default values. Identification of gene co-expression modules was conducted using hierarchical average-linkage clustering. The dynamic tree-cut algorithm was used to identify modules and genes in the same branch that could be assigned to different modules (20). Genes with high intramodular connectivity were considered as intramodular hub genes. The clinicopathologic characteristics investigated in the study are patient age, sex, clinical stage [Ann Arbor staging system; (21)], radiation therapy, ethnicity, survival status and follow-up time and were obtained from the TCGA database. Module-trait associations were estimated using the correlation between the module eigengene and clinical traits, which enables the identification of modules highly correlated with clinical features.
Protein-protein interaction (PPI) network construction and analysis
A PPI network was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (22). Visualization and calculation of degree value for each node was performed using Cytoscape software v3.7.2 (23). Degree centrality was defined as the number of connections one node has and was also analyzed using Cytoscape software. Hub nodes which have the highest degree of centrality connect most adjacent proteins in the PPI network (22). Furthermore, Molecular Complex Detection (MCODE) (24) was used to detect hub clustering modules in the PPI network with default parameters in Cytoscape. GO and KEGG pathway enrichment analyses were also conducted for genes in significant modules.
Copy number variation (CNV) analyses
Focal CNVs and gene-level CNVs of 48 DLBCL samples were detected using the GISTIC algorithm (25) and were downloaded from TCGA database (9). Focal CNVs were considered statistically significant at the cut-off value of q<0.25. The gene-level copy-number alterations of the top 20 frequently altered driver genes were clustered using the heatmap.2 function of gplots package in R (26).
Bootstrap model validation of survival analyses
In order to confirm the associations of driver gene expression with overall survival rate in patients with DLBCL, bootstrap methodology (27) was used for validation. Bootstrapping methodology randomly selected 80% of samples with replacement from the original dataset as a ‘training’ set to determine the median values for driver genes. The original dataset was then used as a ‘testing’ set in which the patients were divided into high and low-expression groups according to the median values. The log-rank test was used to compare the difference in survival rates between the high- and low-expression groups using the R package of survival V3.1–11 (28). This process was repeated 1,000 times, generating 1,000 P-values for each driver gene and the frequency of P<0.05 was counted for each driver gene.
Statistical analysis
The difference in mutation density between groups were compared by Wilcoxon rank-sum test. Linear regression model was used to characterize the associations between clinical features, driver genes and overall survival rate. To establish the association of driver gene expression with overall survival rate of patients with DLBCL, patients were assigned to the ‘high-expression’ group if they exhibited gene expression levels greater than the median values of driver gene expression and to the ‘low-expression’ group if they exhibited expression levels lower than the median value. Kaplan-Meier survival analysis was performed, and survival curves generated. The log-rank test was used to compare the difference in survival rates between the high- and low-expression groups using the R package of survival v3.1–11 (28). P<0.05 was considered to indicate a statistically significant difference.
Results
Somatic mutations in patients with DLBCL
In total, 16,918 somatic mutations were detected in patients with DLBCL (n=48). Somatic mutations were comprised of 9,623 missense, 6,230 silent, 188 splice-site, 353 nonsense, 9 RNA and 515 indels. Of the 515 indels, 332 caused reading frame shifts, and 135 deletions and 48 insertions were located in open reading frames (Fig. S1A). C>T/G>A, A>G/T>C and C>A/G>T were the 3 predominant transitions, with mutation rates of 52.9, 17.5 and 8.5% respectively (Fig. S1B). Indels accounted for 3.2% of variants in DLBCL (Fig. S1B). The somatic mutation density ranged between 0.68–131.29 mutations/megabase (Mb) with an average mutation density of 9.64 mutations/Mb (data not shown). To understand the cause of the mutation density variation, mutation statuses in the DNA mismatch-repair (MMR) pathway genes mutL homolog 1 (MLH1), mutL homolog 3 (MLH3), mutS homolog 2 (MSH2), mutS homolog 3 (MSH3), mutS homolog 6 (MSH6) and PMS1 homolog 2 (PMS2) were analyzed. This revealed that 11 patients with DLBCL had mutations in one of the MMR genes and the average mutation density in patients harboring an MMR mutation was significantly higher compared with wild-type MMR (23.97 mutations/Mb vs. 5.40 mutations/Mb; P<0.01; Fig. S1C). Notably, the DLBCL with the highest mutation density (131.29 mutations/Mb) had 1 missense mutation in PMS2 (data not shown).
Prediction of driver genes and pathways
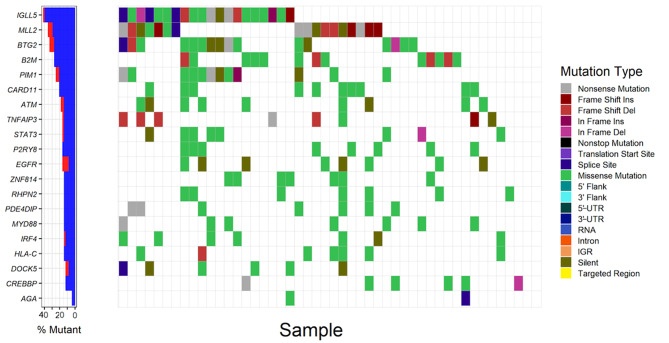
Overall, 8,672 genes were mutated in ≥ one DLBCL sample. There were 12, 47, 109 and 59 driver genes predicted by OncodriveCLUST, OncodriveFM, iCAGES and DrGaP respectively (Tables SI–IV). Combining these 4 sets of driver genes, a total of 208 unique driver genes were detected using all 4 tools. Zinc finger protein 814 (ZNF814), major histocompatibility complex, class I, C (HLA-C), CD79B, rhophilin Rho GTPase binding protein 2 (RHPN2), MYD88 and EZH2 were the common genes identified by OncodriveCLUST and OncodriveFM. Suppressor of cytokine signaling 1 (SOCS1), TP53, signal transducer and activator of transcription 6 (STAT6), actin beta (ACTB), protein tyrosine phosphatase non-receptor type 6 (PTPN6) and LYN proto-oncogene (LYN) were common to OncodriveFM and iCAGES. Fas cell surface death receptor (FAS), inhibitor of nuclear factor kappa B kinase subunit beta (IKBKB), tumor necrosis factor (TNF) and TP53 were common driver genes detected by iCAGES and DrGaP. TP53, BTG anti-proliferation factor 2 (BTG2) and ubiquitin conjugating enzyme E2 A (UBE2A) were common to OncodriveFM and DrGaP, MLH1 was the overlapping driver gene found between OncodriveCLUST and iCAGES. TP53 was the only driver gene predicted by OncodriveFM, iCAGES and DrGaP. Among the 208 driver genes; immunoglobulin lambda like polypeptide 5 (IGLL5), myeloid/lymphoid or mixed-lineage leukemia 2 (MLL2), BTG anti-proliferation factor 2 (BTG2), beta-2-microglobulin (B2M) and Pim-1 proto-oncogene, serine/threonine kinase (PIM1) were the top five recurrently-mutated genes in patients with DLBCL with mutation rates of 41.7, 35.4, 33.3, 27.1, 25.0, respectively (Fig. 1; Table SV). The majority of driver genes were mutated at a low frequency in DLBCL with an average mutation rate of 6.1% (Table SV). By comparing the list of driver genes with curated ONGene and TSGene databases, numerous known oncogenes were identified in the current study, such as signal transducer and activator of transcription 3 (STAT3), epidermal growth factor receptor (EGFR), as well as tumor suppressor genes, such as ATM serine/threonine kinase (ATM), phosphatase and tensin homolog (PTEN). In addition to the list of driver genes, DrGaP also identified 31 driver pathways in DLBCL, including the MAPK signalling pathway, cytokine-cytokine receptor interaction, cell cycle, apoptosis, p53 signalling pathway, pathways in cancer, pancreatic cancer, the Wnt signalling pathway and chronic myeloid leukemia (data not shown).
Figure 1.
Analysis of somatic mutations of the top 20 frequently mutated driver genes in patients with DLBCL (n=48). The left panel demonstrates the mutation rates of the 20 driver genes, blue and red denote synonymous and non-synonymous mutations respectively. The right panel demonstrates the distribution of mutations with different classes of functions in patient samples. UTR, untranslated region; IGR, intergenic region; DLBCL, diffuse large B cell lymphoma; Ins, insertion; Del, deletion; IGLL5, Immunoglobulin lambda like polypeptide 5; MLL2, myeloid/lymphoid or mixed-lineage leukemia 2; BTG2, BTG anti-proliferation factor 2; B2M, beta-2-microglobulin; PIM1, Pim-1 proto-oncogene; CARD11, caspase recruitment domain family member 11; ATM, ATM serine/threonine kinase; TNFAIP3, TNF alpha induced protein 3; STAT3, signal transducer and activator of transcription 3; P2YR8, P2Y receptor family member 8; EGFR, epidermal growth factor receptor; ZNF184, zinc finger protein 184; RHPN2, rhophilin Rho GTPase binding protein 2; PDE4DIP, phosphodiesterase 4D interacting protein; MYD88, MYD88 innate immune signal transduction adaptor; IRF4, interferon regulatory factor 4; HLA-C, major histocompatibility complex class I C; DOCK5, dedicator of cytokinesis 5; CREBBP, CREB binding protein; AGA, aspartylglucosaminidase.
GO term and KEGG pathway enrichment analyses
GO term and KEGG pathway enrichment analyses were performed for 208 driver genes using DAVID. GO enrichment analysis indicated that driver genes were significantly overrepresented in 35 biological processes (Benjamini-adjusted P-value <0.05; Table SVI). The main GO biological processes exhibited a wide spectrum of functional processes, including ‘IκB kinase/NF-κB signaling’, ‘extracellular matrix organization’ and ‘regulation of phosphatidylinositol 3-kinase signaling’ DAVID also revealed driver genes were significantly enriched in 88 KEGG pathways, including ‘acute myeloid leukemia’, ‘melanoma’, ‘colorectal cancer’, ‘non-small cell lung cancer’, ‘T cell receptor signaling pathway’, ‘antigen processing and presentation’, ‘the mTOR signaling pathway’, ‘apoptosis’ and ‘cell cycle’ (Benjamini-adjusted P-value <0.05; Table SVII).
Co-expression network analysis
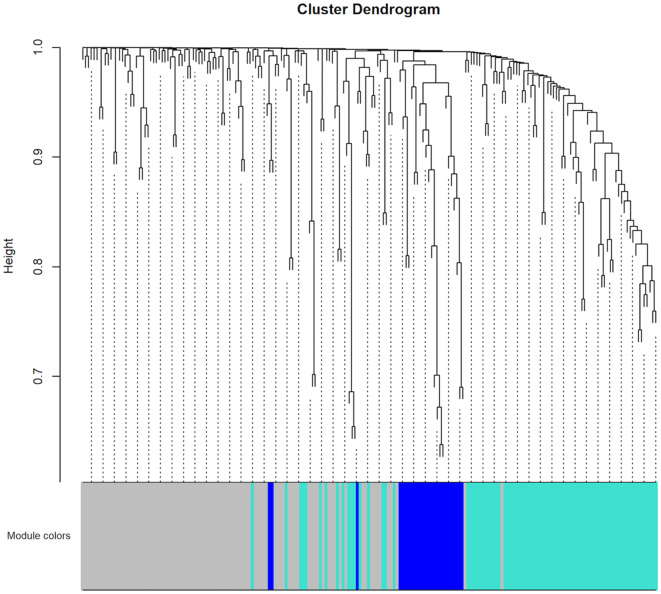
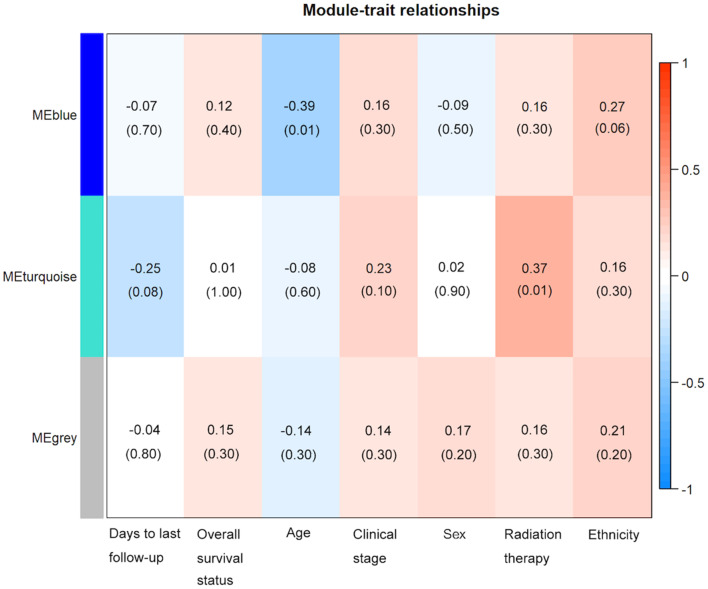
To construct the co-expression network of the 208 driver genes, WGCNA was used based on the expression correlation between driver genes in 48 DLBCL samples. WGCNA analysis identified 3 distinct co-expression modules in DLBCL. These co-expression modules are demonstrated in different colors with 94, 83 and 26 genes in the grey, turquoise and blue modules respectively (Fig. 2). The module-trait association analysis indicated that the turquoise module was positively correlated with radiation therapy and the blue module was negatively correlated with patient age (P<0.05; Fig. 3). Mitogen-activated protein kinase 8 (MAPK8) was the hub gene in the turquoise module. Notch receptor 3 (NOTCH3) was the hub gene in the blue module.
Figure 2.
Hierarchical clustering results of co-expression modules in the patients with DLBCL (n=48). WGCNA was applied to perform the hierarchical clustering. These co-expression modules are demonstrated in different colors with 83 and 26 genes in the turquoise and blue modules, respectively. The grey color represents no genes in any modules.
Figure 3.
Module-trait associations between mutated genes and clinical features in patients with DLBCL, established using weighted gene correlation network analysis. Heatmap presents the correlation between module eigengenes and clinical traits. The corresponding correlation coefficients were presented above the parentheses and P-values were shown within the parentheses. P<0.05 was considered to be statistically significant. The bar on the right demonstrates the degree of correlation between module eigengenes and clinical traits, with red and blue representing positive and negative correlation, respectively. ME, Module Eigengene.
Protein-protein interaction (PPI) network construction and analysis
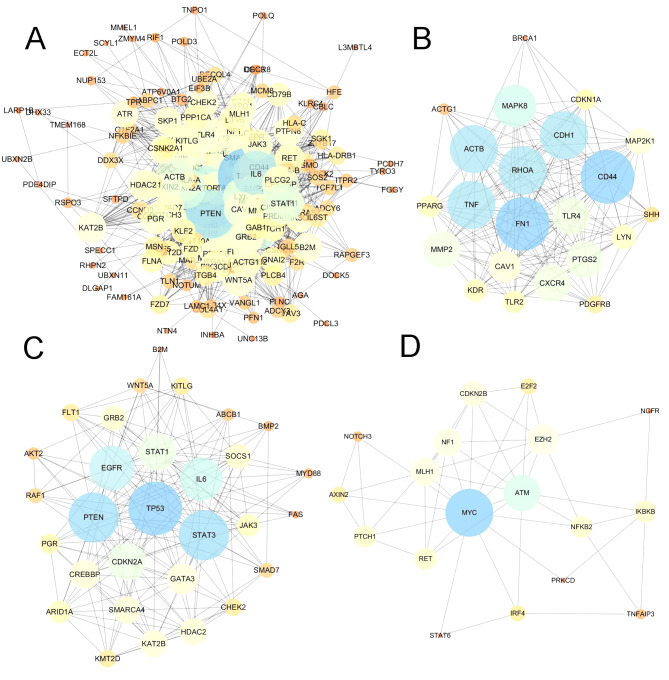
In addition to the co-expression network of driver genes, the present study also characterized the interactions of driver genes at the protein level. STRING was used to construct a PPI network for driver genes. The PPI network comprised 208 nodes and 2,041 edges, with an average node degree of 19.6 (Fig. 4A). The PPI network had significantly more interactions than expected for a random set of proteins of similar size (PPI enrichment; P<0.0001). The nodes which have high degrees possess intensive interactions with other nodes and may serve as key nodes in the PPI network. A total of 9 candidate hub nodes, the degree of which was >4 times the corresponding median values were identified, namely, TP53, MYC, EGFR, PTEN, interleukin 6 (IL6), signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinase 8 (MAPK8), tumor necrosis factor (TNF) and cadherin 1 (CDH1) (Fig. 4A). Furthermore, module analysis was performed to obtain the top 3 modules with high scores using MCODE (Fig. 4B-D). The 9 candidate hub nodes were contained in the three modules. In relation to GO enrichment analysis, genes in module 1 (Fig. 4B) were significantly correlated with 208 GO terms, including ‘positive regulation of apoptosis’, ‘programmed cell death’, ‘cell migration’ and ‘response to hypoxia.’ Genes in module 2 (Fig. 4C) were primarily enriched in ‘regulation of apoptotic process’, ‘cell proliferation’, ‘MAPK cascade’ and ‘phosphatidylinositol-mediated signaling’. Genes in module 3 (Fig. 4D) were not significantly enriched in any GO terms. With respect to KEGG pathway enrichment analysis, the genes in module 1 were enriched in ‘leukocyte transendothelial migration’, ‘melanoma’ and ‘Toll-like receptor signaling pathway’. The genes in module 2 mainly were predominantly enriched in ‘chronic myeloid leukemia’, ‘acute myeloid leukemia’, ‘p53 signaling pathway’ and the ‘hypoxia-inducible factor 1 signaling pathway’. The genes in module 3 were significantly implicated in the ‘cell cycle’, ‘microRNAs in cancer’, ‘small-cell lung cancer’ and the ‘NF-κB signaling pathway’.
Figure 4.
PPI network and module clustering analyses identified several hub proteins in the PPI network. (A) PPI network of all driver genes. (B) Module 1 (MCODE score=15.81). (C) Module 2 (MCODE score=13.17). (D) Module 3 (MCODE score=5.65). MCODE was used to detect hub clustering modules in the PPI network, with default parameters in Cytoscape. Deep node color and increased node size were associated with increased degree value. PPI, protein-protein interaction; MCODE, Molecular Complex Detection.
Copy number variation (CNV) analyses
Focal CNVs of 48 patients with DLBCL were obtained from TCGA. Significant focal gains and deletions (q<0.25) were identified at 40 loci (14 amplifications and 26 deletions) in 93.8% (45/48) of DLBCL samples. Among them, deletions at 6q14.1 and 9p21.3, and amplifications at 1q24.2, 2p16.1 and 7p22.3 were the top 5 most frequent CNVs in DLBCL, with occurrence rates of 35.4 (17/48), 35.4 (17/48), 33.3 (16/48), 33.3 (16/48) and 33.3% (16/48), respectively (Fig. S2). PR/SET domain 1 (PRDM1), cyclin dependent kinase inhibitor 2A (CDKN2A), cyclin dependent kinase inhibitor 2B (CDKN2B), TNF alpha induced protein 3 (TNFAIP3) and R-spondin 3 (RSPO3) were the top five most frequently deleted driver genes in DLBCL, while actin beta (ACTB), BTG anti-proliferation factor 2 (BTG2), placenta expressed transcript 1 (PLET1), CARD11 and DIX domain containing 1 (DIXDC1) were the top five most frequently amplified driver genes in DLBCL (Fig. S3).
Prognosis of patients with DLBCL
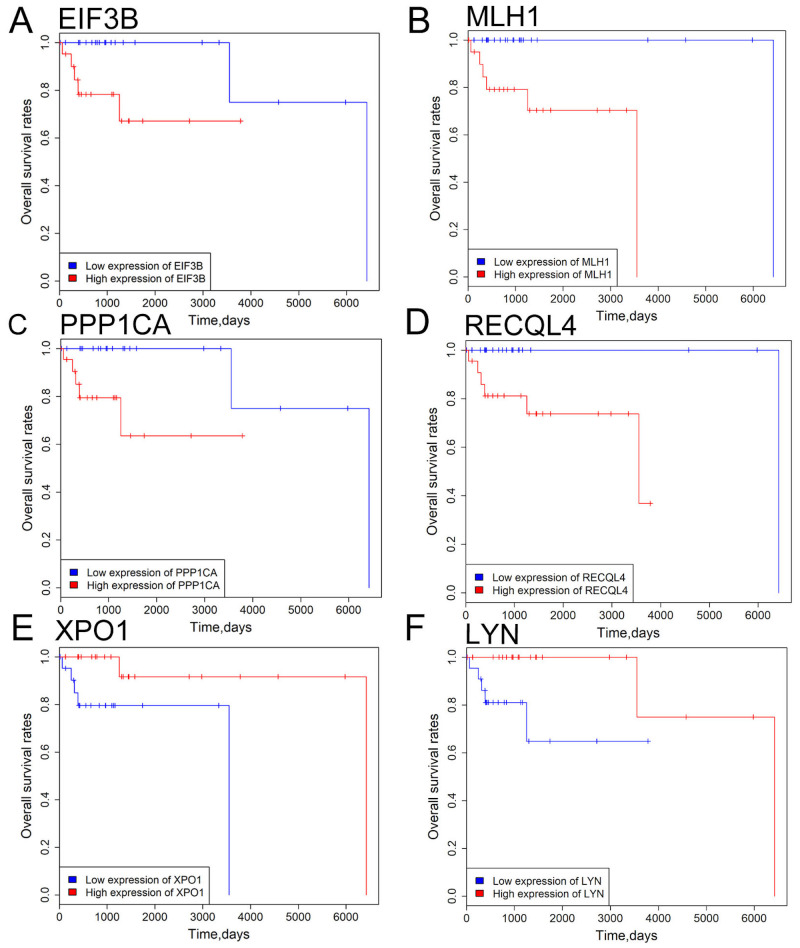
Linear regression model analysis demonstrated overall survival rate was not significantly associated with patient age, clinical stage, radiation therapy, sex and ethnicity in DLBCL (all P>0.05; Table SVIII). To evaluate the association of driver gene expression with patient survival, patients with DLBCL were divided into low- and high-expression groups based on the median expression values of the driver genes. Kaplan-Meier survival analysis indicated that patients with high eukaryotic translation initiation factor 3 subunit B (EIF3B), mutL homolog 1(MLH1), protein phosphatase 1 catalytic subunit alpha (PPP1CA) and RecQ like helicase 4 (RECQL4) expression levels exhibited improved overall survival rate compared with those with low EIF3B, MLH1, PPP1CA and RECQL4 expression levels (Fig. 5). Patients with high exportin 1 (XPO1) and LYN expression exhibited a less favorable prognosis compared with patients with low XPO1 and LYN expression (all P<0.05; Fig. 5). To further verify the aforementioned findings, driver genes were evaluated for their associations with overall survival rate using Kaplan-Meier survival analysis with 1,000 bootstrap resampling. EIF3B, MLH1, PPP1CA, RECQL4, XPO1 and LYN were significantly associated with overall survival rate in patients with DLBCL across the 1,000 bootstrapped samples. EIF3B, MLH1, PPP1CA, RECQL4 and XPO1 exhibited a P-value <0.05 in >60% of the 1,000 testing datasets (EIF3B, 82.6%; MLH1, 94.1%; PPP1CA, 83.2%; RECQL4, 93%; and XPO1, 64.8%), while LYN had a P-value <0.05 in only 29.2% of the testing datasets (data not shown). The current results indicate that EIF3B, MLH1, PPP1CA, RECQL4 and XPO1 may represent potential prognostic biomarkers for patients with DLBCL in the future.
Figure 5.
Survival analysis of 6 driver genes in patients with DLBCL. (A) EIF3B, (B) MLH1, (C) PPP1CA, (D) RECQL4 expression, (E) XPO1 and (F) LYN expression levels were significantly associated with overall survival rate of DLBCL patients. Red and blue curves represent high- and low-expression groups, respectively. P<0.05 was considered to be statistically significant. EIF3B, eukaryotic translation initiation factor 3 subunit B; MLH1, mutL homolog 1; PPP1CA, protein phosphatase 1 catalytic subunit alpha; RECQL4, RecQ like helicase 4; XPO1, exportin 1; LYN, LYN proto-oncogene.
Discussion
Cancer is initiated by the accumulation of driver mutations in cancer genes, which confers a proliferation advantage to cancer cells (29). The average mutation rate is 9.64 mutations/Mb in DLBCL, which is higher compared with the mutation rate in other hematopoietic malignancies, such as chronic lymphocytic leukemia and other leukemias (29,30), and multiple myeloma (31). In the present study, the mutation density varied considerably across DLBCL samples with increased mutation rates in MMR-mutant samples. Moreover, the DLBCL sample with the highest mutation density had one missense mutation in PMS2, which is a key component of the mismatch repair system to correct DNA mismatches and small indels that occur during DNA replication and homologous recombination (32). The results of the present study suggested that mutation density variation is, to a large extent, attributable to mutations in the DNA mismatch repair genes in DLBCL.
The widely applied approach for the detection of driver genes identifies significantly mutated genes in a cohort of cancer samples as compared with the background mutation rate (33,34). In the present study, 4 computational tools, OncodriveCLUST, OncodriveFM, iCAGES and DrGaP were used to detect driver genes using somatic mutations of patients with DLBCL (n=48). MLL2, TP53, CD79B, B2M, CARD11 and EZH2 were predicted as driver genes in DLBCL in the present study, which is consistent with previously published reports (5,6). By comparing the list of driver genes with curated oncogene (15) and tumor suppressor gene (16) databases, numerous known oncogenes were identified in the current study, such as EGFR, STAT3, as well as tumor suppressor genes, such as ATM, PTEN. Notably, in the present study a large fraction of driver genes had low mutation frequencies and were first reported as driver genes in DLBCL, such as CSNK2A1, RECQL4, LARP1B and GAB1. Therefore, the combination of the 4 tools enabled the detection of recurrently and rarely mutated driver genes. For instance, DrGap detected a new driver gene IGLL5, which was not identified via the other 3 computational tools. This may be due to DrGaP predicting driver genes and driver signaling pathways according to a different algorithm. DrGaP integrates biological knowledge of the mutational process in tumors, including the length of protein-coding regions, transcript isoforms, variation in mutation types, differences in background mutation rates, redundancy of the genetic code and multiple mutations in one gene. DrGaP use a Poisson process to model the random nature of somatic mutations, a Bayesian model to estimate background mutation rates and a likelihood ratio test to test the significance of driver genes and pathways. The newly identified driver genes in the present study provide promising candidates for functional validation in future studies.
By performing WGCNA analysis in the present study, 3 co-expression modules were detected, the turquoise module was positively associated with radiation therapy and the blue module was negatively associated with patient age. MAPK8 was the hub gene in the turquoise module. NOTCH3 was the hub gene indicating that these genes were correlated with other genes at the mRNA expression level. Therefore, they may have key roles in the co-expression network. The PPI network analysis also identified 9 candidate hub nodes, namely, TP53, MYC, EGFR, PTEN, IL6, STAT3, MAPK8, TNF and CDH1, and 3 modules. The 9 nodes identified in the present study were major hub nodes and the 3 modules may represent the key biological characteristics in the PPI network.
Finally, in the present study 5 driver genes were significantly associated with the overall survival rate of patients with DLBCL, including EIF3B, MLH1, PPP1CA, RECQL4 and XPO1. Of the five genes, XPO1 has been reported to be oncogene and a negative prognostic factor in mantle cell lymphoma (35), lung adenocarcinoma (36) and gastric cancer (37). XPO1 encodes a protein which functions as the trafficker of a wide range of proteins, including tumor suppressors, growth regulatory, proinflammatory and antiapoptotic proteins (38). XPO1 serves oncogenic and anti-apoptotic roles in transformed cells and is upregulated in mantle cell lymphoma (35), lung adenocarcinoma (36) and gastric cancer (37). In concordance with the findings of the present study, upregulated expression of XPO1 is associated with poor prognosis in gastric carcinoma (37), acute myeloid leukemia (39), pancreatic cancer (40) and lung adenocarcinoma (31). The results obtained in the present study, combined with previously published reports (35–39), indicate that XPO1 may exert oncogenic functions and represent a negative prognostic factor in cancers.
Expression analysis of EIF3B, MLH1, PPP1CA, RECQL4 and XPO1 may be valuable in clinical settings. Cytological or surgical specimens of DLBCL exhibiting high expression of EIF3B, MLH1, PPP1CA, RECQL4 and low expression of XPO1 may be associated with a favorable clinical outcome, which needs to be verified in large-scale and more vigorous future studies.
Despite enhancing the understanding of pathogenesis of DLBCL, the present study is not without limitations. For example, there was a lack of functional validation for the novel driver genes. Furthermore, the overall survival rate-associated genes were not validated in an independent DLBCL dataset, due to lack of publicly available DLBCL data. Overall, the present study discovered a set of driver genes and driver pathways in DLBCL, and demonstrated that the driver genes, such as EIF3B, MLH1, PPP1CA, RECQL4 and XPO1 may serve as potential prognostic biomarkers for patients with DLBCL.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- DLBCL
diffuse large B cell lymphoma
- TCGA
The Cancer Genome Atlas
- iCAGES
Integrated Cancer Genome Score
- GO
Gene ontology
- WGCNA
weighted gene correlation network analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- STRING
Search Tool for the Retrieval of Interacting Genes and Proteins
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- CNV
copy number variation
Funding
The present study was funded by the Science and Technology Bureau of Yinzhou (grant no. 20191JCGY020007).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available in the figshare repository (https://figshare.com/s/268d9f525ceb5d172b60).
Authors' contributions
YL designed the study. RP, KS and LC downloaded somatic mutation, RNA-seq data, CNVs and clinical data from the TCGA database. KS, LC, ZF, TW and RP predicted driver genes and pathways, performed WGCNA co-expression, PPI network, CNV and survival analyses. LC, ZF, RP and KS participated in the writing and revision of the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by world health organization subtypes. CA Cancer J Clin. 2016;66:443–459. doi: 10.3322/caac.21357. [DOI] [PubMed] [Google Scholar]
- 3.Fisher RI, Gaynor ER, Dahlberg S, Oken MM, Grogan TM, Mize EM, Glick JH, Coltman CA, Jr, Miller TP. Comparison of a standard regimen (CHOP) with three intensive chemotherapy regimens for advanced non-Hodgkin's lymphoma. N Engl J Med. 1993;328:1002–1006. doi: 10.1056/NEJM199304083281404. [DOI] [PubMed] [Google Scholar]
- 4.Pfreundschuh M, Schubert J, Ziepert M, Schmits R, Mohren M, Lengfelder E, Reiser M, Nickenig C, Clemens M, Peter N, et al. Six versus eight cycles of bi-weekly CHOP-14 with or without rituximab in elderly patients with aggressive CD20+ B-cell lymphomas: A randomised controlled trial (RICOVER-60) Lancet Oncol. 2008;9:105–116. doi: 10.1016/S1470-2045(08)70002-0. [DOI] [PubMed] [Google Scholar]
- 5.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, Leppa S, Pasanen A, Meriranta L, Karjalainen- Lindsberg ML, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494.e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ld W, Dw P, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorecta l cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 9.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. Cell-of- origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Cunningham F, Rios D, McLaren WM, Smith J, Pritchard B, Spudich GM, Brent S, Kulesha E, Marin- Garcia P, et al. Ensembl variation resources. BMC Genomics. 2010;11:293. doi: 10.1186/1471-2164-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012;40:e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C, Guo Y, Yang H, He Z, Liu X, Wang K. iCAGES: Integrated CAncer GEnome score for comprehensively prioritizing driver genes in personal cancer genomes. Genome Med. 2016;8:135. doi: 10.1186/s13073-016-0390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua X, Xu H, Yang Y, Zhu J, Liu P, Lu Y. DrGaP: A powerful tool for identifying driver genes and pathways in cancer sequencing studies. Am J Hum Genet. 2013;93:439–451. doi: 10.1016/j.ajhg.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Sun J, Zhao M. ONGene: A literature-based database for human oncogenes. J Genet Genomics. 2016;44:2016–2018. doi: 10.1016/j.jgg.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhao M, Sun J, Zhao Z. TSGene: A web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:970–976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carbone PP, Kaplan HS, Musshoff K, Smithers DW, Tubiana M. Report of the committee on Hodgkin's disease staging classification. Cancer Res. 1971;31:1860–1861. [PubMed] [Google Scholar]
- 22.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45((D1)):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warnes G, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Mächler MM, Steffen AM. gplots: Various R programming tools for plotting data. https://www.researchgate.net/publication/303186599_gplots_Various_R_programming_tools_for_plotting_data. [2005];R package version. 2 [Google Scholar]
- 27.Stine R. An introduction to bootstrap methods: Examples and ideas. Sociol Methods Res. 1989;18:243–291. doi: 10.1177/0049124189018002003. [DOI] [Google Scholar]
- 28.Fox J. Cox proportional-hazards regression for survival data the cox proportional-hazards model. An R and S-PLUS companion to applied regression. 2002;2002:1–18. [Google Scholar]
- 29.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/S0959-437X(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, Mooney TB, Callaway MB, Dooling D, Mardis ER, et al. MuSiC: Identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimura M, Ishizawa J, Ruvolo V, Dilip A, Quintás-Cardama A, McDonnell TJ, Neelapu SS, Kwak LW, Shacham S, Kauffman M, et al. Induction of p53-mediated transcription and apoptosis by exportin-1 (XPO1) inhibition in mantle cell lymphoma. Cancer Sci. 2014;105:795–801. doi: 10.1111/cas.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao W, Lu C, Chen L, Keohavong P. Overexpression of CRM1: A characteristic feature in a transformed phenotype of lung carcinogenesis and a molecular target for lung cancer adjuvant therapy. J Thorac Oncol. 2015;10:815–825. doi: 10.1097/JTO.0000000000000485. [DOI] [PubMed] [Google Scholar]
- 37.Zhou F, Qiu W, Yao R, Xiang J, Sun X, Liu S, Lv J, Yue L. CRM1 is a novel independent prognostic factor for the poor prognosis of gastric carcinomas. Med Oncol. 2013;30:726. doi: 10.1007/s12032-013-0726-1. [DOI] [PubMed] [Google Scholar]
- 38.Ishizawa J, Kojima K, Hail N, Jr, Tabe Y, Andreeff M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol Ther. 2015;153:25–35. doi: 10.1016/j.pharmthera.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kojima K, Kornblau SM, Ruvolo V, Dilip A, Duvvuri S, Davis RE, Zhang M, Wang Z, Coombes KR, Zhang N, et al. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013;121:4166–4174. doi: 10.1182/blood-2012-08-447581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang WY, Yue L, Qiu WS, Wang LW, Zhou XH, Sun YJ. Prognostic value of CRM1 in pancreas cancer. Clin Invest Med. 2009;32:E315. doi: 10.25011/cim.v32i6.10668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available in the figshare repository (https://figshare.com/s/268d9f525ceb5d172b60).