Abstract
We have applied a combination of venomics, in vivo neutralization assays, and in vitro third-generation antivenomics analysis to assess the preclinical efficacy of the monospecific anti-Macrovipera lebetina turanica (anti-Mlt) antivenom manufactured by Uzbiopharm® (Uzbekistan) and the monospecific anti-Vipera berus berus antivenom from Microgen® (Russia) against the venom of Dagestan blunt-nosed viper, Macrovipera lebetina obtusa (Mlo). Despite their low content of homologous (anti-Mlt, 5–10%) or para-specific (anti-Vbb, 4–9%) F(ab')2 antibody fragments against M. l. obtusa venom toxins, both antivenoms efficiently recognized most components of the complex venom proteome's arsenal, which is made up of toxins derived from 11 different gene families and neutralized, albeit at different doses, key toxic effects of M. l. obtusa venom, i.e., in vivo lethal and hemorrhagic effects in a murine model, and in vitro phospholipase A2, proteolytic and coagulant activities. The calculated lethality neutralization potencies for Uzbiopharm® anti-Mlt and anti-Vbb Microgen® antivenoms were 1.46 and 1.77 mg/mL, indicating that 1 mL of Uzbiopharm® and Microgen® antivenoms may protect mice from 41 to 50 LD50s of Mlo venom, respectively. The remarkable degree of conservation of immunogenic determinants between species of the clades of European and Oriental viper, which evolved geographically segregated since the early Miocene, suggests an eventual window of opportunity for the treatment of envenomings by Eurasian snakes. Clearly, the rational use of heterologous antivenoms requires establishing their para-specificity landscapes. This paper illustrates the analytical power of combining in vitro and in vivo preclinical quantitative assays toward this goal.
Graphical abstract
Highlights
-
•
Efficacy against M. l. obtusa venom by two antivenoms was investigated.
-
•
Quantification of lethality neutralizing antibodies was assessed.
-
•
Anti-Vipera berus antivenom showed paraspecificity against M. l. obtusa venom.
-
•
This study provides hints as how to improve the potency of the antivenoms sampled.
1. Introduction
Old World vipers (subfamily Viperinae within family Viperidae) are a group of venomous snakes endemic to Europe, Africa and Asia. Also known as true adders or viperines, these Eurasian snakes (extant genera Eristicophis, Pseudocerastes, Vipera, Macrovipera, Montivipera and Daboia) had their roots in a basal segregation of the Vipera sensu lato group (Laurenti, 1768) on three landmasses separated by the Mediterranean and Paratethys Seas, Europe, the Middle East and North Africa, during the early Miocene period (23.3–16.3 million years ago) (Rögl and Steininger, 1983; Szyndlar and Rage, 1999; Lenk et al., 2001, Garrigues et al., 2005). Present-day Old World snakes are distributed in a wide variety of habitats from North Africa to just within the Arctic Circle and from Great Britain to Pacific Asia (Mallow et al., 2003, Phelps, 2010).
During the last decades the phylogeny of the Vipera sensu lato polyphyletic group has undergone constant revision and divisions by a number of authors. Three major clades have been identified (Lenk et al., 2001, Garrigues et al., 2005), the European vipers (De Smedt, 2006, Kreiner, 2007); the oriental vipers, represented by i) the blunt-nosed Macrovipera lebetina subspecies lebetina (Linnaeus, 1758), turanica (Chernov, 1940) (Terentiev and Chernov, 1940), obtusa (Dwigubsky, 1832), cernovi (Chikin and Shcherbak, 1992), and transmediterranea (Nilson and Andrén, 1988); M. schweizerei (Werner, 1935); and the recently described Macrovipera razii sp. n., with at least seven known representatives from central and southern Iran (Oraie et al., 2018), within the genus Macrovipera (Reuss, 1927, Herrmann et al., 1992); ii) the Mountain vipers of the Montivipera xanthina-raddei complex (Nilson et al.,1999); and iii) a group of Asian and North African vipers within genus Daboia (Gray, 1842).
The Dagestan blunt-nosed viper Macrovipera lebetina obtusa (Dwigubsky, 1832) is endemic to Asia. Having by far the widest range in central Asia, this large stout-bodied species, which can reach lengths of up to 1.7 m, is found in dry and well vegetated rocky mountainous areas between 1000 and 2500 m elevation from central Turkey through Syria, Lebanon, Iraq, northern Jordan, the Caucasus region (incl. Armenia), Azerbaijan, Dagestan, western and northwestern Iran, southern Afghanistan, Pakistan and the Kashmir region (Mallow et al., 2003, Oraie et al., 2018). In Pakistan, M. l. obtusa is restricted to the western highlands, and is allopatric with Daboia russelii in the Indus River valley (Khan, 1983). Crepuscular and nocturnal, but often abroad during daylight on overcast days, M.l. obtusa climb and forage in bushes. Adults feed primarily on small mammals, whereas young take mainly lizards (Phelps, 2010).
The venom of the Dagestan blunt-nosed viper, a WHO category 2 species (Warrell, 2010), is highly potent. A mean dry venom yield of 48 mg per snake and intravenous (i.v.) LD50 of 12–18 μg/18 g mouse body weight have been reported (Latifi, 1984, Kurtović et al., 2014). Human envenomings by M. l. obtusa cause life-threating systemic hemodynamic disturbances, reduced functionality of the kidneys, and ischemia at the bite site (Schweiger, 1983, Göçmen et al., 2006, Sharma et al., 2008). Acute kidney injury is not common and if happens, is due to hypotension, and deposit of hemoglobin, myoglobin, and fibrin in the renal tubules causing acute tubular necrosis (Burdmann et al., 1993). However, information on the epidemiology of envenomings by M. l. obtusa across its distribution is scarce (Chippaux, 2012, Dehghani et al., 2014, Zamani et al., 2016) or non-existent.
Treatment of snakebites envenomings is critically dependent on the availability of effective antivenoms. This study was designed to assess a comparative preclinical efficacy of the monospecific anti-M. lebetina turanica (anti-Mlt) antivenom manufactured by Uzbiopharm® (Uzbekistan) and the monospecific anti-Vipera berus berus antivenom from Microgen® (Russia) to neutralize key toxic effects of M. l. obtusa venom, i.e., lethal, defibrinogenetic, hemorrhagic, phospholipase A2 activity, proteolytic, and coagulant, by combination of in vivo neutralization assays and in vitro third-generation antivenomics analysis.
2. Materials and methods
2.1. Venom and antivenoms
Venom from Macrovipera lebetina obtusa (Mlo) was pooled from 10 adult individuals between 80 and 110 cm in length and older than 3 years, of both sexes, collected, during the day in spring and autumn and in summer nights, in different regions of the Republic of Dagestan (Russian Federation). Venom was air-dried at room temperature and stored at −8 °C until used. Monospecific anti-M. lebetina turanica antivenom was manufactured by Uzbiopharm® (Tashkent, Uzbekistan) (batch 23, expiry date March 2020; 98 ± 4 mg F(ab')2/mL, 9.4 mL/ampoule). The monospecific anti-Vipera berus berus antivenom was from Microgen® (Moscow, Russia) (Al-Shekhadat et al., 2019) (batches C43 and C37, expiry dates June 2020 and January 2021, respectively; 85–110.5 mg F(ab')2/mL, 1.1–1.3 mL/ampoule).
2.2. Animals
CD-1 mice (18–20 g body weight) were used throughout the study. The protocols involving the use of mice were approved by the Institutional Committee for the Care and Use of Laboratory Animals (CICUA) of Universidad de Costa Rica (Act 82–2008, date of the approval: 18 September 2008).
2.3. Proteomic characterization of Macrovipera lebetina obtusa venom
Crude air-dried at room temperature venom was dissolved in 0.05% trifluoroacetic acid (TFA) and 5% acetonitrile (ACN) to a final concentration of 15 mg/mL. Insoluble material was removed by centrifugation in an Eppendorf centrifuge at 13,000×g for 10 min at room temperature, and the proteins contained in 40 μL (600 μg) were separated by RP-HPLC using a Agilent LC 1100 High Pressure Gradient System equipped with a Teknokroma Europa C18 (25 cm × 4 mm, 5 μm particle size, 300 Å pore size) column and a DAD detector. The column was developed at a flow rate of 1.0 mL/min with a linear gradient of 0.1% TFA in MilliQ® water (solution A) and 0.1% TFA in acetonitrile (solution B), isocratic (5% B) for 5 min, followed by 5–25% B for 10 min, 25–45% B for 60 min, and 45–70% B for 10 min. Protein detection was carried out at 215 nm with a reference wavelength of 400 nm. Fractions were collected manually across the entire elution range, dried in a vacuum centrifuge (Savant™, ThermoFisher Scientific), and redissolved in MilliQ® water. Molecular masses of the purified proteins were estimated by non-reduced and reduced SDS-PAGE (on 15% polyacrylamide gels).
For SDS-PAGE analysis RP-HPLC sample aliquots were mixed with ¼ volume of 4x sample buffer (0.25M Tris-HCl pH 6.8, 8% SDS, 30% glycerol, 0.02% bromophenol blue, with or without 10% 2-mercaptoethanol) and heated at 85 °C for 15 min, run under reducing conditions, and the gels were stained with Coomassie Brilliant Blue G-250. Protein bands of interest were excised and subject to in-gel disulphide bond reduction (10 mM dithiothreitol, 30 min at 65 °C) and sulphydryl group alkylation (50 mM iodoacetamide, 2h in the dark at room temperature), followed by overnight digestion with sequencing-grade trypsin (66 ng/μL in 25 mM ammonium bicarbonate, 10% ACN; 0.25 μg/sample), using a Genomics Solution ProGest™ Protein Digestion Workstation. Tryptic digests were dried in a vacuum centrifuge (SPD SpeedVac®, ThermoSavant), redissolved in 14 μL of 5% ACN containing 0.1% formic acid, and 7 μL submitted to LC-MS/MS. Tryptic peptides were separated by nano-Acquity UltraPerformance LC® (UPLC®) using a BEH130 C18 (100 μm × 100mm, 1.7 μm particle size) column in-line with a Waters SYNAPT G2 High Definition Mass Spectrometry System. The flow rate was set to 0.6 μL/min and the column was developed with a linear gradient of 0.1% formic acid in water (solution A) and 0.1% formic acid in ACN (solution B), isocratically 1% B for 1 min, followed by 1–12% B for 1min, 12–40% B for 15min, 40–85% B for 2min. Doubly and triply charged ions were selected for CID-MS/MS. Fragmentation spectra were interpreted i) manually (de novo sequencing), ii) using the on-line form of the MASCOT Server (version 2.6) at http://www.matrixscience.com against the last update (Release 234 of October 15th, 2019) of NCBI non-redundant database, and iii) processed in Waters Corporation's ProteinLynx Global SERVER 2013 version 2.5.2. (with Expression version 2.0). The following search parameters were used: Taxonomy: bony vertebrates; Enzyme: trypsin (two missed cleavage allowed); MS/MS mass tolerance was set to ± 0.6 Da; carbamidomethyl cysteine and oxidation of methionine were selected as fixed and variable modifications, respectively. All matched MS/MS data were manually checked. Peptide sequences assigned by de novo MS/MS were matched to homologous proteins available in the NCBI non-redundant protein sequences database using the online BLASTP program (Altschul et al., 1990) at https://blast.ncbi.nlm.nih.gov/Blast.cgi. The relative abundances of the chromatographic peaks obtained by reverse-phase HPLC fractionation of the whole venom were calculated as “percentage of total peptide bond concentration in the peak” by dividing the peak area by the total area of the chromatogram (Calvete, 2014, Eichberg et al., 2015). For chromatographic peaks containing single components (by SDS-PAGE and/or MS), this figure is a good estimate of the % by weight (g/100 g) of the pure venom component (Calderón-Celis et al., 2017). When more than one venom protein was present in a reverse-phase fraction, their proportions (percentage of total protein band area) were estimated by densitometry of Coomassie-stained SDS-polyacrylamide gels using MetaMorph® Image Analysis Software (Molecular Devices). Conversely, the relative abundances of different proteins contained in the same SDS-PAGE band were estimated based on the mean relative ion intensity of the three most abundant peptide ions associated with each protein by MS/MS analysis. The analytical variability associated with this label-free approach has been estimated within a relative error of 15% (Silva et al., 2006). The relative abundances of the protein families present in the venom were calculated as the ratio of the sum of the percentages of the individual proteins from the same toxin family to the total area of venom protein peaks in the reverse-phase chromatogram.
2.4. Toxic and enzymatic venom activities in mice and their neutralization assays
2.4.1. Lethality
For the estimation of the Median Lethal Dose (LD50), groups of five CD-1 mice received by the intraperitoneal (i.p.) route increasing doses of venom (15 μg–240 μg per mouse) dissolved in a volume of 0.5 mL 0.12 M NaCl, 0.04 M phosphate, pH 7.2 (PBS). Deaths occurring within 48 h were recorded and the LD50 was estimated by probits as the minimum amount of venom causing the death of 50% of the mice injected, with 95% confidence limits (Finney, 1947, Al-Shekhadat et al., 2019). For the neutralization of lethality, mixtures containing a fixed dose of venom and various dilutions of antivenom were prepared, and incubated at 37 °C for 30 min. Aliquots of 0.5 mL of each mixture, containing a dose of venom corresponding to 4 LD50s, were then injected intraperitoneally (i.p.) into groups of five mice. Mixtures corresponded to various ratios of mg venom/mL antivenom. A control group was injected with 4 LD50s of venom incubated with PBS instead of antivenom. Deaths occurring during 48 h were recorded, and the neutralizing ability of antivenom was expressed as the Median Effective Dose (ED50), i.e. the venom/antivenom ratio at which half of the population of injected mice is protected, estimated by Probits. (Finney, 1947).
Antivenom potency (P) is the amount of venom (mg) completely neutralized per mL of antivenom. P was calculated using formula P = [(n-1)/ED50] × LD50, where “n" is the number of median lethal doses (LD50s) used as challenge dose to determine the antivenoms median effective dose, ED50. For the calculation of P, LD50 and ED50 are expressed, respectively, as (mg venom/mouse) and (mL of antivenom that protect 50% of the mice population inoculated with n x LD50). In the calculation of P, (n-1) x LD50 is used instead of the total amount of venom, n x LD50, because at the endpoint of the neutralization assay, one LD50 remains unneutralized and causes the death of 50% of mice (Araujo et al., 2008, Morais et al., 2010).
2.4.2. Hemorrhagic activity
To assess the hemorrhagic activity of venoms, groups of three CD-1 mice received an intradermal (i.d.) injection, in the ventral abdominal region, of 0.1 mL of PBS containing several dilutions of venom (from 0.12 μg to 2.0 μg). Mice were sacrificed by CO2 inhalation 2 h after injection, the skin was removed, and the area of the hemorrhagic lesion in the inner side of the skin was measured. The Minimum Hemorrhagic Dose (MHD) corresponds to the dose of venom that induces a hemorrhagic area of 10 mm diameter (Gutiérrez et al., 1985). For the assessment of the neutralizing capacity of antivenoms, mixtures containing a fixed dose of venom and various dilutions of antivenom were prepared, and incubated at 37 °C for 30 min (Gutiérrez et al., 1985). Then, aliquots of 0.1 mL of each mixture, containing a dose of venom corresponding to 10 Minimum Hemorrhagic Doses (MHDs), were injected i. d. into groups of three mice, as described. Mixtures corresponded to various ratios of mg venom/mL antivenom. A control group of mice was injected with the same dose of venom incubated with PBS instead of antivenom. Mice were sacrificed as described 2 h after injection, and the area of the hemorrhagic lesion was measured. Neutralizing ability was expressed as the Median Effective Dose (ED50), corresponding to the ratio venom/antivenom at which the diameter of the hemorrhagic spot is reduced by 50% when compared to the diameter of the hemorrhagic lesion in mice injected with venom incubated with no antivenom (Gutiérrez et al., 1985).
2.4.3. Defibrinogenating activity
Various amounts of venom dissolved in 200 μL of PBS were injected i. v. to groups of three mice following the method described by Gené and coworkers (1989). One hour after injection, mice were bled from the orbital plexus under ether anaesthesia, the blood was placed in dry glass tubes and left undisturbed for 2 h at 22–25 °C. Thereafter, the tubes were gently tilted and the presence, or absence, of clots was recorded. The Minimum Defibrinogenating Dose (MDD) corresponded to the minimum dose of venom that rendered blood unclottable in all mice tested.
2.5. In vitro toxic and enzymatic venom activities and their neutralization assays
2.5.1. Coagulant activity
Coagulant activity of Mlo venom was determined based on the turbidimetric assay described (O'Leary and Isbister, 2010) as modified by Sánchez et al. (2018). Briefly, different amounts of venom, dissolved in 100 μL TBS (25 mM Tris-HCl, 137 mM NaCl, 3.4 mM KCl, pH 7.4) were added in triplicate to wells in a 96-well plate and incubated for 5 min at 37 °C in a microplate reader (Cytation 3 Imaging Reader, BioTek). Then, 4 μL of 0.4 M CaCl2 was added to 100 μL of human citrated plasma previously incubated at 37 °C, and this mixture was added immediately to each venom-containing well using a multichannel pipette. Samples were mixed for 5 s shaking step, and the absorbance at 340 nm was monitored every 30 s over 15 min. The increase in absorbance reflects the formation of a clot. Coagulant activity was expressed as the Minimum Coagulant Dose (MCD), corresponding to the minimum dose of venom that induces a change in absorbance of 0.01 units within 1 min. Control consisted of plasma incubated with TBS alone. For the study of neutralization, 100 μL of mixtures containing a fixed dose of venom and various dilutions of antivenom were prepared, and incubated at 37 °C for 30 min (Gené et al., 1989). Then, each mixture, containing a venom dose corresponding to 2 Minimum Coagulant Doses (MCDs), were tested for coagulant activity as described above. A control group was included, corresponding to plasma incubated with venom that was previously incubated with PBS instead of antivenom. Changes in absorbance were recorded and neutralization was expressed as Effective Dose (ED), corresponding to the ratio of venom/antivenom in which the change in absorbance is prolonged three times as compared to plasma incubated with venom alone.
2.5.2. Phospholipase A2 (PLA2) activity
The titrimetric method described by Dole (1956) and Gutiérrez et al. (1986) was followed, using egg yolk phospholipids as substrate. Various venom doses were prepared in triplicate for each venom, and 100 μL of each solution was placed in 1 mL of egg yolk diluted 1:5 in a solution of 0.1 M Tris, 0.01 M CaCl2 and 1% Triton X-100 (pH 8.5). Mixtures comprising various ratios of mg venom/mL antivenom were incubated at 37 °C for 30 min. Free fatty acids were extracted and titrated. Activity was expressed as μEq fatty acid released per mg protein per min. For neutralization, mixtures were prepared containing a fixed amount of venom (25 μg) and variable dilutions of antivenom. Controls included venom incubated with PBS instead of antivenom. After an incubation of 30 min at 37 °C, the PLA2 activity of the mixtures was assessed as described. Neutralization was expressed as Median Effective Dose (ED50), corresponding to the ratio of venom/antivenom in which PLA2 activity was reduced by 50% as compared to the activity of venom alone.
2.5.3. Proteinase activity
Proteinase activity was assessed on azocasein following the method described by Gutiérrez et al. (2008). Briefly, various amounts of venom dissolved in 20 μL of PBS, were added to 100 μL of substrate (10 mg/mL azocasein dissolved in 25 mM Tris, 150 mM NaCl, 5 mM CaCl2, pH 7.4), followed by incubation at 37 °C for 90 min. Reactions were stopped by the addition of 200 μL of 5% trichloroacetic acid. Then, 150 μL of each supernatant were mixed with 150 μL of 0.5M NaOH, and the absorbance at 450 nm was recorded. Proteolytic activity was expressed as units/mg venom, one unit corresponding to a change in absorbance of 0.2/min. For neutralization, mixtures were prepared containing a fixed amount of venom (8 μg) and variable dilutions of antivenom. The antivenom ED50 was expressed as the ratio of μL of antivenom/mg venom (or mg venom/mL antivenom) at which the response of the control of venom is reduced 50% (Gutiérrez, 2018).
2.5.4. Third-generation antivenomics
Third-generation antivenomics (Pla et al., 2017, Calvete et al., 2018) was applied to compare the specific immunoreactivity of the anti-Mlt antivenom (Uzbiopharm®, Uzbekistan) and the paraspecific immunoreactivity of the anti-Vbb antivenom (Microgen®, Russia) (Al-Shekhadat et al., 2019) towards the venom of M. lebetina obtusa from Republic of Dagestan (Russian Federation). To this end, the antivenoms were dialyzed against MilliQ® water, lyophilized, and 122 mg (anti-Mlt) and 116 mg (anti-Vbb) of total lyophilizate weight were reconstituted in 3 mL of 0.2 M NaHCO3, 0.5 M NaCl, pH 8.3 (coupling buffer). The concentrations of these antivenom stock solutions were determined spectrophotometrically using an extinction coefficient for a 1 mg/mL concentration (ε0.1%) at 280 nm of 1.36 (mg/mL)−1 cm−1 (Howard and Kaser, 2014). Antivenom affinity columns were prepared in batch. To this end, 3 mL of CNBr-activated Sepharose™ 4B matrix (Ge Healthcare, Buckinghamshire, UK) packed in a ABT column (Agarose Bead Technologies, Torrejón de Ardoz, Madrid) and washed with 15x matrix volumes of cold 1 mM HCl, followed by two matrix volumes of coupling buffer to adjust the pH of the column to 8.0–9.0. CNBr-activated instead of N-hydroxysuccinimide (NHS)-activated matrix was employed because NHS released during the coupling procedure absorbs strongly at 280 nm, thus interfering with the measurement of the concentration of antibodies remaining in the supernatant of the coupling solution. The antivenom stock solutions in 3 mL of coupling buffer were incubated with 3 mL CNBr-activated matrix for 4 h at room temperature. Antivenom coupling yields, estimated measuring A280nm before and after incubation with the matrix, were 80.2 mg (anti-Mlt) and 85.5 mg (anti-Vbb) (F(ab')2 (i.e. 26.7 mg anti-Mlo and 28.5 mg anti-Vbb/mL CNBr-activated Sepharose™ 4B matrix). After the coupling, remaining active matrix groups were blocked with 3 mL of 0.1 M Tris-HCl, pH 8.5 at room temperature for 4 h. Affinity columns, each filled with 280 μL (anti-Vbb) or 300 μL (anti-Mlt) of affinity matrix containing 8 mg of immobilized F(ab')2 molecules, were alternately washed with three matrix volumes of 0.1 M acetate containing 0.5 M NaCl, pH 4.0–5.0, and three matrix volumes of 0.1 M Tris-HCl, pH 8.5. This procedure was repeated 6 times. The columns were then equilibrated with 3 vol of working buffer (PBS, 20 mM phosphate buffer, 135 mM NaCl, pH 7.4) and incubated with increasing amounts (100–1200 μg of total venom proteins) of M. l. obtusa dissolved in ½ matrix volume of PBS, and the mixtures incubated for 1 h at 25 °C in an orbital shaker. As specificity controls, 300 μL of CNBr-activated Sepharose™ 4B matrix, without (mock) or with 8 mg of immobilized control (naïve) horse IgGs, were incubated with venom and developed in parallel to the immunoaffinity columns. The non-retained eluates of columns incubated with 100–300, 600, 900, and 1200 μg of venom were recovered, respectively, with 3x, 5x, 7x, and 9x matrix volume of PBS, and the immunocaptured proteins were eluted, respectively, with 3x (100–300 μg) and 6x (600–1200 μg) matrix volume of 0.1M glycine-HCl, pH 2.7 buffer, and brought to neutral pH with 1M Tris-HCl, pH 9.0. The entire fractions eluted in 100–300 μg, ½ of the fractions recovered in 600 μg, of the non-retained fractions and ½ of the retained fractions recovered in 900 μg, ¼ of the non-retained fractions and ½ of the retained fractions recovered in 1200 μg, were concentrated in a Savant SpeedVac™ vacuum centrifuge (ThermoFisher Scientific, Waltham, MA USA) to 45 μL, 40 μL of which were then fractionated by reverse-phase HPLC using an Agilent LC 1100 High Pressure Gradient System (Santa Clara, CA, USA) equipped with a Discovery® BIO Wide Pore C18 (15 cm × 2.1 mm, 3 μm particle size, 300 Å pore size) column and a DAD detector. The column was developed at a flow rate of 0.4 mL/min with a linear gradient of 0.1% TFA in MilliQ® water (solution A) and 0.1% TFA in acetonitrile (solution B), isocratic (5% B) for 1 min, followed by 5–25% B for 5 min, 25–45% B for 35 min, and 45–70% B for 5 min. Protein detection was carried out at 215 nm with a reference wavelength of 400 nm. The fraction of non-immunocaptured molecules was estimated as the relative ratio of the chromatographic areas of the toxin recovered in the non-retained (NR) and retained (R) affinity chromatography fractions using the equation %NRi = 100-[(Ri/(Ri + NRi)) x 100], where Ri corresponds to the area of the same protein “i" in the chromatogram of the fraction retained and eluted from the affinity column. However, for some toxins that were poorly recovered in the column-retained fraction owing to their high binding affinity to the immobilized antivenom likely preventing their elution from the column (Calvete et al., 2015), the percentage of non-immunocaptured toxin“i” (% NRtoxin“i”) was calculated as the ratio between the chromatographic areas of the same peak recovered in the non-retained fraction (NRtoxin“i”) and in a reference venom (Vtoxin“i”) containing the same amount of total protein that the parent venom sample and run under identical chromatographic conditions, using the equation %NRtoxin“i” = (NRtoxin“i”/Vtoxin“i”) x 100. The percentage of antivenom anti-toxin F(ab')2 molecules was calculated by dividing [(1/2 maximal amount (in μmoles) of total venom proteins bound per antivenom vial) x molecular mass (in kDa) of antibody (F(ab')2, 110 kDa) molecule] by the [total amount of antibody (F(ab')2) (in mg) per antivenom vial (Calvete et al., 2018, Sanz et al., 2018, Al-Shekhadat et al., 2019). Binding saturation was computed by extrapolation from data modelled in Excel to degree 2 polynomial functions.
3. Results and discussion
3.1. The Dagestan blunt-nosed viper, Macrovipera lebetina obtusa, venom proteome
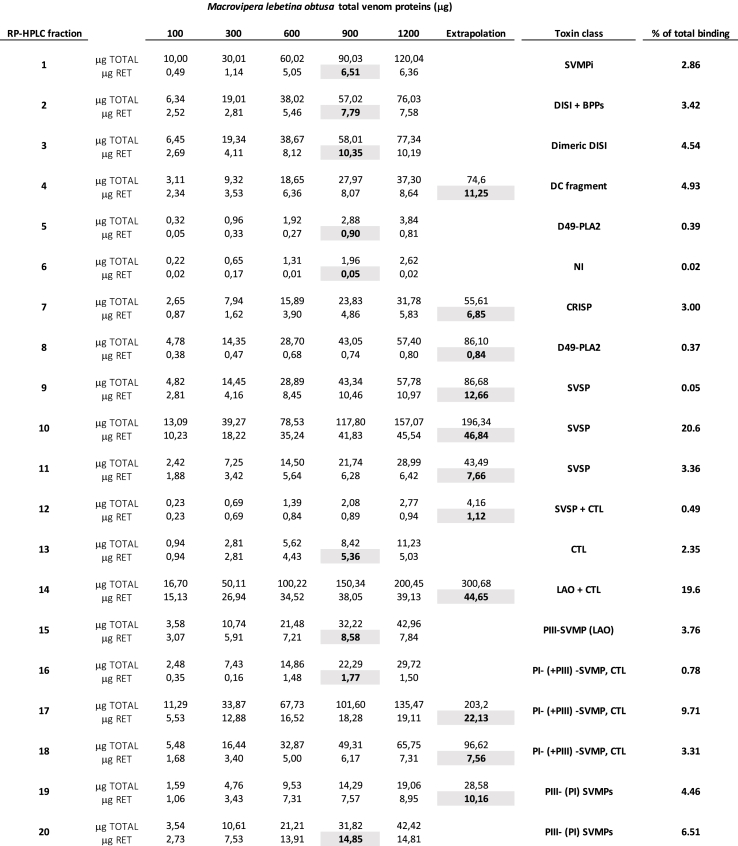
The venom proteome of M. l. obtusa (Dagestan, Russia) was characterized and quantified by applying the previously described (Calvete, 2014; Eichberg et al., 2015) two-step pre-MS decomplexation protocol (reverse-phase HPLC and SDS-PAGE) followed by peptide-centric bottom-up analysis of tryptic digests of the electrophoretically-separated protein bands eluted in the different reverse-phase chromatographic fractions. Supplementary Table S1 shows the details of the MS/MS assignment, quantification and database matching of the proteins eluted in the 40 reverse-phase chromatographic fractions (Fig. 1A) and further resolved by SDS-PAGE of each chromatographic fraction (Fig. 1B). The venom proteome of the Dagestan blunt-nosed viper comprised a complex arsenal of peptides and proteins derived from precursor proteins encoded by genes belonging to 11 different families (Fig. 1C).
Fig. 1.
Proteomic analyses of venom of the Dagestan blunt-nosed viper, Macrovipera lebetina obtusa. Panel A, reverse-phase chromatographic separation of the venom proteins. Panel B, SDS–PAGE profile of the chromatographic fractions analyzed under non-reduced (upper gels) and reduced (lower gels) conditions. Panel C, pie chart displaying the relative occurrence (in percentage of total venom proteins) of toxins from different protein families in the venom proteome. SVMPi, tripeptide inhibitors of snake venom metalloproteinases; BPP, Bradykinin potentiating peptides; DISI, disintegrin; DC, disintegrin-like/cysteine-rich fragment of SVMP of class PIII; D49-PLA2, D49 phospholipases A2; CRISP, cysteine-rich secretory protein; SVSP, snake venom serine proteinase; PI- and PIII-SVMP, metalloproteinases of class PI and PIII, respectively; CTL, C-type lectin-like protein; 5′NT, 5′-nucleotidase; LAO, L-amino acid oxidase; PDE, phosphodiesterase; Hyal, hyaluronidase. na, not assigned venom component.
Endogenous tripeptide inhibitors of SVMPs (SVMPi) and bradykinin-potentiating peptides (BPPs) are released into the venom proteome by the proteolytic processing of a common precursor (Graham et al., 2005, Cidade et al., 2006). These peptide components comprise, respectively, 4.8% and 5.6% of the Dagestan blunt-nosed viper venom proteome (Fig. 1C). SVMPi regulate the proteolytic activities of SVMPs in a reversible manner under physiological conditions, thus protecting glandular tissues and venom factors from the proteolytic activity of SVMPs stored at high concentration in an inactive but competent state for many months in the lumen of the venom gland of many Viperidae snakes (Huang et al., 1998, Huang et al., 2002, Munekiyo and Mackessy, 2005, Wagstaff et al., 2008). Bradykinin-potentiating peptides are inhibitors of the angiotensin I-converting enzyme, which enhance the hypotensive effect of the circulating bradykinin, contributing to cardiovascular shock in the snake's prey or human victim (Ferreira et al., 1970, Greene et al., 1972, Luft, 2008). Among the venom proteins, snake venom Zn2+-dependent metalloproteinases of the PI and PIII classes, which comprise, respectively, 14.6% and 9.4% of the venom proteome (Fig. 1C), along with proteolytic product comprising the C-terminal disintegrin-like and cysteine-rich domains of PIII-SVMPs (DC-fragments, 0.6%), and medium-sized and dimeric disintegrins released from PII-SVMP precursors (~3.5% of Mlo venom proteome), (Fox and Serrano, 2005, Juárez et al., 2008, Carbajo et al., 2015), represent the most abundantly expressed (~28% of the total venom proteome) gene family in the Dagestan blunt-nosed viper venom proteome (Fig. 1C). Other major venom toxin families are, in order of relative abundance, venom serine proteinases (SVSP, 23.4%) (Swenson and Markland, 2005, Serrano, 2013), phospholipases A2 (PLA2, 13.6%) (Koludarov et al., 2019), C-type lectin-like proteins (CTL, 8.7%) (Clemetson, 2010, Arlinghaus and Eble, 2012), and proteoforms of obtustatin (short DISI, 7.4%), a short disintegrin that express the lysine-threonine-serine (KTS) α1β1 integrin inhibitory tripeptide in its active loop (https://www.ncbi.nlm.nih.gov/protein/P83469) (Marcinkiewicz et al., 2003) (Fig. 1C). The remaining 18.4% of the venom proteome is constituted by 6 toxin classes, none of which exceeds 2% relative abundance (Fig. 1C). This set of minor toxins include homo and hetero dimeric disintegrins VLO4 [P0C6A8] and VLO5 [P0C6A9, P0C6B0] encoded by short-coding messages (Okuda et al., 2002, Calvete et al., 2003, Sanz et al., 2008); cysteine-rich secretory protein (CRISP); 5′-nucleotidase (5′NT); L-amino acid oxidase (LAO); phosphodiesterase (PDE); and hyaluronidase (Hyal) (Sanz et al., 2008, Siigur et al., 2019).
The complex toxin arsenal of M. lebetina obtusa venom may prevent blood coagulation (serine proteinases) and platelet aggregation (disintegrins, C-type lectin-like proteins, and L-amino acid oxidase); exert hemolytic and myotoxic effects (phospholipases A2); and disrupt the extracellular matrix of the vascular subendothelium (Zn2+-dependent metalloproteinases). These pharmacological activities may account for the severe pain at the bite site, nausea and vomiting, swelling, necrosis and systemic disseminated hemostatic manifestations reported in human envenomings inflicted by M. l. obtusa (Corkill, 1932, Sharma et al., 2008, Warrell, 2010, Zamani et al., 2016).
3.2. Toxic and enzymatic activities of M. L. obtusa venom and their neutralization by Uzbiopharm® and Microgen® antivenoms
In the murine model, the venom of M. l. obtusa (Dagestan, Russia) was lethal at a Median Lethal Dose (LD50) of 34.95 (95% CI, 18.98–54.93) μg/mouse (Table 1). The venom also exhibited Minimum Hemorrhagic Dose (MHD) of 0.18 ± 0.10 μg/mouse, but it did not show defibrinogenating activity when up to 20 μg were injected intravenously, the highest non-lethal dose tested (Table 1). In in vitro assays, the venom showed phospholipasic, proteolytic, and coagulant activities (Table 1). Both, the monospecific anti-Mlt antivenom from Uzbiopharm® and the Microgen® monospecific anti-Vbb antivenom were capable of neutralizing, with similar efficacy, venom lethality (Table 1). The calculated lethality neutralization potencies (P, the amount (mg) of venom completely neutralized per mL of antivenom) for Uzbiopharm® anti-Mlt and anti-Vbb Microgen® antivenoms were 1.46 and 1.77. These figures indicate that 1 mL of Uzbiopharm® and Microgen® antivenoms may protect mice from 41 to 50 LD50s of Mlo venom, respectively. For comparison, the ED50 of a monospecific F(ab')2 antivenom produced at Razi Institute (Tehran, Iran) was 2.2 ± 0.4 mg/mL (Latifi et al., 1978, Latifi, 1984). This value is similar to the ED50s determined for the Uzbiopharm® (1.93 (95%CI 1.18–2.90) mg/mL) and Microgen® (2.36 (95%CI 1.03–3.39) mg/mL) antivenoms (Table 1). However, since the authors did not report the number of LD50 used in the determination of the ED50, it is not possible to calculate the potency of Razi antivenom. A new polyvalent equine F(ab')2 antivenom (Inoserp Europe) designed to treat envenoming by snakes in the Eurasian region, generated against a mixture of venoms from the European vipers V. ammodytes, V. aspis, V. berus, V. latastei, Montivipera xanthina, Macrovipera schweizeri, M. l. obtusa, M. l. cernovi, and M. l. turanica (García-Arredondo et al., 2019), showed an intravenous ED50, against 5 LD50s of M. l. obtusa (Azerbaijan) venom (LD50 = 16.32 (15.73–16.93) μg/mouse), of 3.47 (95% CI 3.38–3.56) mg/mL, which translates into a calculated P of 2.13 mg/mL (Supplementary Table S2). This figure indicates that 1 mL of Inoserp Europe may protect mice against 130.5 and 61 LD50s of Mlo from Azerbaijan and Dagestan venom, respectively. However, given the marked difference between the LD50s reported for Mlo venom from Iran (i.v., 9.4–22.5 μg/mouse of 16–18 g body weight (Latifi, 1984); i,v., 18.4 ± 1.4 μg/mouse of 16–18 g body weight (Kurtović et al., 2014); Azerbaijan (i.v., 15.73–16.93 μg/mouse of 18–20 g body weight) (García-Arredondo et al., 2019) and Dagestan (i.p., 18.98–54.93 μg/mouse of 18–20 g body weight) (this work), differences in mean body weight of mice, and different administration route, a comparison between antivenoms produced by Razi Institute, Inoserp Europe, Uzbiopharm® and Microgen® should be interpreted cautiously.
Table 1.
Toxic effects of Macrovipera lebetina obtusa venom (V) in mice and their neutralization by the monospecific anti-Mlt antivenom (AV) from Uzbiopharm® and the monospecific anti-Vbb Microgen® antivenom.a 95% confidence limits are expressed in parentheses. Other results are presented as mean ± S.D; NA, not analyzed.
| Toxic activity | Neutralization by antivenom |
||||
|---|---|---|---|---|---|
|
Uzbiopharma Ltd. (Uzbekistan) |
Microgen (Russia) |
||||
| μL AV/mg V | mg V/mL AV | μL AV/mg V | mg V/mL AV | ||
| Lethality (i.p.) [mg/mouse] | 34,95 (18,98–54,93)a | 518 (345–847)a | 1,93 (1,18–2,90)a | 424 (295–971)a | 2,36 (1,03-3,39)a |
| Hemorrhagic (MHD) [mg/mouse] | 0,18 ± 0,10 | 2168 ± 325 | 0,47 ± 0,06 | 478 ± 14 | 2,09 ± 0,06 |
| PLA2 activity [μeq/mg/min] | 9,84 ± 0,34 | 639 ± 63 | 1,57 ± 0,15 | 217 ± 13 | 4,61 ± 0,27 |
| Proteinase [U/mg] | 2,13 ± 0,21 | 872 ± 68 | 1,15 ± 0,01 | 994 ± 12 | 1,01 ± 0,08 |
| Procoagulant (MCD) [μg/mouse] | 0,20 ± 0,00 | 264 ± 23 | 3,80 ± 0,31 | 1979 ± 90 | 0,51 ± 0,02 |
| Defibrinogenic (MDD) | Null up to 20 μg | NA | NA | NA | NA |
Uzbiopharm® anti-Mlt and Microgen® anti-Vbb antivenoms were also effective neutralizing the in vivo and in vitro toxic effects of Dagestan blunt-nosed viper venom. However, whereas the homolog antivenom from Uzbiopharm® performed better blocking the procoagulant and proteolytic activities, heterologous Microgen® antivenom was more effective reversing the venom's hemorrhagic and PLA2 activities (Table 1).
3.3. Third-generation antivenomics: quantification of venom-specific antivenom antibodies
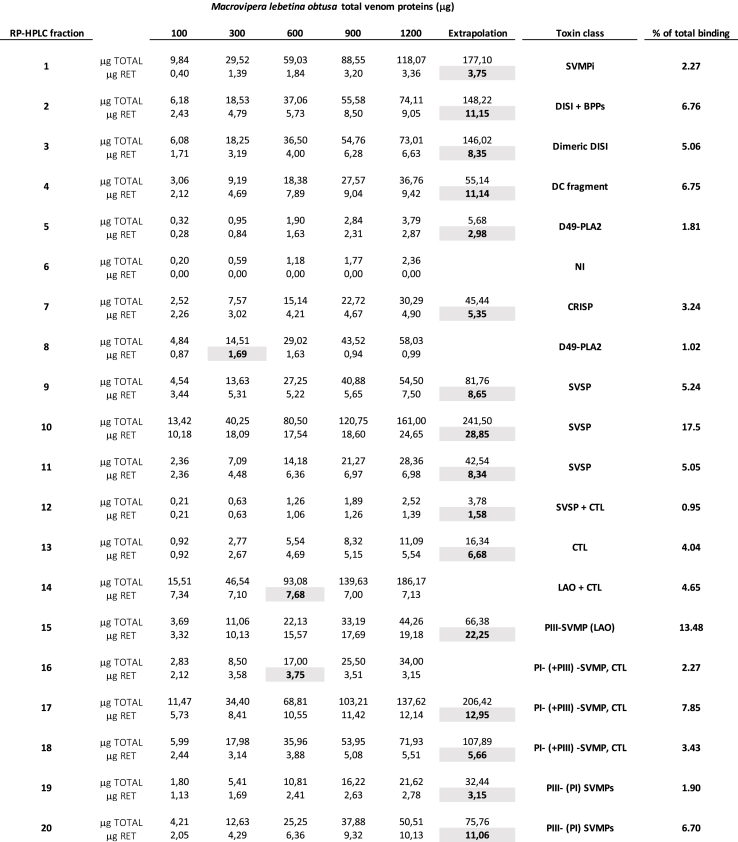
A third-generation antivenomics (3GA) approach was applied to quantify, in a concentration-dependent and toxin-resolved manner, the immunorecognition landscape of the Uzbiopharm® and Microgen® antivenoms towards M. l. obtusa (Dagestan, Russia) venom. The results displayed, respectively in Table 2, Table 3 and Supplementary Tables S3 and S4, show that excepting for the poorly-immunogenic peptides eluted in the first fractions of the reverse-phase chromatogram, both antivenoms efficiently immunocaptured all the major venom proteins (Fig. 2, panels A and B).
Table 2.
Concentration-dependent immunoretained (RET) Macrovipera lebetina obtusa (Mlo) venom proteins by Uzbiopharm® (Uzbekistan) anti-Mlt antivenom affinity column. Maximal binding for each RP-HPLC fraction is highlighted in bold face and grey background.
Table 3.
Concentration-dependent immunoretained (RET) Macrovipera lebetina obtusa (Mlo) venom proteins by Microgen® (Russia) anti-Vbb antivenom affinity column. Maximal binding for each RP-HPLC fraction is highlighted in bold face and grey background.
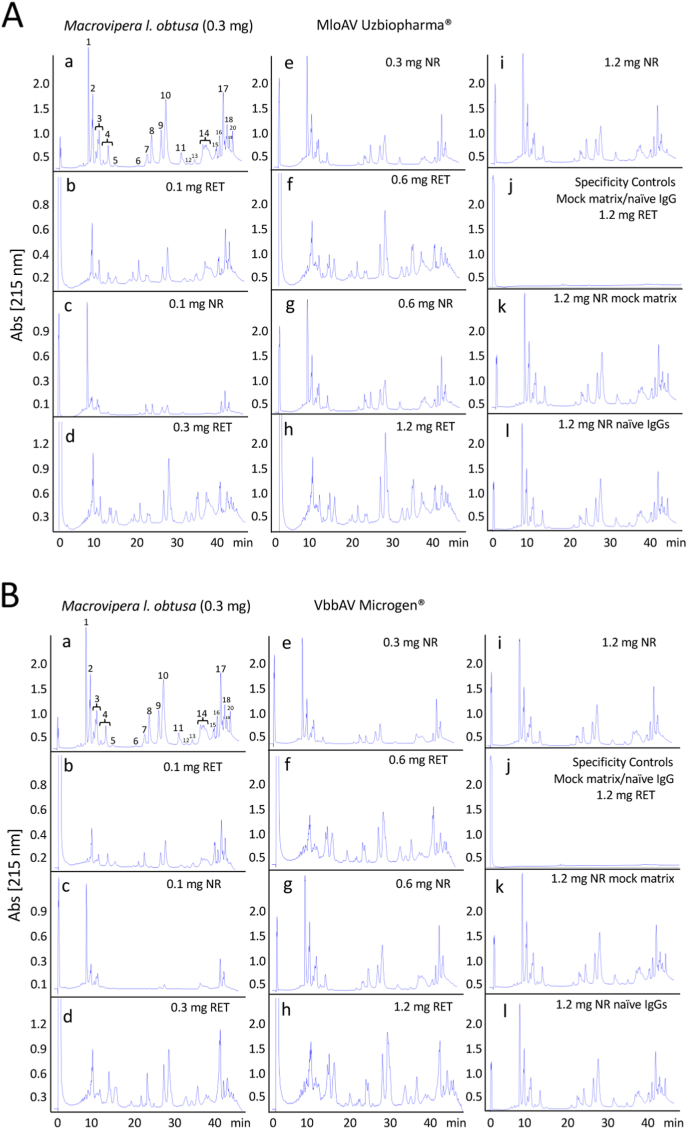
Fig. 2.
Comparative immunorecognition ability of the anti-Mlt (Uzbiopharm®) and anti-Vbb (Microgen®) antivenoms towards Macrovipera lebetina obtusa venom toxins. Third-generation antivenomic analyses of anti-Mlt (panel A) and anti-Vbb (panel B) antivenoms challenged with increasing concentration of M. l. obtusa venom. Reverse-phase chromatographic analysis of whole M. l. obtusa venom (panels a) and of the non-retained and the immunoretained fractions recovered from affinity column (8 mg immobilized antivenom F(ab’)2 molecules) incubated with increasing amounts (100–1200 μg) of venom from M. l. obtusa from Dagestan (Russian Federation) are displayed in panels b through i. Panels j–l show reverse-phase HPLC separations of the retained and non-retained venom fractions on mock matrix and naïve equine IgG affinity columns, respectively.
Affinity columns containing 8 mg anti-Mlt of F(ab')2 antibodies of Uzbiopharm® antivenom had maximal binding capacity of 227.88 μg M. lebetina obtusa venom proteins (e.g. 28.49 mg venom/g antivenom). Since the protein concentration of the batch of anti-Mlt used was 98 mg/mL and a vial contained 9.4 mL, the maximal binding capacity per vial was 26.24 mg M. l. obtusa venom proteins. Considering an average molecular mass for Mlo toxins of 29.8 kDa (calculated as Σ (% i x Mi), where % i is the relative abundance of toxin “i” and Mi its molecular mass in Da), 26.24 mg of M. l. obtusa venom proteins equals 0.88 μmoles of venom molecules. Assuming that one or both antigen-binding sites of an F(ab')2 molecule were occupied at maximal antigen binding capacity, one vial of Uzbiopharm® anti-Mlt antivenom contained, respectively 0.88 μmoles (96.85 mg) or 0.44 μmoles (48.42 mg) of toxin-binding molecules. This figures correspond, respectively, to 10.5% and 5.25% of the total Uzbiopharm® anti-Mlt F(ab')2 molecules.
Affinity columns containing 8 mg of immobilized Microgen® antivenom F(ab')2 molecules had capacity to immunoretain 165.02 μg of M. l. obtusa venom proteins (20.63 mg venom/g antivenom). Bearing in mind that the vials of this antivenom are manufactured with 1.3 mL of 85 mg F(ab')2/mL (110.5 mg F(ab')2/vial), the 3GA outcome indicated for Microgen® antivenom a maximal binding capacity of 2.68 mg (0.088 μmoles) M. l. obtusa venom proteins per vial. This result translates into an estimate of between 4.32% (both Ag binding site occupied) and 8.68% (one Ag binding site occupied) of total vial's F(ab')2 molecules bearing paraspecificity towards M. l. obtusa toxins.
3.4. Quantification of lethality neutralizing through combination of in vivo and in vitro preclinical data
The ability to bind toxins is a necessary but not sufficient molecular property to endow an antivenom with clinical utility. The percentage of lethality neutralizing antibodies can be calculated as the ratio between the antivenom potency and its maximum toxin binding capacity, both terms expressed as mg venom/vial. This parameter was (13.72/26.24) x 100 = 52.3% for Uzbiopharm® anti-Ml antivenom and (2.3/2.68) x 100 = 85.8% for Microgen® anti-Vbb antivenom. This means that 52.3% of the 5.25–10.5% Uzbiopharm® anti-Mlt F(ab')2 molecules that recognize M. l. obtusa venom toxins (i.e. 2.75–5.49% of the total Uzbiopharm® anti-Mlt F(ab')2 molecules) represent clinically relevant antibodies. For Microgen® anti-Vbb, 3.71–7.45% of the total antivenom F(ab')2 molecules are potential live-saving antibodies. If the preclinical data are confirmed in clinical studies, the treatment of a M. l. obtusa bite, which can deliver up to 90 mg of venom (Latifi, 1984), would nominally require up to 42 (Inoserp Europe), 51 (Microgen® anti-Vbb) or 62 (Uzbiopharm® anti-Ml) mL of antivenom.
4. Concluding remarks
This paper illustrates the analytical power of combining venomics, third-generation antivenomics, and neutralization assays of the lethal and toxic venom activities to assess the preclinical efficacy of antivenoms. Here, we show that the monospecific anti-Vipera berus berus antivenom from Microgen® (Russia) displays remarkable paraspecificity towards the toxins of Dagestan blunt-nosed viper venom. Analogously, Kurtović and co-workers have also reported that the Vipera ammodytes-specific antivenom (European viper venom antiserum, Institute of Immunology, Zagreb, Croatia) is preclinically effective in neutralizing lethal toxicity and hemorrhagic activity of venoms of Armenian mountain snakes – Montivipera raddei and M. l. obtusa. (Kurtović et al., 2014). This evidence points to a notable degree of conservation of immunogenic determinants between European and Oriental vipers since their geographic segregation during the early Miocene (23.3–16.3 million years ago) (Rögl and Steininger, 1983; Szyndlar and Rage, 1999; Lenk et al., 2001, Garrigues et al., 2005).
Kurtović et al. (2014) reported a protective efficacy for different batches of Zagreb V. ammodytes-specific antivenom (R, defined as the number of LD50 doses that can be neutralized per mL of undiluted serum) of 47.8 ± 20 to 135 ± 21.5 Mlo i. v. LD50s/mL. This range overlaps in its lower limit with the neutralizing capacity of the Microgen® anti-Vbb antivenom. However, the potency of the anti-Vbb antivenom could nominally be improved 11.5–23 times by affinity chromatographic purification of its F(ab')2 antibodies showing paraspecificity against M. l. obtusa (Dagestan) venom toxins.
Acknowledgements
This research was partly funded by grant BFU2017-89103-P (Ministerio de Ciencia, Innovación y Universidades, Madrid, Spain), by Vicerrectoría de Investigación, Universidad de Costa Rica (741-A0-804), and by LLC “Innova plus” (Saint-Petersburg, Russia).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.toxcx.2020.100035.
Contributor Information
Davinia Pla, Email: dpla@ibv.csic.es.
Sarai Quesada-Bernat, Email: squesada@ibv.csic.es.
Yania Rodríguez, Email: yrodriguez@ibv.csic.es.
Andrés Sánchez, Email: andres.sanbre@gmail.com.
Mariángela Vargas, Email: mariangela.vargasarroyo@ucr.ac.cr.
Mauren Villalta, Email: mcvillalta@gmail.com.
Susana Mesén, Email: susanamesen@gmail.com.
Álvaro Segura, Email: alvaro.seguraruiz@ucr.ac.cr.
Denis O. Mustafin, Email: denis.o.mustafin@gmail.com.
Yulia A. Fomina, Email: fomina.ua@innovaplus.ru.
Ruslan I. Al-Shekhadat, Email: ceo@innovaplus.ru.
Juan J. Calvete, Email: jcalvete@ibv.csic.es.
Ethical statement
International ethical guidelines for scientific papers were followed in the preparation of this manuscript.
Author contribution
DP, SQ-B, and YR performed the biochemical and proteomic characterizations. SQ-B carried out the antivenomics assays. AnS, MAV, MV, SM, and ÁlS performed the in vivo assays. DOM, YAF, and RIA-S, participated in the discussion of the results, carried out a critical review of the work. JJC, ÁlS and RIA-S were responsible for the conception of the work. JJC, ÁlS supervised the experimental work. JJC drafted the manuscript. All the authors participated in the discussion of the results, read and approved the final manuscript.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Al-Shekhadat R.I., Lopushanskaya K.S., Segura Á., Gutiérrez J.M., Calvete J.J., Pla D. Vipera berus berus venom from Russia: venomics, bioactivities and preclinical assessment of Microgen antivenom. Toxins. 2019;11:E90. doi: 10.3390/toxins11020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo H.P., Bourguignon S.C., Boller M.A., Dias A.A., Lucas E.P., Santos I.C., Delgado I.F. Potency evaluation of antivenoms in Brazil: the national control laboratory experience between 2000 and 2006. Toxicon. 2008;51:502–514. doi: 10.1016/j.toxicon.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Arlinghaus F.T., Eble J.A. C-type lectin-like proteins from snake venoms. Toxicon. 2012;60:512–519. doi: 10.1016/j.toxicon.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Burdmann E.A., Woronik V., Prado E.B., Abdulkader R.C., Saldanha L.B., Barreto O.C., Marcondes M. Snakebite-induced acute renal failure: an experimental model. Am. J. Trop. Med. Hyg. 1993;48:82–88. doi: 10.4269/ajtmh.1993.48.82. [DOI] [PubMed] [Google Scholar]
- Calvete J.J., Moreno-Murciano M.P., Theakston R.D., Kisiel D.G., Marcinkiewicz C. Snake venom disintegrins: novel dimeric disintegrins and structural diversification by disulphide bond engineering. Biochem. J. 2003;372:725–734. doi: 10.1042/BJ20021739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete J.J., Gutiérrez J.M., Sanz L., Pla D., Lomonte B. Antivenomics: a proteomics tool for studying the immunoreactivity of antivenoms. In: Kool J., Niessen W.M., editors. Analyzing Biomolecular Interactions by Mass Spectrometry. first ed. Wiley-VCH Verlag GmbH & Co.; 2015. pp. 227–239. [Google Scholar]
- Calvete J.J., Rodríguez Y., Quesada-Bernat S., Pla D. Toxin-resolved antivenomics-guided assessment of the immunorecognition landscape of antivenoms. Toxicon. 2018;148:107–122. doi: 10.1016/j.toxicon.2018.04.015. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. Next-generation snake venomics: protein-locus resolution through venom proteome decomplexation. Expert Rev. Proteomics. 2014;11:315–329. doi: 10.1586/14789450.2014.900447. [DOI] [PubMed] [Google Scholar]
- Calderón-Celis F., Cid-Barrio L., Encinar J.R., Sanz-Medel A., Calvete J.J. Absolute venomics: absolute quantification of intact venom proteins through elemental mass spectrometry. J. Proteomics. 2017;164:33–42. doi: 10.1016/j.jprot.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Carbajo R.J., Sanz L., Pérez A., Calvete J.J. NMR structure of bitistatin – a missing piece in the evolutionary pathway of snake venom disintegrins. FEBS J. 2015;282:341–360. doi: 10.1111/febs.13138. [DOI] [PubMed] [Google Scholar]
- Chikin Y.A., Shcherbak N.N. Vestnik Zoologii; Kiev: 1992. New subspecies of Vipera lebetina chernovi ssp. nov.. (Reptilia, Viperidae) from Middle Asia [in Russian] pp. 45–49. [Google Scholar]
- Chippaux J. Epidemiology of snakebites in Europe: a systematic review of the literature. Toxicon. 2012;59:86–99. doi: 10.1016/j.toxicon.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Cidade D.A., Simão T.A., Dávila A.M., Wagner G., Junqueira-de-Azevedo I.L., Ho P.L., Bon C., Zingali R.B., Albano R.M. Bothrops jararaca venom gland transcriptome: analysis of the gene expression pattern. Toxicon. 2006;48:437–461. doi: 10.1016/j.toxicon.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Clemetson K.J. Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon. 2010;56:1236–1246. doi: 10.1016/j.toxicon.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Corkill N.L. An inquiry into snake bite in Iraq. Indian J. Med. Res. 1932;20:599–696. [Google Scholar]
- De Smedt . JDS Verlag; Halblech, Germany: 2006. J. The Vipers of Europe; p. 340. [Google Scholar]
- Dehghani R., Mehrpour O., Shahi M.P., Jazayeri M., Karrari P., Keyler D., Zamani N. Epidemiology of venomous and semi-venomous snakebites (ophidia: Viperidae, colubridae) in the kashan city of the isfahan province in Central Iran. J. Res. Med. Sci. 2014;19:33–40. [PMC free article] [PubMed] [Google Scholar]
- Dole V.P. A relation between non-esterified fatty acids in plasma and the metabolism of glucose. J. Clin. Invest. 1956;35:150–154. doi: 10.1172/JCI103259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwigubsky I.A. University of Moscow Press; 1832. Experience of the Natural History of All Animals of the Russian Empire; p. 48. 1832. [Google Scholar]
- Eichberg S., Sanz L., Calvete J.J., Pla D. Constructing comprehensive venom proteome reference maps for integrative venomics. Expert Rev. Proteomics. 2015;12:557–573. doi: 10.1586/14789450.2015.1073590. [DOI] [PubMed] [Google Scholar]
- Ferreira S.H., Bartelt D.C., Greene L.J. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry. 1970;9:2583–2593. doi: 10.1021/bi00815a005. [DOI] [PubMed] [Google Scholar]
- Finney D.J. Macmillan; Oxford, UK: 1947. Probit Analysis. A Statistical Treatment of the Sigmoid Response Curve. [Google Scholar]
- Fox J.W., Serrano S.M. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- García-Arredondo A., Martínez M., Calderón A., Saldívar A., Soria R. Preclinical assessment of a new polyvalent antivenom (Inoserp Europe) against several species of the subfamily Viperinae. Toxins. 2019;11:E149. doi: 10.3390/toxins11030149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues T., Dauga C., Ferquel E., Choumet V., Failloux A.-B. Molecular phylogeny of Vipera Laurenti, 1768 and the related genera Macrovipera (Reuss, 1927) and Daboia (Gray, 1842), with comments about neurotoxic Vipera aspis aspis populations. Mol. Phylogenet. Evol. 2005;35:35–47. doi: 10.1016/j.ympev.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Gené J.A., Roy A., Rojas G., Gutiérrez J.M., Cerdas L. Comparative study on coagulant, defibrinating, fibrinolytic and fibrinogenolytic activities of Costa Rican crotaline snake venoms and their neutralization by a polyvalent antivenom. Toxicon. 1989;27:841–848. doi: 10.1016/0041-0101(89)90096-2. [DOI] [PubMed] [Google Scholar]
- Göçmen B., Arikan H., Ozbel Y., Mermer A., Ciçek K. Clinical, physiological and serological observations of a human following a venomous bite by Macrovipera lebetina lebetina (Reptilia: serpentes) Turk. Parazitoloji Derg. 2006;30:158–162. [PubMed] [Google Scholar]
- Graham R.L.J., Graham C., McClean S., Chen T., O′Rourke M., Hirst D., Theakston D., Shaw C. Identification and functional analysis of a novel bradykinin inhibitory peptide in the venoms of New World Crotalinae pit vipers. Biochem. Biophys. Res. Commun. 2005;338:1587–1592. doi: 10.1016/j.bbrc.2005.10.130. [DOI] [PubMed] [Google Scholar]
- Gray J.E. Monographic synopsis of the vipers, or the family Viperidae. Zool. Miscellany. 1842;2:68–71. [Google Scholar]
- Greene L.-J., Camargo A.C., Krieger E.M., Stewart J.M., Ferreira S.H. Inhibition of the conversion of angiotensin I to II and potentiation of bradykinin by small peptides present in Bothrops jararaca venom. Circ. Res. 1972;31(Suppl. 2):62–71. [PubMed] [Google Scholar]
- Gutiérrez J.M., Gené J.A., Rojas G., Cerdas L. Neutralization of proteolytic and hemorrhagic activities of Costa Rican snake venoms by a polyvalent antivenom. Toxicon. 1985;23:887–893. doi: 10.1016/0041-0101(85)90380-0. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Lomonte B., Chaves F., Moreno E., Cerdas L. Pharmacological activities of a toxic phospholipase A isolated from the venom of the snake Bothrops asper. Comp. Biochem. Physiol. 1986;84C:159–164. doi: 10.1016/0742-8413(86)90183-0. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Sanz L., Escolano J., Fernández J., Lomonte B., Angulo Y., Rucavado A., Warrell D.A., Calvete J.J. Snake venomics of the lesser Antillean pit vipers Bothrops caribbaeus and Bothrops lanceolatus: correlation with toxicological activities and immunoreactivity of a heterologous antivenom. J. Proteome Res. 2008;7:4396–4408. doi: 10.1021/pr8003826. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M. Preclinical assessment of the neutralizing efficacy of snake antivenoms in Latin America and the Caribbean: a review. Toxicon. 2018;146:138–150. doi: 10.1016/j.toxicon.2018.02.053. [DOI] [PubMed] [Google Scholar]
- Herrmann H.W., Joger U., Nilson G. Phylogeny and systematics of viperinae snakes III: resurrectiion of the genus Macrovipera (Reuss, 1927) as suggested by biochemical evidence. Amphibia-Reptilia. 1992;13:375–392. [Google Scholar]
- Howard G.C., Kaser M.R. second ed.second ed. CRC Press, Taylor & Francis Group; Boca Raton (FL: 2014. Making and Using Antibodies: A Practical Handbook. [Google Scholar]
- Huang K.F., Hung C.C., Wu S.H., Chiou S.H. Characterization of three endogenous peptide inhibitors for multiple metalloproteinases with fibrinogenolytic activity from the venom of Taiwan habu (Trimeresurus mucrosquamatus) Biochem. Biophys. Res. Commun. 1998;248:562–568. doi: 10.1006/bbrc.1998.9017. [DOI] [PubMed] [Google Scholar]
- Huang K.F., Chiou S.H., Ko T.P., Wang A.H. Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by x-ray crystallography and synthetic inhibitor analogues. Eur. J. Biochem. 2002;269:3047–3056. doi: 10.1046/j.1432-1033.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- Juárez P., Comas I., González-Candelas F., Calvete J.J. Evolution of snake venom disintegrins by positive Darwinian selection. Mol. Biol. Evol. 2008;25:2391–2407. doi: 10.1093/molbev/msn179. [DOI] [PubMed] [Google Scholar]
- Khan M.S. Venomous terrestrial snakes of Pakistan. Snake. 1983;15:101–105. [Google Scholar]
- Koludarov I., Jackson T.N.W., Pozzi A., Mikheyev A.S. Family saga: reconstructing the evolutionary history of a functionally diverse gene family reveals complexity at the genetic origins of novelty. 2019. bioRxiv 583344. [DOI]
- Kreiner G. Edition Chimaira; Frankfurt am Main, Germany: 2007. The Snakes of Europe; p. 317. [Google Scholar]
- Kurtović T., Lang Balija M., Ayvazyan N., Halassy B. Paraspecificity of Vipera ammodytes-specific antivenom towards Montivipera raddei and Macrovipera lebetina obtusa venoms. Toxicon. 2014;78:103–112. doi: 10.1016/j.toxicon.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Latifi M., GAMS C., F d . vol. 8. Academic Press; London: 1978. Commercial production of anti-snakebite serum (antivenom) p. 561. (Biology of the Reptilia). [Google Scholar]
- Latifi M. Variation in yield and lethality of venoms from Iranian snakes. Toxicon. 1984;22:373–380. doi: 10.1016/0041-0101(84)90081-3. [DOI] [PubMed] [Google Scholar]
- Laurenti J.N. Specimen medicum, exhibens synopsin reptilium emendatam cum experimentis circa venena et antidota reptilium austriacorum. Joan. Thom. Nob. de Trattnern. 1768:214. Vienna(+ Plates I-V) [Google Scholar]
- Lenk P., Kalyabina S., Wink M., Joger U. Evolutionary relationships among the true vipers (Reptilia: Viperidae) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2001;19:94–104. doi: 10.1006/mpev.2001.0912. [DOI] [PubMed] [Google Scholar]
- Linnaeus C. Systema naturæ per regna tria naturæ, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Tomus I. Editio decima, reformata. Laurentii Salvii, Holmiæ. 1758:824. [Google Scholar]
- Luft F.C. The Bothrops legacy: vasoactive peptides from Brazil. Renin Rep. 2008;10:57–64. doi: 10.3317/jraas.2008.009. [DOI] [PubMed] [Google Scholar]
- Mallow D., Ludwig D., Nilson G. Krieger Publishing Company; Malabar, Florida: 2003. True Vipers: Natural History and Toxinology of Old World Vipers; p. 359. [Google Scholar]
- Morais V., Ifran S., Berasain P., Massaldi H. Antivenoms: potency or median effective dose, which to use? J. Venom. Anim. Toxins Incl. Trop. Dis. 2010;16:191–193. [Google Scholar]
- Munekiyo S.M., Mackessy S.P. Presence of peptide inhibitors in rattlesnake venoms and their effects on endogenous metalloproteases. Toxicon. 2005;45:255–263. doi: 10.1016/j.toxicon.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Nilson G., Andrén C. Vipera lebetina transmediterranea, a new subspecies of viper from North Africa, with remarks on the taxonomy of V. lebetina and V. mauritanica (Reptilia: Viperidae) Bonn. Zool. Beitrage. 1988;39:371–379. [Google Scholar]
- Nilson G., Tuniyev B., Andrén C., Orlov N., Joger U., Herrmann H.-W. Taxonomic position of the Vipera xanthina complex. Kaupia. 1999;8:99–102. [Google Scholar]
- Okuda D., Koike H., Morita T. A new gene structure of the disintegrin family: a subunit of dimeric disintegrin has a short coding region. Biochemistry. 2002;41:14248–14254. doi: 10.1021/bi025876s. [DOI] [PubMed] [Google Scholar]
- O' Leary M., Isbister G.K. A turbidimetric assay for the measurement of clotting times of procoagulant venoms in plasma. J. Pharmacol. Toxicol. Methods. 2010;61:27–31. doi: 10.1016/j.vascn.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Oraie H., Rastegar-Pouyani E., Khosravani A., Moradi N., Akbari A., Sehhatisabet M.E., Shafiei S., Stümpel N., Joger U. Molecular and morphological analyses have revealed a new species of blunt-nosed viper of the genus Macrovipera in Iran. Salamandra. 2018;54:233–248. [Google Scholar]
- Phelps T. Edition Chimaira; Frankfurt am Main, Germany: 2010. Old World Vipers. A Natural History of the Azemiopinae and Viperinae; p. 558. [Google Scholar]
- Pla D., Rodríguez Y., Calvete J.J. Third generation antivenomics: pushing the limits of the in vitro preclinical assessment of antivenoms. Toxins. 2017;9:E158. doi: 10.3390/toxins9050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss T. Sechs europäische giftschlangengattungen. Zool. Anz. 1927;73(5/8):124–129. [Google Scholar]
- Rögl F., Steininger F.F. Vom Zerfall der Thetys zu Mediterran und Paratethys. Ann. Naturhist. Mus. Wien 85/A (1983) 1983:135–163. [Google Scholar]
- Sánchez A., Herrera M., Villalta M., Solano D., Segura Á., Lomonte B., Gutiérrez J.M., León G., Vargas M. Proteomic and toxinological characterization of the venom of the South African Ringhals cobra Hemachatus haemachatus. J. Proteomics. 2018;181:104–117. doi: 10.1016/j.jprot.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Sanz L., Ayvazyan N., Calvete J.J. Snake venomics of the Armenian mountain vipers Macrovipera lebetina obtusa and Vipera raddei. J. Proteomics. 2008;71:198–209. doi: 10.1016/j.jprot.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Sanz L., Quesada-Bernat S., Chen P.Y., Lee C.D., Chiang J.R., Calvete J.J. Translational venomics: third-generation antivenomics of anti-siamese russell's viper, Daboia siamensis, antivenom manufactured in taiwan CDC's vaccine center. Trav. Med. Infect. Dis. 2018;3:E66. doi: 10.3390/tropicalmed3020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano S.M. The long road of research on snake venom serine proteinases. Toxicon. 2013;62:19–26. doi: 10.1016/j.toxicon.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Sharma L.R., Lal V., Simpson I.D. Snakes of medical significance in India: the first reported case of envenoming by the levantine Viper (Macrovipera lebetina) Wilderness Environ. Med. 2008;19:195–198. doi: 10.1580/07-WEME-CR-175.1. [DOI] [PubMed] [Google Scholar]
- Siigur J., Aaspõllu A., Siigur E. Biochemistry and pharmacology of proteins and peptides purified from the venoms of the snakes Macrovipera lebetina subspecies. Toxicon. 2019;158:16–32. doi: 10.1016/j.toxicon.2018.11.294. [DOI] [PubMed] [Google Scholar]
- Silva J.C., Gorenstein M.V., Li G.Z., Vissers J.P., Geromanos S.J. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Swenson S., Markland F.S., Jr. Snake venom fibrin(ogen)olytic enzymes. Toxicon. 2005;45:1021–1039. doi: 10.1016/j.toxicon.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Schweiger M. Die Folgen eines schweres Bisses von Vipera lebetina obtusa; Seine medizinische Behandlung und Spätfolgen. Herpetofauna. 1983;26:14–16. [Google Scholar]
- Terentiev P.V., Chernov S.A. Second Revised and Enlarged Edition; Leningrad, Uchpedgiz: 1940. A Field Guide to Amphibians and Reptiles of the USSR. [Google Scholar]
- Wagstaff S.C., Favreau P., Cheneval O., Laing G.D., Wilkinson M.C., Miller R.L., Stöcklin R., Harrison R.A. Molecular characterisation of endogenous snake venom metalloproteinase inhibitors. Biochem. Biophys. Res. Commun. 2008;365:650–656. doi: 10.1016/j.bbrc.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Warrell D.A. WHO Library Cataloguing-in-Publication data; 2010. Guidelines for the Management of Snake-Bites. 978-92-9022-377-4. [Google Scholar]
- Werner F. Reptilien der Ägäischen inseln. Sitzber. Akad. Wiss. Wien, Abt., I. 1935;144(1–2):81–117. [Google Scholar]
- Zamani N., Modir-Fallah Rad L., Soltaninejad K., Shadnia S. A retrospective study on snakebite victims in a tertiary referral center. Iranian J. Toxicol. 2016;10:47–50. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.