Just listen to your patient, he is telling you the diagnosis.
Sir William Osler (1849–1919),
Canadian physician and professor of medicine
What will the future of rare diseases look like?
We picture an 18-year-old young man from a village somewhere in Europe. His name is Omer. Omer has been suffering from disease episodes since his early childhood: bursts of fever, fatigue and abdominal pain. He visited many doctors, but none of them was able to establish a satisfactory treatment. He went through a diagnostic and therapeutic odyssey typical for rare disease patients. Fatigue has led him into social isolation. He is perceived as a malingerer.1
Omer uses today’s technology to find help
He begins to Google. Due to the unspecific nature of his approach, the search does not bring up specific results, though. Helpful medical knowledge is not accessible to him this way. Consequently, he looks for help through social media. He is targeted for an advertisement of a symptom checker app because of his previous searches and app downloads in his Ad-ID. He reads ‘start a symptom assessment!’ and decides to instal the app.
Omer starts the symptom assessment in his mother tongue. The chatbot’s underlying ontology is designed to translate professional medical concepts into common language. It initially asks about his basic demographics, ethnicity, pre-existing conditions, current symptoms and their specific attributes. The dialogue reveals that Omer has lately also shown a red, hot, swollen, sharply bordered painful eruption in the calf.
Based on its internal probabilistic disease models, the chatbot’s reasoning algorithms compile a first list of possible underlying diseases, among them several autoimmune diseases and periodic fever syndromes. The chatbot realises the complex situation and the need for a deeper anamnesis. It continues assessing the course and quality of Omer’s symptoms. It asks about the onset, intensity and duration of his fevers thereby identifying the presence of repeating disease episodes. In reaction, the chatbot encourages Omer to track his symptoms over time to increase the value of timing-related information.
By asking detailed questions regarding the possible diseases, the reasoning algorithm rules some of them in and some of them out. For example, the reported absence of weight loss renders tuberculosis less likely. Omer is invited to upload pictures of his skin manifestations aiming to classify them based on a specific diagnostic algorithm. In the consequence, the absence of an urticarial rash reduces Omer’s probability of having Muckle-Wells syndrome. The chatbot asks for red flag symptoms to exclude emergencies. In the end, it establishes a well-founded list of differentials that it uses as hypotheses for the following steps—some periodic fever syndromes are still on it.
Its working hypotheses lead the chatbot to ask about Omer’s previous diseases and treatments. Omer reports a previous appendectomy and the use of painkillers. The app considers the previous appendectomy as hint towards Familial Mediterranean fever (FMF) but at the same time realises the need for more information.
Therefore, explaining the purpose in clear words and following the ethical recommendations of the European Union (EU),2 the chatbot asks for access to health-relevant data stored elsewhere:
Activity data tracked by the smartphone to retrieve information about the disease frequency and periodicity.
His current and past location to adjust for regional disease incidences.
Insurance notes from his medical record to confirm his appendectomy and previous treatments.
Omer consents.
Given the possibility of genetic disorders that can manifest in typical facial phenotypes, the chatbot suggests to perform a selfie face scan, using the smartphone’s three-dimensional sensor. Facial dysmorphism is not found and the app recalculates the probabilities of different genetic disorders.
Seeing Omer’s long medical history, the chatbot considers a genetic component and asks for access to his genetic profile—not only to look for gene mutations but also for single nucleotide polymorphisms. Thereby, it aims to personalise disease probabilities to make the diagnostic process more efficient. However, Omer denies to have such a profile. The chatbot subsequently starts a more thorough family history assessment which uncovers that his uncle is experiencing similar fevers.
Because the collected evidence seems sufficient the chatbot decides to end the assessment at this point. The resulting probability for FMF is high enough, with regards to FMF disease criteria3:
Recurrent febrile episodes.
Signs of peritonitis during episodes.
Possibly Erysipelas-like skin manifestations.
Appropriate ethnicity.
Age <20 years.
(Suspected) family history of FMF.
Based on all the collected data, the app provides an assessment report containing a list of the possible underlying diseases ranked by their calculated probabilities.
The reasoning algorithm recognises that securing the diagnosis of FMF needs professional support. Therefore, it compiles a sequence of diagnostic steps:
A physical examination and an ultrasound scan to evaluate for lymphadenopathy and splenomegaly.
An interferon-gamma release assay to rule out tuberculosis.
A blood test to evaluate presence of signs of inflammation and autoimmune diseases.
A urine sample to test for proteinuria as a sign of renal amyloidosis.
A genetic test to confirm a Mediterranean fever (MEFV) gene mutation.
The app creates a status-quo report in which all previous findings, working hypothesis, remaining differentials and the next diagnostic steps are presented. It asks Omer to send it to a doctor of his choice. However, Omer denies to have an appropriate doctor. Because of this, the app retrieves experts for this disease from a repository and suggests the nearest specialist. Omer agrees to contact the expert in the nearest city 90 km away and to forward the assessment report. The chatbot now sees that the expert’s clinic has a standardised consumer-to-clinical handover application programming interface. After an automated check of the data has confirmed the legitimacy of the request, it delivers Omer's case data in a structured, ontology-based form.
The expert spots Omer’s request in the inbox of his clinical decision support system (CDSS). Beside a time-saving and well-structured description of the status-quo, Omer’s app provides as well information on why certain tests are needed and what their outcomes would mean for the diagnostic progress. Moreover, it visualises the underlying reasoning in an understandable and traceable way according to the rules of the International Telecommunication Union4 to make the machine’s arguments as verifiable as possible. Underlying affected molecular pathways are presented, too.
After having examined Omer’s report, the expert agrees on FMF being a possible explanation. Referring to the recommendations for diagnosis of FMF,5 the app actively suggests a specific genetic test. To save Omer the long way to the city, the specialist sets up a video consultation with him to explain the situation and the diagnostic process to follow: Omer receives a sequence of home test kits for (dried) blood, urine and genetic testing via mail and returns them.
The genetic test confirms a biallelic pathogenic mutation in the MEFV gene. The CDSS recommends colchicine treatment following the recommendations of the European League Against Rheumatism.6 In a video consultation the expert explains the results and medication to Omer, sending him an electronic prescription via the app. Omer picks up the medication in his local pharmacy the next day.
Omer’s treatment success and the potential side effects are continuously monitored, making use of patient-reported outcome measures, symptom tracking and data from sensors. Urine tests to monitor for amyloidosis are performed at home. The chatbot regularly interacts with Omer to ask about his well-being and compliance.
Once, the app notifies the expert due to changes in the tracked parameters suggestive of therapy failure. After a tele-consultation, medication is changed to an anti-IL-1 treatment. The following outcome measures improve and Omer remains free of attacks.
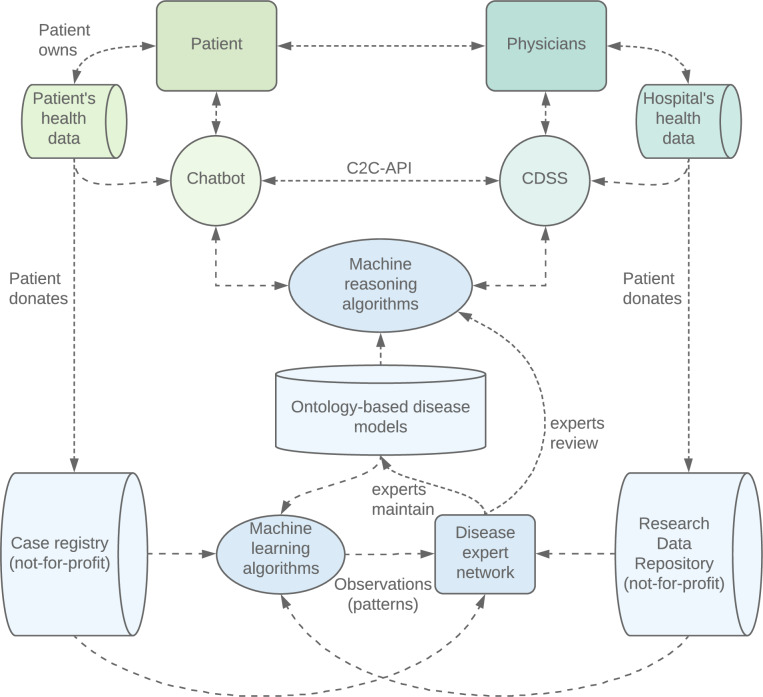
Omer’s data are stored in a standardised ontology-based format in his personal health wallet. He is thankful for the help he has perceived and decides to donate his data to the medical community. His case data are added to an international, not-for-profit registry. Insights from Omer’s case are analysed in conjunction with other cases, are checked by an expert panel and returned into the app’s knowledge base to enable a learning system that improves with the knowledge added by every case (figure 1).
Figure 1.
The system consists of a consumer part (green), a clinical part (turquoise) and a hybrid intelligent system (blue) that are able to communicate. The hybrid intelligent system consists of a network of different parts, including case registry, research data repository, knowledge models and algorithms. The framework is designed to enable patients and physicians to exchange health and research data, access valid medical knowledge and use reviewed reasoning technology while ensuring the quality of the components, auditability and a learning system. C2C-API: consumer-to-clinical handover application programming interface; CDSS: clinical decision support system
Is something like this already possible? Almost
There are symptom checker apps that perform symptom assessments to compile lists of likely diseases or to give safe next step advice.7 8 One of them is combining knowledge models of diseases with probabilistic reasoning that deliver promising results also in the field of rare diseases.9 There is an increasing number of deep learning artificial intelligence algorithms10 that support specific diagnostic tasks like the classification of specific skin conditions11 12 or the recognition of facial phenotypes of genetic disorders based on images.13 There is polygenic risk assessment.14 There are expert repositories that allow to search for the right specialist.15 EU recommendations exist for electronic health record formats that will enable patients and physician to share health data securely.16 CDSS to help experts to diagnose rare diseases have been developed.9 17 Video consultations,18 19 remote self-testing kits20 and electronic prescriptions21 are a reality. Dry-blood sampling is on its way.22 Remote patient monitoring with sensors23 is in use. Digital therapeutics are a reality,24 disease telemonitoring exists25 and digital tracking of outcome measures has shown positive effects.26 Most information about rare diseases is already collected in databases that use common standards to describe disease phenotypes and are connected to databases of genetic variants.27
But now, to unfold the full potential of these technologies, and at the same time avoiding their risks, the pieces have to be integrated in a seamless, easy-to-use and ethical platform patients and physicians can trust (figure 1). To ensure trust, traceability of the system processes will be crucial to overcome the limitations of non-transparent systems and guarantee accountability in the event of incorrect diagnosis or treatment complications. A system that is transparent in its decision making is needed, so that patients and physicians will be able to act autonomously and responsibly. Convenience should not be enough when it comes to personal health. Trustworthiness of data handling as well as medical quality of knowledge-base and algorithms are essential. Here the authors see clinical and scientific experts to take responsibility and set up a not-for-profit repository that provides ontology-based up-to-date knowledge. The repository will function as a gold-standard basis for hybrid intelligent systems regulated by risk-managing methodologies similar to medical device regulation. Machine learning algorithms will analyse the outcome data and point out interesting patterns to experts who will explore the biomedical causality behind these observations according to proven scientific standards.
In this future, disease experts will collaboratively provide their knowledge and machines will participate in the sphere of proven knowledge and feed back their observations to the experts. Then, machines and humans will truly work and think together—and it will usher in the age of AI.
Glossary of terms
| Chatbot | A computer programme that conducts a conversation in natural language via text or voice |
| Clinical decision support system (CDSS) | An expert system that supports physicians by facilitating the diagnostic reasoning process |
| Consumer-to-clinical handover application programming interface | A communication protocol between a health chatbot application and a CDSS to facilitate the transfer of case information |
| Hybrid intelligent system | A software system that combines methods from multiple fields of artificial intelligence |
| Knowledge models | A representation of (medical) knowledge in machine readable form |
| Machine reasoning algorithm | A predefined set of steps to draw conclusions from input data |
| Machine learning algorithm | A mathematical model that learns how to give a prediction/output based on input data through a training process |
| Ontology | A formal specification of a shared, usually hierarchically organised conceptualisation to represent the knowledge of a subject area, for example from the medical domain |
| Research data repository | A subject-specific database designed to store, organise, use, share and archive research datasets |
| Symptom checker | A software designed to determine the possible causes for user-reported symptoms to provide advice on next steps or treatment |
Footnotes
Handling editor: Josef S Smolen
MCH and SR contributed equally.
Contributorship:MCH and SR wrote the manuscript, contributed equally and share first authorship. MK collected background Information and edited the manuscript. ADW wrote parts of the manuscript and performed medical supervision. All authors read and approved the final manuscript. SR: doctoral student at Department of Nephrology and Hypertension, Hannover Medical School, Hannover, Germany
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: MCH: cofounder Ada Health. SR: employee Ada Health.
Patient consent for publication: Not required.
Provenance and peer review: Commissioned; externally peer reviewed.
References
- 1. Erbis G, Schmidt K, Hansmann S, et al. . Living with autoinflammatory diseases: identifying unmet needs of children, adolescents and adults. Pediatr Rheumatol 2018;16:81 10.1186/s12969-018-0300-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ethics guidelines for trustworthy AI [Internet], 2019. Available: https://ec.europa.eu/digital-single-market/en/news/ethics-guidelines-trustworthy-ai [Accessed 6 Jan 2020].
- 3. Livneh A, Langevitz P, Zemer D, et al. . Criteria for the diagnosis of familial Mediterranean fever. Arthritis Rheum 1997;40:1879–85. 10.1002/art.1780401023 [DOI] [PubMed] [Google Scholar]
- 4. Salathé EPFL M, Wiegand Fraunhofer H, Wenzel Fraunhofer H, et al. . White paper of the Focus Group on Artificial Intelligence for Health (FG-AI4H) [Internet], 2020. Available: https://www.itu.int/go/fgai4h [Accessed 8 Jan 2020].
- 5. Giancane G, Ter Haar NM, Wulffraat N, et al. . Evidence-Based recommendations for genetic diagnosis of familial Mediterranean fever. Ann Rheum Dis 2015;74:635–41. 10.1136/annrheumdis-2014-206844 [DOI] [PubMed] [Google Scholar]
- 6. Ozen S, Demirkaya E, Erer B, et al. . EULAR recommendations for the management of familial Mediterranean fever. Ann Rheum Dis 2016;75:644–51. 10.1136/annrheumdis-2015-208690 [DOI] [PubMed] [Google Scholar]
- 7. Semigran HL, Linder JA, Gidengil C, et al. . Evaluation of symptom checkers for self diagnosis and triage: audit study. BMJ 2015;351:h3480 10.1136/bmj.h3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fraser H, Coiera E, Wong D. Safety of patient-facing digital symptom checkers. The Lancet 2018;392:2263–4. 10.1016/S0140-6736(18)32819-8 [DOI] [PubMed] [Google Scholar]
- 9. Ronicke S, Hirsch MC, Türk E, et al. . Can a decision support system accelerate rare disease diagnosis? evaluating the potential impact of ADA DX in a retrospective study. Orphanet J Rare Dis 2019;14:69 10.1186/s13023-019-1040-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Topol EJ. High-Performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44–56. 10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 11. Han SS, Kim MS, Lim W, et al. . Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol 2018;138:1529–38. 10.1016/j.jid.2018.01.028 [DOI] [PubMed] [Google Scholar]
- 12. Esteva A, Kuprel B, Novoa RA, et al. . Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–8. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gurovich Y, Hanani Y, Bar O, et al. . Identifying facial phenotypes of genetic disorders using deep learning. Nat Med 2019;25:60–4. 10.1038/s41591-018-0279-0 [DOI] [PubMed] [Google Scholar]
- 14. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 2018;19:581–90. 10.1038/s41576-018-0018-x [DOI] [PubMed] [Google Scholar]
- 15. se-atlas: Kartierung von Versorgungseinrichtungen und Patientenorganisationen für Menschen mit Seltenen Erkrankungen [Internet]. Available: https://www.se-atlas.de/ [Accessed 6 Jan 2020].
- 16. Recommendation on a European Electronic Health Record exchange format [Internet], 2019. Available: https://ec.europa.eu/digital-single-market/en/news/recommendation-european-electronic-health-record-exchange-format [Accessed 9 Jan 2020].
- 17. Svenstrup D, Jørgensen HL, Winther O. Rare disease diagnosis: a review of web search, social media and large-scale data-mining approaches. Rare Dis 2015;3:e1083145 10.1080/21675511.2015.1083145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ignatowicz A, Atherton H, Bernstein CJ, et al. . Internet videoconferencing for patient-clinician consultations in long-term conditions: a review of reviews and applications in line with guidelines and recommendations. Digit Health 2019;5:205520761984583. 10.1177/2055207619845831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mold F, Hendy J, Lai Y-L, et al. . Electronic consultation in primary care between providers and patients: systematic review. JMIR Medical Informatics 2019;7:e13042 10.2196/13042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tidy EJ, Shine B, Oke J, et al. . Home self-testing kits: helpful or harmful? Br J Gen Pract 2018;68:360–1. 10.3399/bjgp18X698021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Porterfield A, Engelbert K, Coustasse A. Electronic prescribing: improving the efficiency and accuracy of prescribing in the ambulatory care setting. Perspect Health Inf Manag 2014;11:1g. [PMC free article] [PubMed] [Google Scholar]
- 22. Vázquez-Morón S, Ardizone Jiménez B, Jiménez-Sousa MA, et al. . Evaluation of the diagnostic accuracy of laboratory-based screening for hepatitis C in dried blood spot samples: a systematic review and meta-analysis. Sci Rep 2019;9:7316 10.1038/s41598-019-41139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noah B, Keller MS, Mosadeghi S, et al. . Impact of remote patient monitoring on clinical outcomes: an updated meta-analysis of randomized controlled trials. npj Digit Med 2018;1 10.1038/s41746-017-0002-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makin S. The emerging world of digital therapeutics. Nature 2019;573:S106–9. 10.1038/d41586-019-02873-1 [DOI] [PubMed] [Google Scholar]
- 25. Paré G, Jaana M, Sicotte C. Systematic review of home telemonitoring for chronic diseases: the evidence base. J Am Med Inform Assoc 2007;14:269–77. 10.1197/jamia.M2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basch E, Deal AM, Kris MG, et al. . Symptom monitoring with patient-reported outcomes during routine cancer treatment: a randomized controlled trial. J Clin Oncol 2016;34:557–65. 10.1200/JCO.2015.63.0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Köhler S, Vasilevsky NA, Engelstad M, et al. . The human phenotype ontology in 2017. Nucleic Acids Res 2017;45:D865–76. 10.1093/nar/gkw1039 [DOI] [PMC free article] [PubMed] [Google Scholar]