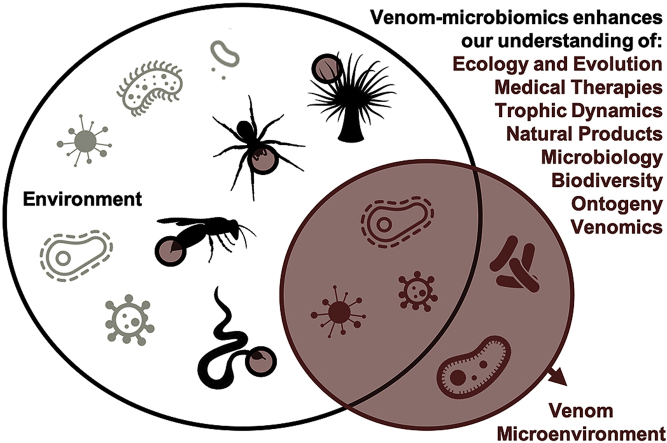
Abstract
Venom is a known source of novel antimicrobial natural products. The substantial, increasing number of these discoveries have unintentionally culminated in the misconception that venom and venom-producing glands are largely sterile environments. Culture-dependent and -independent studies on the microbial communities in venom microenvironments reveal the presence of archaea, algae, bacteria, fungi, protozoa, and viruses. Venom-centric microbiome studies are relatively sparse to date with the adaptive advantages that venom-associated microbes might offer to their hosts, or that hosts might provide to venom-associated microbes, remaining largely unknown. We highlight the potential for the discovery of venom microbiomes within the adaptive landscape of venom systems. The considerable number of convergently evolved venomous animals, juxtaposed with the comparatively few known studies to identify microbial communities in venom, provides new possibilities for both biodiversity and therapeutic discoveries. We present an evidence-based argument for integrating microbiology as part of venomics (i.e., venom-microbiomics) and introduce iVAMP, the Initiative for Venom Associated Microbes and Parasites (https://ivamp-consortium.github.io/), as a growing collaborative consortium. We express commitment to the diversity, inclusion and scientific collaboration among researchers interested in this emerging subdiscipline through expansion of the iVAMP consortium.
Keywords: Bacteria, Coevolution, Holobiont, Microbiome, Symbiont, Virus
Graphical abstract
Highlights
-
•
Venom-microbiome studies as an integrative field of venomics and microbiology.
-
•
Argument for multi-omics-based discovery through a microenvironment framework.
-
•
Introduction of a venom-microbiome research consortium (iVAMP).
1. Text
While scientific research in toxinology and microbiology has persisted for centuries, a cursory search of the literature reveals less than 150 studies overlap between these two fields despite each significantly advancing as a result of next generation sequencing technology (Fig. 1, Supplemental Table 1, Supplemental Code). The integration of genomics (Moran and Gurevitz, 2006), transcriptomics (Pahari et al., 2007), and proteomics (Fry, 2005) into the study of venom has contributed to new toxin discovery and associated biological activity (Oldrati et al., 2016, Calvete, 2017). Over the past 15 years, microbiome research has yielded breakthroughs in our knowledge of unculturable microbial “dark matter” (Bernard et al., 2018), the origins of life (Spang et al., 2017), and human health (Arnold et al., 2016, Clavel et al., 2016). Providing ecological and evolutionary context has enhanced both microbiology (Boughner and Singh, 2016, Hird, 2017) and venomics (Prashanth et al., 2016, Sunagar et al., 2016, Calvete, 2017). We thus propose viewing venom as a microenvironment that occupies a unique niche in which microbes may adapt as a critical perspective for investigating the dynamics of venom-microbe interactions.
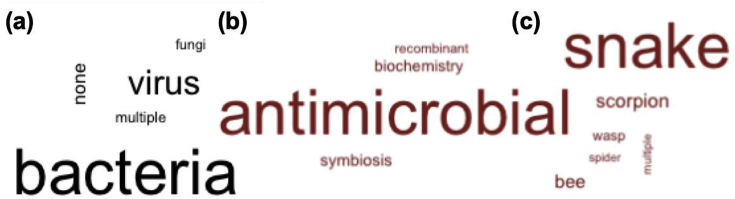
Fig. 1.
Word clouds representative ofSupplemental Table 1content. A breakdown of 140 resultant articles from searching Web of Science for venom-microbe studies. (a) Most articles are either bacteria- or virus-specific, and a subset (16 articles) are not related to studies involving microbes. After removing these articles, investigation of the remaining 126 show (b) approximately 71% focus on venom toxins exhibiting antimicrobial properties with only about 11% focused on venom-microbe interactions. (c) Roughly 57% of the surveyed studies focus on snake venom, and the remaining studies are largely from arthropods.
Researchers in the fields of both venomics and microbiology share common interests in natural products (Katz and Baltz, 2016, Robinson et al., 2017) and adaptive evolution (Phuong et al., 2016, Hird, 2017). With more information on the presence and diversity of venom-associated microbiomes (Table 1), future research efforts can focus on how microbes colonize and thrive in venom glands as a starting point for integrating these fields (McFall-Ngai, 2014, Nunes-Alves, 2015). For example, examining the biology of the host using microscopy (Schlafer and Meyer, 2017) and biomechanics (Yevick and Martin, 2018) could result in translated predictive models (Biggs et al., 2015) for identifying the underlying mechanisms of toxin and metabolite function (Sapp, 2016, Adnani et al., 2017). Determining if and which venom microenvironments are truly sterile, and if microbes contribute to shaping the genetic architecture of the venom gland, will prove critical in our understanding of venom evolution (Conlin et al., 2014) and antimicrobial resistance (Adnani et al., 2017). Correlating microbial community profiles with functional characteristics of venom could provide yet another layer to the venomics field that would deepen our insight on the mechanisms driving venom variation. Identifying microbial species that have adapted to these seemingly extreme environments (Rampelotto, 2013) will open new avenues of research, and emphasizes the need for phylogenetically representative venom host model systems to be bred axenically in vivo to allow researchers to test the functional roles of venom-associated microbes observed in the wild (Fig. 2).
Table 1.
Explicit Sequencing and Next-Generation venom microbiome studies, including published & in progress work within iVAMP (“+” denotes a collaboration formed because of access to the iVAMP network). Next-generation venom microbiome studies are comparatively recent, and few in number. Even so, the diversity of these host and microbial community studies highlight the potential benefits of integrating microbiology and venomics (Webb and Summers, 1990, Peraud et al., 2009, Goldstein et al., 2013, Debat, 2017, Torres et al., 2017, Esmaeilishirazifard et al., 2018).
| Published Studies | Organism | Tissue | Wild/Captive | Approach |
|---|---|---|---|---|
| Webb and Summers, 1990 | Wasp | Venom gland | Captive | Culture, Sanger Sequencing |
| Peraud et al. (2009) | Cone-snail (3 species) | Body, Hepatopancreas, Venom Duct | Wild | Culture, FISH, Sanger Sequencing |
| Goldstein et al. (2013) | Monitor Lizard | Saliva, Gingiva | Captive | Culture, Sanger Sequencing, 16S |
| Simmonds et al. (2016) | Parasitoid Wasp | Venom Gland | Wild | RNAseq/reverse transcriptomics |
| Debat, 2017 | Spiders | Transcriptomes of the Body, Brain, Silk Gland Venom Gland |
Wild | Data-mining (NGS) |
| Torres et al. (2017) | Cone-snail (8 species) | Venom Duct, Muscle, External Duct | Wild | 16S, 454 |
| Esmaeilishirazifard et al. (2018) | Snakes (5 species) Spiders (2 species) | Venom, Oral Cavity | Wild, Captive | Culture, 16S, WGS |
| iVAMP Projects in progress |
Organism |
Tissue |
Wild/Captive |
Approach |
| Colston | Snakes (multiple) | Venom, Venom Glands, Venom Ducts, Oral Cavity, Muscle, Stomach and GIT | Wild, Captive | 16S, RNAseq transcriptomics, Proteomics |
| Harms + Macrander | Lionfish: Pterois volitans | venom glands, venom | Wild (Invasive) | Transcriptomics, Proteomics |
| Keiser + Colston | Spiders: Stegodyphus | venom glands, venom | Wild, Captive | 16S, RNAseq transcriptomics, Proteomics |
| Stiers, Colston | Snake: Crotalus scutulatus | Venom, Venom Glands, Venom Ducts, Oral Cavity, Muscle, Stomach and GIT | Wild, Captive | 16S, RNAseq transcriptomics, Proteomics |
| Ul-Hasan, Nobile, Petras | Cone-snail: Californiconus californicus | Venom, Venom Duct, Hepatopancreas, Shell, Egg | Wild, Captive | 16S and 18S, Proteomics, Metabolomics |
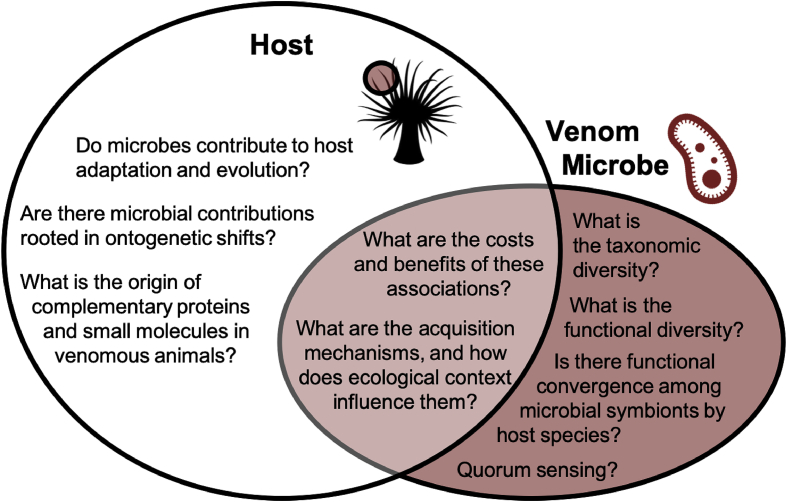
Fig. 2.
Proposed questions for venom-microbiome exploration of the ecology and evolution of venomous hosts and their microbial associates. A Venn diagram displaying the intersections of microbiology and venomics through an ecology and evolution focus. The questions presented are examples of possible areas of investigation to advance the field.
The host-microbe interactions that naturally occur in the venom microenvironment remain largely unknown, and addressing this knowledge gap through directed microbiome sequencing experiments within a wildtype ecosystem framework will strengthen our understanding of animal associated microbes (McFall-Ngai et al., 2013). A variety of microbial studies have found tetrodotoxin-producing bacteria in venomous and poisonous animals (Hwang et al., 1989, Cheng et al., 1995, Pratheepa and Vasconcelos, 2013, Stokes et al., 2014) as well as a number of viruses with RNA genomes residing in venom (Debat, 2017). These studies contrast with the notion of the venom microenvironment as largely sterile in that the primary research on venom-gland derived toxin compounds focuses on antimicrobial properties (Fig. 1). However, (1) compounds derived from or contained within venom that demonstrate antimicrobial activity against clinical and/or reference strains (Almeida et al., 2018) may not reflect what occurs against wild-type strains that co-evolved within venom glands (Reis et al., 2018), and (2) cultured microbes can produce compounds in a lab setting that they may not produce in nature (McCoy and Clapper, 1979, Simmons et al., 2008, Peraud et al., 2009, Catalán et al., 2010, Quezada et al., 2017b, Quezada et al., 2017a, Quezada et al., 2017b, Silvestre et al., 2005, Yu et al., 2011). The captive environment, which is already known to affect the host venom profile (Willemse et al., 1979, Freitas-de-Sousa et al., 2015), may also influence microbial composition of the oral and venom microbiomes (Hyde et al., 2016), which has led to a call for microbiome studies to utilize wild-collected samples (Colston and Jackson, 2016, Hird, 2017). Studying the venom microbiome, and considering the adaptive traits of microbes under selection in an ecological context as it occurs in the wild, clarifies the evolutionary pressures for these antimicrobial compounds found in venom (Fig. 2). In vitro, in vivo, and natural venom microbiome experiments alongside culture-dependent and -independent techniques contribute to our understanding of mutual symbioses, with room for predictive modeling to identify novel niches for microbial adaptation and competition (Bull et al., 2010, Zhu et al., 2018).
An initial search shows approximately 100 papers per year have consistently been published on venom antimicrobial peptides (PubMed search term - antimicrobial AND peptide AND venom 14th Mar 2019) for the past 5 years. The few venom-microbiome studies in the literature to date (Table 1) indicate a clear need for an expansion of the subdiscipline of venom-microbiome research, and this has led to the formation of an international, collaborative cohort of researchers referred to as the Initiative for Venom Associated Microbes and Parasites (or iVAMP, https://ivamp-consortium.github.io/). A major goal of the iVAMP consortium is to provide a platform for the scientific community to openly discuss areas of interest to the field. Fig. 2 outlines some examples of ongoing questions that may be of interest to iVAMP researchers. By emphasizing representation through practice, this consortium supports working with and for communities from which we sample rather than taking from them. Involving scientists across the globe through initiatives like iVAMP extends beyond the requirements of legislation, such as the Nagoya Protocol (Buck and Hamilton, 2011), to ensure that science is accessible to the public and inclusive of all parties involved. Overall, the approach taken by this initiative expands suggested practices (Weber and Schell Word, 2001; Cheng et al., 2018) for the benefit of scientific innovation and discovery.
As an organization, iVAMP has explicit goals and approaches for furthering the fields of microbiome research and venomics (Fig. 2) as well as specific aims for conducting ethical, inclusive, reproducible science. In doing so, our practices seek to prevent counterproductive competition and instead embrace interdisciplinary, collaborative scientific research. The broad scientific disciplines covered by iVAMP members provide a network that allows researchers access to a variety of technical platforms and key resources that otherwise may not be available in individual labs. This is especially important for those researchers who may want to enter the venomics field, but lack accessibility to the necessary resources or instrumentation. Expansion of knowledge on microbes living in the many diverse venom host microenvironments additionally contributes to currently absent aspects of holobiont and coevolutionary theory (Faure Denis et al., 2018). Through iVAMP, researchers set an open-access tone for the subdiscipline of venom-microbiomics that will be useful well into the future.
Acknowledgements
We thank the conference organizers of Evolution, the Gordon Research Conference, and the Society for Integrative and Comparative Biology for contributing to environments conducive to a major source of these collaborations. We also thank the Toxicon editor-in-chief, Prof. Glenn King, for encouraging this contribution and the two anonymous reviewers for helpful suggestions that improved this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.toxcx.2019.100016.
Funding
We acknowledge support from the affiliated institutions of the authors.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adnani N., Rajski S.R., Bugni T.S. Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep. 2017;34:784–814. doi: 10.1039/c7np00009j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.R., Mendes B., Lancellotti M., Marangoni S., Vale N., Passos Ó., Ramos M.J., Fernandes P.A., Gomes P., Da Silva S.L. A novel synthetic peptide inspired on Lys49 phospholipase A2 from Crotalus oreganus abyssus snake venom active against multidrug-resistant clinical isolates. Eur. J. Med. Chem. 2018;149:248–256. doi: 10.1016/j.ejmech.2018.02.055. [DOI] [PubMed] [Google Scholar]
- Arnold J.W., Roach J., Azcarate-Peril M.A. Emerging technologies for gut microbiome research. Trends Microbiol. 2016;24:887–901. doi: 10.1016/j.tim.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard G., Pathmanathan J.S., Lannes R., Lopez P., Bapteste E. Microbial dark matter investigations: how microbial studies transform biological knowledge and empirically sketch a logic of scientific discovery. Genome Biol. Evol. 2018;10:707–715. doi: 10.1093/gbe/evy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs M.B., Medlock G.L., Kolling G.L., Papin J.A. Metabolic network modeling of microbial communities. WIREs Syst. Biol. Med. 2015;7:317–334. doi: 10.1002/wsbm.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughner L.A., Singh P. Microbial Ecology: where are we now? Postdoc J. 2016;4:3–17. doi: 10.14304/SURYA.JPR.V4N11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Hamilton C. The Nagoya Protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization to the convention on biological diversity. RECIEL. 2011;20:47–61. [Google Scholar]
- Bull J.J., Jessop T.S., Whiteley M. Deathly drool: evolutionary and ecological basis of septic bacteria in Komodo Dragon mouths. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete J.J. Venomics: integrative venom proteomics and beyond. Biochem. J. 2017;474:611–634. doi: 10.1042/BCJ20160577. [DOI] [PubMed] [Google Scholar]
- Catalán A., Espoz M.C., Cortés W., Sagua H., González J., Araya J.E. Tetracycline and penicillin resistant Clostridium perfringens isolated from the fangs and venom glands of Loxosceles laeta: its implications in loxoscelism treatment. Toxicon. 2010;56:890–896. doi: 10.1016/j.toxicon.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Cheng C.A., Hwang D.F., Tsai Y.H., Chen H.C., Jeng S.S., Noguchi T., Ohwada K., Hasimoto K. Microflora and tetrodotoxin-producing bacteria in a gastropod, Niotha clathrata. Food Chem. Toxicol. 1995;33:929–934. doi: 10.1016/0278-6915(95)00061-6. [DOI] [PubMed] [Google Scholar]
- Cheng H., Dove N.C., Mena J.M., Perez T., Ul-Hasan S. The Biota Project: a case study of a multimedia, grassroots approach to scientific communication for engaging diverse audiences. Integr. Comp. Biol. 2018;58:1294–1303. doi: 10.1093/icb/icy091. [DOI] [PubMed] [Google Scholar]
- Clavel T., Lagkouvardos I., Hiergeist A. Microbiome sequencing: challenges and opportunities for molecular medicine. Expert Rev. Mol. Diagn. 2016;16:795–805. doi: 10.1080/14737159.2016.1184574. [DOI] [PubMed] [Google Scholar]
- Colston T.J., Jackson C.R. Microbiome evolution along divergent branches of the vertebrate tree of life: what is known and unknown. Mol. Ecol. 2016;25:3776–3800. doi: 10.1111/mec.13730. [DOI] [PubMed] [Google Scholar]
- Conlin P.L., Chandler J.R., Kerr B. Games of life and death: antibiotic resistance and production through the lens of evolutionary game theory. Curr. Opin. Microbiol., Antimicrobials. 2014;21:35–44. doi: 10.1016/j.mib.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Debat H.J. An RNA virome associated to the golden orb-weaver spider Nephila clavipes. Front. Microbiol. 2017;8:2097. doi: 10.3389/fmicb.2017.02097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis Faure, Simon Jean-Christophe, Thierry Heulin. Holobiont: a conceptual framework to explore the eco-evolutionary and functional implications of host–microbiota interactions in all ecosystems. New Phytol. 2018;218:1321–1324. doi: 10.1111/nph.15199. [DOI] [PubMed] [Google Scholar]
- Esmaeilishirazifard E., Usher L., Trim C., Denise H., Sangal V., Tyson G.H., Barlow A., Redway K., Taylor J.D., Kremmyda-Vlachou M., Loftus T.D., Lock M.M.G., Wright K., Dalby A., Snyder L.A.S., Wuster W., Trim S., Moschos S.A. 2018. Microbial Adaptation to Venom Is Common in Snakes and Spiders. bioRxiv 348433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-de-Sousa L.A., Amazonas D.R., Sousa L.F., Sant'Anna S.S., Nishiyama M.Y., Serrano S.M.T., Junqueira-de-Azevedo I.L.M., Chalkidis H.M., Moura-da-Silva A.M., Mourão R.H.V. Comparison of venoms from wild and long-term captive Bothrops atrox snakes and characterization of Batroxrhagin, the predominant class PIII metalloproteinase from the venom of this species. Biochimie. 2015;118:60–70. doi: 10.1016/j.biochi.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Fry B.G. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein E.J.C., Tyrrell K.L., Citron D.M., Cox C.R., Recchio I.M., Okimoto B., Bryja J., Fry B.G. Anaerobic and aerobic bacteriology of the saliva and gingiva from 16 captive komodo dragons (Varanus komodoensis): new implications for the “bacteria as venom” model. J. Zoo Wildl. Med. 2013;44:262–272. doi: 10.1638/2012-0022R.1. [DOI] [PubMed] [Google Scholar]
- Hird S.M. Evolutionary biology needs wild microbiomes. Front. Microbiol. 2017;8:725. doi: 10.3389/fmicb.2017.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.F., Arakawa O., Saito T., Noguchi T., Simidu U., Tsukamoto K., Shida Y., Hashimoto K. Tetrodotoxin-producing bacteria from the blue-ringed octopus Octopus maculosus. Mar. Biol. 1989;100:327–332. [Google Scholar]
- Hyde E.R., Navas-Molina J.A., Song S.J., Kueneman J.G., Ackermann G., Cardona C., Humphrey G., Boyer D., Weaver T., Mendelson J.R., McKenzie V.J., Gilbert J.A., Knight R. The oral and skin microbiomes of captive komodo dragons are significantly shared with their habitat. mSystems. 2016;1 doi: 10.1128/mSystems.00046-16. e00046-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Baltz R.H. Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- McCoy R.H., Clapper D.R. The oral flora of the South Texas tarantula, Dugesiella anax (araneae: theraphosidae) J. Med. Entomol. 1979;16:450–451. doi: 10.1093/jmedent/16.5.450. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M.J. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu. Rev. Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M., Hadfield M.G., Bosch T.C.G., Carey H.V., Domazet-Lošo T., Douglas A.E., Dubilier N., Eberl G., Fukami T., Gilbert S.F., Hentschel U., King N., Kjelleberg S., Knoll A.H., Kremer N., Mazmanian S.K., Metcalf J.L., Nealson K., Pierce N.E., Rawls J.F., Reid A., Ruby E.G., Rumpho M., Sanders J.G., Tautz D., Wernegreen J.J. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y., Gurevitz M. When positive selection of neurotoxin genes is missing. The riddle of the sea anemone Nematostella vectensis. FEBS J. 2006;273:3886–3892. doi: 10.1111/j.1742-4658.2006.05397.x. [DOI] [PubMed] [Google Scholar]
- Nunes-Alves C. Vibrio genes involved in squid colonization. Nat. Rev. Microbiol. 2015;13:3. [Google Scholar]
- Oldrati V., Arrell M., Violette A., Perret F., Sprüngli X., Wolfender J.-L., Stöcklin R. Advances in venomics. Mol. Biosyst. 2016;12:3530–3543. doi: 10.1039/c6mb00516k. [DOI] [PubMed] [Google Scholar]
- Pahari S., Mackessy S.P., Kini R.M. The venom gland transcriptome of the Desert Massasauga Rattlesnake (Sistrurus catenatus edwardsii): towards an understanding of venom composition among advanced snakes (Superfamily Colubroidea) BMC Mol. Biol. 2007;8:115. doi: 10.1186/1471-2199-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraud O., Biggs J.S., Hughen R.W., Light A.R., Concepcion G.P., Olivera B.M., Schmidt E.W. Microhabitats within venomous cone snails contain diverse actinobacteria. Appl. Environ. Microbiol. 2009;75:6820–6826. doi: 10.1128/AEM.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuong M.A., Mahardika G.N., Alfaro M.E. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genomics. 2016;17:401. doi: 10.1186/s12864-016-2755-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashanth J.R., Dutertre S., Jin A.H., Lavergne V., Hamilton B., Cardoso F.C., Griffin J., Venter D.J., Alewood P.F., Lewis R.J. The role of defensive ecological interactions in the evolution of conotoxins. Mol. Ecol. 2016;25:598–615. doi: 10.1111/mec.13504. [DOI] [PubMed] [Google Scholar]
- Pratheepa V., Vasconcelos V. Microbial diversity associated with tetrodotoxin production in marine organisms. Environ. Toxicol. Pharmacol. 2013;36:1046–1054. doi: 10.1016/j.etap.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Quezada M., Licona-Cassani C., Cruz-Morales P., Salim A.A., Marcellin E., Capon R.J., Barona-Gómez F. Diverse cone-snail species harbor closely related Streptomyces species with conserved chemical and genetic profiles, including polycyclic tetramic acid macrolactams. Front. Microbiol. 2017;8:2305. doi: 10.3389/fmicb.2017.02305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada M., Shang Z., Kalansuriya P., Salim A.A., Lacey E., Capon R.J. Waspergillamide A, a nitro depsi-tetrapeptide diketopiperazine from an Australian mud dauber wasp-associated Aspergillus sp. (CMB-W031) J. Nat. Prod. 2017;80:1192–1195. doi: 10.1021/acs.jnatprod.6b01062. [DOI] [PubMed] [Google Scholar]
- Rampelotto P.H. Extremophiles and extreme environments. Life. 2013;3:482–485. doi: 10.3390/life3030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis P.V.M., Boff D., Verly R.M., Melo-Braga M.N., Cortés M.E., Santos D.M., Pimenta A.M. de C., Amaral F.A., Resende J.M., de Lima M.E. LyeTxI-b, a synthetic peptide derived from Lycosa erythrognatha spider venom, shows potent antibiotic activity in vitro and in vivo. Front. Microbiol. 2018;9:667. doi: 10.3389/fmicb.2018.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.D., Undheim E.A.B., Ueberheide B., King G.F. Venom peptides as therapeutics: advances, challenges and the future of venom-peptide discovery. Expert Rev. Proteom. 2017;14:931–939. doi: 10.1080/14789450.2017.1377613. [DOI] [PubMed] [Google Scholar]
- Sapp J. The symbiotic self. Evol. Biol. 2016;43:596–603. [Google Scholar]
- Schlafer S., Meyer R.L. Confocal microscopy imaging of the biofilm matrix. J. Microbiol. Methods. 2017;138:50–59. doi: 10.1016/j.mimet.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Silvestre F.G., Castro C.S. de, Moura J.F. de, Giusta M.S., Maria M.D., Álvares É.S.S., Lobato F.C.F., Assis R.A., Gonçalves L.A., Gubert I.C., Chávez-Olórtegui C., Kalapothakis E. Characterization of the venom from the Brazilian brown spider Loxosceles similis Moenkhaus, 1898 (araneae, Sicariidae) Toxicon. 2005;46:927–936. doi: 10.1016/j.toxicon.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Simmonds T.J., Carillo D., Burke G.R. Characterization of a venom gland-associated rhabdovirus in the parasitoid wasp Diachasmimorpha longicaudata. J. Insect Physiol. 2016;91–92:48–55. doi: 10.1016/j.jinsphys.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Simmons T.L., Coates R.C., Clark B.R., Engene N., Gonzalez D., Esquenazi E., Dorrestein P.C., Gerwick W.H. Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4587–4594. doi: 10.1073/pnas.0709851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Stairs C.W., Lombard J., Eme L., Ettema T.J.G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 2017;15.:711–723. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- Stokes A.N., Ducey P.K., Neuman-Lee L., Hanifin C.T., French S.S., Pfrender M.E., Iii E.D.B., Jr E.D.B. Confirmation and distribution of tetrodotoxin for the first time in terrestrial invertebrates: two terrestrial flatworm species (Bipalium adventitium and Bipalium kewense) PLoS One. 2014;9 doi: 10.1371/journal.pone.0100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K., Morgenstern D., Reitzel A.M., Moran Y. Ecological venomics: how genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteom., Proteomics in Evolutionary Ecology. 2016;135:62–72. doi: 10.1016/j.jprot.2015.09.015. [DOI] [PubMed] [Google Scholar]
- Torres J.P., Tianero M.D., Robes J.M.D., Kwan J.C., Biggs J.S., Concepcion G.P., Olivera B.M., Haygood M.G., Schmidt E.W. Stenotrophomonas-like bacteria are widespread symbionts in cone snail venom ducts. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.01418-17. e01418-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.A., Summers M.D. Venom and viral expression products of the endoparasitic wasp Campoletis sonorensis share epitopes and related sequences. Proc. Natl. Acad. Sci. U.S.A. 1990;87:4961–4965. doi: 10.1073/pnas.87.13.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.R., Schell Word C. The communication process as evaluative context: what do nonscientists hear when scientists speak? Bioscience. 2001;51:487–495. [Google Scholar]
- Willemse G.T., Hattingh J., Karlsson R.M., Levy S., Parker C. Changes in composition and protein concentration of puff adder (Bitis arietans) venom due to frequent milking. Toxicon. 1979;17:37–42. doi: 10.1016/0041-0101(79)90253-8. [DOI] [PubMed] [Google Scholar]
- Yevick H.G., Martin A.C. Quantitative analysis of cell shape and the cytoskeleton in developmental biology. Wiley Interdiscip. Rev. Dev. Biol. 2018;7:e333. doi: 10.1002/wdev.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu V.C.-H., Yu P.H.-F., Ho K.-C., Lee F.W.-F. Isolation and identification of a new tetrodotoxin-producing bacterial species, Raoultella terrigena, from Hong Kong marine puffer fish Takifugu niphobles. Mar. Drugs. 2011;9:2384–2396. doi: 10.3390/md9112384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Cusumano A., Bloem J., Weldegergis B.T., Villela A., Fatouros N.E., van Loon J.J., Dicke M., Harvey J.A., Vogel H., Poelman E.H. Symbiotic polydnavirus and venom reveal parasitoid to its hyperparasitoids. Proc. Natl. Acad. Sci. U.S.A. 2018;115:5205–5210. doi: 10.1073/pnas.1717904115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.