Abstract
Snake venom may vary in composition and toxicity across the geographic distribution of a species. In the case of the three species of the Neotropical rattlesnakes Crotalus simus, C. culminatus and C. tzabcan recent research has revealed that their venoms can contain a neurotoxic component (crotoxin homologs), but is not always the case. In the present work, we detected and quantified crotoxin homologs in venom samples from three species distributed across Mexico, to describe variation at the individual and subspecific levels, using slot blot and ELISA immunoassays. We found that all C. simus individuals analyzed had substantial percentages of crotoxin homologs in their venoms (7.6–44.3%). In contrast, C. culminatus lacked them completely and six of ten individuals of the species C. tzabcan had low percentages (3.0–7.7%). We also found a direct relationship between the lethality of a venom and the percentage of crotoxin homologs it contained, indicating that the quantity of this component influences venom lethality in the rattlesnake C. simus.
Keywords: Crotalus simus, Crotoxin, Monoclonal antibody, Polyclonal antibodies, Quantification of crotoxin homologs
Highlights
-
•
Monoclonal antibodies were produced that specifically recognized crotoxin homologs in venoms of Crotalus species.
-
•
Crotoxin homologs were quantified in three species of Crotalus: C. simus, C. culminatus and C. tzabcan.
-
•
All specimens of C. simus contained crotoxin homologs at different levels, while C. culminatus venoms lacked them completely.
-
•
In C. tzabcan, some venoms possess and other lack crotoxin homologs.
1. Introduction
Viperid venoms are composed of a large number of inorganic and organic molecules, the latter include proteins, some of which are responsible for generating physiopathology when envenomation occurs (Gutiérrez, 2002, Gutiérrez et al., 2009). Sixty-three protein families have been reported in viperid venoms (Tasoulis and Isbister, 2017). However, it is known that the most important families are snake venom metalloproteases (SVMPs), snake venom serine proteases (SVSPs) and phospholipases A2 (PLA2) (Calvete, 2017, Durban et al., 2017, Lomonte et al., 2012, Tasoulis and Isbister, 2017). Together, these families usually represent more than 70% of the protein composition. Viperid venoms can be generally classified into type I (low lethal potency, high enzymatic activity) and type II (high lethal activity, low enzymatic activity) (Mackessy, 2008) and there are venoms that behave as intermediate, like some populations of C. s. scutulatus and C. simus (high lethality and high enzymatic activity) (Borja et al., 2018, Castro et al., 2013).
One of the families of proteins with greatest diversity of toxic activities is PLA2s, where two groups have been described based on the presence or absence of enzymatic activity (Gutiérrez et al., 2008, Gutiérrez and Lomonte, 2013, Kini, 2003, Kini, 2005, Kini and Evans, 1987, Kini and Evans, 1989, Lomonte et al., 2003, Lomonte and Rangel, 2012). Among those with catalytic activity is a group of neurotoxins that have been called crotoxin homologs because they are similar to crotoxin, a PLA2 that was first purified and crystallized from the venom of the South American rattlesnake C. durissus terrificus (Slotta and Fraenkel-Conrat, 1938). Crotoxin is a β-neurotoxin that inhibits the release of the neurotransmitter acetylcholine at the neuromuscular junction, producing possibly lethal, neurotoxic effects (Faure et al., 1991, Faure et al., 1993, Faure et al., 1994, Rangel-Santos et al., 2004, Slotta and Fraenkel-Conrat, 1938). The protein is a heterodimeric complex united by non-covalent bonds, consisting of a basic PLA2 (CB) (M.W. 14,350 Da, pI 8.2) with neurotoxic and enzymatic activity and a non-toxic acidic protein called crotapotin (CA), which consists of three disulfide-linked polypeptide chains (M.W. 9490 Da, pI 3.4), whose function is to direct CB to its target site (Faure et al., 1993, Faure et al., 1994, Faure et al., 2011, Gutiérrez, 2002). CA increases the lethal potential of CB and each complex may include 4 isoforms, whose identities directly influence the toxicity of the venom (Canziani et al., 1983, Faure et al., 1991, Faure et al., 1993).
The venoms of C. d. durissus, C. d. terrificus and C. d. ruruima, for example, have been reported to have more than 50% of crotoxin (Calvete et al., 2010). More recently, crotoxin homologs have been reported in the venoms of other species of the genus Crotalus, including some populations of C. scutulatus scutulatus (Mojave toxin) named as “type A”; and the population lacking of this toxin is named “type B” (Borja et al., 2014, Borja et al., 2018, Cate and Bieber, 1978, Dobson et al., 2018, Massey et al., 2012, Strickland et al., 2018). Other examples are C. viridis concolor (concolor toxin) (Mackessy et al., 2003), C. tigris (Mojave-like toxin) (Minton and Weinstein, 1984, Weinstein and Smith, 1990), C. vegrandis (vegrandis toxin) (Chen et al., 2004) and C. horridus (canebrake toxin) (Glenn et al., 1994). Similar components have been described in the venoms of a few non-Crotalus viperids, like Bothriechis nigroviridis (nigroviriditoxin) (Lomonte et al., 2015) and Ophryacus sphenophrys (sphenotoxin) (Neri-Castro et al., 2019). Neurotoxic venoms tend to have median lethal doses (LD50) from 10 to 100 times lower when compared to venoms without neurotoxic components (Borja et al., 2018, Castro et al., 2013, Glenn et al., 1982, Mackessy, 2008, Mackessy, 2010a, Mackessy, 2010b, Rivas et al., 2017, Strickland et al., 2018). Populations with presence and absence of crotoxin homologs have been described in venoms of C. s. scutulatus and C. lepidus and research indicates that this can also be the case for C. tzabcan (Borja et al., 2014, Borja et al., 2018, Castro et al., 2013, Durban et al., 2017, Rivas et al., 2017, Saviola et al., 2017).
The rattlesnake Crotalus simus is distributed from Mexico to Costa Rica and typically inhabits semiarid regions, including tropical dry forest, chaparral, tropical deciduous forest and pastures. Previously, C. simus was classified as part of the C. durissus group, which includes snakes from North, Central, and South America. Campbell and Lamar (2004) then separated the C. durissus complex into three species: C. totonacus, C. durissus and C. simus. They further divided C. simus into three subspecies: C. s. simus, C. s. culminatus and C. s. tzabcan (Campbell and Lamar, 2004). Finally, in 2005, Wüster (Wüster et al., 2005) and collaborators proposed to elevate the three subspecies described by Campbell to species level: a) C. simus, which is distributed along the Atlantic slope, from central Veracruz in Mexico to western Honduras, and along the Pacific coast from the Isthmus of Tehuantepec to central Costa Rica; b) C. culminatus, which is found from southern Michoacan to the Isthmus of Tehuantepec and c) C. tzabcan, which consists of populations from the Yucatan Peninsula and northern Belize and Guatemala.
The venoms of these three species of the C. simus complex were recently characterized by Castro et al. (2013) who reported intraspecific variation in their biological and biochemical activities. They observed that there were marked differences in venom lethality among them three species: C. simus venoms showed the lowest median lethal doses (LD50 0.18–0.65 μg/g), and C. culminatus the highest (LD50 3.42–15.9 μg/g), whereas C. tzabcan showed intermediate lethality (LD50 0.47–8.21 μg/g). Also, proteomic analysis showed that crotoxin homologs made up 14.3% of the total C. simus venom, while C. culminatus venom lacked them completely (Castro et al., 2013) and C. tzabcan contained 3% (Durban et al., 2017).
Variation in venom composition over a species' geographic distribution is an integral part of intraspecific variation. Clinically, these geographical differences can have a great impact, because envenomations could present different symptomatology depending on the region (Mackessy, 2008). It is also important to know whether there is a geographic variation in venoms used as immunogens, because this might affect whether antivenom quality remains the same for different antivenom batches (Gutiérrez et al., 2017). The wide distributional range of the rattlesnake C. simus makes this species a good model for venom variation studies. This research would also increase our knowledge regarding the species' biology and provide information to improve the selection of venoms used for immunization of animals during antivenom production (Calvete, 2017, Chaves et al., 1992, Gutiérrez et al., 2010b, Gutiérrez et al., 2009, Lomonte et al., 2008).
The aim of the present work was to detect and quantify crotoxin homologs in the venom of different organisms of the three species of the C. simus complex: C. simus, C. culminatus and C. tzabcan.
2. Materials and methods
2.1. Venoms
Crotalus simus venom samples used were collected under permit from the Secretaría de Medio Ambiene y Recursos Naturales (SEMARNAT). Other venom samples came from the collection UMA TSAAB KAN (identification number UMA-IN-0183-YUC-10). C. scutulatus scutulatus pools of type A venom, used as a positive control for the presence of crotoxin homologs and type B venom, used as a negative control, were from the National Natural Toxins Research Center (NNTRC, Kingsville, Texas, USA) and include material from two or more individuals. C. durissus terrificus venom, used to purify the sub B of crotoxin and as a positive control, was from captive snakes kept at the Instituto Carlos Malbrán, Buenos Aires, Argentina. We assigned identification numbers to all venom samples. All the individual venoms from C. simus, C. culminatus and C. tzabcan are part of the IBt-UNAM venom bank, (for more details see Castro et al., 2013).
2.2. Protein quantification
Venoms were quantified using the bicinchoninic acid method (BCA). We followed the methods described in the manual of the commercial Pierce™ BCA Protein Assay kit.
2.3. Electrophoresis
Samples of 15 μg of each venom were analyzed using SDS-PAGE 15% under denaturing and reducing (2-mercaptoethanol) conditions. Gels were dyed using Coomasie 0.2% R-250. We included molecular weight markers (Biolabs) in each gel (Laemmli, 1970).
2.4. Crotoxin purification
Crotoxin was purified from C. durissus terrificus venom by size-exclusion chromatography (Aird et al., 1990). We used a glass column 196 cm in length and 0.9 cm in diameter, packed with Sephadex G-75 (Sigma). The buffer used was 20 mM ammonium acetate with 6 M urea, pH 4.7, with a flow of 16 mL/h. Fraction 3 was lyophilized and then passed through an ion-exchange chromatographic column (Mono Q 5/5 μm) on an FPLC system (AccesoLab) as previously described by Rangel-Santos et al. (2004), to yield highly purified crotoxin B subunit (CB).
2.5. Production and purification of polyclonal antibodies (pAbs)
Three rabbits from the New Zealand strain were immunized over three months in intervals of fifteen days, with increasing doses of CB (10–300 μg/rabbit) alternating with incomplete Freund's adjuvant and alum (aluminum hydroxide and magnesium hydroxide, Thermo Fisher). The serum of the three rabbits was mixed and the antibodies were purified by affinity chromatography in a Sepharose 4B resin activated with cyanogen bromide (Sigma) and coupled to CB (5 mg).
2.6. Production of monoclonal antibodies (mAb)
Two groups of five mice from the Balb/C strain were immunized via intraperitoneal injection with CB. We started with 1 μg of toxin with incomplete Freund's adjuvant, and immunized once a week, intercalating with alum, until we reached 5 μg of toxin over a 2-months period. Spleen lymphocytes from the immunized mice were fused with murine myeloma cells from the cell line SP2/0 Ag14 ATCC. Antibodies from an established hybridoma were purified using affinity chromatography in a Sepharose column coupled to protein A (Invitrogen™) (Valdés et al., 2001).
2.7. Detection of crotoxin homologs by slot blot
The slot blot analysis was performed with 50 μg of venom in PBS (native conditions) on a polyvinylidene fluoride membrane (Merck Millipore), using a Hoefer chamber (Amersham Pharmacia Biotech). We used the monoclonal antibody for detection antibody (1 μg/mL), and goat anti-mouse (diluted 1:4000) coupled to alkaline phosphatase (Millipore) as the secondary antibody. Finally, we used BCIP/NBT buffer (Invitrogen) to obtain a colorimetric reaction.
2.8. Detection of crotoxin homologs by western blot
We used a Western blot to analyze 2 μg of venom under reducing conditions (2-mercaptoethanol) on a nitrocellulose membrane (Merck Millipore). For the recognition antibody, we used our polyclonal rabbit anti-crotoxin antibody, using a concentration of 5 μg/mL. As a secondary antibody, we used goat anti-rabbit IGg (diluted 1:4000) coupled to alkaline phosphatase (Millipore). Again, we obtained a colorimetric reaction using BCIP/NBT Invitrogen reagents, according to the manufacturer's protocols.
2.9. Enzyme-linked immunosorbent assay (ELISA)
We measured the antibody titres generated in mice or rabbit using indirect ELISA and sandwich-type ELISA to quantify the percentage of crotoxin homologs in C. simus venoms, as explained below:
Indirect ELISA: 96-well plates (Nunc MaxiSorp®) were sensitized with 100 μL of 5 μg/mL CB in 0.05 M carbonate-bicarbonate buffer (pH 9.5), incubating for 2 h at 37 °C. Then, the wells were washed three times with TBST pH 8 (50 mM Tris/HCl, 150 mM NaCl, 0.5% Tween 20). Next, the wells were blocked with gelatin (50 mM Tris/HCl pH8, 0.5% gelatin, 0.2% Tween 20) at 37 °C for 2 h. We repeated the washing and then incubated for 1 h at 37 °C with the serum sample of interest in 1:3 consecutive dilutions. The wells were washed again and then incubated for 1 h at 37 °C with the secondary antibody diluted 1:4000 (anti-mouse or anti-rabbit, depending on the analysis), coupled with horseradish peroxidase (HRP) (Merck Millipore). Finally, the plate was incubated for 10 min with ABTS (2,2′-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt-colorimetric method) for detection.
Sandwich-type ELISA: we used the same buffers as in the indirect ELISA and all incubations were done at 37 °C. 96-well plates (Nunc MaxiSorp®) were sensitized with 100 μL of 1 μg/mL 4F6 monoclonal antibody for 1 h. After washing, the wells were blocked for 2 h with gelatin (200 μL of 0.5% gelatin in 50 mM Tris, 0.2% Tween 20, pH 8.0). We then generated a standard curve with CB at a starting concentration of 2 μg/mL and 1:3 consecutive dilutions. Sample venoms were placed in known concentrations diluted in the same way and incubated for 1 h. Next, we added 100 μL of 1 μg/mL polyclonal rabbit anti-crototoxin antibody to each well, incubated for 1 h, washed and incubated with goat anti-rabbit antibody coupled to HRP (diluted 1:4000) (Millipore) for another hour. Finally, the colorimetric reaction was obtained using ABTS, as before. Absorbances were quantified in an ELISA reader (Magellan R) at 405 nm. To determine the concentration of crotoxin homologs in sample venoms, the values were interpolated from the CB standard curve, fitted to a sigmoidal dose-response (variable slope) non-linear regression, using the software GraphPad Prism v. 6.0 (GraphPad Software).
3. Results
3.1. Detection of crotoxin by polyclonal antibodies from rabbit
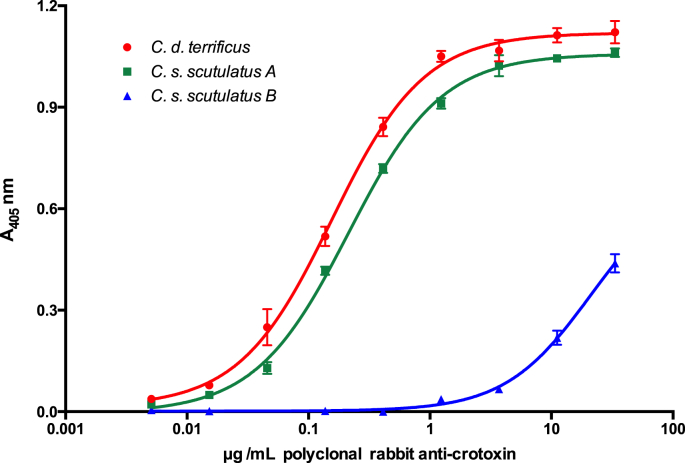
As expected, the obtained polyclonal antibodies recognized CB (14 kDa) from C. durissus terrificus venom (Fig. 1). Also, Fig. 2 shows an indirect ELISA, demonstrating recognition of C. d. terrificus and C. s scutulatus type A venoms, used as positive controls (C+), but not of type B venom of C. s. scutulatus, used as a negative control (C-). Therefore, polyclonal antibody recognizes 134 times more C. d. terrificus and 96 times more C. s. scutulatus type A compared to the venom of C. s. scutulatus type B.
Fig. 1.
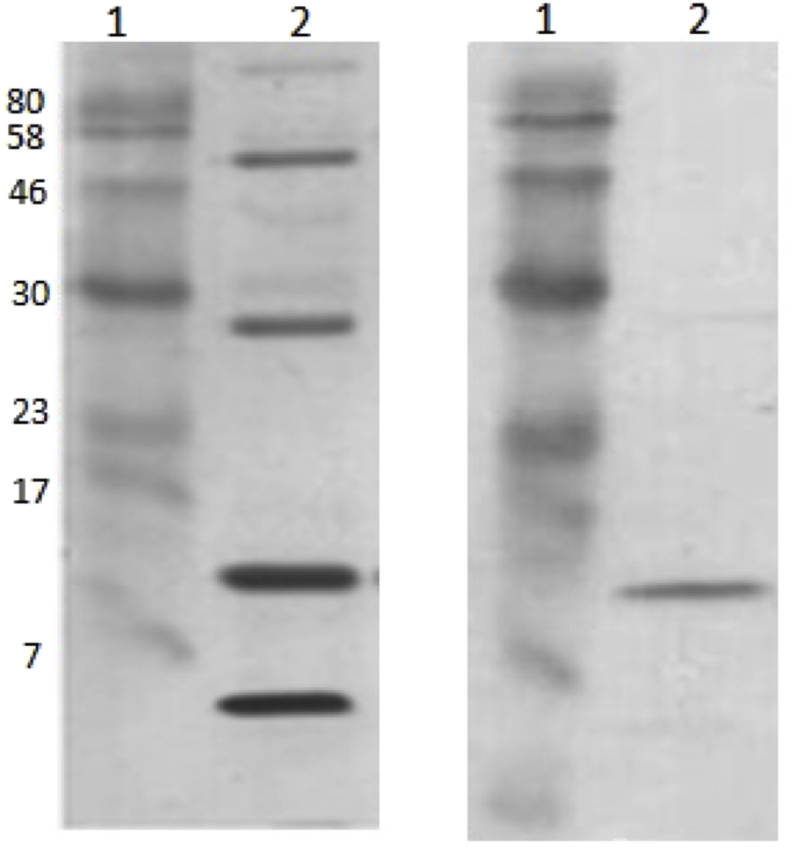
SDS-PAGE 15% (left) and Western blot (right) of C. durissus terrificus venom, incubation with 5 μg/mL antibodies obtained from the immunopurification of polyclonal rabbit serum. 1. Molecular weight marker; 2. C. durissus terrificus venom 20 μg (SDS-PAGE) and 2 μg (Western blot).
Fig. 2.
Recognition curves of polyclonal rabbit anti-crotoxin with (red circle) C. d. terrificus venom (C+), (green square) C. s. scutulatus type A (C+), and (blue triangle) C. s. scutulatus type B (C-). The titers are 0.15 ± 0.006 μg/mL for C. d. terrificus, 0.21 ± 0.01 μg/mL for C. s. scutulatus type A and 20.1 ± 7.8 01 μg/mL for C. s. scutulatus type B. Bars represent mean ± SD of triplicates.
3.2. Detection of crotoxin by monoclonal antibody 4F6
Regarding the production of monoclonal antibodies (mAb), we found one with a concentration of 240 μg/mL in ascites fluid, which we labelled 4F6. This antibody recognized the two positive controls (C. d. terrificus and C. s. scutulatus type A), while presenting no cross-reactivity with the PLA2s from C. s. scutulatus type B venom (Fig. 3).
Fig. 3.
Recognition curve of monoclonal antibody 4F6 with venom from (red circle) C. d. terrificus (C+), (green square) C. s. scutulatus type A (C+), and (blue triangle) for C. s. scutulatus type B (C). The titers are 1.09 ± 0.01 μg/mL for C. d. terrificus and 1.97 ± 0.23 μg/mL for C. s. scutulatus type A. Bars represent mean ± SD of triplicates.
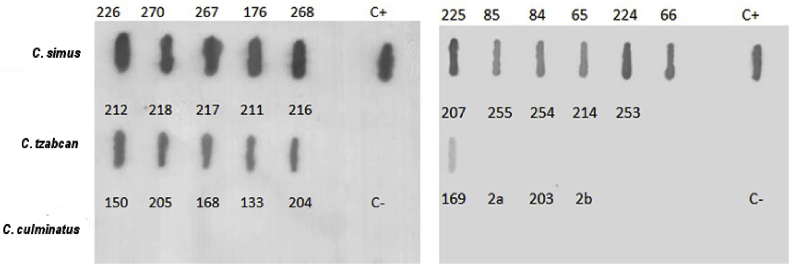
Detection of crotoxin homologs was carried out using the slot blot technique under native venom conditions (non-reducing). The mAb 4F6 recognized these components in the venom of the 11 C. simus samples analyzed and showed no cross-reactivity with the negative control, nor with any protein from C. culminatus venoms. Furthermore, the mAb recognized these neurotoxins in 6 samples from C. tzabcan (samples: IBt 212, IBt 218, IBt 217, IBt 211, IBt 216, and IBt 207), but not in 4 samples from the same species (samples: IBt 255, IBt 254, IBt 214, and IBt 253) (Fig. 4).
Fig. 4.
Slot blots of native venom (50 μg) of different individuals from the species C. simus, C. tzabcan, and C. culminatus; C. d. terrificus (C+); and C. s. scutulatus type B (C-). Incubation with mAb 4F6 anti-crotoxin (2 μg/mL).
3.3. Quantitative analysis of crotoxin homologs
In accordance with the observations made using the slot blot technique, no crotoxin homologs were found using ELISA in any of the venoms from the species C. culminatus (Table 1).
Table 1.
Median lethal dose (μg/g mouse) and percentage of crotoxin homologs found in venoms of C. simus, C. culminatus and C. tzabcan.
| Species | Sample ID | Locality | LD50 |
Crotoxin homologs |
|
|---|---|---|---|---|---|
| (μg/g mouse) | Detectiona | % ± SD | |||
| C. simus | IBt 226 | La Tinaja, Veracruz | 0.18 (0.15–0.21) | + | 38.5 ± 0.7 |
| IBt270 | Actopan, Veracruz | 0.20 (0.18–0.21) | + | 21.6 ± 0.73 | |
| IBt267 | Puente Nacional, Veracruz | 0.21 (0.20–0.21) | + | 26.3 ± 0.88 | |
| IBt 176 | Santo Domingo, Oaxaca | 0.20 (0.20–0.21) | + | 14.5 ± 0.4 | |
| IBt 268 | Actopan, Veracruz | 0.26 (0.15–0.27) | + | 26.2 ± 0.4 | |
| IBt 225 | Tinajas, Veracruz | 0.26 (0.26–0.26) | + | 44.3 ± 0.24 | |
| IBt 085 | Chiapa de Corzo, Chiapas | 0.26 (0.21–0.30) | + | 7.6 ± 0.5 | |
| IBt 084 | Chiapa de Corzo, Chiapas | 0.30 (0.26–0.32) | + | 6.3 ± 0.42 | |
| IBt 065 | Copainalá, Chiapas | 0.31 (0.30–0.31) | + | 8.4 ± 0.8 | |
| IBt 224 | Playas del Conchal, Veracruz | 0.32 (0.31–0.32) | + | 39.7 ± 0.8 | |
| IBt 066 | Chiapa de Corzo, Chiapas | 0.65 (0.61–0.67) | + | 13.7 ± 0.4 | |
| C. tzabcan | IBt 212 | Solidaridad, Quintana Roo | 0.80 (0.71–0.91) | + | 7.7 ± 0.45 |
| IBt 218 | Chetumal, Quintana Roo | 0.91 (0.89–0.93) | + | 4.4 ± 0.34 | |
| IBt 217 | Chetumal, Quintana Roo | 1.08 (0.96–1.20) | + | 3.0 ± 0.16 | |
| IBt 211 | Solidaridad, Quintana Roo | 1.19 (1.08–1.31) | + | 4.6 ± 0.74 | |
| IBt 216 | Chetumal, Quintana Roo | 1.79 (1.71–1.87) | + | 5.0 ± 0.43 | |
| IBt 207 | Solidaridad, Quintana Roo | 1.99 (1.84–2.16) | + | 7.3 ± 0.45 | |
| IBt 255 | Mérida, Yucatán | 4.68 (4.42–5.00) | – | 0 | |
| IBt 254 | Mérida, Yucatán | 7.21 (5.29–8.79) | – | 0 | |
| IBt 214 | Solidaridad, Quintana Roo | 7.89 (7.47–8.31) | – | 0 | |
| IBt 253 | Mérida, Yucatán | 8.21 (8.00–8.42) | – | 0 | |
| C. culminatus | IBt 150 | Puente de Ixtla, Morelos | 3.42 (3.37–3.47) | – | 0 |
| IBt 205 | Chilpancingo, Guerrero | 3.47 (3.37–3.63) | – | 0 | |
| IBt 168 | Tepalcingo, Morelos | 7.95 (7.79–8.10) | – | 0 | |
| IBt 133 | Alpuyeca, Morelos | 8.53 (8.42–8.68) | – | 0 | |
| IBt 204 | Tlaltizapán, Morelos | 9.31 (9.10–9.58) | – | 0 | |
| IBt 169 | Tepalcingo, Morelos | 9.84 (9.86–10.0) | – | 0 | |
| Csim2A | Villa de Ayala, Morelos | 15.0 (14.3–15.8) | – | 0 | |
| IBt 203 | Tlaltizapán, Morelos | 15.5 (15.0–16.0) | – | 0 | |
| Csim2B | Villa de Ayala, Morelos | 15.9 (14.7–17.3) | – | 0 | |
Detection by slot blot, venoms with presence for crotoxin homologs (+), venoms with absence of crotoxin homologs (−).
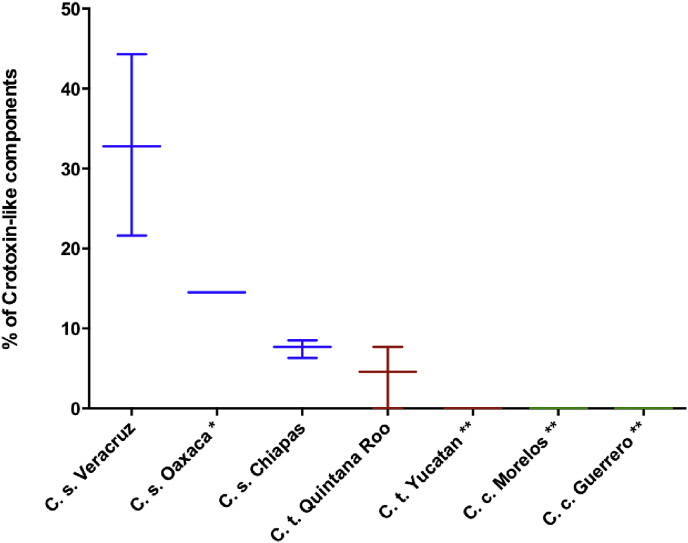
The limit of quantification (LoQ) of our ELISA was 3.5 ng/mL. With this technique, we determined that the venom of the positive control (C. d. terrificus) contained 44.8%, type A venom from C. s. scutulatus contained an average of 23.8%, and we did not detect these components at all in type B venom from that same species. Table 1 shows the mean percentage of crotoxin homologs in venoms from the three species of C. simus, according to their geographic distributions. Venom from individuals of C. simus collected in the state of Veracruz had the highest mean percentage of these neurotoxins (32.8%), followed by the single sample from Oaxaca (14.5%), whereas venoms from individuals collected in Chiapas and Quintana Roo had the lowest average percentage of crotoxin homologs (9%). Individual IBt 225 stood out as having the venom with the highest concentration of these proteins (44.3%) (Table 1). We did not find them at all in the venoms of four C. tzabcan individuals (IBt 255, IBt 254, IBt 214, and IBt 253), one of which was collected in Quintana Roo and the rest in Yucatán (Table 1).
4. Discussion
4.1. Polyclonal and monoclonal antibodies
The recognition of C. d. terrificus venom by previously purified polyclonal antibodies (under reducing conditions) was evaluated by western blot. We only observed recognition of the band corresponding to the basic subunit of crotoxin. Additionally, we carried out an ELISA to evaluate our antibodies' ability to recognize native proteins from C. d. terrificus venom and from C. s. scutulatus “A” venom as positive controls. We also tested type B venom from C. s. scutulatus as a negative control. We observed that the antibodies recognized PLA2s from both positive controls. Yet, the antibodies recognized non-neurotoxic PLA2s from C. s. scutulatus population B with low intensity, 134 times lower than C. d. terrificus venom and 96 times less than C. s. scutulatus type A. The basic subunit of crotoxin has approximately 50% sequence identity with non-neurotoxic PLA2s (Aird et al., 1986). However, this identity refers to the linear sequence so the probability of cross-recognition is likely lower given that many epitopes are a result of three-dimensional structure (Sela et al., 1967). Additionally, although a great variety of antigenic sites are presented to the animal immune system during immunization, there are particular epitopes antigens that may substantially stimulate the production of antibodies, some of which may be associated with the active site of a toxin (Oshima-Franco et al., 1999). A great variety of cross recognition levels have been found with antibodies to heterologous and homologous PLA2s, including lack of cross-reactivity among certain species within the same family. These differences reflect variation in the antigenic sites of enzymes belonging to the same family (Nair et al., 1980). For example, polyclonal antibodies obtained against non-neurotoxic PLA2s did not recognize the basic subunit of Mojave toxin, showing that, despite high sequence identity, the two classes of PLA2s are antigenically different (Rael et al., 1986).
We evaluated the recognition pattern of the obtained mAb 4F6 using indirect ELISA. It showed cross-reactivity with Mojave toxin, but it did not recognize non-neurotoxic PLA2s from type B C. s. scutulatus venom. Rael and collaborators observed the same phenomenon in 1986, when they obtained a mAb that recognized the basic subunit of crotoxin and showed no cross-reactivity with non-neurotoxic PLA2s from the same venom (Rael et al., 1986). They therefore concluded that, despite high sequence identity between neurotoxic and non-neurotoxic PLA2s, it is probable that the three-dimensional configuration of CB is different from that of other PLA2s. It is also worth mentioning that the mAb from Rael et al. recognized several isoforms of crotoxin. Their work did not evaluate whether the antibody showed recognition of the different isoforms that may exist in the venoms of C. d. terrificus and C. simus. However, it is very possible that such recognition occurs, since the sequence identity between the crotoxin isoforms of C. d. terrificus, C. simus (Costa Rican and Mexico populations), and Mojave neurotoxin is between 98% and 100% (Calvete et al., 2012, Durban et al., 2017, Faure et al., 1991, Faure et al., 1994, Massey et al., 2012), suggesting that the structural epitopes of all three toxins are similar.
4.2. Qualitative analysis by slot blot
Due to the wide geographic distribution and high levels of genetic divergence among species of the C. simus complex, these species represents an interesting model to study variation in venom composition (Castro et al., 2013). We did not observe cross-reactivity with non-neurotoxic PLA2s from type B C. s. scutulatus venom, nor was there reactivity with venom proteins of C. culminatus. These results are consistent with the proteomic results of Castro et al. (2013). Additionally, we determined that all C. simus individuals studied and six of the ten C. tzabcan individuals studied had crotoxin homologs in their venom. These experiments were not performed with polyclonal antibodies because they showed ELISA recognition (low) to the venom of C. s. scutulatus type B whose venom lacks crotoxin. In slot blot experiments the results are qualitative (presence or absence) so antibodies that have cross reactivity to non-neurotoxic PLA2s, even if it is low, could give unreliable results, while monoclonal antibodies are specific for crotoxin homologs.
4.3. Quantitative analysis of crotoxin homologs
The purpose of quantifying crotoxin homologs in venoms from three species was to relate these quantities to the median lethal dose of each venom, as proposed by Calvete et al. (2012), who report that the species with the highest percentage of crotoxin have low LD50s. We thus worked with the same adult individuals from which Castro et al. (2013) had already obtained intravenous LD50 values (Table 1), as well as biological and biochemical activities and proteomes of pools from C. simus and C. culminatus venoms. As a result of our quantification, we observed that venom from C. simus individuals had the highest average concentration of crotoxin homologs (7.6%–44.3%), followed by 6 individuals of C. tzabcan (3.0%–7.7%). However, 4 C. tzabcan individuals and all of the 9 individuals of C. culminatus we evaluated lacked the neurotoxin, which is consistent with the slot blot results.
In 2010, Calvete and collaborators performed a proteomic and biological analysis of the venom of the species C. simus and the C. durissus complex, which are distributed in Central and South America, respectively. They proposed that the concentration of crotoxin in venom is directly related with its lethal activity (Calvete et al., 2010). In general, the venoms that have crotoxin homologs have higher lethal potency than those that lack them. However, there are cases where the percentage of these components do not appear to be the only factor increasing lethality, for example, the venoms of individuals IBt 225 and IBt 085 both had LD50 values of 5 μg/g mouse, but they had very different percentages of crotoxin homologs: 44.3% and 7.6%, respectively. There may be several reasons for this discrepancy, one of which has to do with the ratio of CB and CA in each venom, given that the relative levels of CA may increase the lethality (Canziani et al., 1983). Another potential explanation may involve the isoforms that each venom contains; these differ slightly in their molecular structure, as well as in their chromatographic properties and electrophoretic mobility. The isoforms of crotoxin homologs CB from different species (i.e. C. s. scutulatus, C. d. terrificus, C. oreganus concolor and C. tzabcan) often differ in a small number of amino acid residues (typically 1 to 5) and, in some cases, these differences modify the enzymatic and pharmacological properties of the toxin. The LD50 of different isoforms ranges from 0.07 to 0.45 μg/g (Faure and Bon, 1988, Faure et al., 1993). Finally, variation in non-neurotoxic components of the venoms, like SVMP, can also influence venom lethality.
In our study, we also found that the lethality (LD50) of snake venoms not containing crotoxin homologs (those of C. culminatus and four individuals of C. tzabcan) is 26–100 times lower when compared to the lethality of individuals that have this protein. Previously, the same relationship was reported in populations of the snake C. s. scutulatus, where type B venom (which does not contain crotoxin) has 20 times lower lethality in comparison with type A venom (Glenn et al., 1982). The causes for this difference in the presence of neurotoxins have been poorly addressed and some authors attribute the variation in venom composition to local adaptations, which confer advantages such as specialization on different prey species. However, recent research has described that variation between neurotoxic and haemorrhagic specimens within the same species is due to very marked differences in haplotypes of PLA2 and SVMPs complex genes (Dowell et al., 2018).
4.4. Intraspecific variation
The variation in venom toxicity we observed also exhibited a geographic pattern, as has been described in several snake species (Borja et al., 2013, Borja et al., 2018, Glenn et al., 1982, Glenn et al., 1994,; Gutiérrez et al., 2010a, Salazar et al., 2007, Strickland et al., 2018, Sunagar et al., 2014). We found that Crotalus simus from eastern Mexico, in the state of Veracruz had the highest average concentration of crotoxin homologs (14.5%–44.3%), while those from Chiapas presented lower proportions (6.3%–13.7%). Taxonomic studies suggest that the populations of the state of Veracruz is a different species from those of South (Chiapas) and Central America (Wüster et al., 2005), however, this has not yet been clarified due to the low number of samples analyzed in the aforementioned study. Six individuals of C. tzabcan from the state of Quintana Roo had low neurotoxin concentrations (3%–7.7%), in contrast, three individuals from the state of Yucatán and one from Quintana Roo did not present crotoxin homologs (Fig. 5 and Table 2). This is interesting because we now know that there are positive and negative individuals for crotoxin homologs. However, with the few samples we have we cannot delimit populations or correlate with external factors, so a larger number of samples should be studied along this species distribution.
Fig. 5.
Intervals of crotoxin homologs percentage for different individuals per state of origin. Horizontal lines represent mean of all data and error bars represent minimum and maximum percentages. *Data of a single individual. **crotoxin quantifications were zero.
Table 2.
Percentage of crotoxin homologs (mean and range) in venoms from individuals of the three species of Crotalus simus, according to their geographic distributions.
| Species | Locality | n | % Crotoxin (Mean) | % Crotoxin (Range) |
|---|---|---|---|---|
| C. simus | Veracruz | 6 | 32.8 | 21.6–44.3 |
| Oaxaca | 1 | 14.5 | 14.5 | |
| Chiapas | 4 | 7.7 | 6.3–8.5 | |
| Total | 11 | 22.0 | 6.3–44.3 | |
| C. tzabcan | Quintana Roo | 7 | 4.6 | 0–7.7 |
| Yucatán | 3 | 0 | 0 | |
| Total | 10 | 3.2 | 0–7.7 | |
| C. culminatus | Morelos | 8 | 0 | 0 |
| Guerrero | 1 | 0 | 0 | |
| Total | 9 | 0 | 0 |
In the states of Morelos and Guerrero, we did not find neurotoxin in any of the samples belonging to C. culminatus but this species also includes populations in the state of Oaxaca, where it overlaps with the distribution of C. simus. It is thus important to increase our sampling to determine whether there are populations of C. simus without crotoxin homologs, or individuals of C. culminatus with neurotoxic venoms. This phenomenon, as previously mentioned, has been reported in the subspecies C. s. scutulatus, where there is a marked geographic variation in venom composition (Glenn et al., 1982).
There are two possible reasons why venom may vary across a species' geographic distribution. The first occurs in nearby or sympatric populations, as in the case of C. scutulatus, where there are two divergent populations without significant morphological differences, but which differ by the presence or absence of Mojave toxin. There is evidence that populations with type A and type B venoms were historically isolated, however, there is currently no physical barrier between these populations, and, in fact, intermediate populations have been described (Borja et al., 2018, Glenn et al., 1982, Massey et al., 2012, Schield et al., 2018, Strickland et al., 2018, Wilkinson et al., 2018, Zancolli et al., 2018). The second type of venom variation has been seen in isolated populations. Small, isolated populations tend to have homogeneous venoms. In contrast, large populations tend to conserve many venom components when isolated but also show variation in the total spectrum of venom, related to the time since isolation (Chippaux et al., 1991). The particular case of the C. simus complex has not been previously studied. Nevertheless, previous researchers have proposed that Mexico is the center of diversification for rattlesnakes, and that their diversity, also reflected in the composition and toxicity of their venoms, is a consequence of physiographic and climatic fluctuations over the past 50 million years (Flores-Villela, 1993, Wüster et al., 2005). The species of the genus Crotalus have been proposed to, originally, possess the genes for subunits A and B of crotoxin homologs (because the ancestor that arrived through the north of America had them) and then some were lost over the course of time (Calvete et al., 2010, Calvete, 2017, Dowell et al., 2016). On the other hand, the intraspecific variation in the case of C. simus of Costa Rica and Mexico is ontogenetic, where the concentrations of crotoxin are greater in neonates than in adults (Calvete et al., 2010, Castro et al., 2013, Durban et al., 2017, Durban et al., 2013, Lomonte et al., 1983), Still, it is necessary to carry out studies with a greater number of samples of both juvenile and adult specimens of various species, to further analyze the regulation mechanisms that have generated the described variation.
5. Conclusions
In the present study, we obtained important information regarding the presence/absence of crotoxin homologs across the geographic distribution of the C. simus, C. culminatus and C. tzabcan, describing a significant interspecific as well as intraspecific variation in venom composition. This type of analysis may be used to identify and quantify crotoxin homologs in venoms of other species of snakes, which may help to predict their neurotoxic properties and improve hospital treatment of patients bitten by snakes. On the other hand, the identification of species or populations of viperids with neurotoxins will help to evaluate antivenoms and eventually improve them. The ELISA technique used here, has important advantages such as the analysis of multiple samples at the same time, and being relatively cheap and fast.
Ethical statement
All animal work was performed according to the guidelines approved by the bioethics committee of the Instituto de Biotecnología, UNAM.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Edgar Neri Castro is a doctoral student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and a scholarship recipient from Consejo Nacional de Ciencia y Tecnología, México (CONACYT) with registration number 254145. This project was partially financed by Dirección General de Asuntos del Personal Académico of UNAM (DGAPA-PAPIIT: IN207218) and CONACYT (project 221343). The authors thank the following herpetariums: UMA TSÁAB KAAN (SEMARNAT registration number UMA-IN-0183-YUC-10) and Deval Animal (DGVS-CR-IN-0957-D.F./07) for assistance in the extraction of venoms. Also, we thank IBt-Animal House personnel (Elizabeth Mata and Gabriela Cabeza) for their help with experimental animals, and Felipe Olvera for his technical skills.
References
- Aird S.D., Kaiser I.I., Lewis R.V., Kruggel W.G. A complete amino acid sequence for the basic subunit of crotoxin. Arch. Biochem. Biophys. 1986;249:296–300. doi: 10.1016/0003-9861(86)90005-6. [DOI] [PubMed] [Google Scholar]
- Aird S.D., Yates J.R., Martino P.A., Shabanowitz J., Hunt D.F., Kaiser I.I. The amino acid sequence of the acidic subunit B-chain of crotoxin. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1990;1040:217–224. doi: 10.1016/0167-4838(90)90079-U. [DOI] [PubMed] [Google Scholar]
- Borja M., Lazcano D., Martínez-Romero G., Morlett J., Sánchez E., Cepeda-Nieto A.C., Garza-García Y., Zugasti-Cruz A. Intra-specific variation in the protein composition and proteolytic activity of venom of Crotalus lepidus morulus from the Northeast of Mexico. Copeia. 2013;2013:702–716. doi: 10.1643/OT-13-005. [DOI] [Google Scholar]
- Borja M., Castañeda G., Espinosa J., Neri E., Carbajal A., Clement H., García O., Alagon A. Mojave rattlesnake ( Crotalus scutulatus scutulatus ) with Type B venom from Mexico. Copeia. 2014;2014:7–13. doi: 10.1643/OT-12-041. [DOI] [Google Scholar]
- Borja M., Neri-Castro E., Castañeda-Gaytán G., Strickland J.L., Parkinson C.L., Castañeda-Gaytán J., Ponce-López R., Lomonte B., Olvera-Rodríguez A., Alagón A., Pérez-Morales R. Biological and proteolytic variation in the venom of Crotalus scutulatus scutulatus from Mexico. Toxins (Basel) 2018;10:1–19. doi: 10.3390/toxins10010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete J.J., Sanz L., Cid P., de la Torre P., Flores-Díaz M., Dos Santos M.C., Borges A., Bremo A., Angulo Y., Lomonte B., Alape-Girón A., Gutiérrez J.M. Snake venomics of the Central American rattlesnake Crotalus simus and the south American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in south America. J. Proteome Res. 2010;9:528–544. doi: 10.1021/pr9008749. [DOI] [PubMed] [Google Scholar]
- Calvete J.J., Pérez A., Lomonte B., Sánchez E.E., Sanz L. Snake venomics of Crotalus tigris: the minimalist toxin arsenal of the deadliest neartic rattlesnake venom. Evolutionary clues for generating a pan-specific antivenom against crotalid type II venoms. J. Proteome Res. 2012;11:1382–1390. doi: 10.1021/pr201021d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete J.J. Venomics: integrative venom proteomics and beyond. Biochem. J. 2017;474:611–634. doi: 10.1042/BCJ20160577. [DOI] [PubMed] [Google Scholar]
- Campbell J., Lamar W.W. Cornell University Press; Ithaca, New York, USA: 2004. The Venomous Reptiles of the Western Hemisphere. [Google Scholar]
- Canziani G., Seki C.I., Vidal J.C.I. 1983. The Mechanism of Inhibition of Phospholipase Activity of Crotoxin by Crotoxin a Complex Is the Major Toxin from a South American Rattlesnake ( Crotalus durissus Terrificus ) Venom . This Complex Consists of a Moderately Toxic Phosp 21. [Google Scholar]
- Castro E.N., Lomonte B., del Carmen Gutiérrez M., Alagón A., Gutiérrez J.M. Intraspecies variation in the venom of the rattlesnake Crotalus simus from Mexico: different expression of crotoxin results in highly variable toxicity in the venoms of three subspecies. J. Proteom. 2013;87:103–121. doi: 10.1016/j.jprot.2013.05.024. [DOI] [PubMed] [Google Scholar]
- Cate R.L., Bieber A.L. Purification and characterization of mojave (Crotalus scutulatus scutulatus) toxin and its subunits. Arch. Biochem. Biophys. 1978;189:397–408. doi: 10.1016/0003-9861(78)90227-8. [DOI] [PubMed] [Google Scholar]
- Chaves F., Gutiérrez J., Brenes F. Pathological and biochemical changes induced in mice after intramuscular injection of venom from newborn specimens of the snake Bothrops asper (Terciopelo) Toxicon. 1992;30:1099–1109. doi: 10.1016/0041-0101(92)90055-A. [DOI] [PubMed] [Google Scholar]
- Chen Y.-H., Wang Y.-M., Hseu M.-J., Tsai I.-H. Molecular evolution and structure–function relationships of crotoxin-like and asparagine-6-containing phospholipases A 2 in pit viper venoms. Biochem. J. 2004;381:25–34. doi: 10.1042/BJ20040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux J.P., Williams V., White J. Snake venom variability: methods of study, results and interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- Dobson J., Yang D.C., op den Brouw B., Cochran C., Huynh T., Kurrupu S., Sánchez E.E., Massey D.J., Baumann K., Jackson T.N.W., Nouwens A., Josh P., Neri-Castro E., Alagón A., Hodgson W.C., Fry B.G. Rattling the border wall: pathophysiological implications of functional and proteomic venom variation between Mexican and US subspecies of the desert rattlesnake Crotalus scutulatus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2018;205:62–69. doi: 10.1016/j.cbpc.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell N.L., Giorgianni M.W., Kassner V.A., Selegue J.E., Sanchez E.E., Carroll S.B. The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr. Biol. 2016;26:2434–2445. doi: 10.1016/j.cub.2016.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell N.L., Giorgianni M.W., Griffin S., Kassner V.A., Selegue J.E., Sanchez E.E., Carroll S.B. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr. Biol. 2018:1–11. doi: 10.1016/j.cub.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J., Pérez A., Sanz L., Gómez A., Bonilla F., Rodríguez S., Chacón D., Sasa M., Angulo Y., Gutiérrez J.M., Calvete J.J. Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genomics. 2013;14:1–17. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban J., Sanz L., Trevisan-Silva D., Neri-Castro E., Alagón A., Calvete J.J. Integrated venomics and venom gland transcriptome analysis of juvenile and adult Mexican rattlesnakes Crotalus simus, C. tzabcan, and C. culminatus revealed miRNA-modulated ontogenetic shifts. J. Proteome Res. 2017;16:3370–3390. doi: 10.1021/acs.jproteome.7b00414. [DOI] [PubMed] [Google Scholar]
- Faure G., Bon C. Crotoxin, a phospholipase A2 neurotoxin from the South American rattlesnake Crotalus durissus terrificus: purification of several isoforms and comparison of their molecular structure and of their biological activities. Biochemistry. 1988;27:730–738. doi: 10.1021/bi00402a036. [DOI] [PubMed] [Google Scholar]
- Faure G., Saliou B., Bon C., Guillaume J.L., Camoin L. Multiplicity of acidic subunit isoforms of crotoxin, the phospholipase A2 neurotoxin from Crotalus durissus terrificus venom, results from posttranslational modifications. Biochemistry. 1991;30:8074–8083. doi: 10.1021/bi00246a028. [DOI] [PubMed] [Google Scholar]
- Faure G., Harvey A.L., Thomson E., Saliou B., Radvanyi F., Bon C. Comparison of crotoxin isoforms reveals that stability of the complex plays a major role in its pharmacological action. Eur. J. Biochem. 1993;214:491–496. doi: 10.1111/j.1432-1033.1993.tb17946.x. [DOI] [PubMed] [Google Scholar]
- Faure G., Choumet V., Bouvhier C., Camoin L., Guillaume J.L., Monegier B., Vuilhorgne M., Bon C. The origin of the diversity of crotoxin isoforms in the venom of Crotalus durissus terrificus. Eur. J. Biochem. 1994;223:161–164. doi: 10.1111/j.1432-1033.1994.tb18978.x. [DOI] [PubMed] [Google Scholar]
- Faure G., Xu H., Saul F.A. Crystal structure of crotoxin reveals key residues involved in the stability and toxicity of this potent heterodimeric β-neurotoxin. J. Mol. Biol. 2011;412:176–191. doi: 10.1016/j.jmb.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Flores-Villela O. Herpetofauna of Mexico: distribution and endemism. Biol. Divers. Mex. Orig. Distrib. 1993:253–280. [Google Scholar]
- Glenn J.L., Straight R.C., Wolfe M.C., Hardy D.L. 1982. Geographical Variation in Crotalus Scutulatus Scutulatus (Mojave Rattlesnake) Venom Properties. [DOI] [PubMed] [Google Scholar]
- Glenn J.L., Straight R.C., Wolt T.B. Regional variation in the presence of canebrake toxin in Crotalus horridus venom. Comp. Biochem. Physiol. Part C Pharmacol. 1994;107:337–346. doi: 10.1016/1367-8280(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M. Comprendiendo los venenos de serpientes: 50 Años de investigaciones en América Latina. Rev. Biol. Trop. 2002;50:377–394. [PubMed] [Google Scholar]
- Gutiérrez J.M., Alberto Ponce-Soto L., Marangoni S., Lomonte B. Systemic and local myotoxicity induced by snake venom group II phospholipases A2: comparison between crotoxin, crotoxin B and a Lys49 PLA2 homologue. Toxicon. 2008;51:80–92. doi: 10.1016/j.toxicon.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Lomonte B., León G., Alape-Girón A., Flores-Díaz M., Sanz L., Angulo Y., Calvete J.J. Snake venomics and antivenomics: proteomic tools in the design and control of antivenoms for the treatment of snakebite envenoming. J. Proteom. 2009;72:165–182. doi: 10.1016/j.jprot.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Sanz L., Flores-Díaz M., Figueroa L., Madrigal M., Herrera M., Villalta M., León G., Estrada R., Borges A., Alape-Girón A., Calvete J.J. Impact of regional variation in Bothrops asper snake venom on the design of antivenoms: integrating antivenomics and neutralization approaches. J. Proteome Res. 2010;9:564–577. doi: 10.1021/pr9009518. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Williams D., Fan H.W., Warrell D.A. Snakebite envenoming from a global perspective: towards an integrated approach. Toxicon. 2010;56:1223–1235. doi: 10.1016/j.toxicon.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Lomonte B. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon. 2013;62:27–39. doi: 10.1016/j.toxicon.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Solano G., Pla D., Herrera M., Segura Á., Vargas M., Villalta M., Sánchez A., Sanz L., Lomonte B., León G., Calvete J.J. Preclinical evaluation of the efficacy of antivenoms for snakebite envenoming: state-of-the-art and challenges ahead. Toxins (Basel) 2017;9:1–22. doi: 10.3390/toxins9050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini R.M., Evans H.J. Structure-function relationships of phospholipases. The anticoagulant region of phospholipases A2. J. Biol. Chem. 1987;262:14402–14407. doi: 10.1016/0041-0101(88)90060-8. [DOI] [PubMed] [Google Scholar]
- Kini R.M., Evans H.J. A model to explain the pharmacological effects of snake venom phospholipases A2. Toxicon. 1989;27:613–635. doi: 10.1016/0041-0101(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Kini R.M. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Kini R.M. Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon. 2005;45:1147–1161. doi: 10.1016/j.toxicon.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Gené J.A., Gutiérrez J., Cerdas L. Estudio comparativo de los venenos de serpiente cascabel (Crotalus durissus durissus) de ejemplares adultos y recien nacidos. Toxicon. 1983;21:379–384. doi: 10.1016/0041-0101(83)90094-6. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Angulo Y., Calderón L. An overview of lysine-49 phospholipase A2 myotoxins from crotalid snake venoms and their structural determinants of myotoxic action. Toxicon. 2003;42:885–901. doi: 10.1016/j.toxicon.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Escolano J., Fernández J., Sanz L., Angulo Y., Gutiérrez J.M., Calvete J.J. Snake venomics and antivenomics of the arboreal neotropical pitvipers Bothriechis lateralis and Bothriechis schlegelii. J. Proteome Res. 2008;7:2445–2457. doi: 10.1021/pr8000139. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Rangel J. Snake venom Lys49 myotoxins: from phospholipases A2 to non-enzymatic membrane disruptors. Toxicon. 2012;60:520–530. doi: 10.1016/j.toxicon.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Rey-Suárez P., Tsai W.C., Angulo Y., Sasa M., Gutiérrez J.M., Calvete J.J. Snake venomics of the pit vipers Porthidium nasutum, Porthidium ophryomegas, and Cerrophidion godmani from Costa Rica: toxicological and taxonomical insights. J. Proteom. 2012;75:1675–1689. doi: 10.1016/j.jprot.2011.12.016. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Mora-Obando D., Fernández J., Sanz L., Pla D., María Gutiérrez J., Calvete J.J. First crotoxin-like phospholipase A2 complex from a New World non-rattlesnake species: nigroviriditoxin, from the arboreal Neotropical snake Bothriechis nigroviridis. Toxicon. 2015;93:144–154. doi: 10.1016/J.TOXICON.2014.11.235. [DOI] [PubMed] [Google Scholar]
- Mackessy S.P., Williams K., Ashton K.G. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: a case of venom paedomorphosis? Copeia. 2003;2003:769–782. doi: 10.1643/HA03-037.1. [DOI] [Google Scholar]
- Mackessy S.P. Venom composition in rattlesnakes: trends and biological significance. In: Hayes W.K., Beaman K.R., Cardwell M.D., Bush S.P., editors. The Biology of Rattlesnakes. Loma Linda, California. 2008. pp. 495–510. [Google Scholar]
- Mackessy S.P. Evolutionary trends in venom composition in the Western Rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon. 2010;55:1463–1474. doi: 10.1016/j.toxicon.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Mackessy S.P. The Field of reptile toxinology: snakes, lizards, and their venoms. In: Mackessy S.P., editor. Handbook of Venoms and Toxins of Reptiles. CRC Press; Boca Raton, Florida, USA: 2010. pp. 3–23. [Google Scholar]
- Massey D.J., Calvete J.J., Sánchez E.E., Sanz L., Richards K., Curtis R., Boesen K. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from southern Arizona. J. Proteom. 2012;75:2576–2587. doi: 10.1016/j.jprot.2012.02.035. [DOI] [PubMed] [Google Scholar]
- Minton S.A., Weinstein S.A. Protease activity and lethal toxicity of venoms from some little known rattlesnakes. Toxicon. 1984;22:828–830. doi: 10.1016/0041-0101(84)90169-7. [DOI] [PubMed] [Google Scholar]
- Nair C., Nair B.C., Elliott W.B. Immunological comparison of phospholipases A2 present in rattlesnake (genus Crotalus) venoms. Toxicon. 1980;18:675–680. doi: 10.1016/0041-0101(80)90098-7. [DOI] [PubMed] [Google Scholar]
- Neri-Castro E., Lomonte B., Valdés M., Ponce-López R., Bénard-Valle M., Borja M., Strickland J.L., Jones J.M., Grünwald C., Zamudio F., Alagón A. Venom characterization of the three species of Ophryacus and proteomic profiling of O. sphenophrys unveils Sphenotoxin, a novel Crotoxin-like heterodimeric β-neurotoxin. J. Proteom. 2019;192:196–207. doi: 10.1016/j.jprot.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Oshima-Franco Y., Hyslop S., Prado-Franceschi J., Cruz-Höfling M.A., Rodrigues-Simioni L. Neutralizing capacity of antisera raised in horses and rabbits against Crotalus durissus terrificus (South American rattlesnake) venom and its main toxin, crotoxin. Toxicon. 1999;37:1341–1357. doi: 10.1016/S0041-0101(98)00246-3. [DOI] [PubMed] [Google Scholar]
- Rael E.D., Salo R.J., Zepeda H. Monoclonal antibodies to Mojave toxin and use for isolation of cross-reacting proteins in Crotalus venoms. Toxicon. 1986;24:661–668. doi: 10.1016/0041-0101(86)90029-2. [DOI] [PubMed] [Google Scholar]
- Rangel-Santos A., Dos-Santos E.C., Lopes-Ferreira M., Lima C., Cardoso D.F., Mota I. A comparative study of biological activities of crotoxin and CB fraction of venoms from Crotalus durissus terrificus, Crotalus durissus cascavella and Crotalus durissus collilineatus. Toxicon. 2004;43:801–810. doi: 10.1016/j.toxicon.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Rivas E., Neri-Castro E., Bénard-Valle M., Hernánez-Dávila A.I., Zamudio F., Alagón A. General characterization of the venoms from two species of rattlesnakes and an intergrade population (C. lepidus x aquilus) from Aguascalientes and Zacatecas, Mexico. Toxicon. 2017;138:191–195. doi: 10.1016/j.toxicon.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Salazar A.M., Rodríguez-Acosta A., Girón M.E., Aguilar I., Guerrero B. A comparative analysis of the clotting and fibrinolytic activities of the snake venom (Bothrops atrox) from different geographical areas in Venezuela. Thromb. Res. 2007;120:95–104. doi: 10.1016/j.thromres.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Saviola A.J., Gandara A.J., Bryson R.W., Mackessy S.P. Venom phenotypes of the rock rattlesnake (Crotalus lepidus) and the ridge-nosed rattlesnake (Crotalus willardi) from México and the United States. Toxicon. 2017;138:119–129. doi: 10.1016/j.toxicon.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Schield D.R., Adams R.H., Card D.C., Corbin A.B., Jezkova T., Hales N.R., Meik J.M., Perry B.W., Spencer C.L., Smith L.L., García G.C., Bouzid N.M., Strickland J.L., Parkinson C.L., Borja M., Castañeda-Gaytán G., Bryson R.W., Flores-Villela O.A., Mackessy S.P., Castoe T.A. Cryptic genetic diversity, population structure, and gene flow in the Mojave rattlesnake (Crotalus scutulatus) Mol. Phylogenet. Evol. 2018;127:669–681. doi: 10.1016/j.ympev.2018.06.013. [DOI] [PubMed] [Google Scholar]
- Sela M., Schechter I., Borek F. Antibodies to sequential and conformational determinants. Cold Spring Harb. Symp. Quant. Biol. 1967;32:537–545. [Google Scholar]
- Slotta K.H., Fraenkel-Conrat H.L. Schlangengifte, III. Mitteil.: reinigung und krystallisation des Klapperschlangen-Giftes. Berichte der Dtsch. Chem. Gesellschaft (A B Ser.) 1938;71:1076–1081. doi: 10.1002/cber.19380710527. [DOI] [Google Scholar]
- Strickland J.L., Mason A.J., Rokyta D.R., Parkinson C.L. Phenotypic variation in Mojave Rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins (Basel) 2018:1–23. doi: 10.3390/toxins10040135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K., Undheim E.A.B., Scheib H., Gren E.C.K., Cochran C., Person C.E., Koludarov I., Kelln W., Hayes W.K., King G.F., Antunes A., Fry B.G. Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J. Proteom. 2014;99:68–83. doi: 10.1016/j.jprot.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Tasoulis T., Isbister G.K. A Review and database of snake venom proteomes. Toxins (Basel) 2017;9 doi: 10.3390/toxins9090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés R., Ibarra N., González M., Alvarez T., García J., Llambias R., Pérez C.A., Quintero O., Fischer R. CB.Hep-1 hybridoma growth and antibody production using protein-free medium in a hollow fiber bioreactor. Cytotechnology. 2001;35:145–154. doi: 10.1023/A:1017921702775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein S.A., Smith L.A. Preliminary fractionation of tiger rattlesnake ( Crotalus tigris ) venom. Toxicon. 1990;28:1447–1455. doi: 10.1016/0041-0101(90)90158-4. [DOI] [PubMed] [Google Scholar]
- Wilkinson J.A., Glenn J.L., Straight R.C., Sites J.W. 2018. Distribution and Genetic Variation in Venom A and B Populations of the Mojave Rattlesnake ( Crotalus Scutulatus Scutulatus in Arizona Published by : Allen Press on behalf of the Herpetologists ’ League Stable; pp. 54–68.https://www.jstor.org/stable/3892815.REF.47 [Google Scholar]
- Wüster W., Ferguson J.E., Quijada-Mascareñas J.A., Pook C.E., Salomão M.D.G., Thorpe R.S. Tracing an invasion: landbridges, refugia, and the phylogeography of the neotropical rattlesnake (serpentes: viperidae: Crotalus durissus) Mol. Ecol. 2005;14:1095–1108. doi: 10.1111/j.1365-294X.2005.02471.x. [DOI] [PubMed] [Google Scholar]
- Zancolli A.G., Calvete J.J., Cardwell M.D., Greene H.W., Hayes K., Hegarty M.J., Herrmann H., Holycross A.T., Dominic I., Mulley J.F., Sanz L., Travis Z.D., Whorley J.R., Catharine E., Wüster W. When one phenotype is not enough-divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. bioRxiv. 2018:1–21. doi: 10.1101/413831. [DOI] [PMC free article] [PubMed] [Google Scholar]